Abstract
Porcine circovirus type 2 (PCV2) is the pivotal pathogen causing porcine circovirus-associated diseases. In this study, 62 PCV2 isolates were identified from seven farms in southern China from 2013 to 2015 and phylogenetic trees were reconstructed based on whole-genome sequences or the cap gene. In this investigation, PCV2b was the main genotype in circulation throughout these farms. Furthermore, an emerging mutant (PCV2b-1C), isolated from PCV2-vaccinated farms, was the predominant strain prevalent on these farms. In addition, we isolated a new cluster that may represent evolution of the virus through recombination of PCV2b-1A/1B and PCV2b-1C. Finally, we discuss evidence that antigenicity and surface structure variation of the capsid resulted from mutation of the C-terminal loop (Loop CT) of the PCV2b-1C Cap in silico.
Introduction
Porcine circoviruses (PCVs), the smallest, non-enveloped animal viruses of the genus Circovirus in the family Circoviridae, have a circular ssDNA genome (Cheung, 2006). There are two known main genotypes, porcine circovirus type 1 (PCV1) and PCV2, with high levels of nucleotide homology and a common genomic organization (Segalés et al., 2013). The first isolation of PCV1 was from a porcine kidney cell line (PK15; ATCC CCL-33) as a contaminant and non-pathogenic agent in swine (Tischer et al., 1986, 1974). Furthermore, PCV2 causes post-weaning multisystemic wasting syndrome, porcine dermatitis and nephropathy syndrome, respiratory distress, acute pulmonary oedema, and other PCV-associated diseases (PCVADs) (Chae, 2005; Segalés, 2012; Segalés et al., 2005), now present in every major swine-producing country and causing great economic losses in the global swine industry (Alarcon et al., 2013; Gillespie et al., 2009).
PCV2 has a circular genome of 1766–1768 nt (Wei et al., 2013) with three major ORFs. Whereas ORF1 encodes replication-associated proteins (Rep and Rep′) essential for replication of viral DNA (Cheung, 2003; Finsterbusch & Mankertz, 2009), ORF2 (cap gene) encodes the major structural protein of the capsid (molecular mass 27.8 kDa), the main antigenic determinant of the virus (Nawagitgul et al., 2000; Pogranichnyy et al., 2000). Finally, ORF3, embedded in ORF1, encodes a protein that is not essential for PCV2 replication, but has potential apoptotic activity (Juhan et al., 2010; Liu et al., 2005).
Based on genome sequence analysis of PCV2 isolates, PCV2 was divided into three main genotypes (PCV2a, PCV2b and PCV2c) with a threshold distance of 0.02 (Cortey et al., 2011; Grau-Roma et al., 2008). In addition, PCV2a was further subdivided into five clusters (2A, 2B, 2C, 2D and 2E) and PCV2b into three clusters (1A, 1B and 1C); PCV2c, which has only been reported in Denmark, has three isolates in GenBank (An et al., 2007; Olvera et al., 2007; Wang et al., 2013). Based on sequence analysis of the cap gene of PCV2 recovered from various countries, virus isolates have also been divided into three main genotypes (PCV2a, PCV2b and PCV2c) with a threshold distance of 0.035 (Cortey et al., 2011; Segalés et al., 2008), and PCV2b was subdivided into two clusters (1A/1B and 1C) (An et al., 2007; Cai et al., 2012). Previous studies indicated that PCV2 is continuously evolving through a series of point mutations in ORF2 and genome recombination between PCV2a and PCV2b (Cai et al., 2012; Mu et al., 2012).
Recently, PCV2b has become the predominant genotype worldwide (Beach & Meng, 2012; Trible & Rowland, 2012). Of note, a newly emerging mutant of PCV2b (designated PCV2b-1C) has been frequently isolated from PCV2-vaccinated farms in the USA, Korea and Germany (Eddicks et al., 2015; Opriessnig et al., 2013; Reiner et al., 2015; Seo et al., 2014; Xiao et al., 2012). An extra lysine present at the C terminus (CT) of PCV2 Cap is a unique feature of this mutant; furthermore, the presence of this additional amino acid increases virus titre in sera (Opriessnig et al., 2014b) and pathogenicity (Guo et al., 2012).
Infection with PCV2 is a huge burden to swine production, even in farms that vaccinate against the agent, throughout southern China – an important swine-breeding region. However, the genotype prevalence and genetic variation of PCV2 in this region from 2013 to 2015 remain unknown. Currently, there is no specific treatment available for PCV2 infection and PCVADs. Vaccination has proved to be the best way to prevent PCV2 infection in field. Thus, investigation of the antigens and capsid structure variations of the virus is critical to improve diagnosis, new vaccine design and our understanding of PCV2 pathogenesis. Therefore, the main objectives of this study were to characterize the prevalence, genetic variation and phylogenetic characteristics of PCV2 isolates in southern China from 2013 to 2015. In particular, alterations in antigenicity and surface structure of the capsid due to altered Cap amino acids are evaluated and discussed.
Results
Preparation of PCV2 genomic DNA and sequence analysis
Based on PCR, 62/160 (38.8 %) samples were apparently PCV2-positive; therefore, these 62 samples were subjected to genomic DNA amplification (Table S1, available in the online Supplementary Material) and sequence analysis. Based on complete genomic DNA sequence analysis, pairwise similarities of the 62 isolates ranged from 94.8 to 99.9 %.
Phylogenetic analysis of the 62 PCV2 isolates
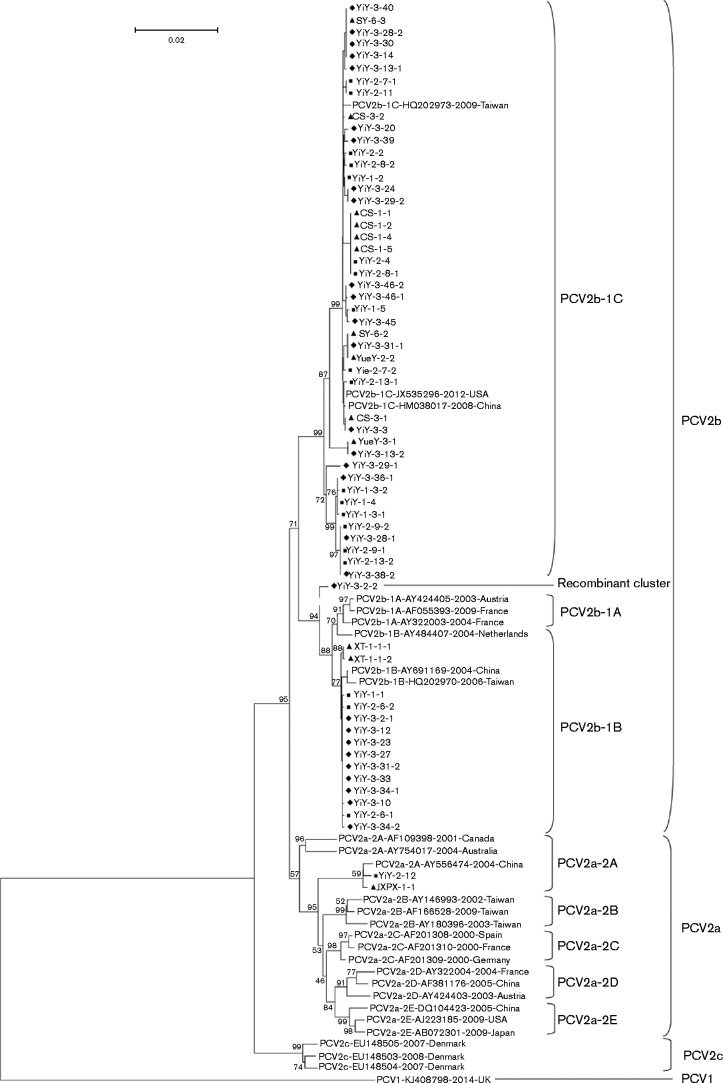
A phylogenetic tree was reconstructed using complete genomic DNA sequences of the 62 PCV2 isolates along with 27 PCV2 and one PCV1 reference strains deposited in GenBank (Fig. 1). Of the 62 PCV2 isolates, two and 59 readily fitted into the clades of PCV2a and PCV2b, respectively, indicating that PCV2b was the predominant genotype circulating in southern China from 2013 to 2015 in our investigation. Of the 59 PCV2b isolates, 23.7 % (14/59) belonged to cluster 1B and 76.3 % (45/59) belonged to cluster 1C, although no PCV2b isolate was obtained in cluster 1A (Fig. 1), suggesting that PCV2b-1C was an emerging predominant cluster. Notably, one isolate (YiY-3-2-2) was classified into a new cluster in a distinct branch located between PCV2b-1A/1B and PCV2b-1C within phylogenetic trees, based on either the complete genome (Fig. 1) or the cap gene (Fig. 2a), whereas this isolate was indistinguishable from the others in the PCV2b-1B cluster in the phylogenetic trees based on the rep gene (Fig. 2b). Therefore, we inferred that variations of the cap gene likely reflected evolution of PCV2. In addition, based on analysis of genomic sequences of all 62 PCV2 isolates, PCV2a had higher homology (95.7–96.2 %) with PCV2b-1A/1B when compared with PCV2b-1C (94.8–95.2 %). Furthermore, similarity between PCV2b-1A/1B and PCV2b-1C isolates ranged from 96.2 to 99.9 %, whereas homologies within PCV2b-1C and PCV2b-1A/1B clusters were 98.2–99.9 and 99.7–99.9 %, respectively (Table 1).
Fig. 1.
Phylogenetic tree of the 62 PCV2 isolates in our study with 28 reference strains from PCV1 (KJ408798), PCV2a (2A: AF109398, AY754017 and AY556474; 2B: AY180396, AY146993, AF166528; 2C: AF201308, AF201309, AF201310; 2D: AY322004, AY424403, AF381176; 2E: DQ104423, AJ223185, AB072301), PCV2b (1A: AY322003, AY424405, AF055393; 1B: AY484407, AY691169, HQ202970; 1C: HQ202973, HM038017, JX535296) and PCV2c (EU148503, EU148504, EU148505) deposited in GenBank. The phylogenetic tree was reconstructed based on complete genomic sequences of isolates described above. mega5.05 was used to reconstruct a neighbour-joining tree using a Kimura two-parameter model. Reliabilities of the tree were assessed (1000 bootstrap replications). ▴, 13 PCV2 isolates in 2013; ▪, 20 PCV2 isolates in 2014; ♦, 29 PCV2 isolates in 2015. Bar, 0.02 substitutions per site.
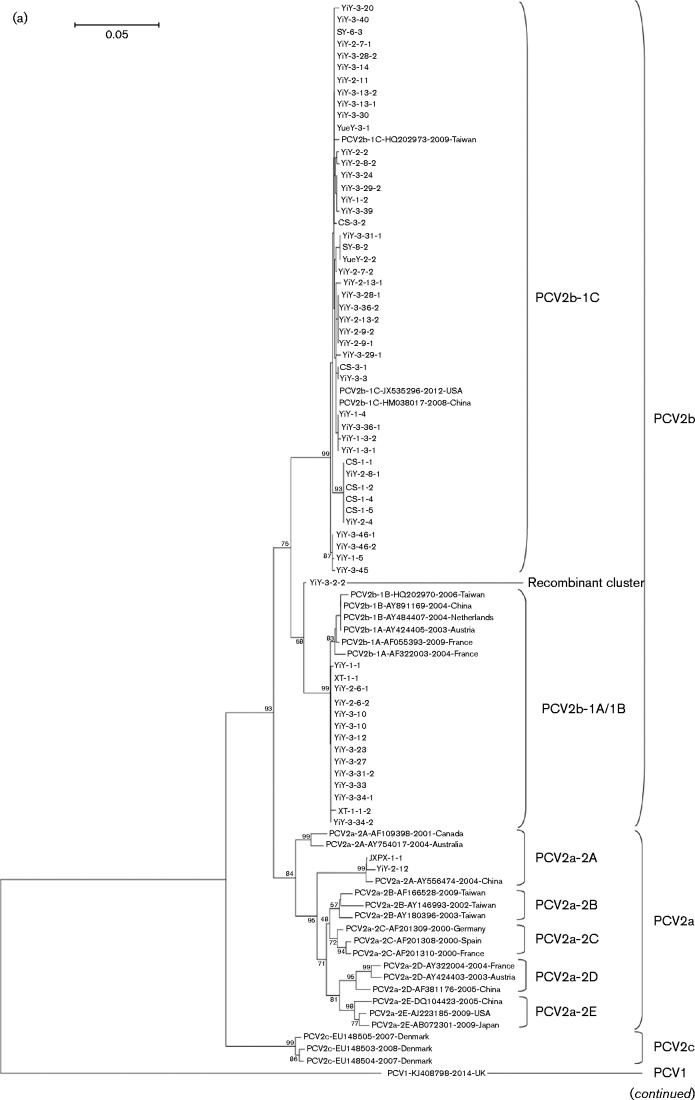
Fig. 2.
Phylogenetic trees of the 62 PCV2 isolates with 28 reference strains were reconstructed based on either the (a) cap or (b) rep genes. Bars, 0.05 (a) and 0.02 (b) substitutions per site.
Table 1. Homology of complete genomic sequence amongst our isolates and PCV2 reference strains.
| Strain (GenBank accession no.) | Identity of strains (%) | |||||
|---|---|---|---|---|---|---|
| LG (HM038034) (PCV2a) | Isolates in our study (PCV2a) | WuHan (FJ598044) (PCV2b-1A/1B) | Isolates in our study (PCV2b-1A/1B) | BDH (HM038017) (PCV2b-1C) | Isolates in our study (PCV2b-1C) | |
| LG (HM038034) (PCV2a) | 100 | 96.5 | 95.6 | 95.6–95.8 | 95.5 | 95.2–95.5 |
| Isolates in our study (PCV2a) | 99.5 | 95.6–95.7 | 95.7–96.2 | 94.9–95.0 | 94.8–95.2 | |
| WuHan (FJ598044) (PCV2b-1A/1B) | 100 | 99.1–99.3 | 96.1 | 96.0–97.1 | ||
| Isolates in our study (PCV2b-1A/1B) | 99.7–99.9 | 96.3–96.4 | 96.2–99.9 | |||
| BDH (HM038017) (PCV2b-1C) | 100 | 98.5–99.9 | ||||
| Isolates in our study (PCV2b-1C) | 98.2–99.9 | |||||
Recombination analysis
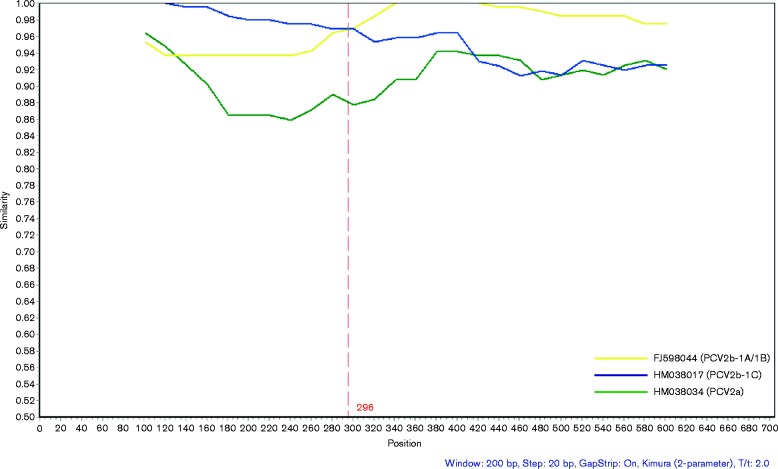
Based on phylogenetic analyses, the new isolate (YiY-3-2-2) was identified as a distinct cluster with bootstrap support values of >50 %, indicating evolution traces of the isolate in both phylogenetic trees were reliable (Figs 1 and 2a). As this isolate was located between PCV2b-1A/1B and PCV2b-1C within the phylogenetic tree, the cap gene of the isolate was compared with the strains from clusters of PCV2b-1A/1B (strain WH; GenBank accession number FJ598044), PCV2b-1C (strain rBDH; GenBank accession number HM038017) and PCV2a (strain LG; GenBank accession number HM038034), previously reported in China (Guo et al., 2012; Zhai et al., 2014). This recombination analysis revealed that a potential breakpoint might be located at position 296 (position in alignment of cap gene; Fig. 3). The new gene cluster (YiY-3-2-2) had higher sequence similarity to PCV2b-1C (HM038017) before the breakpoint and shared greater sequence similarity compared with PCV2b-1A/1B (FJ598044) after the breakpoint, albeit with a lower sequence similarity as compared with PCV2a (HM038034) (Fig. 3). Nucleotide sequences before the breakpoint of the isolate (YiY-3-2-2) had 99.0 and 94.9 % identity with PCV2b-1C (HM038017) and PCV2b-1A/1B (FJ598044), respectively, whereas nucleotide sequences after the breakpoint of the isolate had 98.8 and 94.3 % identity with PCV2b-1A/1B (FJ598044) and PCV2b-1C (HM038017). A similar result was observed when we randomly used three distinct strains of PCV2b-1A/1B (GenBank accession number HQ713495), PCV2b-1C (GenBank accession number JX535296) and PCV2a (GenBank accession number DQ397521), recovered from the USA as reference sequences for the recombination analysis of this isolate (YiY-3-2-2) (Fig. S1a). As controls, the cap genes of two new isolates (XT-1-1-1 and CS-1-1) in our study were substituted for the queries in the recombination analyses. The results indicated no recombination event in either of these two cases (Fig. S1b, c), despite both of the control query isolates showing high homology to the recombination isolate of YiY-3-2-2 (98.3 and 96 %). Therefore, we inferred that this isolate (YiY-3-2-2) was the product of a recombination event between the genomes of PCV2b-1A/1B and PCV2b-1C. Analysis of the breakpoint position at Cap indicated that the recombination event might create a novel surface combination between clusters 1C and 1A/1B on the capsid of the recombination cluster (Fig. S2). Three-dimensional structure analysis suggested that the variable amino acid residues were all distributed on the capsid surface (Fig. S2b). Before the breakpoint, amino acid residues of this recombination isolate exhibited 100 % identity to Cap of cluster 1C (Fig. S2a), and the distinct amino acid residues between the clusters 1C and 1A/1B were located in or close to the Loop BC-decorated fivefold axes on the capsid (green in Fig. S2b). However, after the breakpoint the amino acid residues of this recombination isolate were much closer to the traditional PCV2b strains (PCV2b-1A/1B) in which an extra lysine is absent at the CT of Cap (Fig. S2a).
Fig. 3.
The new cluster of YiY-3-2-2 was a product of genome recombination between PCV2b-1A/1B and PCV2b-1C. The cap gene similarity was compared between the cap gene of the new cluster (YiY-3-2-2) and those of PCV2 reference strains of PCV2b-1A/1B (GenBank accession number FJ598044), PCV2b-1C (GenBank accession number HM038017) and PCV2a (GenBank accession number HM038034) using SimPlot molecular recombination software. The y-axis indicates identity within a sliding window (200 bp wide) centred on the position plotted, with a step size between plots of 20 bp. GapStrip, on; Kimura (two-parameter); T/t = 2.0. The red vertical line indicates a potential breakpoint.
Analysis of Cap amino acid sequences
To investigate Cap amino acid variations amongst all 61 PCV2 isolates, these Cap amino acid sequences were aligned. The sequence conservation found amongst aligned amino acid sequences was 87.6–100 %. Seven conserved regions of Cap (each represented by ≥ 10 residues) were mapped at residues 1–33, 92–120, 135–150, 152–168, 170–189, 192–205 and 216–231, whereas variable amino acid residues were detected at positions 34, 37, 47, 53, 59–60, 63, 68, 77, 86, 88–91, 121, 131, 133–134, 151, 169, 190–191, 206, 210, 215 and 232–234 (Table 2). Of these, residues at positions 47, 53, 59–60, 169, 190–191, 206 and 232–234 were located at three conformational epitopes, recognized by neutralizing antibodies (Lekcharoensuk et al., 2004). Furthermore, 16 amino acid residues (47S, 59A, 60T/S, 63T, 77D, 86T, 88K, 89I, 91I, 131P, 133S/T, 151P, 190S, 191K, 206K and 232K) were identified to differentiate clusters of PCV2a from others (PCV2b-1A/1B and 1C); three and 11 amino acid residues were unique in the clusters of PCV2b-1A/1B (89R, 210E and 233S/P) and PCV2b-1C (34Y/H, 37H/R, 53I, 68N, 89L, 90T, 121T, 134N, 169R/G, 215I and 234K), respectively (Table 2).
Table 2. Variable amino acid residues in the putative capsid protein of ORF2 amongst 61 PCV2 isolates and 40 reference strains.
Of the 61 PCV2 isolates, only two isolates were PCV2a, 14 were PCV2b-1A/1B and 45 were PCV2b-1C.
| Position* | PCV2a | PCV2b-1A/1B | PCV2b-1C | |||
|---|---|---|---|---|---|---|
| Isolates in our study | Reference strains† | Isolates in our study | Reference strains‡ | Isolates in our study | Reference strains§ | |
| 34 | H | 1Q/24H | H | H | 6Y/39H | H |
| 37 | R | R | R | R | 1H/44R | R |
| 47 | S | 1S/4A/20T | T | T | T | T |
| 53 | F | 1I/24F | F | F | I | 3F/9I |
| 59 | A | 1R/24A | K | 1A/9R | K | 1R/2A/9K |
| 60 | 1S/1T | 2S/23T | T | T | T | 1S/11T |
| 63 | T | 5T/6S/14R | R | 2R/8K | R | 2S/10R |
| 68 | A | A | A | A | N | 3A/9N |
| 77 | D | 5N/20D | N | N | N | N |
| 86 | T | T | S | S | S | S |
| 88 | K | K | P | P | P | P |
| 89 | I | I | R | R | L | L |
| 90 | S | S | S | S | T | T |
| 91 | I | I | V | V | V | V |
| 121 | S | 10S/15T | S | S | T | T |
| 131 | P | 4I/9P/12T | T | T | T | T |
| 133 | 1S/1T | 6S/19A | A | A | A | A |
| 134 | T | 1P/24T | T | T | N | 2T/10N |
| 151 | P | 5T/20P | T | T | T | 3P/9T |
| 169 | S | 3G/22S | S | S | 16R/29G | R |
| 190 | S | 4T/21S | T | A | T | 2S/10T |
| 191 | K | 1K/4G/10R/10A | G | G | G | 2A/10G |
| 206 | K | 4I/21K | I | I | I | 3K/9I |
| 210 | D | D | E | E | D | D |
| 215 | V | V | V | V | I | I |
| 232 | K | 1N/24K | N | 1K/9N | N | 1K/11N |
| 233 | P | P | 1S/13P | P | P | P |
| 234 | – | – | – | – | K | K |
Amino acid positions in Cap. Bold indicates amino acid residues located in loops; otherwise, the amino acid residues are located in β-strands, except for amino acid residues 34 and 37 that are located in the nuclear localization signal domain.
PCV2a reference sequences: GenBank accession numbers AB072302, AF117753, AF109398, AY754017, AY556474 (2A); AY180396, AF364094, AY146993, HQ202944, AF166528 (2B); AY256455, AF201308, AF201309, AF201310, AY256459 (2C); AY322004, AF201306, AY424403, AY288135, AF381176 (2D); DQ104423, AY699793, AF264040, AJ223185, AB072301 (2E).
PCV2b-1A/1B reference sequences: GenBank accession numbers AY322003, AY424405, DQ141322, HQ202966, AF055393 (1A); AY484407, AY691169, AY556475, AY678532, HQ202970 (1B).
PCV2b-1C reference sequences: GenBank accession numbers HQ202972, HM038017, AY713470, AY484410, JX535296, HQ395022, HQ395025, HQ395029, HQ3950038, HQ3950043, HQ3950052, HQ3950061.
– , Dashes indicate amino acid residues at this position are absent among these PCV2 genotypes.
Effects of Loop CT variations/mutations on the surface structure and the antigenicity of the PCV2 capsid
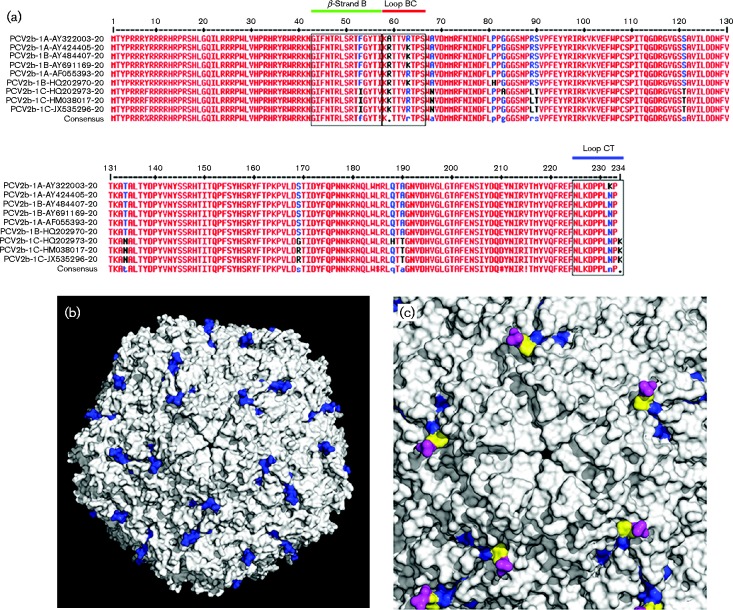
A typical feature of the newly emerging mutant of PCV2b-1C was demonstrated by the presence of an extra lysine at the CT of Cap (Guo et al., 2012). Based on the crystal structure of PCV2 Cap, the CT consisted of a loop, lacking a typical secondary structure; therefore, we designated this loop as Loop CT. In traditional PCV2b isolates, Loop CT contained nine amino acid residues (Fig. 4a). Based on the three-dimensional structure, Loop CT loops were exposed on the exterior surface after 60 Caps self-assembled into the capsid (blue; Fig. 4b). Notably, the last two amino acid residues (232NP233) of this loop were missed in the crystal structure, but it was clear that residues of 230PL231 at the end of Loop CT projected from the capsid surface (yellow and magenta; Fig. 4c) (Khayat et al., 2011). Therefore, we inferred the missing two amino acid residues (232NP233) should extend further from the capsid surface. Furthermore, the extra amino acid residue (234K), exclusively present at the end of Loop CT of PCV2b/1C, was also located on the capsid surface and could enhance/change surface patterns of the capsid as compared with traditional PCV2b isolates. Based on crystal structure analysis, we inferred that Loop CT (blue) was adjacent to and evenly distributed around the Loop BC-decorated fivefold axes (red) on the capsid surface, although it kept a distance from Loop BC and β-strand B (green) in both the primary and three-dimensional structures of the Cap monomer (Figs 4a and 5a, b). In the capsid, Loop CT arranged closely to Loop BC and β-strand B of neighbouring subunits (Fig. 5c), and some of the surface composed by β-strand B was below and partly obscured by Loop BC and Loop CT (Fig. 5d). Previous studies suggested Loop BC, Loop CT and β-strand B contained critical conformational epitopes, capable of being recognized by PCV2 neutralizing antibodies (Huang et al., 2011; Lekcharoensuk et al., 2004; Liu et al., 2013; Shang et al., 2009). Therefore, alteration of amino acid composition in Loop CT might have changed the antigenic epitope(s) of the PCV2 capsid.
Fig. 4.
Primary amino acid sequence alignment of PCV2b Cap and surface structure of PCV2 capsid. (a) Comparison of Cap amino acid residues amongst nine PCV2b reference isolates (GenBank accession numbers AY322003, AY424405, AF055393, AY484407, AY691169, HQ202970, HQ202973, HM038017 and JX535296). (b, c) The surface structure of a PCV2b capsid was generated based on the crystal structure of PCV2CS (Protein Data Bank ID: 3R0R; Khayat et al., 2011) and displayed using PyMOL. The amino acid residues of Loop CT are labelled in blue (b) and the two amino acid residues (230PL231) at the end of Loop CT are marked in magenta (c). Note that the last two amino acid residues (232NP233) of Cap were absent in the crystal structure.
Fig. 5.
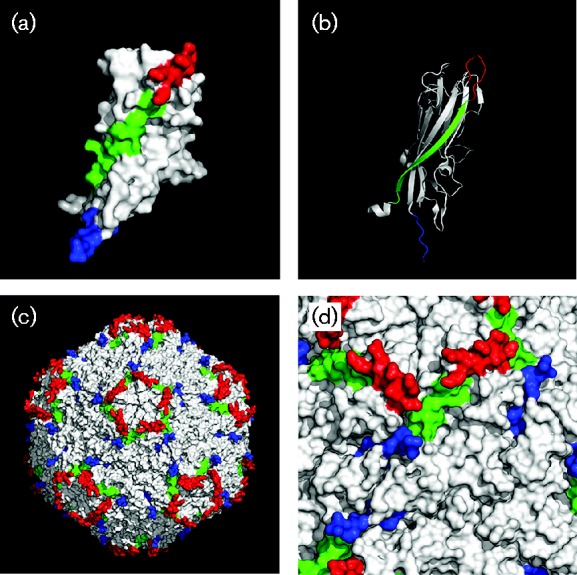
Localization and orientation of β-strand B, Loop BC and Loop CT in the crystal structures of both the PCV2 Cap monomer and capsid. Localization of β-strand B (green), Loop BC (red) and Loop CT (blue) in the Cap monomer is displayed in a (a) surface model or (b) a backbone model. After 60 Cap monomers self-assembled into a capsid, the orientations of β-strand B, Loop BC and Loop CT on the surface are displayed on (c) a global scale or (d) an enlarged scale. Note that the last two amino acid residues (232NP233) of Cap were absent in the crystal structure.
Discussion
PCV2b has become one of the prevalent genotypes worldwide over the last decade (de Castro et al., 2012; Manokaran et al., 2008; Pérez et al., 2011; Reiner et al., 2015; Wang et al., 2013; Wiederkehr et al., 2009). In this regard, a genotypic shift of PCV2 was reported from 2001 to 2011 in Taiwan and 77.1 % of isolates belonged to the PCV2b genotype (Wang et al., 2013). Wei et al. (2013) reported that 61/65 isolates fitted into the PCV2b genotype from 2011 to 2012 in southern China. Furthermore, cluster 1C (45/61) was the predominant cluster in that study (Wei et al., 2013).
Based on our investigation, we inferred that PCV2b was the main genotype circulating in southern China from 2013 to 2015, accounting for >95 % (59/62) of PCV2 isolates (Fig. 1). In particular, an emerging PCV2b-1C with an extra lysine present at the CT of Cap was the predominant genotype, accounting for 72.6 % of the positive samples in our study. This mutant has also been frequently isolated from PCV2-vaccinated farms in various countries (Eddicks et al., 2015; Opriessnig et al., 2013; Reiner et al., 2015; Seo et al., 2014; Xiao et al., 2012). Moreover, these PCV2b-1C isolates in our study do not exhibit 100 % genome identity to any strain deposited in GenBank, which strongly suggests PCV2 is still evolving within cluster 1C of PCV2. In our study, the homology (94.8–95.2 %) between PCV2b-1C isolates and PCV2a was less than the homology between PCV2b-1A/1B and PCV2a, which ranged from 95.7 to 96.2 % (Table 1). Thus, despite the current evidence suggesting that PCV2a-based vaccines can provide cross-protection from infection by PCV2b (Ma et al., 2007; Opriessnig et al., 2014a; Segalés, 2012), ongoing evolution of PCV2b poses a future challenge regarding efficacy of PCV2a-based commercial vaccines.
As a non-enveloped DNA virus, the PCV2 capsid is responsible for engagement of factors (clues) in host cells to translocate and release the viral genome to the correct site for DNA replication (Suomalainen & Greber, 2013). Therefore, alteration of the capsid may affect viral life cycle, antigenicity and neutralizing antibody recognition. Previous studies on epitope mapping of PCV2, along with the crystal structure analysis of Cap in our study, revealed Loop CT, Loop BC and β-strand B contained a few critical epitopes recognized by neutralizing antibodies (Huang et al., 2011; Lekcharoensuk et al., 2004; Liu et al., 2013; Shang et al., 2009). The amino acid residues (231LNP233) of Loop CT constitute a linear epitope and also participate in formation of a conformational epitope (Lekcharoensuk et al., 2004; Shang et al., 2009). Removal of the last three residues of Loop CT resulted in failed recognition by one neutralizing antibody to PCV2 capsid (Shang et al., 2009). The region from amino acid residue 47 to 57 of Cap was one of the conformational epitopes recognized by the neutralizing antibody (6H2) (Lekcharoensuk et al., 2004). Notably, replacement of Loop CT with the corresponding one from PCV1 Cap will weaken the recognition capacity of the neutralizing antibody against the PCV2 capsid (Lekcharoensuk et al., 2004), indicating the mutation in Loop CT had an adverse effect on neutralizing antibody binding to the conformational epitope. Structural analysis of PCV2 Cap demonstrated that the amino acid residues 47–57 form the first β-strand (B) in the viral jelly roll (Fig. 5b) (Khayat et al., 2011). We concluded the exterior surface constituted by β-strand B was located beneath Loop BC and Loop CT, and partly in the shadows of both loops (Fig. 5d). Therefore, variations in Loop BC or Loop CT could affect recognition of antibodies to the epitope(s) in β-strand B. Furthermore, an additional amino acid at the end of Loop CT may impair antibody recognition of the epitope by steric hindrance. Therefore, the extra lysine of PCV2 Cap may not only affect the linear epitope (231LNP233) recognized by antibodies, but also increase impediments of antibodies recognizing epitopes around Loop CT. In addition, the extra, positively charged lysine projecting from the capsid surface may enhance initial, non-specific interaction of the virus with negatively charged heparan sulphate or chondroitin sulphate B on the cell membrane during PCV2 infection. It was reported that heparan sulphate and chondroitin sulphate B are attachment receptors for PCV2 entry into its host cells, and that blocking the interaction of PCV2 with these receptors dramatically impairs PCV2 internalization (Misinzo et al., 2006). However, apparently no heparin-binding motif of PCV2 Cap has been reported, although analysis of the amino acid sequence of PCV2 Cap revealed a putative heparin-binding motif (98IRKVKV103) and the crystal structure indicates the motif is hidden inside the capsid. Recently, a PCV2 mutant with an additional lysine at the end of Loop CT had significantly more PCV2 DNA in sera compared with traditional PCV2b strains (Opriessnig et al., 2014b). Therefore, infectivity of this mutant may be stronger than traditional strains in vivo, although the association between infectivity and the extra lysine remains to be determined.
Five isolates were identified as a recombinant cluster that arose from an inter-genotypic recombination between PCV2a and PCV2b strains within ORF2 (Cai et al., 2012). The new isolate (YiY-3-2-2) in our study was present in a distinct branch located between PCV2b-1A/1B and PCV2b-1C in phylogenetic trees, based on either complete genomes (Fig. 1) or cap genes (Fig. 2a). Recombination assay revealed a new recombination event occurred between PCV2b-1A/1B and PCV2b-1C within ORF2, which yielded a novel PCV2 strain (Fig. 3). Interestingly, the recombination isolate forms a combinatorial surface pattern on its capsid since the surface regions around the icosahedral fivefold axes are the same as those of PCV2b-1C, whereas other parts are similar to the traditional PCV2b (PCV2b-1A/1B). In particular, the combination between the icosahedral fivefold axes and Loop CT on the capsid surface is novel. Therefore, the capsid of the recombination isolate displays a surface pattern around the icosahedral fivefold axes derived from PCV2b-1C along with a Loop CT-determining surface pattern from PCV2b-1A/1B. The potential effects of the new surface combination on antigenicity of the capsid remain to be determined. Generally, co-infection with distinct PCV2 isolates in the same pig is a precondition for genome recombination and two distinct isolates were recovered from one pig in our study (Table S1).
Alignment of Cap sequences amongst the 61 isolates with 40 reference strains deposited in GenBank revealed a few amino acid residues that were highly divergent (Table 2). Specifically, amino acids at positions 47, 53, 59–60, 169, 190–191, 206 and 232–234 were located at three putative conformational epitopes (Lekcharoensuk et al., 2004). Furthermore, amino acid residues at positions 59–60 participated in the formation of conformational neutralizing epitopes (Huang et al., 2011; Liu et al., 2013). Mutation of A59R in Cap of PCV2a (CL, LG and JF2) strains completely inhibited immunoreactivity of three PCV2a strains with mAb 8E4 (Huang et al., 2011), but mutations of R59A and A60T in Cap of the YJ strain increased neutralization and recognition by mAb 8E4 (Liu et al., 2013). Structural analysis demonstrated these amino acid residues were all located on the capsid surface (Loop BC); therefore, continuous mutations at these crucial positions may help the virus avoid the host immune system, thereby promoting virus propagation under field conditions.
In conclusion, the mutated PCV2b strain (PCV2b-1C) has been the predominant genotype in southern China from 2012 to 2015. In particular, based on three-dimensional structure and in silico analyses of antigenicity and surface structure variation of the capsid, we inferred that amino acid mutations in the capsid surface may have potential effects on antigenic patterns and neutralizing antibody recognition of the virus.
Methods
Sample collection, viral DNA extraction and PCV2 detection
Field samples were collected during 2013–2015 from seven commercial farms in southern China, including Jiangxi, Hunan, Jiangsu and Hubei provinces. Tissue samples (spleen, lymphoid node, lung, kidney and liver) (n = 160) from pigs diagnosed with doubtful PCVADs were tested by PCR. Viral genomic DNA was extracted from fresh or frozen tissue samples using a TIANamp Genomic DNA kit (Tiangen) following the manufacturer's protocol. All samples were screened for PCV2 by PCR using primer pairs pPCV2-cap-F-ZY009 and pPCV2-cap-R-ZY010 (Table S2). Thermal cycling conditions were 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 60 °C and 45 s at 72 °C, and a final elongation step of 10 min at 72 °C. Finally, PCR products were separated with 1 % agarose gel electrophoresis and positive samples confirmed based on the presence of DNA bands (∼780 bp).
PCV2 genomic DNA amplification and sequence determination
In total, 62 PCV2-positive DNA samples were used for PCV2 genomic amplification (Table S2) and sequence analysis. The PCV2 genomic DNAs were PCR-amplified using the primer pairs pPCV2-FL-F-KpnI-ZY001 and pPCV2-FL-R-HindIII-ZY002 (Table S2). Two restriction sites (KpnI and HindIII) were separately introduced into the 5′-terminal ends of the primers for genomic DNA, inserting into pSP72 vector (Promega) in order to obtain the PCV2 complete genome sequence. The PCR mixtures (50 μl) contained 25 μl 2 × EasyTaq PCR SuperMix (TransGen Biotech), 0.4 μM each primer and 100 ng extracted DNA. The PCR was performed in a thermocycler under the following conditions: 5 min at 94 °C, followed by 25 cycles of 30 s at 94 °C, 30 s at 60 °C and 2 min at 72 °C, and a final step of 10 min at 72 °C. Resultant PCR products were purified using a Gel Extraction kit (Omega) following the manufacturer's instructions, and then both the vector (pSP72) and insert of purified PCV2 genomic DNA were doubly digested with KpnI and HindIII at 37 °C for 1 h. After digestion, products were purified using a Cycle-Pure kit (Omega). The ligation mixture contained 5 U T4 DNA ligase (Thermo), 100 ng PCV2 genomic DNA fragment and 20 ng vector, and was incubated at 22 °C for 30 min. The ligation product was then transformed into Trans5α chemically competent cells (TransGen Biotech) and plasmid DNA extracted using a Plasmid DNA Mini kit (ProCell) according to the manufacturer's instructions. Positive plasmid-containing inserts were identified by restriction analysis with KpnI and HindIII. Finally, positive plasmids were sent for DNA sequencing at a commercial facility (GenScript) using universal primer T7 and two primers (pPCV2-FL-Seq001 and pPSP72-BSP6-Seq002 in Table S1). Resulting DNA fragments were assembled using dnaman software (Lynnon).
Bioinformatics analysis
A neighbour-joining tree was constructed using mega5.05 with a Kimura two-parameter model. Reliability of the neighbour-joining tree was calculated using 1000 bootstrap replicates. Sequence similarity was analysed and aligned with the clustal w method of the MegAlign program of dnastar version 7.10 (Lasergene). PCV2 cap genes of the putative recombinant isolate and reference strains were further analysed with SimPlot version 3.5.1 recombination analysis software to determine nucleotide identity.
Three-dimensional mapping of amino acids in PCV2 virus-like particles
Three-dimensional structures of the PCV2 capsid and Cap were displayed with PyMOL version 1.7.4.4 (www.pymol.org). Structure data for the capsid and Cap were downloaded from the Protein Data Bank (ID: 3R0R; www.rcsb.org) (Khayat et al., 2011).
Supplementary Data
Supplementary Data
References
- Alarcon P., Rushton J., Wieland B. (2013). Cost of post-weaning multi-systemic wasting syndrome and porcine circovirus type-2 subclinical infection in England – an economic disease model Prev Vet Med 110 88–102 10.1016/j.prevetmed.2013.02.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D. J., Roh I. S., Song D. S., Park C. K., Park B. K. (2007). Phylogenetic characterization of porcine circovirus type 2 in PMWS and PDNS Korean pigs between 1999 and 2006 Virus Res 129 115–122 10.1016/j.virusres.2007.06.024 . [DOI] [PubMed] [Google Scholar]
- Beach N. M., Meng X. J. (2012). Efficacy and future prospects of commercially available and experimental vaccines against porcine circovirus type 2 (PCV2) Virus Res 164 33–42 10.1016/j.virusres.2011.09.041 . [DOI] [PubMed] [Google Scholar]
- Cai L., Ni J., Xia Y., Zi Z., Ning K., Qiu P., Li X., Wang B., Liu Q., other authors (2012). Identification of an emerging recombinant cluster in porcine circovirus type 2 Virus Res 165 95–102 10.1016/j.virusres.2012.01.008 . [DOI] [PubMed] [Google Scholar]
- Chae C. (2005). A review of porcine circovirus 2-associated syndromes and diseases Vet J 169 326–336 10.1016/j.tvjl.2004.01.012 . [DOI] [PubMed] [Google Scholar]
- Cheung A. K. (2003). The essential and nonessential transcription units for viral protein synthesis and DNA replication of porcine circovirus type 2 Virology 313 452–459 10.1016/S0042-6822(03)00373-8 . [DOI] [PubMed] [Google Scholar]
- Cheung A. K. (2006). Rolling-circle replication of an animal circovirus genome in a theta-replicating bacterial plasmid in Escherichia coli J Virol 80 8686–8694 10.1128/JVI.00655-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortey M., Olvera A., Grau-Roma L., Segalés J. (2011). Further comments on porcine circovirus type 2 (PCV2) genotype definition and nomenclature Vet Microbiol 149 522–523 10.1016/j.vetmic.2010.11.009 . [DOI] [PubMed] [Google Scholar]
- de Castro A. M., Cruz T. F., Salgado V. R., Kanashiro T. M., Ferrari K. L., Araujo J. P., Jr, Brandão P. E., Richtzenhain L. J. (2012). Detection of porcine circovirus genotypes 2a and 2b in aborted foetuses from infected swine herds in the State of São Paulo, Brazil Acta Vet Scand 54 29 10.1186/1751-0147-54-29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddicks M., Fux R., Szikora F., Eddicks L., Majzoub-Altweck M., Hermanns W., Sutter G., Palzer A., Banholzer E., Ritzmann M. (2015). Detection of a new cluster of porcine circovirus type 2b strains in domestic pigs in Germany Vet Microbiol 176 337–343 10.1016/j.vetmic.2015.01.013 . [DOI] [PubMed] [Google Scholar]
- Finsterbusch T., Mankertz A. (2009). Porcine circoviruses – small but powerful Virus Res 143 177–183 10.1016/j.virusres.2009.02.009 . [DOI] [PubMed] [Google Scholar]
- Gillespie J., Opriessnig T., Meng X. J., Pelzer K., Buechner-Maxwell V. (2009). Porcine circovirus type 2 and porcine circovirus-associated disease J Vet Intern Med 23 1151–1163 10.1111/j.1939-1676.2009.0389.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Roma L., Crisci E., Sibila M., López-Soria S., Nofrarias M., Cortey M., Fraile L., Olvera A., Segalés J. (2008). A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence Vet Microbiol 128 23–35 10.1016/j.vetmic.2007.09.007 . [DOI] [PubMed] [Google Scholar]
- Guo L., Fu Y., Wang Y., Lu Y., Wei Y., Tang Q., Fan P., Liu J., Zhang L., other authors (2012). A porcine circovirus type 2 (PCV2) mutant with 234 amino acids in capsid protein showed more virulence in vivo, compared with classical PCV2a/b strain PLoS One 7 e41463 10.1371/journal.pone.0041463 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. P., Lu Y. H., Wei Y. W., Guo L. J., Liu C. M. (2011). Identification of one critical amino acid that determines a conformational neutralizing epitope in the capsid protein of porcine circovirus type 2 BMC Microbiol 11 188 10.1186/1471-2180-11-188 . [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Juhan N. M., LeRoith T., Opriessnig T., Meng X. J. (2010). The open reading frame 3 (ORF3) of porcine circovirus type 2 (PCV2) is dispensable for virus infection but evidence of reduced pathogenicity is limited in pigs infected by an ORF3-null PCV2 mutant Virus Res 147 60–66 10.1016/j.virusres.2009.10.007 . [DOI] [PubMed] [Google Scholar]
- Khayat R., Brunn N., Speir J. A., Hardham J. M., Ankenbauer R. G., Schneemann A., Johnson J. E. (2011). The 2.3-angstrom structure of porcine circovirus 2 J Virol 85 7856–7862 10.1128/JVI.00737-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekcharoensuk P., Morozov I., Paul P. S., Thangthumniyom N., Wajjawalku W., Meng X. J. (2004). Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2 J Virol 78 8135–8145 10.1128/JVI.78.15.8135-8145.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chen I., Kwang J. (2005). Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis J Virol 79 8262–8274 10.1128/JVI.79.13.8262-8274.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Huang L., Wei Y., Tang Q., Liu D., Wang Y., Li S., Guo L., Wu H., Liu C. (2013). Amino acid mutations in the capsid protein produce novel porcine circovirus type 2 neutralizing epitopes Vet Microbiol 165 260–267 10.1016/j.vetmic.2013.03.013 . [DOI] [PubMed] [Google Scholar]
- Ma C. M., Hon C. C., Lam T. Y., Li V. Y., Wong C. K., de Oliveira T., Leung F. C. (2007). Evidence for recombination in natural populations of porcine circovirus type 2 in Hong Kong and mainland China J Gen Virol 88 1733–1737 10.1099/vir.0.82629-0 . [DOI] [PubMed] [Google Scholar]
- Manokaran G., Lin Y. N., Soh M. L., Lim E. A., Lim C. W., Tan B. H. (2008). Detection of porcine circovirus type 2 in pigs imported from Indonesia Vet Microbiol 132 165–170 10.1016/j.vetmic.2008.04.021 . [DOI] [PubMed] [Google Scholar]
- Misinzo G., Delputte P. L., Meerts P., Lefebvre D. J., Nauwynck H. J. (2006). Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells J Virol 80 3487–3494 10.1128/JVI.80.7.3487-3494.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu C., Yang Q., Zhang Y., Zhou Y., Zhang J., Martin D. P., Xia P., Cui B. (2012). Genetic variation and phylogenetic analysis of porcine circovirus type 2 infections in central China Virus Genes 45 463–473 10.1007/s11262-012-0789-7 . [DOI] [PubMed] [Google Scholar]
- Nawagitgul P., Morozov I., Bolin S. R., Harms P. A., Sorden S. D., Paul P. S. (2000). Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein J Gen Virol 81 2281–2287 10.1099/0022-1317-81-9-2281 . [DOI] [PubMed] [Google Scholar]
- Olvera A., Cortey M., Segalés J. (2007). Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality Virology 357 175–185 10.1016/j.virol.2006.07.047 . [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Xiao C. T., Gerber P. F., Halbur P. G. (2013). Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2 Vet Microbiol 163 177–183 10.1016/j.vetmic.2012.12.019 . [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Gerber P. F., Xiao C. T., Mogler M., Halbur P. G. (2014a). A commercial vaccine based on PCV2a and an experimental vaccine based on a variant mPCV2b are both effective in protecting pigs against challenge with a 2013 U.S. variant mPCV2b strain Vaccine 32 230–237 10.1016/j.vaccine.2013.11.010 . [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Xiao C. T., Gerber P. F., Halbur P. G., Matzinger S. R., Meng X. J. (2014b). Mutant USA strain of porcine circovirus type 2 (mPCV2) exhibits similar virulence to the classical PCV2a and PCV2b strains in caesarean-derived, colostrum-deprived pigs J Gen Virol 95 2495–2503 10.1099/vir.0.066423-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez L. J., de Arce H. D., Cortey M., Domínguez P., Percedo M. I., Perera C. L., Tarradas J., Frías M. T., Segalés J., other authors (2011). Phylogenetic networks to study the origin and evolution of porcine circovirus type 2 (PCV2) in Cuba Vet Microbiol 151 245–254 10.1016/j.vetmic.2011.03.022 . [DOI] [PubMed] [Google Scholar]
- Pogranichnyy R. M., Yoon K. J., Harms P. A., Swenson S. L., Zimmerman J. J., Sorden S. D. (2000). Characterization of immune response of young pigs to porcine circovirus type 2 infection Viral Immunol 13 143–153 10.1089/vim.2000.13.143 . [DOI] [PubMed] [Google Scholar]
- Reiner G., Hofmeister R., Willems H. (2015). Genetic variability of porcine circovirus 2 (PCV2) field isolates from vaccinated and non-vaccinated pig herds in Germany Vet Microbiol 180 41–48 10.1016/j.vetmic.2015.07.026 . [DOI] [PubMed] [Google Scholar]
- Segalés J. (2012). Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis Virus Res 164 10–19 10.1016/j.virusres.2011.10.007 . [DOI] [PubMed] [Google Scholar]
- Segalés J., Allan G. M., Domingo M. (2005). Porcine circovirus diseases Anim Health Res Rev 6 119–142 10.1079/AHR2005106 . [DOI] [PubMed] [Google Scholar]
- Segalés J., Olvera A., Grau-Roma L., Charreyre C., Nauwynck H., Larsen L., Dupont K., McCullough K., Ellis J., other authors (2008). PCV-2 genotype definition and nomenclature Vet Rec 162 867–868 10.1136/vr.162.26.867 . [DOI] [PubMed] [Google Scholar]
- Segalés J., Kekarainen T., Cortey M. (2013). The natural history of porcine circovirus type 2: from an inoffensive virus to a devastating swine disease? Vet Microbiol 165 13–20 10.1016/j.vetmic.2012.12.033 . [DOI] [PubMed] [Google Scholar]
- Seo H. W., Park C., Kang I., Choi K., Jeong J., Park S. J., Chae C. (2014). Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea Arch Virol 159 3107–3111 10.1007/s00705-014-2164-6 . [DOI] [PubMed] [Google Scholar]
- Shang S. B., Jin Y. L., Jiang X. T., Zhou J. Y., Zhang X., Xing G., He J. L., Yan Y. (2009). Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2 Mol Immunol 46 327–334 10.1016/j.molimm.2008.10.028 . [DOI] [PubMed] [Google Scholar]
- Suomalainen M., Greber U. F. (2013). Uncoating of non-enveloped viruses Curr Opin Virol 3 27–33 10.1016/j.coviro.2012.12.004 . [DOI] [PubMed] [Google Scholar]
- Tischer I., Rasch R., Tochtermann G. (1974). Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines Zentralbl Bakteriol Orig A 226 153–167 . [PubMed] [Google Scholar]
- Tischer I., Mields W., Wolff D., Vagt M., Griem W. (1986). Studies on epidemiology and pathogenicity of porcine circovirus Arch Virol 91 271–276 10.1007/BF01314286 . [DOI] [PubMed] [Google Scholar]
- Trible B. R., Rowland R. R. (2012). Genetic variation of porcine circovirus type 2 (PCV2) and its relevance to vaccination, pathogenesis and diagnosis Virus Res 164 68–77 10.1016/j.virusres.2011.11.018 . [DOI] [PubMed] [Google Scholar]
- Wang C., Pang V. F., Lee F., Huang T. S., Lee S. H., Lin Y. J., Lin Y. L., Lai S. S., Jeng C. R. (2013). Prevalence and genetic variation of porcine circovirus type 2 in Taiwan from 2001 to 2011 Res Vet Sci 94 789–795 10.1016/j.rvsc.2012.11.019 . [DOI] [PubMed] [Google Scholar]
- Wei C., Zhang M., Chen Y., Xie J., Huang Z., Zhu W., Xu T., Cao Z., Zhou P., other authors (2013). Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012 Infect Genet Evol 17 87–92 10.1016/j.meegid.2013.03.041 . [DOI] [PubMed] [Google Scholar]
- Wiederkehr D. D., Sydler T., Buergi E., Haessig M., Zimmermann D., Pospischil A., Brugnera E., Sidler X. (2009). A new emerging genotype subgroup within PCV-2b dominates the PMWS epizooty in Switzerland Vet Microbiol 136 27–35 10.1016/j.vetmic.2008.10.028 . [DOI] [PubMed] [Google Scholar]
- Xiao C. T., Halbur P. G., Opriessnig T. (2012). Complete genome sequence of a novel porcine circovirus type 2b variant present in cases of vaccine failures in the United States J Virol 86 12469 10.1128/JVI.02345-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai S. L., Chen S. N., Xu Z. H., Tang M. H., Wang F. G., Li X. J., Sun B. B., Deng S. F., Hu J., other authors (2014). Porcine circovirus type 2 in China: an update on and insights to its prevalence and control Virol J 11 88 10.1186/1743-422X-11-88 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data