Abstract
The stress hormone ABA not only regulates stress response, but is also required for plant development and growth. Some evidences indicate that ABA plays a pivotal role in the ripening process of non climacteric as well as climacteric fruits. In a recent study, we showed that the tomato (Solanum lycopersicum) transcription factor SlZFP2 fine tunes ABA biosynthesis during fruit development through direct suppression of ABA biosynthetic genes and it also regulates fruit ripening through transcriptional suppression of the ripening regulator CNR. This indicates that SlZFP2 likely modulates the cross-talk between ABA and ethylene in regulation of fruit development and ripening in tomato. Gene expression analysis using ABA deficient mutants sit and flc as well as the SlZFP2 RNAi lines of high fruit ABA production showed that ethylene biosynthetic genes LeACS1A, LeACS1 and LeACO1 were positively regulated by ABA during early fruit growth. We reason that ABA promotes basal ethylene biosynthesis in system 1 during fruit growth and likely plays a minor role in ripening regulation after the onset of ripening process.
Keywords: fruit ripening, overexpression, RNA interference, S.lycopersicum, transcription factor
Abbreviations
- SlZFP2
Solanum lycopersicum zinc finger protein 2
- HA-SlZFP2
an expression cassette of hemagglutinin-SlZFP2 fusion protein
- ACS
1-aminocyclopropane-1-carboxylic acid synthase
- ACO
ACC oxidase
- CNR
COLORLESS NON-RIPENING
- NOT
NOTABILIS
- SIT
SITIENS
- FLC
FLACCA
- SlAO1
Solanum lycopersicum aldehyde oxidase 1
- ABA
abscisic acid
- dpa
days post anthesis
- RNAi
RNA interference
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads.
ABA is well known for its roles in seed maturation and germination, in addition to its pivotal roles in stress response.1 At transcriptional level, ABA biosynthesis is regulated by stresses and also developmental processes, for example, during fruit development.2,3 Recently, it has been hypothesized that ABA may be involved in ripening regulation of non-climacteric and climacteric fruits.4-6 Understanding the transcriptional regulation of ABA biosynthesis during fruit development is required to dissect the role of ABA in regulation of ripening process. In a recent publication, we characterized the role of the transcription factor SlZFP2, encoding a single C2H2 zinc finger protein, in tomato fruit development.7 In that study, we found that constitutive expression of HA-SlZFP2 under 35S promoter repressed ABA biosynthesis in leaves and fruits, whereas silencing its expression increased ABA production in young fruits at 5 and 10 dpa. We also revealed that SlZFP2 regulates fruit ripening through transcriptional repression of the ripening regulator CNR. Thus, the SlZFP2 pathway likely modulates crosstalk between ABA biosynthesis and the regulatory network of fruit ripening in tomato.
In tomato, there are two ABA peak levels during fruit development and ripening.2,6 ABA level is high in anthesis ovaries, and then declines rapidly after pollination.8,9 By monitoring the ABA contents in developing fruits, we also found ABA production decreases to relatively low level around 5 dpa, whereas during the cell expansion phase of fruit development ABA production resumes gradually and reaches its second highest level at mature green stage. Through biochemical and gene expression analysis, we have demonstrated that SlZFP2 suppresses ABA biosynthesis through direct binding to the promoters of the ABA biosynthetic genes NOT, FLC, SIT and SlAO1. Since SlZFP2 is mainly expressed during fruit development, it likely plays an important role in maintenance of the dynamic ABA production post pollination. Indeed, SlZFP2 expression negatively correlates with ABA level during fruit development, for example, SlZFP2 expression was relatively low in anthesis ovaries and 20 dpa fruits when high ABA production was observed (Fig. 1). Moreover, we found SlZFP2 expression was downregulated in the young fruits of ABA deficient mutants sit and flc, indicating that there is a feedback regulation on SlZFP2 expression by ABA during fruit development. Similarly, its Arabidopsis homolog AtZFP2 can be induced by ABA in seedlings.10 These results suggest that ABA activates SlZFP2 and the latter in turn represses ABA biosynthesis during fruit development.
Figure 1.
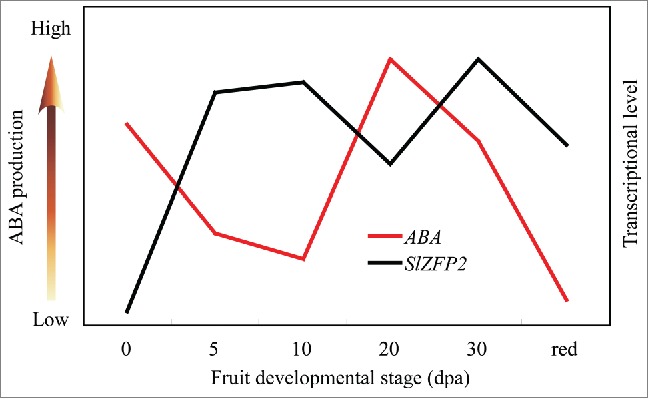
Changes of ABA production and SlZFP2 expression during fruit development and ripening. The figure was draw from our previous published data in (Weng et al. 2015).7 Transcript levels of SlZFP2 during fruit development and ripening were determined by qRT-PCR, and the maximal levels of SlZFP2 expression and ABA content were set at 1.
Besides its role in fine tuning ABA biosynthesis during fruit development, our study has also revealed that SlZFP2 regulates fruit ripening because overexpressing SlZFP2 or HA-SlZFP2 delayed fruit ripening for 5–7 days, whereas silencing its expression by RNAi accelerated fruit ripening. Fruit ripening in tomato is mainly mediated by ethylene, which its production is transcriptionally regulated by several transcription factors.11,12 Among those ripening regulators, CNR inhibits fruit ripening through AP2a mediated negative regulation of ethylene biosynthesis and signaling.13-15 In HA-SlZFP2 overexpression and RNAi lines, CNR was respectively repressed and upregulated during ripening process, demonstrating that SlZFP2 regulates ripening process through CNR pathway. Since downregulation of SlZFP2 led to elevated CNR expression in fruits as early as 15 dpa, SlZFP2 likely functions to prevent CNR expression before the onset of ripening process.
However, the action of SlZFP2 on fruit ripening is more likely through indirect impact on ethylene production because overexpression of this transcription factor only resulted in increased expression of ethylene biosynthetic genes LeACS6, LeACO1 and LeACO3 in ripe fruits at B10 stage (breaker plus 10 days). Their expression was not impacted at the onset of ripening process by overexpression or RNAi-mediated repression of SlZFP2. Thus, the gene expression analysis suggests that elevated or repressed ABA biosynthesis by manipulating SlZFP2 expression has little impact on ethylene production at the onset of ripening process.
Ethylene is the predominant plant hormone regulating climacteric-fruit ripening. In tomato, two systems of ethylene biosynthesis have been proposed, which basal ethylene production is maintained in system 1 during fruit growth and later its production is increased drastically in system 2 during ripening.4 In system 1, LeACS1A, LeACS6, LeACO1, 3 and 4 are responsible for the basal ethylene production.4,16-18 Apparently, SlZFP2 does not directly regulate the induction of ethylene biosynthesis in system 2. However, we found LeACO3 and LeACO4 expression was increased significantly in the 2 dpa fruits of the representative SlZFP2 RNAi line 207 through transcriptome analysis by RNA-seq. The other ethylene biosynthetic genes LeACS1A, LeACS2 and LeACO1, although the LeACS1A and LeACS2 were expressed at low levels, were also expressed at higher levels in the young fruits (Fig. 2A). This suggests that SlZFP2 may regulate ethylene biosynthesis in system 1. Thus, the problematic fruit set observed in these SlZFP2 RNAi lines can be explained by elevated ethylene biosynthesis. Our observation is consistent with early studies that ABA promotes flower and fruit abscission through ethylene biosynthesis.19,20 Given the significant increase in ABA content in SlZFP2 RNAi fruits, SlZFP2 likely regulates ethylene biosynthesis during early fruit growth through ABA pathway. To test the possibility, we analyzed the expression of ethylene biosynthetic genes in ABA deficient mutants sit and flc. We found that LeACO1 was downregulated in both the 5 and 10 dpa fruits of the 2 mutants; LeACS6 expression was also repressed at 5 dpa (Fig. 2B and C). The result further supports the role of ABA in promoting ethylene biosynthesis in system 1. Similarly, ABA inhibits root growth also through enhancing ethylene biosynthesis in Arabidopsis.21 Therefore, if there is an indispensible role for ABA in regulation of fruit ripening, it will possibly lie on its positive effect on basal ethylene production.
Figure 2.
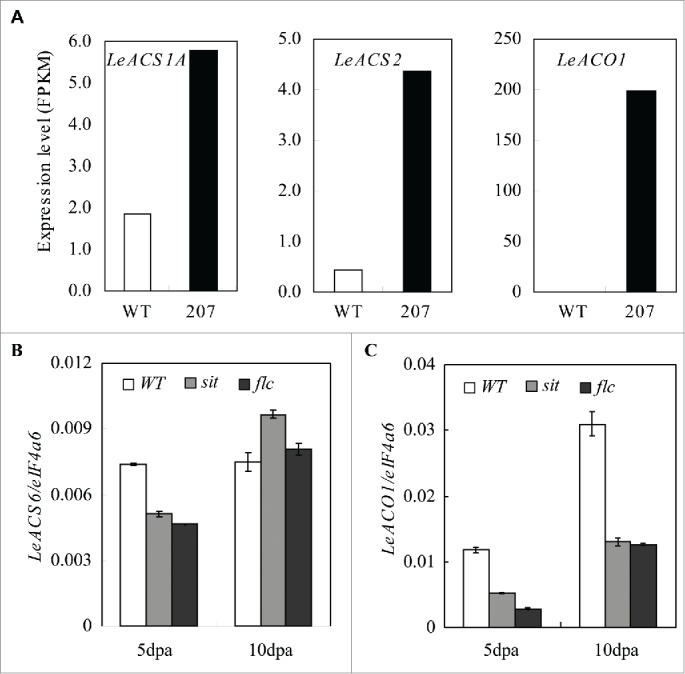
Expression of several ethylene biosynthetic genes in early growth fruits of ABA deficient and overproducing mutants Expression of LeACS1A, LeACS2 and LeACO1 (A) was upregulated in the 2 dpa fruits of the SlZFP2 RNAi line 207, whereas, LeACS6 (B) and LeACO1 (C) was downregulated in the 5 and 10 dpa fruits of the ABA deficient mutants sit and flc. Expression of LeACS6 and LeACO1 was determined by qRT-PCR in 3 biological replicates, and the error bars represent standard deviations of the means. The expression values in (A) were from our previous RNA-seq data (Weng et al 2015).7
Collectively, SlZFP2 plays at least two roles in regulation of fruit development and ripening (Fig. 3). First, SlZFP2, likely induced by high ABA at anthesis, represses ABA biosynthesis after anthesis, and in turn the decrease in ABA level limits ethylene production during fruit set and early fruit growth. Fine-tuning ABA biosynthesis likely helps to maintain ethylene production at its basal level in system 1 for normal fruit growth. Second, SlZFP2 also prevents CNR expression before the onset of ripening process. However, it remains to be determined whether or not the ABA biosynthesis regulated by SlZFP2 interconnects with the CNR-mediated ripening regulation.
Figure 3.
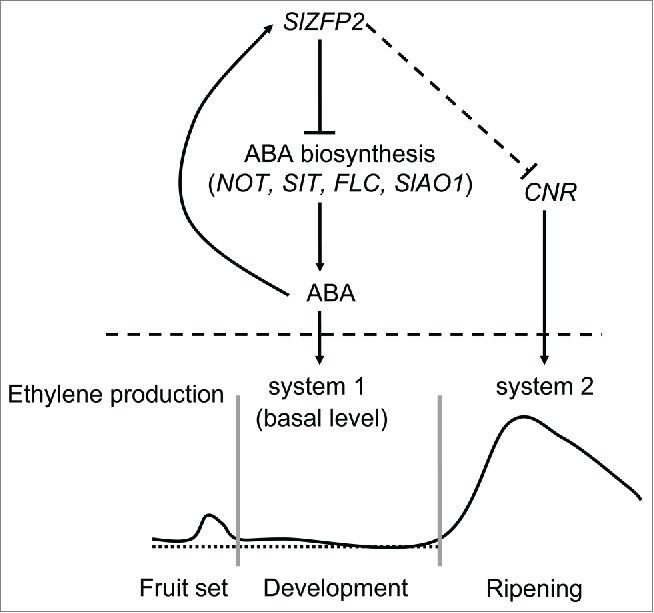
A proposed model for SlZFP2 action on fruit development and ripening During fruit set and development, SlZFP2 acts as a transcription repressor to fine tune ABA biosynthesis through direct binding to the promoters of NOT, SIT, FLC and SlAO1. Decreasing ABA biosynthesis by high SlZFP2 expression leads to relatively lower ethylene production which facilitates fruit set and prevents floral organ senescence. In addition, SlZFP2 also prevents the expression of the ripening regulator CNR before the onset of ripening process, either directly or indirectly.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The work was supported by grants 2012AA100105 and 2012CB113900 from MOST (to HX), grant 2009OHTP07 from CAS (to HX) and grant 31301777 from NSF (to WL).
References
- 1.Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. An update on abscisic acid signaling in plants and more. Mol Plant 2008; 1:198-217; PMID:19825533; http://dx.doi.org/ 10.1093/mp/ssm022 [DOI] [PubMed] [Google Scholar]
- 2.Buta JG, Spaulding DW. Changes in indole-3-acetic acid and abscisic acid levels during tomato (Lycopersicon esculentum Mill.) fruit development and ripening. J Plant Growth Regul 1994; 13:163-6; http://dx.doi.org/ 10.1007/BF00196382 [DOI] [Google Scholar]
- 3.Symons GM, Chua YJ, Ross JJ, Quittenden LJ, Davies NW, Reid JB. Hormonal changes during non-climacteric ripening in strawberry. J Exp Bot 2012; 63:4741-50; PMID:22791823; http://dx.doi.org/ 10.1093/jxb/ers147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar R, Khurana A, Sharma AK. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 2014; 65:4561-75; PMID:25028558; http://dx.doi.org/ 10.1093/jxb/eru277 [DOI] [PubMed] [Google Scholar]
- 5.Leng P, Yuan B, Guo Y. The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot 2014; 65:4577-88; PMID:24821949; http://dx.doi.org/ 10.1093/jxb/eru204 [DOI] [PubMed] [Google Scholar]
- 6.McAtee P, Karim S, Schaffer R, David K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci 2013; 4:79; PMID:23616786; http://dx.doi.org/ 10.3389/fpls.2013.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol 2015; 167:931-49; PMID:25637453; http://dx.doi.org/ 10.1104/pp.114.255174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitsch LM, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, Mariani C, Vriezen WH. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta 2009; 229:1335-46; PMID:19322584; http://dx.doi.org/ 10.1007/s00425-009-0913-7 [DOI] [PubMed] [Google Scholar]
- 9.Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 2008; 177:60-76; PMID:18028300 [DOI] [PubMed] [Google Scholar]
- 10.Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 2002; 115:4891-900; PMID:12432076; http://dx.doi.org/ 10.1242/jcs.00175 [DOI] [PubMed] [Google Scholar]
- 11.Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 2011; 45:41-59; PMID:22060040; http://dx.doi.org/ 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- 12.Seymour GB, Ostergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol 2013; 64:219-41; PMID:23394500; http://dx.doi.org/ 10.1146/annurev-arplant-050312-120057 [DOI] [PubMed] [Google Scholar]
- 13.Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA. Transcriptional control of fleshy fruit development and ripening. J Exp Bot 2014; 65:4527-41; PMID:25080453; http://dx.doi.org/ 10.1093/jxb/eru316 [DOI] [PubMed] [Google Scholar]
- 14.Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 2011; 23:923-41; PMID:21398570; http://dx.doi.org/ 10.1105/tpc.110.081273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 2006; 38:948-52; PMID:16832354; http://dx.doi.org/ 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- 16.Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J 1996; 9:525-35; PMID:8624515; http://dx.doi.org/ 10.1046/j.1365-313X.1996.09040525.x [DOI] [PubMed] [Google Scholar]
- 17.Barry CS, Llop-Tous MI, Grierson D. The regulation of 1-aminocyclopropane-1- carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 2000; 123:979-86; PMID:10889246; http://dx.doi.org/ 10.1104/pp.123.3.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van de Poel B, Bulens I, Markoula A, Hertog ML, Dreesen R, Wirtz M, Vandoninck S, Oppermann Y, Keulemans J, Hell R, et al.. Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant Physiol 2012; 160:1498-514; PMID:22977280; http://dx.doi.org/ 10.1104/pp.112.206086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cracker LE, Abeles FB. Abscission: role of abscisic Acid. Plant Physiol 1969; 44:1144-9; PMID:16657181; http://dx.doi.org/ 10.1104/pp.44.8.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riov J, Dagan E, Goren R, Yang SF. Characterization of abscisic Acid-induced ethylene production in citrus leaf and tomato fruit tissues. Plant Physiol 1990; 92:48-53; PMID:16667264; http://dx.doi.org/ 10.1104/pp.92.1.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Chen Z, Gao J, Gong Z. Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J 2014; 79:44-55; PMID:24738778; http://dx.doi.org/ 10.1111/tpj.12534 [DOI] [PubMed] [Google Scholar]


