Abstract
Despite the rare appearance of potent HIV-neutralizing mAbs in infected individuals requiring prolonged affinity maturation, little is known regarding this process in the majority of viremic individuals. HIV-infected individuals with chronic HIV viremia have elevated numbers of nonconventional tissue-like memory (TLM) B cells that predominate in blood over conventional resting memory (RM) B cells. Accordingly, we investigated affinity maturation in these 2 memory B cell populations. Analysis of IgG-expressing TLM B cells revealed a higher number of cell divisions compared with RM B cells; however, TLM B cells paradoxically displayed significantly lower frequencies of somatic hypermutation (SHM). To assess Ab reactivity in TLM and RM B cells, single-cell cloning was performed on HIV envelope CD4–binding site–sorted (CD4bs-sorted) B cells from 3 individuals with chronic HIV viremia. Several clonal families were present among the 127 cloned recombinant mAbs, with evidence of crosstalk between TLM and RM B cell populations that was largely restricted to non-VH4 families. Despite evidence of common origins, SHM frequencies were significantly decreased in TLM-derived mAbs compared with SHM frequencies in RM-derived mAbs. However, both cell populations had lower frequencies of SHMs than did broadly neutralizing CD4bs–specific mAbs. There was a significant correlation between SHM frequencies and the HIV-neutralizing capacities of the mAbs. Furthermore, HIV neutralization was significantly higher in the RM-derived mAbs compared with that seen in the TLM-derived mAbs, and both SHM frequencies and neutralizing capacity were lowest in TLM-derived mAbs with high polyreactivity. Thus, deficiencies in memory B cells that arise during chronic HIV viremia provide insight into the inadequacy of the Ab response in viremic individuals.
The inadequacy of the antibody response in HIV-viremic individuals is explored by investigating HIV-specific mAbs derived from two populations of memory B cells.
Introduction
HIV infection leads to numerous immunologic abnormalities, especially in individuals whose viremia is not well controlled, either naturally or by antiretroviral therapy (ART). B cells are not direct targets for HIV replication; however, direct and indirect consequences of viral replication such as immune activation and lymphopenia lead to numerous B cell abnormalities over the course of infection (1–3). Abnormalities of B cell terminal differentiation occur early after infection, as evidenced by increased frequencies of plasmablasts in the peripheral blood, most of which are not HIV specific, and correlate with hypergammaglobulinemia and the secretion of inflammatory cytokines (4, 5). Abnormalities in B cell maturation are also observed in HIV infection, especially in advanced disease, with increased frequencies of immature/transitional B cells in the peripheral blood associated with CD4+ T cell lymphopenia and increased serum levels of IL-7 (6). HIV infection is also associated with numerous phenotypic and functional abnormalities in the memory B cell compartment (1–3). These abnormalities arise early, intensify during the chronic phase of viremia, and can be reversed by early initiation of ART (7).
Human memory B cells are mostly identified by the expression of the cell-surface marker CD27 in the absence or presence of Ig class switching (8, 9). However, since the primary role of memory B cells is to rapidly respond upon re-encountering the original stimulating antigen (pathogen), features that reflect this role should form the basis of evaluation of the quality of the memory B cell compartment. Two such features include the capacity to generate a repertoire of resting memory B cells that ensures longevity and the capacity to undergo somatic hypermutation (SHM) in association with T cell help (10, 11). In this regard, the accumulation in resting memory (RM) B cells of SHM in the variable regions of Ig heavy and light chains that convey increased affinity for cognate antigen is the most desirable outcome of an effective B cell response (12).
Several populations of memory B cells do not fall within the classical definition characterized by CD27 expression in the absence or presence of Ig class switching. In healthy individuals, these nonclassical memory B cells represent minor constituents among circulating B cells. For example, IgG+ or IgD– memory B cells that do not express CD27 comprise less than 4% of B cells in the peripheral blood (13, 14). However, nonclassical memory B cells can represent major constituents in various disease settings (12, 15). In this regard, at least 3 phenotypically distinct memory B cell populations, on the basis of the expression of CD21 and CD27, have been identified in the peripheral blood of HIV-viremic individuals. RM B cells (CD21hiCD27+) constitute the majority of circulating memory B cells in healthy individuals, yet a minority in chronic HIV-viremic individuals (7). In contrast, the majority of circulating memory B cells in chronically HIV-viremic individuals consist of tissue-like memory (TLM) (CD21loCD27–) and activated memory (AM) (CD21loCD27+) B cells (7). The former B cell population is named for its similarities to tonsillar tissue–derived counterparts in healthy individuals (16). In addition to the low expression levels of CD21 and CD27, TLM B cells and their tonsil-derived counterparts express the putative inhibitory immunoregulatory receptor FCRL4 (16, 17). TLM B cells in HIV-infected individuals have also been shown to express multiple inhibitory receptors and markers associated with homing to sites of inflammation, including CXCR3 and CD11c (17). Similar properties have been observed in pathogen-induced T cell exhaustion (18) and in diseases with immune-activating or -dysregulating effects on B cells, including common variable immunodeficiency, hepatitis C virus infection, malaria, rheumatoid arthritis, Sjogren’s syndrome, systemic lupus erythematosus, and Wiskott-Aldrich syndrome (19–25). Of note, loss-of-function mutations in CTLA4, an inhibitory receptor that regulates T cell responses yet is not expressed on B cells, has nonetheless been associated with severe perturbations in the B cell compartment, including the expansion of TLM B cells (26, 27).
We recently demonstrated that within given HIV-infected individuals, HIV-specific B cells were enriched within AM and TLM B cell populations, whereas B cells specific for non-HIV pathogens such as influenza and tetanus were distributed preferentially within the RM B cell compartment (28). These findings suggested that the abnormalities in B cell responses were not simply a reflection of changes in the distribution of B cell populations, but rather reflected alterations related to HIV per se. However, these observational findings did not address critical questions regarding the quality of responses against HIV and whether the overall response was affected by the enrichment of the response within abnormal B cell populations. Accordingly, the current study was undertaken to investigate the frequencies of SHMs within conventional and nonconventional B cell populations found in the peripheral blood of chronically HIV-viremic individuals and to relate these findings to the neutralization capacities of the HIV-specific mAbs derived from these respective B cell subsets. Our findings demonstrate that HIV-specific mAbs derived from TLM B cells have lower frequencies of SHMs than do those derived from their RM counterparts, and these differences correlated with weaker HIV-neutralizing activity of TLM- versus RM-derived mAbs.
Results
Enhanced proliferative history of IgG+ TLM versus RM B cells.
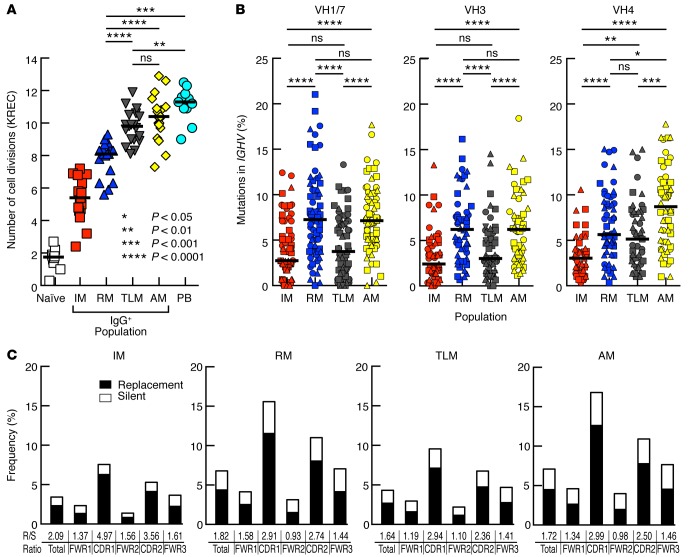
The number of cell divisions undergone in vivo by a defined B cell population can be measured by κ-deleting recombination excision circles (KREC) analysis (29, 30). We previously reported that TLM B cells had undergone fewer cell divisions than did conventional CD27-expressing memory B cells comprising both RM and AM B cells (17). However, we had not taken Ig isotype into consideration. We therefore revisited KREC analyses of Ig-defined B cell populations given 2 recent findings: more cell divisions were observed among Ig class–switched than unswitched memory B cells (29), and our observation that the frequency of class-switched Ig was significantly lower in TLM B cells than in CD27+ memory B cells (31). As expected and shown in Figure 1A, the lowest number of cell divisions occurred in naive B cells, and the highest number of cell divisions was observed in plasmablasts (not sorted on an Ig basis due to an absence of surface IgG expression). When KREC levels were measured among IgG-expressing B cells, the lowest number of cell divisions was found in IgG-expressing CD27–CD21+ B cells (Figure 1A), which we have previously defined as intermediate memory (IM) B cells (28). This is consistent with published findings in healthy individuals (29) and takes into account that CD21lo B cell frequencies are negligible (7) and, as such, are not generally considered in these individuals. Of note, we had previously reported that the frequency of IgG+ IM B cells was very low in HIV-viremic individuals, whereas TLM, RM, and AM B cells each accounted for approximately 30% of the total number of IgG+ circulating B cells (28). Among these 3 major B cell populations, the number of cell divisions undergone by both TLM and AM B cells was significantly greater than RM B cells (Figure 1A). Thus, the nonconventional IgG+ TLM B cells that circulate in the blood of HIV-viremic subjects have an enhanced proliferative history compared with that of conventional IgG+ RM B cells.
Figure 1. Reduced SHMs among Ig heavy chains of TLM B cells despite higher KREC levels.
(A) KREC analysis of B cell populations sorted from the peripheral blood of HIV-infected chronically viremic patients (n = 9–19 per population). Differences between naive and IM and all other populations were significant, although P values are not shown. (B) B cell populations were sorted from the peripheral blood of 3 patients (each identified by a different symbol) from A, and variable regions within the Ig heavy chains (VH) were amplified and sequenced. Mutation frequencies were calculated from nucleotide sequences and the number of clones sequenced per family ranged by population from 62 to 78 for VH1/7, 48 to 65 for VH3, and 45 to 60 for VH4. (C) R/S ratios were determined from the sequences in B. Horizontal bars represent the median values. P values were determined using Friedman/Wilcoxon signed-rank (A) and Kruskal-Wallis and Mann-Whitney U (B) tests. AM, activated memory; IM, intermediate memory; KREC, κ-deleting recombination excision circles; PB, plasmablast; RM, resting memory; R/S, replacement-to-silent; TLM, tissue-like memory.
IgG+ TLM B cells display decreased frequencies of SHMs compared with IgG+ RM B cells.
The accumulation of SHMs in the variable regions of heavy and light chains, a characteristic that defines Ab affinity maturation and clonal selection, has been shown to be tightly linked to cell division (32). Thus, we aimed to determine whether the increased proliferation identified in IgG+ TLM B cells was associated with enhanced accumulation of SHMs. Accordingly, we analyzed the frequencies of SHMs in variable regions of the IgG heavy chain genes within the memory B cell populations described in Figure 1A. Among the VH families analyzed for the 3 individuals reported in Figure 1A, SHM frequencies were significantly higher in AM and RM B cells than in TLM or IM B cell populations for VH1/7 and VH3 (Figure 1B). However, there was no significant difference in SHMs between RM and TLM B cells for the VH4 family. Given the KREC analyses, SHM frequencies would have been predicted to be higher in AM and TLM B cells than in RM B cells. Instead, we observed increased levels of SHMs in conventional IgG+ RM B cells compared with levels in IgG+ TLM memory B cells in 2 of the 3 VH families studied. Finally, we determined the ratios of replacement-to-silent (R/S) mutations for the dataset in Figure 1B. As shown in Figure 1C, R/S ratios for all B cell populations were predictably higher in the complementarity-determining region (CDR) than in the framework regions. VH4 family members had a slightly higher R/S ratio in framework region-1 than did those belonging to non-VH4 families (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/jci.insight.84610DS1). We also found enrichment of usage of the VH4-34 gene among TLM B cells (Supplemental Figure 2A). This VH gene encodes intrinsically self-reactive Abs found in systemic lupus erythematosus and Wiskott-Aldrich syndrome (33, 34) and is associated with HIV-neutralizing activity in HIV infection (35). Despite these unique features, SHM frequencies among VH4-34 sequences were not different compared with those in other VH4 family members in either population (data not shown). We conclude that, contrary to predictions from KREC analyses, IgG+ TLM B cells from HIV-viremic subjects showed overall decreased frequencies of SHM compared with conventional IgG+ RM B cells, despite having undergone more cell divisions.
Characterization of CD4–binding site, HIV-specific mAbs from IgG+ TLM and RM B cells.
Several HIV envelope proteins have been engineered into probes for flow cytometric analyses of antigen-specific B cell responses and demonstrate high sensitivity and specificity for B cells expressing B cell receptors (BCRs) that recognize epitopes within the probe (36, 37). Using such probes to interrogate B cell responses in a large cohort of HIV-infected individuals, we recently demonstrated that HIV-specific B cells are found in higher frequency in the blood in the early stage of infection (28), although they predominantly recognize epitopes, such as the coreceptor (CCR5) of the HIV envelope, that are generally considered non-neutralizing (38). B cell responses that target neutralizing epitopes, such as those against the CD4-binding site (CD4bs) of the HIV envelope, tend to arise several months to years after infection (39). A similar pattern of delayed CD4bs-specific B cell response was observed in our cohort, and the contribution to the overall CD4bs response of TLM B cells was relatively higher when compared with that against the coreceptor response (28).Given these observations, we designed a strategy to single-cell sort CD4bs-specific B cells from IgG+ TLM and RM B cell populations and compare properties of the mAbs cloned from these 2 cell populations. We chose 3 HIV-infected individuals reported in our previous study (28) and in whom the frequency of CD4bs-specific B cells was sufficiently high within both cell populations to perform single-cell sorting (Table 1). These individuals were chronically infected with low-level viremia in the absence of ART, had CD4 counts above 350/μl with an inverted CD4:CD8 ratio, and serological Ab activity that neutralized tier 2 HIV pseudoviruses, albeit with variably restricted profiles of breadth and potency (Supplemental Table 1).
Table 1. Study patients from whom CD4bs-specific B cells were sorted.
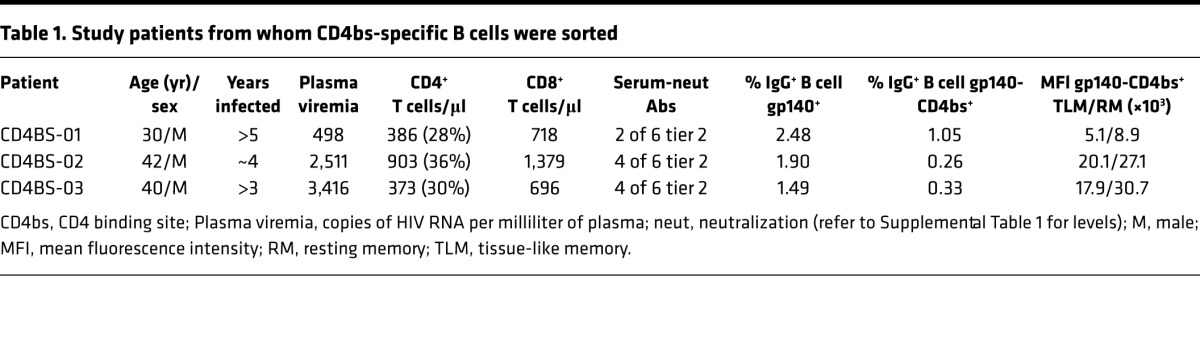
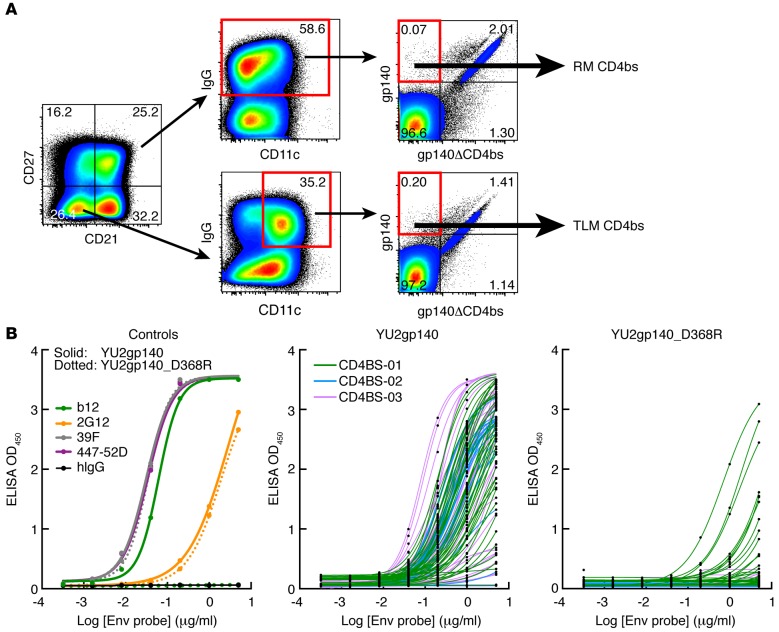
The representative flow plots shown in Figure 2A for 1 of the 3 individuals (CD4BS-01) revealed that the mean fluorescence intensity of CD4bs binding was generally lower in TLM B cells compared with that observed in RM B cells, a feature observed in all 3 individuals (Table 1) and in others (data not shown). These lower intensities were consistent with the relative underrepresentation of HIV-specific TLM B cells when measured by flow cytometry, as previously reported (28). Nonetheless, the number of B cells required for the 2 sorting strategies shown in Figure 2A were similar, and, accordingly, a total of 62 and 65 mAbs were cloned from IgG+ TLM and RM B cells, respectively (Supplemental Table 2; 167 clones total, of which 127 were expressed). The percentage of recombinant mAbs that were specific for the CD4bs of gp140 was evaluated by ELISA and was similar between IgG+ TLM and RM B cell populations (Figure 2B and Supplemental Table 2). Indeed, we found that mAbs specific for the CD4bs sorting antigen represented 87.1% of TLM and 86.1% of RM clones (Figure 2B and Supplemental Table 2). Of the 127 mAbs cloned from IgG+ RM and TLM B cells, 110 were found specific to the CD4bs of HIV envelope protein and are referred to henceforth as CD4bs mAbs. Of note, 18 of the 44 gp140-binding mAbs reconstituted from patient CD4BS-01 retained a relatively low level yet detectable reactivity with the CD4bs mutant (Figure 2B and Supplemental Table 2), an observation that was somewhat unexpected, although possibly related to the lower overall mean fluorescence intensity of HIV-specific B cells in this individual compared with the other 2 (Table 1). In addition, a slightly higher percentage of such mAbs were found in TLM (41.7%) versus RM (31.6%) B cell populations (Supplemental Table 2). Nonetheless, all gp140-specific mAbs retained specificity to the CD4bs, as evidenced by a 50% effective concentration (EC50) binding to the gp140-CD4bs mutant that was at least 2-fold higher than that of binding to WT gp140 and not observed with control mAbs that were not specific to CD4bs (Figure 2B).
Figure 2. mAbs derived from single-cell-sorted RM and TLM B cells are specific to the CD4bs epitope within gp140.
(A) Gating strategy used to single-cell sort CD4bs-binding TLM and RM B cells of CD4BS-01. (B) Evaluation of mAb (n = 110) binding by ELISA to either YU2gp140 (WT protein) or YU2gp140_D368R (CD4bs mutant). Plots on the left are for control mAbs, and those on the right are for mAbs derived from single-cell sorting from the 3 individuals described in Table 1. CD4bs, CD4 binding site; RM, resting memory; TLM, tissue-like memory.
HIV-specific mAbs from IgG+ TLM B cells show decreased SHMs but are clonally related to RM counterparts.
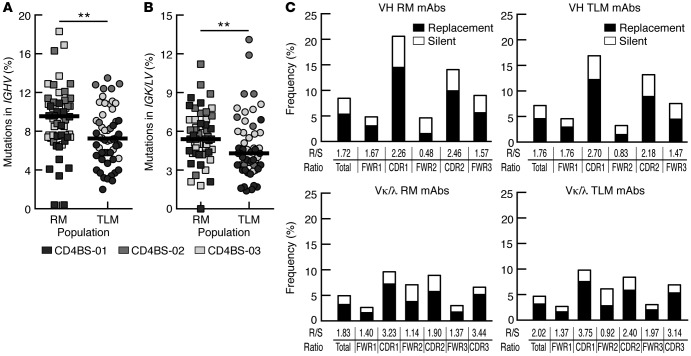
Since total IgG+ TLM B cells displayed lower SHM frequencies than did IgG+ RM B cells, we evaluated whether this difference was also present among CD4bs mAbs. We found that CD4bs mAbs cloned from IgG+ TLM B cells harbored significantly lower frequencies of SHMs in both IGH V (Figure 3A) and IGK/L V (Figure 3B) gene segments compared with frequencies detected in IgG+ RM B cell counterparts. However, HCDR3 lengths were not different between the 2 cell populations; the median for CD4bs mAbs in Supplemental Table 2 was 18 aa for each of the 2 populations. Differences in SHM for both Ig chains from RM- and TLM-derived CD4bs mAbs were mainly attributed to VH4 family members that were found in 20.9% (23 of 110) of CD4bs mAbs in chronically HIV-viremic subjects (Supplemental Figure 2B and Supplemental Table 2); this frequency is similar to that reported in another study (40), although there is a paucity of VH4 gene usage among the highly potent CD4bs mAbs isolated from HIV-infected elite neutralizers (41).
Figure 3. Lower SHM levels among CD4bs mAbs derived from TLM B cells compared with those derived from RM B cells.
SHM frequencies were calculated for (A) heavy- and (B) light-chain variable regions of the CD4bs mAbs (n = 110) derived from TLM and RM B cells of the 3 patients described in Figure 2. Horizontal bars represent median values. (C) R/S ratios were determined for heavy and light chains of all CD4bs mAbs derived from TLM and RM B cells. **P < 0.01, by Mann-Whitney U test. CD4bs, CD4 binding site; RM, resting memory; R/S, replacement-to-silent; SHM, somatic hypermutation; TLM, tissue-like memory.
The observation that SHM frequencies of VH4-encoded CD4bs mAbs were lower among those derived from TLM B cells than those from RM B cells contrasted with the findings depicted in Figure 1, in which differences in SHMs from RM versus TLM B cell populations were restricted to non-VH4 families. To distinguish between the effects of sorting strategies and the effects driven by the specific antigen, heavy-chain SHM frequencies were measured from nonantigen-selected, single-sorted RM and TLM B cells. Differences in SHM frequencies in these B cells were found to mirror those from bulk-sorted cells, in that SHM frequencies were higher among sequences from RM B cells compared with those from TLM B cells for VH1/7 and VH3, but not VH4, families (Supplemental Figure 3). Of note, while VH4-34 gene usage was prevalent among TLM and, to a lesser extent, RM B cells (Supplemental Figure 2A), with a similar trend among single-cell clones (Supplemental Figure 2B), VH4-34 gene usage among CD4bs mAbs was infrequent: 1 mAb derived from CD4BS-01 TLM B cells and 1 clonal family from CD4BS-02 RM B cells (Supplemental Table 2). Thus, differences in VH4 mainly reflect distinctions between B cell populations and effects driven by CD4bs epitope selection.
Analyses of R/S ratios among the CD4bs mAbs from both IgG+ TLM and RM B cells revealed that replacement mutations were normally enriched in IgH and IgK/L CDRs, suggesting that antigenic selection shaped both B cell populations (Figure 3C). In addition, VH4 clones showed R/S ratios that were 1.5- to 2-fold higher than those in non-VH4 family members, revealing an increased selection pressure shaping VH4 responses, despite decreased global SHM frequencies (Supplemental Figure 1B).
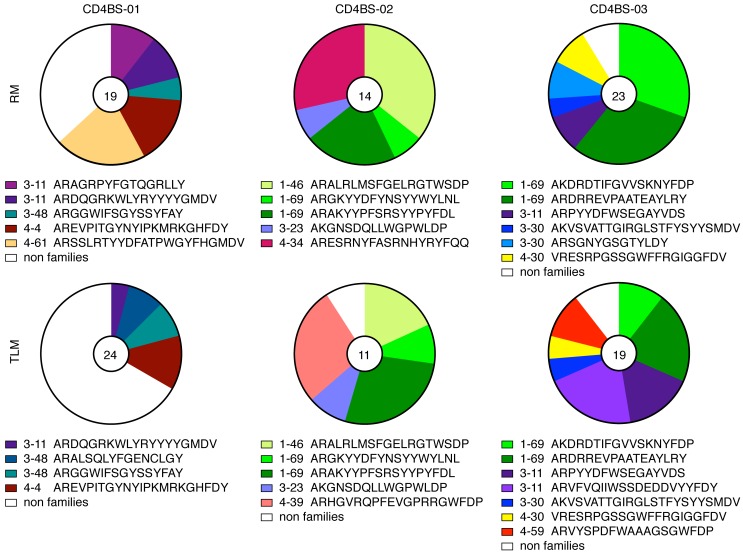
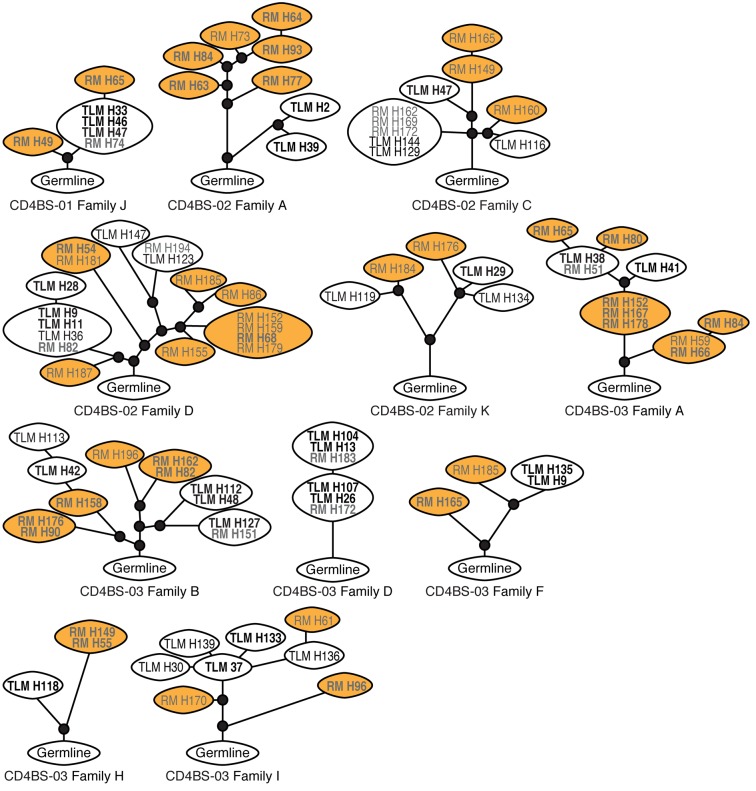
Sequence alignments revealed the presence of several clonally related B cells in all 3 patients studied; these were identified both within and across IgG+ TLM and RM B cells (Figure 4 and Supplemental Figure 4). The most highly represented IgH gene segments were IGHV1-69 and IGHV3-11, both found in multiple clones from 2 of the 3 patients (Supplemental Table 2). Lineage tree analysis was performed to establish clonal evolution within each family, and a total of 11 trees were generated, revealing a mixed pattern of shared and distinct mutations among the families (Figure 5). With the exception of 1 tree, in which clones from the 2 cell populations were fully intermingled (CD4BS-03 family D), all of the other trees contained RM and TLM clones that had clearly evolved separately from an inferred common intermediate (Figure 5). There were several trees in which the division between cell populations was close to the germline sequence, such as in CD4BS-02 family A or in the smaller clonal families CD4BS-03 families F and H. Consistent with the SHM data, RM-derived clones in 6 of the 11 family trees were clearly distinct and more evolved than were corresponding clones derived from TLM B cells (CD4BS-01 family J; CD4BS-02 families A and C and 1 large branch of family D; CD4BS-03 families A and H). Families B and I in CD4BS-03 showed mixed TLM and RM outgrowths, whereas the few remaining families contained RM and TLM clones with similar evolution profiles. Among the 7 clonal expansions specific to either RM or TLM B cells, 4 were VH4 encoded. In contrast, only 2 VH4 families were among the 12 that were found in both B cell populations (Figure 4). Collectively, these observations indicate that CD4bs HIV-specific clonal expansion occurred in either private VH4 clones or, most often, in common precursors of IgG+ TLM and RM B cells, but that among the latter, clonal evolution appeared distinct, with more matured clones of increased SHMs enriched in IgG+ RM B cells.
Figure 4. mAb clonal families within and across derivative B cell populations.
Pie charts of clonal family representation were constructed for each of the 3 sets of CD4bs mAbs derived from TLM and RM B cells. The numbers in the pie charts represent the number of mAbs evaluated. Other color-coordinated identifiers include the heavy-chain family and the clonal family CDR 3 segment. CD4bs, CD4 binding site; CDR, complementarity determining region; RM, resting memory; TLM, tissue-like memory.
Figure 5. Clonal trees generated from Ig heavy-chain sequences show distinct patterns.
Lineage trees were generated from sequences obtained from RM and TLM B cells from the 3 individuals described in Table 1 and Supplemental Table 1. Expressed mAbs are denoted in bold. Clones within ovals contain identical sequences; white ovals denote TLM-only or mixed clones, and orange ovals denote RM-only clones; small black circles identify inferred intermediates from germline sequence at the base of each tree. Branch lengths reflect SHM frequencies. RM, resting memory; SHM, somatic hypermutation; TLM, tissue-like memory.
CD4bs HIV-specific mAbs from IgG+ TLM B cells show lower neutralization capacity than do RM B cells.
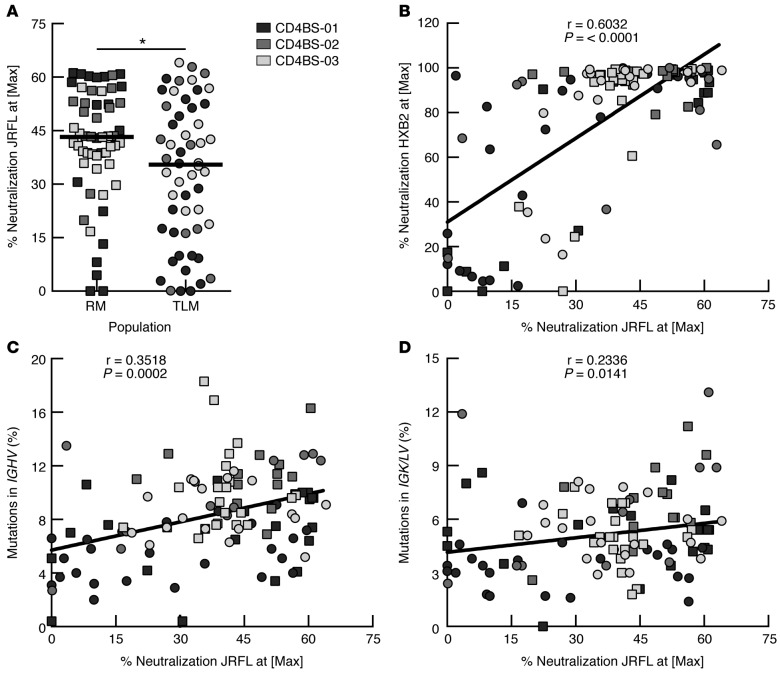
The ability of CD4bs HIV-specific mAbs cloned from IgG+ TLM and RM B cells to neutralize HIV was evaluated in an HIV-1 pseudovirus infectivity assay using a standard TZM-bl assay (42). As shown in Figure 6A, none of the mAbs demonstrated the ability to neutralize the HIV-1 tier 2 JRFL pseudovirus above 80% at the highest concentration of 40 μg/ml, consistent with the low neutralizing potency observed in the serum of these individuals (Supplemental Table 1). In this regard, all 3 patients are typical chronically infected HIV-viremic individuals selected not on the basis of serologic neutralization capacity, but rather for the presence of CD4bs HIV-specific IgG+ TLM and RM B cells (ref. 28 and Table 1). Despite the overall low neutralizing activity, CD4bs mAbs from IgG+ RM B cells had a significantly higher HIV-neutralizing capacity than did those from IgG+ TLM B cells (Figure 6A and Supplemental Figure 5A). However, when evaluated against HXB2, a tier 1 virus known to be highly susceptible to neutralization by CD4bs-specific Abs (43), the majority of CD4bs mAbs displayed strong neutralizing activity, with differences between TLM and RM sources that did not reach statistical significance (Supplemental Figure 5, B and C), yet correlated with activity against JRFL (Figure 6B). Furthermore, there was a significant correlation between HIV-neutralizing capacity and SHM frequencies in the IGH V gene segments (Figure 6C) and, to a lesser yet significant extent, in IGK/L V gene segments (Figure 6D). We conclude that higher SHM frequencies correlate with more efficient neutralization of HIV. However, neutralization by CD4bs mAbs from both TLM and RM B cells is weak in potency and restricted in breadth (evaluated as described in Methods; data not shown), compared with that of potent and broadly neutralizing Abs that display very high SHM frequencies.
Figure 6. CD4bs mAbs derived from TLM B cells have lower HIV-neutralizing capacity than do those from RM B cells.
(A) HIV-neutralizing activities of the CD4bs mAbs (n = 110) were evaluated in TZM-bl cells with JRFL-enveloped HIV pseudotype virus. (B) HIV-neutralizing activities of the CD4bs mAbs were correlated between HXB2- and JRFL-enveloped pseudoviruses. Percentage of neutralization at maximum concentration of mAb (40 μg/ml) of the titration curve was used for the comparisons. Neutralization of JRFL was correlated with SHM percentages in variable segments of heavy (C) and light (D) chains of the corresponding mAbs. Horizontal bars represent median values, and symbols in correlations are the same as those used for the group analyses. *P < 0.05, by Mann-Whitney U test. CD4bs, CD4 binding site; Max, maximum; RM, resting memory; SHM, somatic hypermutation; TLM, tissue-like memory.
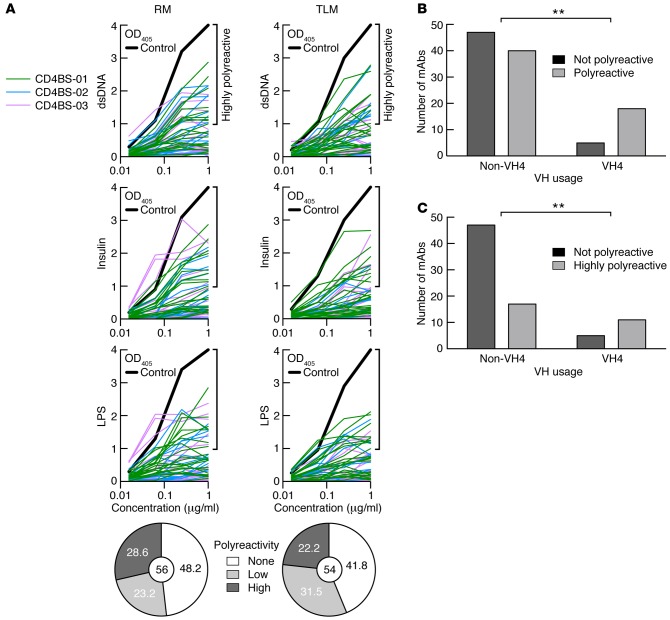
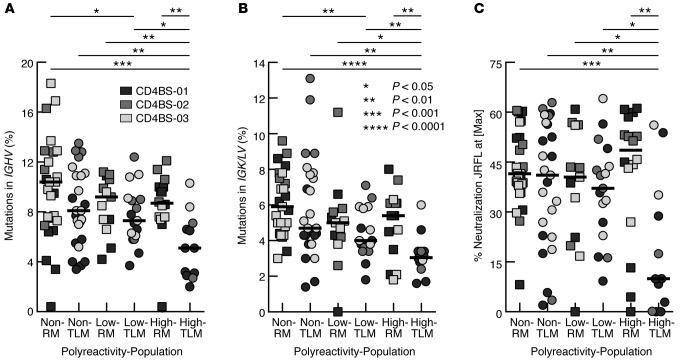
Polyreactivity, as defined by reactivity with dsDNA, insulin, and LPS, has been shown to be associated with many broadly neutralizing anti-HIV Abs and may contribute to their neutralization capacity (44). The assessment of polyreactivity in CD4bs mAbs revealed that the majority of IgG+ RM and TLM B cells expressed polyreactive Abs (51.8% and 53.7% respectively), with similar proportions of low and highly polyreactive Abs (Figure 7A). The number of polyreactive CD4bs mAbs was significantly enriched among those that belonged to the VH4 family when compared with CD4bs mAbs from all other VH families combined (Figure 7B). Furthermore, this difference was largely driven by TLM CD4bs mAbs (Supplemental Figure 2D) and associated with CD4bs mAbs that were defined as highly polyreactive (Figure 7C and Supplemental Figure 2D). TLM-derived highly polyreactive CD4bs mAbs had significantly fewer mutations in both heavy and light chains when compared with any other grouping of CD4bs mAbs by level of polyreactivity and cell population of origin (Figure 8, A and B). However, enrichment of VH4 usage among the highly polyreactive TLM-derived CD4bs mAbs was not significant when compared with the other groups in Figure 8 (data not shown). Finally, consistent with differences in SHMs related to levels of polyreactivity (Figure 8, A and B), the highly polyreactive CD4bs mAbs derived from TLM B cells displayed significantly lower neutralization capacity compared with that of CD4bs mAbs of no or low polyreactivity derived from TLM B cells (Figure 8C).
Figure 7. Enrichment of polyreactivity among VH4-utilizing CD4bs mAbs.
(A) Polyreactivity was evaluated for each mAb (n = 110) by ELISA-based reactivity to dsDNA, insulin, and LPS. Abs were considered polyreactive when they recognized all 3 antigens. Bold lines show ED38-positive control; horizontal lines show the cut-off OD405 for positive reactivity. For each fraction, the frequency of nonpolyreactive (white), low polyreactive (light gray), and highly polyreactive (dark gray) clones is summarized in the pie charts, with the total number of pooled clones isolated from the 3 patients indicated in the centers. Numbers of CD4bs mAbs with no polyreactivity among non-VH4 and VH4 family members compared with (B), all those with polyreactivity, and (C), those with high polyreactivity. **P < 0.01, by Fisher’s exact test. CD4bs, CD4 binding site; RM, resting memory; TLM, tissue-like memory.
Figure 8. Highly polyreactive CD4bs mAbs derived from TLM B cells have the lowest levels of SHMs and HIV-neutralizing capacity.
Frequencies of SHMs for mAbs shown in Figure 3 were analyzed by level of polyreactivity for (A) heavy and (B) light chains. (C) The percentage of HIV neutralization for mAbs shown in Figure 6 was analyzed according to the level of polyreactivity. Horizontal bars represent median values. P values were determined by Kruskal-Wallis and Mann-Whitney U tests. CD4bs, CD4 binding site; RM, resting memory; SHMs, somatic hypermutations; TLM, tissue-like memory.
Taken together, these data indicate that increased SHMs in HIV-specific B cells is associated with an increasing capacity to neutralize the virus, as reported by others (45–49), and the level of CD4bs mAb polyreactivity was a significant determinant of neutralization. Furthermore, those mAbs of high polyreactivity derived from TLM B cells show a distinctly lower capacity to neutralize HIV than all other mAbs, whether derived from RM or TLM B cells (Figure 8C).
Discussion
The present study evaluates a fundamental property of memory B cells, namely the affinity maturation process, in the setting of chronic HIV viremia. We demonstrate that IgG+ TLM B cells, a population of circulating exhausted memory B cells overrepresented in HIV-infected chronically viremic individuals (17), display lower overall frequencies of SHMs when compared with IgG+ RM B cells, despite having undergone more cell divisions in vivo. Decreased SHMs was also a key characteristic of CD4bs mAbs derived from IgG+ TLM when compared with SHMs in RM B cells of HIV-infected, chronically viremic individuals, and these decreased SHM frequencies correlated with reduced HIV-neutralizing capacity. Thus, anti-HIV memory B cells in HIV-viremic individuals obey the clonal theory and affinity maturation model, whereby SHM plays an essential role in the emergence of pathogen-specific, in this case HIV, neutralizing clones (50). However, while TLM B cell–derived CD4bs mAbs were clearly deficient compared with RM B cell–derived CD4bs mAbs, none of the mAbs expressed by either IgG+ RM or TLM B cells from the HIV-viremic individuals studied reached the high frequencies of SHMs that have been associated with potent and broadly neutralizing CD4bs mAbs isolated from a relatively small number of infected individuals (47, 49, 51, 52).
Neutralizing Abs against the CD4bs generally arise during the chronic phase of infection (39, 53–55), with the most potent ones showing a restricted VH1-2 or VH1-46 gene usage or HCDR3-dominated interactions (52). It is noteworthy that none of the CD4bs mAbs described in the present study utilized VH1-2, while 9 utilized VH1-46 and all but 2 originated from RM B cells (Supplemental Table 2). The B cell origins of the most potent and broadly neutralizing mAbs directed against the CD4bs have not been well characterized, although a role for TLM B cells appears unlikely. The VH1-2–restricted mAbs VRC01 and VRC23, as well as the CD4 supersite–targeted mAbs VRC13, VRC15, and CH103, were derived from B cells that expressed CD27, a marker not expressed on TLM B cells (our unpublished observations for VRC01) and personal communication from Nicole Doria-Rose for VRC23 and Rebecca Lynch for VRC13, VRC15, and CH103). Furthermore, with the exception of VRC23, the strategies for identifying these mAbs did not gate for CD27 expression, rather, CD27 happened to be expressed on the selected cells, suggesting that TLM B cells were underrepresented as a source of CD4 supersite–targeted mAbs.
VH4 gene usage among potent and broadly neutralizing CD4bs mAbs is uncommon, with the CH103 lineage (VH4-59) being the exception, as it is characterized by intermediate levels of SHM and HIV neutralization breadth and potency (46). The CH103 lineage is also unique in having been detected early after infection and was found to acquire polyreactivity during its maturation process, an adaption thought to provide an advantage for neutralizing HIV (46), as previously demonstrated (44). In contrast, VH4-utilizing CD4bs mAbs were isolated from both RM and TLM B cells from all 3 individuals in the present study, an observation consistent with a previous report of common VH4 usage among CD4bs mAbs with modest HIV-neutralizing activity (40). However, there was limited crossover of VH4 clones between RM and TLM B cells. Furthermore, although the majority of CD4bs mAbs were polyreactive, the percentage of polyreactivity was the highest among VH4-expressing clones, especially for mAbs with high polyreactivity. In addition, highly polyreactive CD4bs mAbs derived from TLM B cells manifested significantly lower HIV-neutralizing capacity when compared with all other polyreactive and nonpolyreactive CD4bs mAbs (Figure 8C).
Clonal expansions were clearly observed within and across the 2 B cell populations investigated, although VH4 clones appeared more segregated, and RM B cells often displayed increased affinity maturation features compared with those of their TLM-derived counterparts. As described for the CH103 lineage (46), it is likely that a combination of intermediate properties can enhance HIV-neutralizing capacity. Two such examples stand out in our study, both of which were clonally related CD4bs mAbs restricted to RM B cells. The first consisted of 4 members belonging to VH1-46 in patient CD4BS-02 (Figure 4); all possessed a neutralizing capacity that was above the median and a combination of favorable characteristics that included VH usage for targeting the CD4bs (VH1-46), a long HCDR3 (19 aa), and high polyreactivity (Supplemental Table 2). The second consisted of 4 members belonging to VH4-61 in patient CD4BS-01 (Figure 4); all possessed a neutralizing capacity above the median, along with a long HCDR3 (22 aa) and high polyreactivity (Supplemental Table 2). In contrast, only 1 CD4bs mAb of high polyreactivity and HIV-neutralizing activity was found among the TLM population; it was a member of the VH4-30 family isolated from patient CD4BS-03 that was also present in the RM B cell population. Collectively, these findings indicate that, while a certain number of CD4bs-specific clones were shared between IgG+ RM and TLM B cells, there was evidence of more extensive maturation among the former B cell population, in addition to superior clones expressed by RM B cells that were not shared by TLM B cells.
B cells similar to the TLM B cell population have been described in several disease settings (reviewed in ref. 1), including malaria, in which V gene repertoires and parasite-specific mAbs isolated from IgG+ classical memory (equivalent to RM) and atypical memory (equivalent to TLM) B cells have been described recently (56, 57). Few differences were found between the 2 B cell populations, since both contributed to protective B cell immunity; where differences were found, they functionally favored atypical memory B cells, namely, higher levels of SHM and frequencies of polyreactivity (56). It is noteworthy that malaria is associated with seasonal reinfection with Plasmodium falciparum, resulting in cycling periods of clinical disease and remission associated with a gradual increase in memory B cell responses against the parasite (58). In HIV infection, the virus persists with progressive depletion of CD4+ T cells in the majority of individuals not receiving ART, and while the Ab response may evolve over time, the overall frequency of the memory B cell response against HIV is highest in early infection and decreases thereafter (28). TLM B cells arise as a result of the chronic immune-activating effects of persistent HIV viremia, although they upregulate the expression of numerous inhibitory receptors that we have suggested not only inhibit their function, but also protect them from excessive response and death (31), thus perhaps explaining their longevity with occasional turnover, as evidenced by KREC levels, but without the accumulation of functionally beneficial mutations that would be predicted for a population that has undergone extensive cell division (32). Consistent with our current and past observations (17), recent findings in malaria suggest that, despite evidence of common origins between atypical (TLM equivalent) and classical memory B cells, the former arise in response to chronic exposure to the parasite, with features that include expression of several inhibitory receptors and impaired BCR-mediated function (59).
It is not well understood why most HIV-infected individuals fail to mount proper HIV-specific immune responses and produce potent and broadly neutralizing Abs. High-affinity, antigen-specific Abs are normally generated during immune responses in germinal centers (GCs), in which antigen-binding B cells interact with CD4+ Tfh cells that promote B cell activation, proliferation, and differentiation into isotype-switched memory B cells and plasma cells (reviewed in refs. 12, 50). Since Tfh cells are the preferential targets of HIV replication in lymph nodes, it is likely that Tfh cell function will be altered by HIV infection and may lead to aberrant B cell responses (60). In addition, Tfh cells in HIV-infected lymph nodes display dysregulated cytokine patterns that are characterized by decreased IL-4 and increased IFN-γ secretion, whereas IL-21 secretion is affected under certain conditions when compared with Tfh cells in lymph nodes of HIV-negative healthy individuals (ref. 61 and Pantaleo G., personal communication). IL-21 and IL-4 play important roles in B cell activation and proliferation (reviewed in ref. 62). However, in the absence of IL-4 in mice, IL-21 favors the emergence of CD21–/lo B cells that express CXCR3, CD11c, and T-bet (63). Interestingly, TLM B cells in HIV viremia, as well as in common variable immunodeficiency disease, rheumatoid arthritis, and Sjogren’s syndrome, display low levels of CD21 and express CD11c and T-bet and CXCR3 in the case of HIV (refs. 17, 19, 24, and data not shown). These profiles are thus similar to those associated with defective IL-4 production in mice and alterations in Tfh cells in lymphoid tissues of HIV-infected individuals. In addition, IL4R gene expression is diminished in TLM B cells, further reinforcing the effects of altered IL-4 responses (17, 19). The expression of TBX21 encoding T-bet in TLM B cells as well as many IFN-responsive genes is further indicative of abnormal IFN-γ stimulation of these B cells (19). IFN-γ decreases Ab secretion and bias switching to different IgG isotypes (64) and may therefore prevent appropriate immune responses against HIV. Finally, the decreased OX40L and CD40 expression observed in TLM B cells (17, 19, 65) may account for the deficient induction of AID, the enzyme that catalyzes SHMs, whereas proliferation stimulated by Tfh may be preserved, thereby recapitulating the characteristic features of IgG+ TLM B cells in HIV-viremic subjects. Taken together, these observations suggest that deficient HIV-infected Tfh cells may not properly activate B cells recognizing HIV in GCs, leading to the production of TLM B cells with decreased SHMs when cells were likely exposed to IFN-γ in the absence of IL-4.
Recent discussion of heterogeneity among human memory B cells has included TLM B cells in HIV infection and similar B cells in other diseases associated with chronic activation or inflammation (12, 66, 67). Not all features are the same across disease settings, and even within a disease, there is evidence of plasticity and intermediate phenotypes of B cell populations. For example, in HIV disease, AM B cells display hybrid features derived from RM and TLM B cells in addition to unique features of increased activation and susceptibility to apoptosis (reviewed in ref. 1). In this regard, it will be important to examine the contribution of AM B cells to the quality and sustainability of HIV-specific responses. It is also possible that TLM B cells give rise to RM or AM B cells, although this is less likely for RM B cells, given their comparatively fewer cell divisions at the population level (Figure 1A). Nonetheless, such intermediates within the TLM B cell compartment could still hinder affinity maturation, given the inherently low responsiveness to stimulation associated with the expression of multiple inhibitory receptors in this B cell population (31). A similar scenario has been suggested for exhausted T cells (68).
Among B cells that arise or expand in response to chronic activation, some may be polyclonal, reflecting systemic changes induced by the pathogen or condition and affected by factors that are distinct from those of B cells responding to the virus in a more localized environment. Such distinctions may help explain the somewhat disparate observations relative to VH4 gene usage. Chronic immune activation may have global effects on VH families, as shown by deep sequencing in HIV infection (69) and, in particular, on VH4-34 gene usage among TLM B cells in HIV and among similar cells in systemic lupus erythematosus and Wiskott-Aldrich syndrome (33, 34). In contrast, responses against the CD4bs show evidence of constraints on the B cell repertoire, whether it be the VH gene–restricted or HCDR3-dominated properties of mAbs that target the CD4 supersite (52), or the paucity of VH4-34 usage among VH4-based CD4bs mAbs of moderate activity (40). TLM and RM B cells may tackle these constraints differently, especially VH4 family TLM and RM B cells.
In summary, we demonstrate that nonconventional TLM B cells overrepresented in the peripheral blood of chronically infected HIV-viremic individuals show reduced affinity maturation compared with their clonally related conventional RM counterparts, despite evidence of having undergone more cell divisions. We also demonstrate that reduced SHMs are associated with poor HIV neutralization, especially among highly polyreactive Abs derived from TLM B cells. These findings add to our understanding of HIV-induced pathogenesis in chronically infected HIV-viremic individuals as it relates to their dysregulated immune responses.
Methods
Study patients and procedures.
Serum and/or leukapheresis products were collected from 25 chronically infected HIV-viremic individuals who were not receiving ART at the time of the study.
Cell lines, HIV pseudoviruses, and control mAbs.
The following reagents were obtained from the NIH AIDS Reagent Program: the TZM-bl cell line; the plasmid pSG3∆Env to produce HIV pseudoviruses; and the HIV-specific mAbs b12, 2G12, 39F, and 447-52D. Plasmids containing HIV envelopes of HXB2 and JRFL were also obtained from the NIH AIDS Reagent Program and were subcloned into an expression vector, as previously described (70).
Sorting of B cell subpopulations and single HIV-specific B cells.
Peripheral blood mononuclear cells (PBMCs) were obtained by density-gradient centrifugation. Mature (CD10–) B cells were isolated from PBMCs by negative magnetic bead–based selection using a B cell enrichment cocktail that was supplemented with tetrameric anti-CD10 mAb (STEMCELL Technologies). Immunophenotyping to identify suitable subjects for sorting was performed using the following anti-human mAbs: CD27-PerCP-Cy5.5 (O323; eBioscience); CD21-FITC (Bu32) and CD11c Brilliant Violet (BV) 421 (3.9; BioLegend); and CD19-V450 (HIB19), CD20-APC-H7 (L27), and IgG-PE-Cy7 (G18-145; BD Biosciences). HIV envelope CD4bs-specific B cells were identified using 2 trimeric HIV envelope gp140 probes from YU2: gp140-WT and a mutant gp140 probe with a single point mutation in the CD4-binding region (368D/R), as previously described (28). HIV-gp140-WT and CD4bs-mutant probes were conjugated with streptavidin-APC or -PE (Life Technologies), respectively. FACS analyses were performed on a FACSCanto II flow cytometer (BD Biosciences), with data analyses performed using FlowJo software (Tree Star). Sorting of B cell populations and of single HIV-specific B cells into 96-well PCR plates was performed on a modified 3-laser FACSAria instrument (BD Biosciences).
KREC and Ig sequencing.
The KREC assay was performed on DNA of cellular lysates generated from sorted B cell subpopulations, as previously described (30), with modifications (17). For each sorted B cell population, RNA was extracted from 1 × 105 to 4 × 105 cells, and approximately 5 × 105 cell equivalents in cDNA was used to amplify and sequence IgH gene transcripts in each family, as previously described (71). Repertoire analyses, dendrogram trees, and R/S ratios were determined by sequence comparison with the International ImMunoGeneTics information (IMGT) database. B cells were considered to belong to the same clone on the basis of identical V, D, and J gene segment usage and CDR3 length for both heavy- and light-chain Ig genes.
Ab production and binding assays.
Recombinant Abs were generated from single-cell-sorted 96-well PCR plates by reverse transcription PCR (RT-PCR) amplification of the variable regions of the heavy- and light-chain Ig genes and cloning into IgG expression vectors, as previously described (72). Abs were purified using protein G Sepharose 4 Fast Flow beads (GE Healthcare Life Sciences), according to the manufacturer’s recommendations, and concentrations were determined by bead-capture technology using a FACSarray (BD Biosciences). Ab specificity for the CD4bs of gp140 was performed as previously described (28), with the following modifications: ELISA 96-well plates were coated with 100 μl of 2 μg/ml test mAb, followed by 100 μl of 5-fold serial dilutions of biotin-labeled YU2gp140 or YU2gp140_D368R probes starting at 5 μg/ml. Ab polyreactivity, as defined by recognition of all 3 test antigens, dsDNA, insulin, and LPS, was determined as previously described (72). Highly polyreactive Abs were defined as those that displayed ELISA binding values of greater than 1, which corresponds to twice the value of cut-off for positivity.
Neutralization assay.
Neutralization capacity against HIV was measured using a single-round infection assay (73). Pseudotyped HIV bearing JRFL or HXB2 env was incubated with six 4-fold serial dilutions of purified Ab, beginning at 40 μg/ml, for 2 hours before being added to TZM-bl target cells for 48 hours. Duplicate measurements were averaged. Serum HIV neutralization profiles of breadth and potency were determined against the cross-clade panel of pseudoviruses shown in Supplemental Table 1 and performed by Monogram, as described previously (74). Pools of CD4bs mAbs grouped within each individual source by family or population were also evaluated for potency against the same cross-clade panel of pseudoviruses.
Statistics.
Multiple testing was performed by Friedman or Kruskal-Wallis, which, if significant, prompted pair-wise comparisons by Wilcoxon signed-rank or Mann-Whitney U tests. Two-group comparisons were performed by Mann-Whitney U test. Fisher’s exact test was used to analyze the number of polyreactive mAbs relative to VH usage, the number of clones with an IC40, and the number of VH4-34 members within the VH4 family. Correlations were determined by the Spearman test. All analyses were performed with Prism 6 software (GraphPad Software). P values of less than 0.05 were considered statistically significant.
Study approval.
This study was approved by the IRB of the NIAID, NIH. The participants provided informed consent, in accordance with the requirements of the IRB of the NIAID.
Author contributions
EM, ASF, and SM designed the study and wrote the manuscript. AL, JB, LJYK, JH, CCF, LK, WW, CMB, and YW performed the experiments and acquired the data. ORF and KRG recruited the study participants and oversaw the protocol procedures. TWC and YL provided reagents, analyzed data, and assisted with the study design and revisions.
Supplementary Material
Acknowledgments
We thank the patients for their willingness to participate in this study. We thank Shyam Kottilil for assistance with patient recruitment and care; Carl-Magnus Hogerkorp for assistance with preliminary work; and Catherine Rehm and Sara Jones for specimen processing. This work was supported by the Intramural Research Program of the NIAID, NIH and by the NIH/NIAID grant R01AI102766 (to Y. Li).
Footnotes
Leo J.Y. Kim’s present address is: Case Western Reserve University, Cleveland, Ohio, USA.
Jason Ho’s present address is: Boehringer Ingelheim, Ridgefield, Connecticut, USA.
Cody C. Frear’s present address is: Arizona State University, Tempe, Arizona, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(3):e84610. doi:10.1172/jci.insight.84610.
Contributor Information
Eric Meffre, Email: eric.meffre@yale.edu.
Aaron Louie, Email: aar.louie@gmail.com.
Jason Bannock, Email: jason.Bannock@yale.edu.
Jason Ho, Email: jasonho72@gmail.com.
Lela Kardava, Email: kardaval@niaid.nih.gov.
Wei Wang, Email: wewang@niaid.nih.gov.
Yimeng Wang, Email: wangy@ibbr.umd.edu.
Tae-Wook Chun, Email: twchun@nih.gov.
Yuxing Li, Email: yuxingli@umd.edu.
Anthony S. Fauci, Email: afauci@niaid.nih.gov.
Susan Moir, Email: smoir@niaid.nih.gov.
References
- 1.Moir S, Fauci AS. B-cell exhaustion in HIV infection: the role of immune activation. Curr Opin HIV AIDS. 2014;9(5):472–477. doi: 10.1097/COH.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 2.Moir S, Fauci AS. Insights into B cells and HIV-specific B-cell responses in HIV-infected individuals. Immunol Rev. 2013;254(1):207–224. doi: 10.1111/imr.12067. [DOI] [PubMed] [Google Scholar]
- 3.Amu S, Ruffin N, Rethi B, Chiodi F. Impairment of B-cell functions during HIV-1 infection. AIDS. 2013;27(15):2323–2334. doi: 10.1097/QAD.0b013e328361a427. [DOI] [PubMed] [Google Scholar]
- 4.Buckner CM, et al. Characterization of plasmablasts in the blood of HIV-infected viremic individuals: evidence for nonspecific immune activation. J Virol. 2013;87(10):5800–5811. doi: 10.1128/JVI.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83(8):3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malaspina A, et al. Appearance of immature/transitional B cells in HIV-infected individuals with advanced disease: correlation with increased IL-7. Proc Natl Acad Sci U S A. 2006;103(7):2262–2267. doi: 10.1073/pnas.0511094103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moir S, et al. B cells in early and chronic HIV infection: evidence for preservation of immune function associated with early initiation of antiretroviral therapy. Blood. 2010;116(25):5571–5579. doi: 10.1182/blood-2010-05-285528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188(9):1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tangye SG, Liu YJ, Aversa G, Phillips JH, de Vries JE. Identification of functional human splenic memory B cells by expression of CD148 and CD27. J Exp Med. 1998;188(9):1691–1703. doi: 10.1084/jem.188.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tangye SG, Ma CS, Brink R, Deenick EK. The good, the bad and the ugly — TFH cells in human health and disease. Nat Rev Immunol. 2013;13(6):412–426. doi: 10.1038/nri3447. [DOI] [PubMed] [Google Scholar]
- 11.Tangye SG, Tarlinton DM. Memory B cells: effectors of long-lived immune responses. Eur J Immunol. 2009;39(8):2065–2075. doi: 10.1002/eji.200939531. [DOI] [PubMed] [Google Scholar]
- 12.Tarlinton D, Good-Jacobson K. Diversity among memory B cells: origin, consequences, and utility. Science. 2013;341(6151):1205–1211. doi: 10.1126/science.1241146. [DOI] [PubMed] [Google Scholar]
- 13.Wu YC, Kipling D, Dunn-Walters DK. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front Immunol. 2011;2:e84610. doi: 10.3389/fimmu.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fecteau JF, Cote G, Neron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J Immunol. 2006;177(6):3728–3736. doi: 10.4049/jimmunol.177.6.3728. [DOI] [PubMed] [Google Scholar]
- 15.Kaminski DA, Wei C, Qian Y, Rosenberg AF, Sanz I. Advances in human B cell phenotypic profiling. Front Immunol. 2012;3:e84610. doi: 10.3389/fimmu.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrhardt GR, et al. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med. 2005;202(6):783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moir S, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205(8):1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wherry EJ, et al. Molecular signature of CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Isnardi I, et al. Complement receptor 2/CD21- human naive B cells contain mostly autoreactive unresponsive clones. Blood. 2010;115(24):5026–5036. doi: 10.1182/blood-2009-09-243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charles ED, et al. Clonal B cells in patients with hepatitis C virus-associated mixed cryoglobulinemia contain an expanded anergic CD21low B-cell subset. Blood. 2011;117(20):5425–5437. doi: 10.1182/blood-2010-10-312942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss GE, et al. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J Immunol. 2009;183(3):2176–2182. doi: 10.4049/jimmunol.0901297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakhmanov M, et al. Circulating CD21low B cells in common variable immunodeficiency resemble tissue homing, innate-like B cells. Proc Natl Acad Sci U S A. 2009;106(32):13451–13456. doi: 10.1073/pnas.0901984106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi H, Tanoue S, Kaplan DE. Peripheral CD27(–)CD21(–) B-cells represent an exhausted lymphocyte population in hepatitis C cirrhosis. Clin Immunol. 2014;150(2):184–191. doi: 10.1016/j.clim.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saadoun D, et al. Expansion of autoreactive unresponsive CD21–/low B cells in Sjogren’s syndrome-associated lymphoproliferation. Arthritis Rheum. 2013;65(4):1085–1096. doi: 10.1002/art.37828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castiello MC, et al. Wiskott-Aldrich Syndrome protein deficiency perturbs the homeostasis of B-cell compartment in humans. J Autoimmun. 2014;50:42–50. doi: 10.1016/j.jaut.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuehn HS, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345(6204):1623–1627. doi: 10.1126/science.1255904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubert D, et al. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat Med. 2014;20(12):1410–1416. doi: 10.1038/nm.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kardava L, et al. Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. J Clin Invest. 2014;124(7):3252–3262. doi: 10.1172/JCI74351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berkowska MA, et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 2011;118(8):2150–2158. doi: 10.1182/blood-2011-04-345579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med. 2007;204(3):645–655. doi: 10.1084/jem.20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kardava L, et al. Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. J Clin Invest. 2011;121(7):2614–2624. doi: 10.1172/JCI45685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509(7502):637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolhatkar NS, et al. Altered BCR and TLR signals promote enhanced positive selection of autoreactive transitional B cells in Wiskott-Aldrich syndrome. J Exp Med. 2015;212(10):1663–1677. doi: 10.1084/jem.20150585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pugh-Bernard AE, et al. Regulation of inherently autoreactive VH4-34 B cells in the maintenance of human B cell tolerance. J Clin Invest. 2001;108(7):1061–1070. doi: 10.1172/JCI12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobie JJ, et al. 9G4 autoreactivity is increased in HIV-infected patients and correlates with HIV broadly neutralizing serum activity. PLoS One. 2012;7(4):e84610. doi: 10.1371/journal.pone.0035356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundling C, et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med. 2012;4(142):e84610. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheid JF, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343(2):65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonsignori M, et al. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends Microbiol. 2012;20(11):532–539. doi: 10.1016/j.tim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch RM, et al. The development of CD4 binding site antibodies during HIV-1 infection. J Virol. 2012;86(14):7588–7595. doi: 10.1128/JVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 41.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254(1):225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol. 2006;80(3):1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dey B, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5(5):e84610. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467(7315):591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein F, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153(1):126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, et al. Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell. 2015;161(3):470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 51.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou T, et al. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell. 2015;161(6):1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binley JM, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82(23):11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker LM, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6(8):e84610. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muellenbeck MF, et al. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. J Exp Med. 2013;210(2):389–399. doi: 10.1084/jem.20121970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zinocker S, et al. The V gene repertoires of classical and atypical memory B cells in malaria-susceptible West African children. J Immunol. 2015;194(3):929–939. doi: 10.4049/jimmunol.1402168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss GE, et al. The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. PLoS Pathog. 2010;6(5):e84610. doi: 10.1371/journal.ppat.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Portugal S, et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. Elife. 2015:4. doi: 10.7554/eLife.07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perreau M, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210(1):143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cubas RA, et al. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nat Med. 2013;19(4):494–499. doi: 10.1038/nm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tangye SG. Advances in IL-21 biology-enhancing our understanding of human disease. Curr Opin Immunol. 2015;34:107–115. doi: 10.1016/j.coi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 63. Naradikian MS, et al. IL-4 and IL-21 reciprocally regulate T-BET expression in activated B cells. Presented at: Keystone Symposia: The Golden Anniversary of B Cell Discovery; March 23, 2015; Banff, Alberta, Canada. [Google Scholar]
- 64.Snapper CM, Paul WE. Interferon-γ and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236(4804):944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 65.Terrier B, et al. Expansion of functionally anergic CD21-/low marginal zone-like B cell clones in hepatitis C virus infection-related autoimmunity. J Immunol. 2011;187(12):6550–6563. doi: 10.4049/jimmunol.1102022. [DOI] [PubMed] [Google Scholar]
- 66.Kurosaki T, Kometani K, Ise W. Memory B cells. Nat Rev Immunol. 2015;15(3):149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- 67.Rubtsova K, Rubtsov AV, Cancro MP, Marrack P. Age-associated B cells: A T-bet-dependent effector with roles in protective and pathogenic immunity. J Immunol. 2015;195(5):1933–1937. doi: 10.4049/jimmunol.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao M, Prabakaran P, Chen W, Kessing B, Dimitrov DS. Deep sequencing and Circos analyses of antibody libraries reveal antigen-driven selection of Ig VH genes during HIV-1 infection. Exp Mol Pathol. 2013;95(3):357–363. doi: 10.1016/j.yexmp.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moir S, et al. CD40-Mediated induction of CD4 and CXCR4 on B lymphocytes correlates with restricted susceptibility to human immunodeficiency virus type 1 infection: potential role of B lymphocytes as a viral reservoir. J Virol. 1999;73(10):7972–7980. doi: 10.1128/jvi.73.10.7972-7980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moir S, et al. Humans with chronic granulomatous disease maintain humoral immunologic memory despite low frequencies of circulating memory B cells. Blood. 2012;120(24):4850–4858. doi: 10.1182/blood-2012-05-430959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 73.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montezuma-Rusca JM, et al. Bone marrow plasma cells are a primary source of serum HIV-1-specific antibodies in chronically infected individuals. J Immunol. 2015;194(6):2561–2568. doi: 10.4049/jimmunol.1402424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.