Abstract
Reactivation of latent human cytomegalovirus is a significant infectious complication of organ transplantation and current therapies target viral replication once reactivation of latent virus has already occurred. The specific molecular pathways that activate viral gene expression in response to transplantation are not well understood. Our studies aim to identify these factors, with the goal of developing novel therapies that prevent transcriptional reactivation in transplant recipients. Murine cytomegalovirus (MCMV) is a valuable model for studying latency and reactivation of CMV in vivo. We previously demonstrated that transplantation of MCMV-latently infected kidneys into allogeneic recipients induces reactivation of immediate early (IE) gene expression and epigenetic reprogramming of the major immediate early promoter (MIEP) within 48 h. We hypothesize that these events are mediated by activation of signalling pathways that lead to binding of transcription factors to the MIEP, including AP-1 and NF-κB. Here we show that transplantation induces rapid activation of several members of the AP-1 and NF-κB transcription factor family and we demonstrate that canonical NF-κB (p65/p50), the junD component of AP-1, and nucleosome remodelling complexes are recruited to the MIEP following transplantation. Proteomic analysis of recipient plasma and transcriptome analysis of kidney RNA identified five extracellular ligands, including TNF, IL-1β, IL-18, CD40L and IL-6, and three intracellular signalling pathways associated with reactivation of IE gene expression. Identification of the factors that mediate activation of these signalling pathways may eventually lead to new therapies to prevent reactivation of CMV and its sequelae.
Introduction
Human cytomegalovirus (HCMV) is an opportunistic herpesvirus that has the ability to establish a lifelong latent infection and to reactivate. Reactivation of HCMV in transplant recipients is associated with CMV disease, increased risk of acute and chronic allograft rejection, infection with other opportunistic pathogens, graft failure and death (Razonable et al., 2013). Due to the high prevalence of latent infection, more than 75 % of solid organ transplant recipients experience reactivation of latent HCMV (Fishman et al., 2007). Although effective antiviral drugs have reduced the incidence of CMV-related post-transplant complications, use of these drugs is limited by their toxicity and the emergence of resistant strains (Razonable et al., 2013). Thus, a greater understanding of the molecular events controlling reactivation from latency is needed to develop alternative strategies to prevent CMV disease.
Reactivation of HCMV has been associated clinically with conditions that generate an inflammatory milieu, including allograft rejection, sepsis and acute illness (Cook et al., 1998; Döcke et al., 1994; Fietze et al., 1994; Grattan et al., 1989; Heininger et al., 2001; Hibberd et al., 1992; Kalil & Florescu, 2009; Kutza et al., 1998; Lao et al., 1997; Limaye et al., 2008; Mutimer et al., 1997; Portela et al., 1995; Razonable et al., 2001; Reinke et al., 1994a, 1994b) and the inflammatory cytokine TNF has been proposed to drive reactivation through NF-κB-mediated activation of the major immediate early promoter (MIEP) (Döcke et al., 1994; Fietze et al., 1994; Prösch et al., 1995; Stein et al., 1993). However, due to the species specificity of HCMV, it has not been possible to test this hypothesis in the context of organ transplantation. Murine cytomegalovirus (MCMV) is very similar to HCMV in many important ways, including: (i) ability to establish latency and to reactivate; (ii) hierarchical regulation of viral gene expression; (iii) structure, function and organization of the major immediate early (IE) genes (Keil et al., 1987a, b; Meier & Stinski, 2013; Stenberg et al., 1984); (iv) repression of IE gene expression during latency by heterochromatinization of viral genomes (Grzimek et al., 2001; Hummel & Abecassis, 2002; Hummel et al., 2001; Kurz et al., 1999; Liu et al., 2008; 2010; Reeves & Sinclair, 2013; Reeves et al., 2005; Seckert et al., 2013); (v) the presence of similar regulatory elements (e.g. NF-κB, AP-1, Sp1 and CREB/ATF binding sites) in the viral IE enhancers (Liu et al., 2013; Meier & Stinski, 2013); and (vi) (re)activation of the MIEP and/or IE transcription in response to inflammatory mediators or allogeneic stimulation (Döcke et al., 1994; Fietze et al., 1994; Huang et al., 2012; Hummel & Abecassis, 2002; Hummel et al., 2001; Kew et al., 2014; Lee et al., 2004; Liu et al., 2013; O'Connor & Murphy, 2012; Prösch et al., 1995; Reeves & Compton, 2011; Simon et al., 2005; Stein et al., 1993).
Because the IE genes are the first set of genes expressed in lytic infection (Mocarski et al., 2007), and are required to activate the viral replication programme, transcriptional reactivation of IE gene expression is likely a key event in reactivation of the virus. We previously developed a renal transplant model for reactivation of MCMV IE gene expression (Hummel et al., 2001). In this model, transplantation of latently infected kidneys into allogeneic recipients induced IE gene expression and epigenetic reprogramming of the MIEP within 48 h (Hummel et al., 2001; Liu et al., 2013). In addition, allogeneic transplantation induced expression of a lacZ reporter transgene under the control of the HCMV MIEP (MIEP-lacZ) (Hummel et al., 2001). Although TNF was sufficient to induce both MCMV IE gene expression and the HCMV MIEP-lacZ reporter, it was not required to activate IE gene expression in response to allogeneic transplantation in either of these models (Zhang et al., 2008, 2009). We therefore hypothesized that multiple factors may contribute to reactivation of IE gene expression in the complex environment of an allogeneic transplant. The goal of the present study is to identify additional candidates that may contribute to reactivation of IE gene expression in allogeneic transplants, with the long-term goal of developing novel therapeutic approaches to preventing reactivation of the virus. In addition to the TNF/NF-κB and IL-6/mitogen-activated protein kinase (MAPK) signalling pathways, which have been previously implicated in reactivation of HCMV (Döcke et al., 1994; Fietze et al., 1994; Hargett & Shenk, 2010; Huang et al., 2012; Kew et al., 2014; O'Connor & Murphy, 2012; Prösch et al., 1995; Reeves & Compton, 2011; Stein et al., 1993), we have identified novel signalling pathways that have not to our knowledge been previously associated with reactivation of HCMV. Our results suggest that it may be necessary to target multiple signalling pathways to prevent transcriptional reactivation of CMV.
Results
Multiple NF-κB and AP-1 family members are rapidly activated by renal transplantation
The MCMV MIEP is strikingly enriched in NF-κB and AP-1 binding sites (Dorsch-Häsler et al., 1985; Seckert et al., 2013). NF-κB consists of homo- and heterodimeric complexes of p65/RelA, p50, p52, c-rel and RelB subunits (reviewed by Oeckinghaus et al., 2011). Canonical NF-κB (p65/p50) activates transcription of immune response genes in response to inflammatory mediators, including TNF, IL-1β, Toll-like receptor ligands and oxidative stress. Non-canonical NF-κB (p52/RelB) is activated by CD40L, BAFF and lymphotoxin beta. AP-1 is a dimeric complex of Fos, Jun, Maf and ATF family members. Different complexes of this diverse family have varying roles in cellular proliferation, inflammatory immune response and oxidative stress (Eferl & Wagner, 2003; Hernandez et al., 2008; Karin, 1995; Karin & Shaulian, 2001; Shaulian & Karin, 2002).
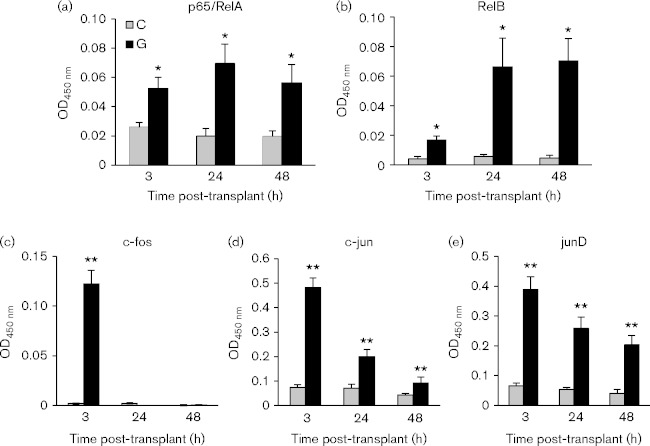
We previously demonstrated that NF-κB p50/p65 and the junD component of AP-1 were activated within 48 h after renal transplantation and that transcription factor activation correlated with reactivation of IE expression (Hummel et al., 2001; Zhang et al., 2008). Here, we have expanded our analysis to include earlier times and additional NF-κB and AP-1 family members (Fig. 1). Latently infected kidneys were transplanted into allogeneic recipients and analysed at 3, 24 or 48 h. Multiple members of the AP-1 family were rapidly activated by transplantation but they differed in the kinetics of subsequent inactivation. c-Fos was strongly, but transiently, activated; junD was also strongly activated at 3 h, and this activation was sustained through 48 h, albeit at lower levels; c-jun was rapidly activated, but was no longer active at 48 h. No activation of JunB, FosB, Fra-1 or Fra-2 was observed at any time point (data not shown). Both the p65/RelA and RelB subunits of NF-κB were activated within 3 h and remained activated at 24 and 48 h, but RelB activity increased between 3 and 24 h, while p65 activity remained constant.
Fig. 1.
Transcription factor activation induced by allogeneic transplantation. Kidneys from latently infected BALB/c mice were transplanted into allogeneic C57BL/6 recipients and harvested 3, 24 or 48 h post-transplant (G), as indicated on the x-axis. The contralateral latent donor kidney (C) was harvested at the time of the transplant as a matching Day 0 control. Nuclear extracts were prepared and analysed by TransAm assays, n = 5/time point. Results are expressed as the mean plus standard error. **P < 0.01; *P < 0.05.
NF-κB and AP-1 family members are differentially regulated in allogeneic transplants
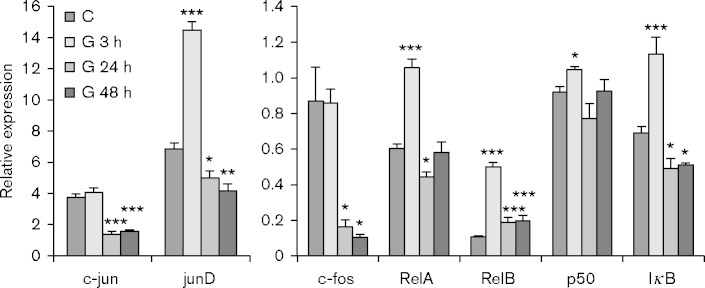
Transcription factor activity is regulated at multiple levels, including RNA transcription and stability, as well as protein localization, stability and post-translational modification. To gain insight into the mechanisms controlling changes in activity of NF-κB and AP-1 family members, we analysed expression of RNAs encoding these proteins (Fig. 2). The results show that expression of p65/RelA was upregulated at 3 h post-transplant, then fell below control levels at 24 h and returned to basal levels by 48 h. Expression of RelB peaked at 3 h, but in contrast to p65/RelA, its expression remained elevated relative to controls at 24 and 48 h. These results suggest that the increased activity of NF-κB family members observed in allogeneic transplants (Fig. 1) may be due in part to increased RNA expression. NF-κB activity is controlled by a complex regulatory circuit (Ruland, 2011). IκBalpha is an inhibitory subunit that sequesters the active p65/p50 complex in the cytoplasm and its expression is induced by NF-κB as part of a negative feedback loop to prevent damage associated with prolonged expression of inflammatory genes controlled by NF-κB. Consistent with these observations, IκB alpha expression was rapidly, but transiently, induced by transplantation (Fig. 2).
Fig. 2.
Expression of transcription factor RNAs in control and kidney allografts. RNA expression was analysed in control (C) kidneys and in grafts harvested 3 (G 3 h), 24 (G 24 h) or 48 h (G 48 h) after transplant and normalized to hypoxanthine phosphoribosyltransferase (HPRT) RNA. Results are expressed as the mean plus standard error. P values were determined by comparison of expression in the graft to pooled controls. ***P < 0.001; **P < 0.01; *P < 0.05; n = 4.
Expression of AP-1 family members was also differentially regulated in allogeneic transplants (Fig. 2). Expression of c-jun and c-fos RNAs were unchanged at 3 h, but were sharply downregulated 24 and 48 h after transplant. The transient increase in activity of c-jun and c-fos at 3 h (Fig. 1) was therefore due to post-transcriptional regulation. In contrast, expression of junD was significantly induced at 3 h post-transplant, and subsequently downregulated.
NF-κB and AP-1 are recruited to the MIEP concomitant with reactivation of IE gene expression
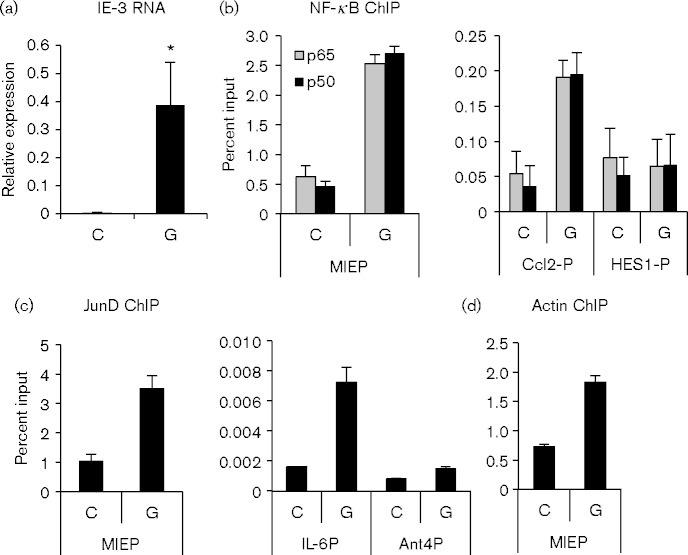
The MCMV MIEP controls expression of two differentially spliced genes, IE-1 and IE-3. IE-3 is the MCMV functional homologue of HCMV IE-2, which is the major transactivator of early gene expression (Angulo et al., 2000; Keil et al., 1987a, 1987b; Martínez et al., 2010; Messerle et al., 1992). We previously showed that allogeneic transplantation induced reactivation of IE gene expression 48 h after transplant, and that RNA polymerase phosphorylated at Ser2 (Pol II pS2) was recruited to both the MIEP and the IE-3 coding region (Hummel et al., 2001; Liu et al., 2013). Here, we show that IE-3 RNA expression was detectable by reverse transcriptase-qPCR at 48 h, but not at 3 or 24 h post-transplant (Fig. 3a and data not shown). To determine whether reactivation of IE gene expression was associated with recruitment of NF-κB and AP-1 to the MIEP, latently infected kidneys were transplanted into allogenic recipients and analysed at 48 h by chromatin immunoprecipitation (ChIP) analyses. We analysed binding of histones, Pol II pS2, NF-κB/RelA and p50 and the junD component of AP-1 to the MIEP. Analysis of Pol II and histone modification was published separately (Liu et al., 2013). These results showed that allogeneic transplantation induced major changes in viral chromatin, including recruitment of pS2-Pol II to the MIEP and the downstream IE-3 exon 5, loss of repressive histone marks and gain of activating marks.
Fig. 3.
Reactivation of IE gene expression correlates with recruitment of transcription factors and remodelling of the MIEP. (a) Expression of IE-3 in matching control (C) and transplanted (G) kidneys 48 h after transplant. MCMV IE-3 RNA expression was normalized to expression of cellular Eef2; n = 5. Error bars show the mean plus standard error. *P < 0.05. (b) ChIP analysis of p65/RelA and p50 NF-κB subunits bound to the MCMV MIEP (left panel) and to the promoters of the cellular Ccl2 and Hes1 genes (right panel) in pooled chromatin from control (C) or 48 h allografts (G). (c) Binding of junD to the MIEP (left panel) and to the promoters of the IL-6 and Ant4 genes (right panel). (d) Binding of actin to the MIEP. The data shown are representative of at least two independent assays, except NF-κB p50 (n = 1). Error bars show sd of technical replicates analysed in triplicate.
Our previous studies showed activation of NF-κB in response to transplantation by analysing binding to oligonucleotides containing consensus NF-κB binding sites (Fig. 1 and Hummel et al., 2001; Zhang et al., 2008). Activation of the transcription factor does not necessarily result in binding to the gene of interest. Here, we show that NF-κB p50 and p65/RelA are in fact bound to the viral MIEP 48 h after transplantation into allogeneic recipients (Fig. 3b). As controls, we show that NF-κB is also bound to the NF-κB-responsive CCL2 promoter (Teferedegne et al., 2006; Wolter et al., 2008), but not to the NF-κB-deficient Hes-1 promoter. In addition, junD was recruited to the MIEP and to the junD-responsive IL-6 promoter (Baccam et al., 2003; Mann et al., 2002; Ndlovu et al., 2009; Smart et al., 2001; Viedt et al., 2002; Zerbini et al., 2003), but not the transcriptionally silent Ant4 promoter (Fig. 3c).
Chromatin remodelling complexes are recruited to the MIEP upon reactivation of IE gene expression
MCMV genomes are highly enriched in histones in latently infected mice, suggesting that the nucleosomes are in a compacted configuration, which is closed to transcription (Liu et al., 2008). Many inducible genes require remodelling of the chromatin by the SWI/SNF complex in order to activate gene expression (Ramirez-Carrozzi et al., 2009). SWI/SNF complexes contain common subunits, including actin, and one of two ATPases, Brahma (BRM) or Brahma-related gene 1 (BRG-1), which confer gene specificity (Kadam & Emerson, 2003; Mohrmann & Verrijzer, 2005). AP-1 family members, including c-jun and c-fos, interact with BRG-1. We therefore analysed recruitment of actin and BRG-1 to the MIEP; there was insufficient material to analyse BRM. Increased binding of actin (Fig. 3d), but not BRG-1 (data not shown), to the MIEP was observed in 48 h allografts. These data indicate that chromatin remodellers may be recruited to the MIEP, although the specific complexes remain to be identified.
Collectively, Figs 1–3 show that allogeneic transplantation induces transcriptional reactivation of IE gene expression and that this correlates with changes in expression, activation and binding of NF-κB and AP-1 transcription factors to the MIEP. In order to develop new strategies that may prevent reactivation of latent CMV, it is important to understand how these transcription factors are activated in the context of transplantation.
Inflammatory mediators are elevated in recipient plasma
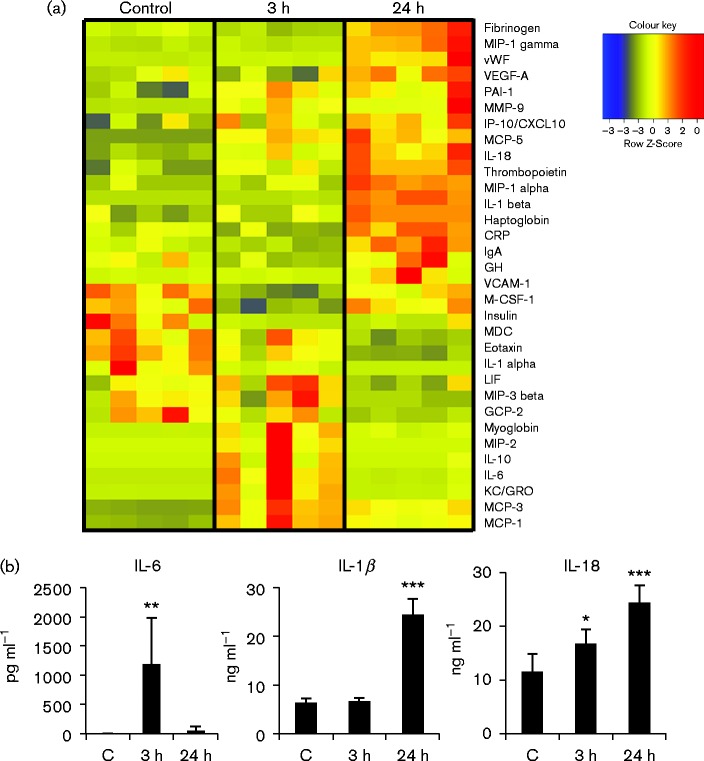
Allogeneic transplantation activates both innate and adaptive inflammatory immune responses at early times post-transplant (Einecke et al., 2005; El-Sawy et al., 2004; Famulski et al., 2007, 2006; Hummel et al., 2009). Transcriptional reactivation of IE gene expression and remodelling of viral chromatin may be mediated by activation of inflammatory signalling pathways induced by interaction of ligands in the plasma, which are elevated in response to allogeneic transplantation, with receptors expressed on cells of the kidney. To identify potential mediators of reactivation, we analysed plasma taken from recipient mice at the time of sacrifice for biomarkers of inflammation. These results showed that several inflammatory cytokines, including IL-1β, IL-18 and IL-6, were elevated relative to controls (Fig. 4). Plasma levels of IL-6 protein were strongly elevated at 3 h, and fell at 24 h (Fig. 4a, b), but were elevated relative to controls at 48 h (Fig. S1, available in the online Supplementary Material). Plasma levels of IL-1 and IL-18 rose more gradually (Fig. 4), but were also elevated relative to controls at 48 h (Fig. S1).
Fig. 4.
Allogeneic transplantation induces release of inflammatory proteins into the plasma. (a) Heat map of differentially regulated plasma proteins in control (C) and recipient mice at 3 and 24 h after transplant, n = 5/group. (b) Graphical representation of the plasma protein values of selected cytokines. Results are expressed as the mean plus sd. ***P < 0.001, **P < 0.01, *P < 0.05.
Inflammatory signalling pathways are activated in renal allografts
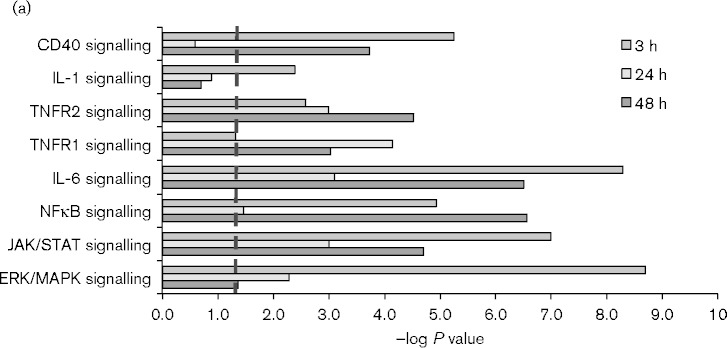
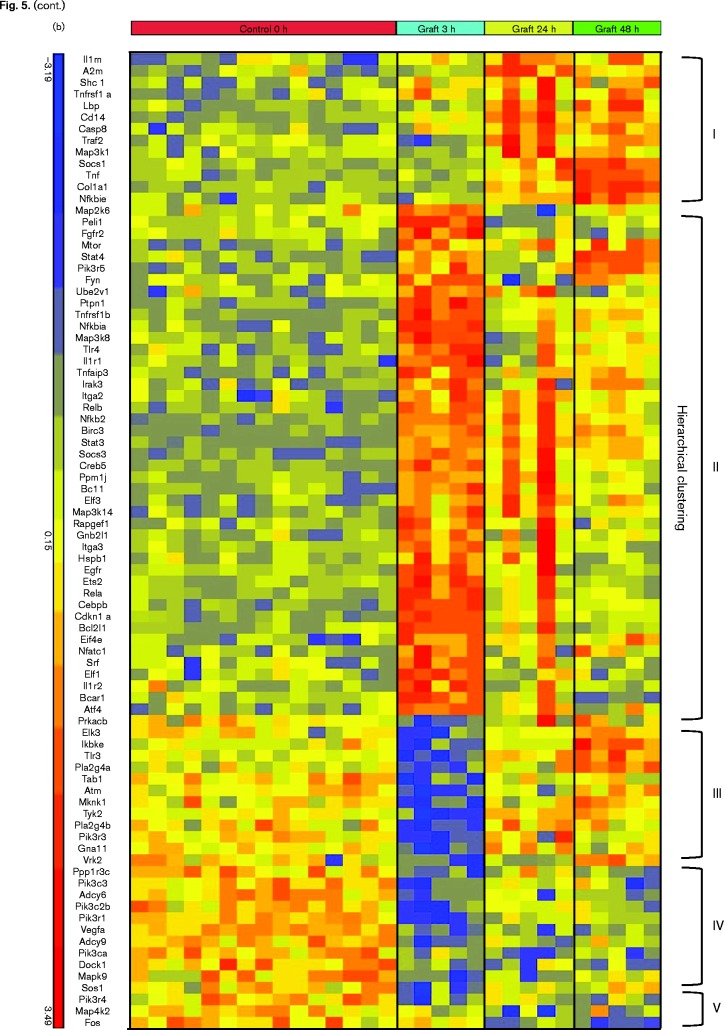
To further investigate the response to allogeneic transplantation, we analysed global changes in gene expression within the transplanted kidney by microarray and Ingenuity Pathway Analysis. As controls, we analysed RNA from the contralateral donor kidneys removed at the time of the transplant. The results (Fig. 5a) showed that TNFR1, TNFR2, IL-1, CD40 and IL-6 signalling, as well as their downstream effector pathways, including NF-κB, JAK/STAT and ERK/MAPK, were differentially regulated at multiple time points. The heat map showing expression of the individual members of these pathways (Fig. 5b) illustrates the complex and rapidly evolving nature of the inflammatory response induced by allogeneic transplantation. Five patterns of expression were apparent in the genes in these signalling pathways: genes in Cluster I were induced 24–48 h after transplant; genes in Cluster II were induced at 3 h and subsequently downregulated; genes in Cluster III were rapidly downregulated and returned to near baseline levels after 24 h; genes in Cluster IV were rapidly downregulated and recovered expression more slowly, while genes in Cluster V were slowly downregulated and remained below baseline at 48 h. Mapk9/JNK2, which phosphorylates and activates c-jun (Jaeschke et al., 2006; Gupta et al., 1996), was among the genes rapidly downregulated at 3 h. Genes induced at 3 h included genes involved in both positive (Tnfrsf1b, IL1R1, TLR4, p65/RelA, Nfkb2/p100, RelB, Map3k14/NIK, IRAK, Ube2v1, Peli1, Bcl10) and negative (Nfkbia/IκB alpha, Tnfaip3/A20) regulation of the NF-κB signalling pathway (Oeckinghaus et al., 2011; Ruland, 2011). Increased expression of these negative regulators may account for the decreased expression of some genes in this pathway observed at 24–48 h. Positive (NF-IL6/CEBPβ, STAT3) and negative (SOCS3, Map2k6) regulators of IL-6 signalling (Akira et al., 1990; Bode et al., 2001; Heinrich et al., 2003) were also strongly upregulated at 3 h. IL-6 expression itself did not meet the threshold for statistical significance in the microarray analysis, due to sample variability, but IL-6 expression was significantly induced in the same RNA samples when analysed by the more quantitative method of RT-qPCR (Fig. 6). Upregulation of TNF expression in response to allotransplantation was also confirmed by RT-qPCR (Fig. 6). In contrast to IL-6, which was upregulated rapidly and transiently, TNF expression increased gradually from 3 to 48 h.
Fig. 5.
Dynamic changes in inflammatory signalling pathways are induced by allogeneic transplantation. (a) Ingenuity Pathway Analysis of selected signalling pathways upregulated by allogeneic transplantation at 3, 24 and 48 h. Pathways with –log P value >1.3 (dashed line) are statistically significant. (b) Heat map of RNA expression of genes within these pathways determined by microarray analysis.
Fig. 6.
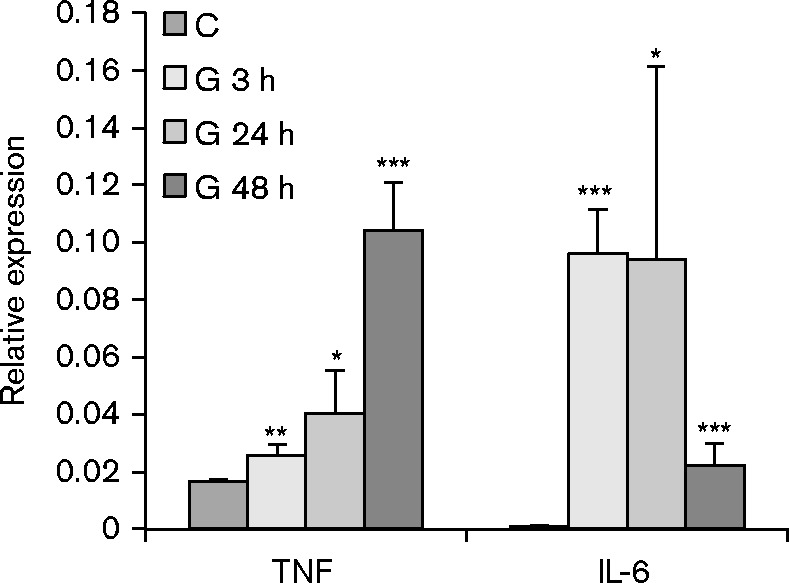
Expression of TNF and IL-6 is induced in kidney allografts. RNA expression was analysed in control (C) kidneys and in grafts harvested 3 (G 3 h), 24 (G 24 h) or 48 h (G 48 h) after transplant and normalized to HPRT RNA. Results are expressed as the mean plus standard error. P values were determined by comparison of expression in the graft to pooled controls. ***P < 0.001; **P < 0.01; *P < 0.05; n = 4.
IL-6 and IL-1β proteins were also elevated in plasma of recipient mice (Fig. 4), indicating a good correspondence between the presence of the ligand in the plasma and the response of the cells in the kidney. CD40 was not included among the plasma analytes, and thus, a correlation between the ligand and receptor signalling could not be determined for this factor. Despite increased expression of TNF RNA in the allografts (Fig. 6) and pathway analysis showing upregulation of TNFR1 and TNFR2 signalling (Fig. 5), TNF protein was not detected in the plasma above threshold levels. This may be due to rapid turnover of TNF protein (plasma t1/2 = 6–7 min) (Beutler et al., 1985), or to dilution of the protein in recipient plasma.
Discussion
CMV genomes are heterochromatinized and the IE genes, which are required for lytic replication, are transcriptionally silent in latent infection (Grzimek et al., 2001; Hummel et al., 2001; Kurz et al., 1997, 1999; Liu et al., 2008, 2010; Seckert et al., 2013; Simon et al., 2005). Activation of IE gene expression, through recruitment of transcription factors that control MIEP activity and remodelling of viral chromatin, is therefore likely required for reactivation of the virus. The MIEP has multiple potential binding sites for AP-1 and NF-κB (Hummel et al., 2001; Seckert et al., 2013). Here, we show that several members of these transcription factor families are rapidly activated and bound to the MIEP following allogeneic transplantation.
Interestingly, individual family members showed dynamic changes in the kinetics of activation/inactivation, as well as changes in RNA expression. c-fos was rapidly and transiently activated following transplantation (Fig. 1). c-fos is an unstable protein whose expression and activity is regulated at multiple levels (Sasaki et al., 2006). Activation of c-jun was also rapidly induced, but in contrast to c-fos, retained some activity relative to controls at 24 h. Loss of c-jun activity may be due in part to the rapid downregulation of Mapk9/JNK2 (Fig. 5b), which activates and phosphorylates c-jun (Jaeschke et al., 2006). In addition, expression of c-jun/c-fos RNA was strongly downregulated by 48 h (Fig. 2). This result is consistent with their roles as early response genes whose RNAs are rapidly expressed in response to stimuli and quickly degraded due to recognition of an AU-rich RNA decay element in the 3′ UTR (Chen & Shyu, 1995). While c-jun and c-fos are well-known oncoproteins with roles in cellular proliferation, junD is atypical of other AP-1 family members in both regulation of gene expression and in biological function (Hernandez et al., 2008). JunD is activated by oxidative stress, including renal ischaemia/reperfusion injury (Kim et al., 2005) and protects cells against apoptosis through upregulation of genes that mitigate oxidative damage (Gerald et al., 2004; Hernandez et al., 2008; Lamb et al., 2003; Pillebout et al., 2003; Tsuji, 2005). As with c-jun and c-fos, junD was rapidly activated by transplantation, but, in contrast to these proteins, junD activity was sustained for 48 h (Fig. 1 and Hummel et al., 2001; Zhang et al., 2008) and downregulation of the RNA relative to controls was less dramatic.
Both p65/RelA and RelB NF-κB family members were rapidly activated by allogeneic transplantation, and remained active at 48 h (Fig. 1), despite downregulation of the RNAs and increased expression of negative regulatory molecules, including Nfkbia (IκBalpha) at 3 h and Nfkbie at 48 h (Figs 2 and 5b). While many studies have focused on canonical signalling pathways leading to activation of p65/RelA, non-canonical pathways leading to activation of RelB have not to our knowledge been previously implicated in reactivation of CMV. The relative roles of different NF-κB family members in regulation of the MIEP requires further investigation. Collectively, the data in Figs 1, 2 and 5 show that MIEP-regulatory transcription factor expression and activity following transplant is regulated by complex and dynamic processes, which are controlled both transcriptionally and post-transcriptionally.
We previously demonstrated that allogeneic transplantation induces transcriptional activation of IE gene expression, and epigenetic reprogramming of MCMV chromatin, including a switch from heterochromatic to euchromatic histones and recruitment of RNA polymerase II to the MIEP (Hummel et al., 2001; Liu et al., 2013). Here we show that transcription factors thought to be important in regulation of IE gene expression, including the canonical NF-κB subunits p50 and p65/RelA and the junD component of AP-1, are bound to the MIEP in response to allogeneic transplantation. To our knowledge, this is the first demonstration that these transcription factors are not only activated by stimuli that induce reactivation; they are in fact recruited to the viral DNA concomitant with reactivation of IE gene expression. Though not definitive, these correlative results strongly suggest that these factors play an important role in activating expression of the IE genes.
In this respect, the MCMV MIEP is similar to other viral enhancers, which mimic inflammatory immune response genes (Kropp et al., 2014). Cellular inflammatory response genes have been divided into two classes, primary and secondary, based on their requirement for new protein synthesis (Hargreaves et al., 2009; Medzhitov & Horng, 2009; Ramirez-Carrozzi et al., 2009, 2006). Primary response genes are rapidly inducible, due to constitutive binding of RNA polymerase to nucleosome-free promoters, and elongation of transcription is the rate-determining step in activating expression of these genes. Conversely, the promoter regions of secondary response genes are covered by nucleosomes, and remodelling of the chromatin is required for binding of RNA polymerase to activate transcription. Our studies show that binding of RNA polymerase to the MIEP is not detectable in latent mice and that, in addition to RNA polymerase and transcription factors, actin, a component of many chromatin-remodelling complexes, is recruited to the MIEP following transplantation. Thus, regulation of the MIEP may be more similar to secondary immune response genes.
Remodelling complexes can alter the configuration of the chromatin through: (i) nucleosome sliding, in which the position of the nucleosome on the DNA changes; (ii) remodelling, in which histones remain bound but the DNA becomes more accessible; (iii) ejection of nucleosomes from the DNA; and (iv) replacement of canonical histones with a variant histone (Mohrmann & Verrijzer, 2005). We previously observed no change in binding of histone H3 to the MIEP in allografts (Liu et al., 2013). These results therefore suggest that remodelling does not occur through eviction.
Current antiviral therapies target viral DNA replication in cells in which reactivation of latent genomes has already occurred. An alternative strategy, which would be less susceptible to viral escape mutants, would be to target cellular pathways that lead to transcriptional reactivation of IE gene expression. A greater understanding of the molecular mechanisms leading to activation of the MIEP is required to realize this goal. TNF expression is upregulated by allogeneic transplantation (Fig. 6 and Hummel et al., 2009, 2001) and TNF is sufficient to activate MCMV IE gene expression and/or reactivation in vivo (Cook et al., 2006; Hummel et al., 2001; Simon et al., 2005). However, our previous studies showed that TNF was not required for reactivation of IE gene expression in response to allogeneic transplantation (Zhang et al., 2009). These results suggest that multiple factors may contribute to transcriptional reactivation of viral gene expression in the context of allogeneic transplantation. To identify additional factors, we performed plasma proteomic analysis and transcriptional profiling of genes that were differentially expressed following allogeneic transplantation. While many pathways are activated in the complex environment of an allogeneic transplant, our studies identified five extracellular ligands that may contribute to reactivation of IE gene expression and reprogramming of viral chromatin. These include TNF, IL-1 and IL-18, which activate canonical NF-κB through engagement of their cell surface receptors, and CD40/CD40L, which activates both canonical and non-canonical NF-κB (Pomerantz & Baltimore, 2002). These pathways also activate AP-1, through activation of JNK-mediated phosphorylation of jun family members (Quezada et al., 2004; Weber et al., 2010; Wullaert et al., 2006). In addition, we found elevated levels of IL-6 in the plasma, increased IL-6 expression and activation of IL-6 signalling pathways in the kidney. Recent studies indicate that IL-6 activates the HCMV MIEP in monocyte-derived dendritic cells through recruitment of CREB (cAMP response element-binding protein) and MSK (mitogen- and stress-activated protein kinase)-mediated histone phosphorylation (Kew et al., 2014; Reeves & Compton, 2011). Although reactivation of the MIEP may occur through different pathways in different models, our results suggest that IL-6 may also have a role in reprogramming of viral chromatin in transplant-induced reactivation of CMV.
The alloimmune response to a transplanted organ is initiated by non-specific injury resulting from mitochondrial damage, oxidative stress and release of ‘danger signals’ from damaged tissue (de Groot & Rauen, 2007; Gallucci et al., 1999; Kono & Rock, 2008). These signals induce maturation of antigen-presenting cells in the graft that migrate to the lymph nodes and activate the adaptive arm of the immune response. NF-κB and AP-1 are activated by ischaemia/reperfusion injury and oxidative stress as well as inflammatory cytokines (Gloire et al., 2006; Kamata et al., 2005; Karin, 1995; Karin & Shaulian, 2001; Kim et al., 2005; Morgan & Liu, 2011; Oeckinghaus et al., 2011; Shaulian & Karin, 2002). In our studies, we observed very rapid activation of NF-κB and AP-1 family members, and biphasic activation of some pathways, including CD40, IL-6, NF-κB and JAK/STAT signalling (Fig. 5a). These observations suggest that different factors may contribute to activation of NF-κB and AP-1 at different times in the rapidly evolving environment of an allogeneic transplant, and that it may be necessary to target mediators of intracellular damage, such as reactive oxygen species, as well as extracellular ligands, to prevent reactivation of IE gene expression. Identification of the signalling pathways that lead to reactivation and the factors that activate these pathways will be the focus of future studies.
Allograft rejection, sepsis, acute illness, IL-6 and TNF have long been implicated in reactivation of HCMV in patients (Cook et al., 1998; Döcke et al., 1994; Fietze et al., 1994; Grattan et al., 1989; Heininger et al., 2001; Hibberd et al., 1992; Kalil & Florescu, 2009; Kutza et al., 1998; Lao et al., 1997; Limaye et al., 2008; Mutimer et al., 1997; Portela et al., 1995; Razonable et al., 2001; Reinke et al., 1994a) and allogeneic stimulation, TNF, IL-6 and lipopolysaccharide have been shown to induce reactivation of HCMV in experimental models (Hargett & Shenk, 2010; Huang et al., 2012; Kew et al., 2014; O'Connor & Murphy, 2012; Reeves & Compton, 2011; Söderberg-Nauclér et al., 1997). Allogeneic transplantation and TNF are sufficient to activate both an HCMV MIEP-lacZ transgene and MCMV IE gene expression (Hummel et al., 2001). Thus, there is considerable convergence between clinical studies of patients infected with HCMV, experimental models of HCMV latency/reactivation and animal studies with MCMV. Identification of the factors that induce reactivation of MCMV IE gene expression may therefore eventually lead to new approaches to prevent reactivation of HCMV and its sequelae.
Methods
Mice and transplants
BALB/c and C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). MCMV (Smith strain) was purchased from the American Type Culture Collection, and propagated in mice by harvesting salivary glands 14 days post-infection. Virus stocks were titrated on confluent monolayers of murine embryo fibroblasts. To establish latency, 3–4-week old female BALB/c mice were infected by intraperitoneal injection with 5 × 105 p.f.u. of MCMV (Smith strain) and housed for 3–6 months in the Northwestern University Center for Comparative Medicine. Donor kidneys from latently infected BALB/c mice were transplanted into recipient C57BL/6 mice as previously described (Zhang et al., 1995), except that recipients were bilaterally nephrectomized at the time of the transplant. The pararenal glands were left intact. The contralateral donor kidneys were frozen in liquid nitrogen at the time of the transplant for use as matching, Day 0 latent controls. Transplanted kidneys were frozen in liquid nitrogen at the time of sacrifice, and recipient plasma was collected for analysis of plasma proteins. Recipients were sacrificed at 3, 24 or 48 h, as indicated in the text. These studies were approved by the Northwestern University Institutional Animal Care and Use Committee and conducted accordingly.
Transcription factor activation
Analysis of transcription factors was performed on nuclear extracts isolated from frozen kidney tissue using TransAm kits as directed by the manufacturer (Active Motif). Nuclear extract (10 or 5 μg) was used to analyse NF-κB or AP-1 family members, respectively.
ChIP analyses
Transplanted kidneys were harvested at 48 h for ChIP analysis. The contralateral donor kidneys were removed at the time of the transplant for use as latent controls. Fresh tissue was finely minced on ice, and the chromatin was fixed in 1 % formaldehyde as previously described (Liu et al., 2008, 2010), and frozen in liquid nitrogen. Due to the very low MCMV DNA copy number in kidneys of latent mice [∼1 copy of MCMV DNA per 10 000 cellular genomes (Li et al., 2012)] and the large amount of chromatin required for each ChIP, chromatin from 5–6 kidneys was pooled for each antibody. Frozen pellets of fixed tissue from 30–50 transplants were resuspended in hypotonic lysis buffer, and processed for immunoprecipitation as previously described (Liu et al., 2008, 2010). Ten per cent of the chromatin was removed for analysis of input DNA. The remaining chromatin was pre-cleared with IgG and incubated overnight at 4 °C with antibodies against target proteins as previously described (Liu et al., 2008, 2010). The following antibodies were used: NF-κBp65 (Abcam, Ab7970-1), NF-κBp50 (Abcam Ab7971), junD (SantaCruz Biotech, sc-74x) and actin (SantaCruz Biotech, sc-7210). qPCR analysis to quantify input and immunoprecipitated DNA were performed as previously described (Liu et al., 2008, 2010).
Plasma protein analysis
Plasma (70 μl) from transplant recipients or untreated C57BL/6 control mice was frozen in liquid nitrogen and analysed for inflammatory proteins at MyriadRBM by Luminex using the Rodent MAPv3.1 platform.
Transcriptome analysis
Frozen tissue was immediately transferred to tubes containing TriZol and 5 mm stainless steel beads (Qiagen) and the tissue was disrupted by mechanical shaking in a TissueLyser (Qiagen) at room temperature for 5 min. RNAs were purified with PureLink RNA Minikits (Ambion), using on-column DNase treatment as directed by the manufacturer. RNA was quantified on a nanodrop spectrophotometer and quality was assessed on an Agilent 2100 bioanalyser. Genome-wide RNA expression was analysed using Affymetrix MoGene-2_1-st-v1 microarrays.
RT-qPCR
cDNAs were synthesized from 2 μg total cell RNA using a High Capacity cDNA Reverse Transcriptase kit (Applied Biosystems) for analysis of cellular gene expression. Gene-specific RNAs were amplified from 10 ng cDNA under standard conditions with the following TaqMan Gene Expression Assays (Applied Biosystems): TNF, Mm00443260_g1; RelA (p65), Mm00501346_m1; Nfkb1 (p50), Mm00476361_m1; RelB, Mm00485664_m1; c-jun, Mm00495062_s1; c-fos, Mm00487425_m1; junD, Mm04208316_s1; Nfkbib/IkBalpha, Mm00456849_m1. Relative gene expression was determined using hypoxanthine phosphoribosyltransferase (HPRT) RNA (Mm01545399_m1) as the internal control. MCMV IE-3 expression was analysed as previously described (Liu et al., 2008).
Statistical analysis
To identify changes in plasma protein expression, the MyriadRBM data were first filtered by excluding analytes in which >75 % of the measurements were below the detection threshold. Fisher's exact test was performed to ensure that none of the excluded proteins was significantly more likely to be undetectable in one group than another. The filtered multi-analyte protein MAP data were first log-transformed for better approximation to normal distribution and then analysed by a linear model, in which the common variance is estimated with the pooled data per protein wise. P values were obtained by performing t-test between the conditions for each protein, and then adjusted by Benjamini-Hochberg false discovery rate procedure for multiple comparisons. Changes in expression were visualized with a heat map generated in R.
For genome-wide RNA expression, microarrays were normalized using Robust Multichip Average (RMA) (Bolstad et al., 2003) and signal filters of Log2 < 5.69 were used to exclude probe sets with low signal intensities. Pairwise class comparisons were carried out using a one-way ANOVA by the Method of Moments (Eisenhart, 1947) in Partek Genomics Suite 6.6. A False Discovery Rate (FDR) of < 5 % was used for all class comparisons. Pathway mapping to biologically significant pathways was done using Ingenuity Pathway Analysis. All pathways were adjusted using the Benjamini-Hochberg correction for multiple testing. The microarray expression data from the study were deposited at the NIH Gene Expression Omnibus (GEO) website under the accession number GSE 72392.
Student's t-test was used to determine statistical significance for expression of individual RNAs and plasma proteins. P < 0.05 was considered statistically significant.
Acknowledgements
The authors thank MyriadRBM for analysis of plasma proteins. These studies were funded by NIH 1R21AI076771 and the Northwestern University Comprehensive Transplant Center.
Supplementary Data
Supplementary Data
References
- Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. (1990). A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family EMBO J 9 1897–1906 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo A., Ghazal P., Messerle M. (2000). The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth J Virol 74 11129–11136 10.1128/JVI.74.23.11129-11136.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam M., Woo S. Y., Vinson C., Bishop G. A. (2003). CD40-mediated transcriptional regulation of the IL-6 gene in B lymphocytes: involvement of NF-kappa B, AP-1, and C/EBP J Immunol 170 3099–3108 10.4049/jimmunol.170.6.3099 . [DOI] [PubMed] [Google Scholar]
- Beutler B. A., Milsark I. W., Cerami A. (1985). Cachectin/tumor necrosis factor: production, distribution, and metabolic fate in vivo J Immunol 135 3972–3977 . [PubMed] [Google Scholar]
- Bode J. G., Ludwig S., Freitas C. A., Schaper F., Ruhl M., Melmed S., Heinrich P. C., Häussinger D. (2001). The MKK6/p38 mitogen-activated protein kinase pathway is capable of inducing SOCS3 gene expression and inhibits IL-6-induced transcription Biol Chem 382 1447–1453 10.1515/BC.2001.178 . [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias Bioinformatics 19 185–193 10.1093/bioinformatics/19.2.185 . [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Shyu A. B. (1995). AU-rich elements: characterization and importance in mRNA degradation Trends Biochem Sci 20 465–470 10.1016/S0968-0004(00)89102-1 . [DOI] [PubMed] [Google Scholar]
- Cook C. H., Yenchar J. K., Kraner T. O., Davies E. A., Ferguson R. M. (1998). Occult herpes family viruses may increase mortality in critically ill surgical patients Am J Surg 176 357–360 10.1016/S0002-9610(98)00205-0 . [DOI] [PubMed] [Google Scholar]
- Cook C. H., Trgovcich J., Zimmerman P. D., Zhang Y., Sedmak D. D. (2006). Lipopolysaccharide, tumor necrosis factor alpha, or interleukin-1beta triggers reactivation of latent cytomegalovirus in immunocompetent mice J Virol 80 9151–9158 10.1128/JVI.00216-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot H., Rauen U. (2007). Ischemia-reperfusion injury: processes in pathogenetic networks: a review Transplant Proc 39 481–484 10.1016/j.transproceed.2006.12.012 . [DOI] [PubMed] [Google Scholar]
- Döcke W. D., Fietze E., Syrbe U., von Baehr R., Volk H. D., Prösch S., Kimel V., Krüger D. H., Zuckermann H., Klug C. (1994). Cytomegalovirus reactivation and tumour necrosis factor Lancet 343 268–269 10.1016/S0140-6736(94)91116-9 . [DOI] [PubMed] [Google Scholar]
- Dorsch-Häsler K., Keil G. M., Weber F., Jasin M., Schaffner W., Koszinowski U. H. (1985). A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus Proc Natl Acad Sci U S A 82 8325–8329 10.1073/pnas.82.24.8325 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R., Wagner E. F. (2003). AP-1: a double-edged sword in tumorigenesis Nat Rev Cancer 3 859–868 10.1038/nrc1209 . [DOI] [PubMed] [Google Scholar]
- Einecke G., Melk A., Ramassar V., Zhu L. F., Bleackley R. C., Famulski K. S., Halloran P. F. (2005). Expression of CTL associated transcripts precedes the development of tubulitis in T-cell mediated kidney graft rejection Am J Transplant 5 1827–1836 10.1111/j.1600-6143.2005.00974.x . [DOI] [PubMed] [Google Scholar]
- Eisenhart C. (1947). The assumptions underlying the analysis of variance Biometrics 3 1–21 10.2307/3001534 . [DOI] [PubMed] [Google Scholar]
- El-Sawy T., Miura M., Fairchild R. (2004). Early T cell response to allografts occurring prior to alloantigen priming up-regulates innate-mediated inflammation and graft necrosis Am J Pathol 165 147–157 10.1016/S0002-9440(10)63283-X . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski K. S., Einecke G., Reeve J., Ramassar V., Allanach K., Mueller T., Hidalgo L. G., Zhu L. F., Halloran P. F. (2006). Changes in the transcriptome in allograft rejection: IFN-gamma-induced transcripts in mouse kidney allografts Am J Transplant 6 1342–1354 10.1111/j.1600-6143.2006.01337.x . [DOI] [PubMed] [Google Scholar]
- Famulski K. S., Broderick G., Einecke G., Hay K., Cruz J., Sis B., Mengel M., Halloran P. F. (2007). Transcriptome analysis reveals heterogeneity in the injury response of kidney transplants Am J Transplant 7 2483–2495 10.1111/j.1600-6143.2007.01980.x . [DOI] [PubMed] [Google Scholar]
- Fietze E., Prösch S., Reinke P., Stein J., Döcke W. D., Staffa G., Löning S., Devaux S., Emmrich F., other authors (1994). Cytomegalovirus infection in transplant recipients The role of tumor necrosis factor. Transplantation 58 675–680 10.1097/00007890-199409000-00007 . [DOI] [PubMed] [Google Scholar]
- Fishman J. A., Emery V., Freeman R., Pascual M., Rostaing L., Schlitt H. J., Sgarabotto D., Torre-Cisneros J., Uknis M. E. (2007). Cytomegalovirus in transplantation – challenging the status quo Clin Transplant 21 149–158 10.1111/j.1399-0012.2006.00618.x . [DOI] [PubMed] [Google Scholar]
- Gallucci S., Lolkema M., Matzinger P. (1999). Natural adjuvants: endogenous activators of dendritic cells Nat Med 5 1249–1255 10.1038/15200 . [DOI] [PubMed] [Google Scholar]
- Gerald D., Berra E., Frapart Y. M., Chan D. A., Giaccia A. J., Mansuy D., Pouysségur J., Yaniv M., Mechta-Grigoriou F. (2004). JunD reduces tumor angiogenesis by protecting cells from oxidative stress Cell 118 781–794 10.1016/j.cell.2004.08.025 . [DOI] [PubMed] [Google Scholar]
- Gloire G., Legrand-Poels S., Piette J. (2006). NF-kappaB activation by reactive oxygen species: fifteen years later Biochem Pharmacol 72 1493–1505 10.1016/j.bcp.2006.04.011 . [DOI] [PubMed] [Google Scholar]
- Grattan M. T., Moreno-Cabral C. E., Starnes V. A., Oyer P. E., Stinson E. B., Shumway N. E. (1989). Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis JAMA 261 3561–3566 10.1001/jama.1989.03420240075030 . [DOI] [PubMed] [Google Scholar]
- Grzimek N. K., Dreis D., Schmalz S., Reddehase M. J. (2001). Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs J Virol 75 2692–2705 10.1128/JVI.75.6.2692-2705.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Barrett T., Whitmarsh A. J., Cavanagh J., Sluss H. K., Dérijard B., Davis R. J. (1996). Selective interaction of JNK protein kinase isoforms with transcription factors EMBO J 15 2760–2770 . [PMC free article] [PubMed] [Google Scholar]
- Hargett D., Shenk T. E. (2010). Experimental human cytomegalovirus latency in CD14+ monocytes Proc Natl Acad Sci U S A 107 20039–20044 10.1073/pnas.1014509107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D. C., Horng T., Medzhitov R. (2009). Control of inducible gene expression by signal-dependent transcriptional elongation Cell 138 129–145 10.1016/j.cell.2009.05.047 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heininger A., Jahn G., Engel C., Notheisen T., Unertl K., Hamprecht K. (2001). Human cytomegalovirus infections in nonimmunosuppressed critically ill patients [see comments] Crit Care Med 29 541–547 10.1097/00003246-200103000-00012 . [DOI] [PubMed] [Google Scholar]
- Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003). Principles of interleukin (IL)-6-type cytokine signalling and its regulation Biochem J 374 1–20 10.1042/bj20030407 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. M., Floyd D. H., Weilbaecher K. N., Green P. L., Boris-Lawrie K. (2008). Multiple facets of junD gene expression are atypical among AP-1 family members Oncogene 27 4757–4767 10.1038/onc.2008.120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd P. L., Tolkoff-Rubin N. E., Cosimi A. B., Schooley R. T., Isaacson D., Doran M., Delvecchio A., Delmonico F. L., Auchincloss H.,, Jr., Rubin R. H. (1992). Symptomatic cytomegalovirus disease in the cytomegalovirus antibody seropositive renal transplant recipient treated with OKT3 Transplantation 53 68–72 10.1097/00007890-199201000-00013 . [DOI] [PubMed] [Google Scholar]
- Huang M. M., Kew V. G., Jestice K., Wills M. R., Reeves M. B. (2012). Efficient human cytomegalovirus reactivation is maturation dependent in the Langerhans dendritic cell lineage and can be studied using a CD14+ experimental latency model J Virol 86 8507–8515 10.1128/JVI.00598-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M., Abecassis M. M. (2002). A model for reactivation of CMV from latency J Clin Virol 25 (Suppl 2) 123–136 10.1016/S1386-6532(02)00088-4 . [DOI] [PubMed] [Google Scholar]
- Hummel M., Zhang Z., Yan S., DePlaen I., Golia P., Varghese T., Thomas G., Abecassis M. I. (2001). Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency J Virol 75 4814–4822 10.1128/JVI.75.10.4814-4822.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M., Kurian S. M., Lin S., Borodyanskiy A., Zhang Z., Li Z., Kim S. J., Salomon D. R., Abecassis M. (2009). Intragraft TNF receptor signaling contributes to activation of innate and adaptive immunity in a renal allograft model Transplantation 87 178–188 10.1097/TP.0b013e3181938971 . [DOI] [PubMed] [Google Scholar]
- Jaeschke A., Karasarides M., Ventura J. J., Ehrhardt A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006). JNK2 is a positive regulator of the cJun transcription factor Mol Cell 23 899–911 10.1016/j.molcel.2006.07.028 . [DOI] [PubMed] [Google Scholar]
- Kadam S., Emerson B. M. (2003). Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes Mol Cell 11 377–389 10.1016/S1097-2765(03)00034-0 . [DOI] [PubMed] [Google Scholar]
- Kalil A. C., Florescu D. F. (2009). Prevalence and mortality associated with cytomegalovirus infection in nonimmunosuppressed patients in the intensive care unit Crit Care Med 37 2350–2358 10.1097/CCM.0b013e3181a3aa43 . [DOI] [PubMed] [Google Scholar]
- Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005). Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases Cell 120 649–661 10.1016/j.cell.2004.12.041 . [DOI] [PubMed] [Google Scholar]
- Karin M. (1995). The regulation of AP-1 activity by mitogen-activated protein kinases J Biol Chem 270 16483–16486 10.1074/jbc.270.28.16483 . [DOI] [PubMed] [Google Scholar]
- Karin M., Shaulian E. (2001). AP-1: linking hydrogen peroxide and oxidative stress to the control of cell proliferation and death IUBMB Life 52 17–24 10.1080/15216540252774711 . [DOI] [PubMed] [Google Scholar]
- Keil G. M., Ebeling-Keil A., Koszinowski U. H. (1987a). Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products J Virol 61 526–533 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil G. M., Ebeling-Keil A., Koszinowski U. H. (1987b). Sequence and structural organization of murine cytomegalovirus immediate-early gene 1 J Virol 61 1901–1908 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew V. G., Yuan J., Meier J., Reeves M. B. (2014). Mitogen and stress activated kinases act co-operatively with CREB during the induction of human cytomegalovirus immediate-early gene expression from latency PLoS Pathog 10 e1004195 10.1371/journal.ppat.1004195 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Varghese T. K., Zhang Z., Zhao L. C., Thomas G., Hummel M., Abecassis M. (2005). Renal ischemia/reperfusion injury activates the enhancer domain of the human cytomegalovirus major immediate early promoter Am J Transplant 5 1606–1613 10.1111/j.1600-6143.2005.00912.x . [DOI] [PubMed] [Google Scholar]
- Kono H., Rock K. L. (2008). How dying cells alert the immune system to danger Nat Rev Immunol 8 279–289 10.1038/nri2215 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp K. A., Angulo A., Ghazal P. (2014). Viral enhancer mimicry of host innate-immune promoters PLoS Pathog 10 e1003804 10.1371/journal.ppat.1003804 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz S., Steffens H. P., Mayer A., Harris J. R., Reddehase M. J. (1997). Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs J Virol 71 2980–2987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz S. K., Rapp M., Steffens H. P., Grzimek N. K., Schmalz S., Reddehase M. J. (1999). Focal transcriptional activity of murine cytomegalovirus during latency in the lungs J Virol 73 482–494 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutza A. S., Muhl E., Hackstein H., Kirchner H., Bein G. (1998). High incidence of active cytomegalovirus infection among septic patients [see comments] Clin Infect Dis 26 1076–1082 10.1086/520307 . [DOI] [PubMed] [Google Scholar]
- Lamb J. A., Ventura J. J., Hess P., Flavell R. A., Davis R. J. (2003). JunD mediates survival signaling by the JNK signal transduction pathway Mol Cell 11 1479–1489 10.1016/S1097-2765(03)00203-X . [DOI] [PubMed] [Google Scholar]
- Lao W. C., Lee D., Burroughs A. K., Lanzani G., Rolles K., Emery V. C., Griffiths P. D. (1997). Use of polymerase chain reaction to provide prognostic information on human cytomegalovirus disease after liver transplantation J Med Virol 51 152–158 . [PubMed] [Google Scholar]
- Lee Y., Sohn W. J., Kim D. S., Kwon H. J. (2004). NF-kappaB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7 Eur J Biochem 271 1094–1105 10.1111/j.1432-1033.2004.04011.x . [DOI] [PubMed] [Google Scholar]
- Li Z., Wang X., Yan S., Zhang Z., Jie C., Sustento-Reodica N., Hummel M., Abecassis M. (2012). A mouse model of CMV transmission following kidney transplantation Am J Transplant 12 1024–1028 10.1111/j.1600-6143.2011.03892.x . [DOI] [PubMed] [Google Scholar]
- Limaye A. P., Kirby K. A., Rubenfeld G. D., Leisenring W. M., Bulger E. M., Neff M. J., Gibran N. S., Huang M. L., Santo Hayes T. K., other authors (2008). Cytomegalovirus reactivation in critically ill immunocompetent patients JAMA 300 413–422 10.1001/jama.2008.697 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. F., Yan S., Abecassis M., Hummel M. (2008). Establishment of murine cytomegalovirus latency in vivo is associated with changes in histone modifications and recruitment of transcriptional repressors to the major immediate-early promoter J Virol 82 10922–10931 10.1128/JVI.00865-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. F., Yan S., Abecassis M., Hummel M. (2010). Biphasic recruitment of transcriptional repressors to the murine cytomegalovirus major immediate-early promoter during the course of infection in vivo J Virol 84 3631–3643 10.1128/JVI.02380-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. F., Wang X., Yan S., Zhang Z., Abecassis M., Hummel M. (2013). Epigenetic control of cytomegalovirus latency and reactivation Viruses 5 1325–1345 10.3390/v5051325 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J., Oakley F., Johnson P. W., Mann D. A. (2002). CD40 induces interleukin-6 gene transcription in dendritic cells: regulation by TRAF2, AP-1, NF-kappa B, and CBF1 J Biol Chem 277 17125–17138 10.1074/jbc.M109250200 . [DOI] [PubMed] [Google Scholar]
- Martínez F. P., Cosme R. S., Tang Q. (2010). Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation J Gen Virol 91 2664–2676 10.1099/vir.0.022301-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Horng T. (2009). Transcriptional control of the inflammatory response Nat Rev Immunol 9 692–703 10.1038/nri2634 . [DOI] [PubMed] [Google Scholar]
- Meier J. L., Stinski M. F. (2013). Major immediate-early enhancer and its gene products. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention, pp. 152–173. Edited by Reddehase M. J. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Messerle M., Bühler B., Keil G. M., Koszinowski U. H. (1992). Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3 J Virol 66 27–36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Shenk T., Pass R. F. (2007). Cytomegaloviruses. In Fields Virology 5th edn, pp. 2702–2772. Edited by Knipe D. M., Howley P. M. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. [Google Scholar]
- Mohrmann L., Verrijzer C. P. (2005). Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes Biochim Biophys Acta 1681 59–73 10.1016/j.bbaexp.2004.10.005 . [DOI] [PubMed] [Google Scholar]
- Morgan M. J., Liu Z. G. (2011). Crosstalk of reactive oxygen species and NF-κB signaling Cell Res 21 103–115 10.1038/cr.2010.178 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutimer D., Mirza D., Shaw J., O'Donnell K., Elias E. (1997). Enhanced (cytomegalovirus) viral replication associated with septic bacterial complications in liver transplant recipients Transplantation 63 1411–1415 10.1097/00007890-199705270-00007 . [DOI] [PubMed] [Google Scholar]
- Ndlovu M. N., Van Lint C., Van Wesemael K., Callebert P., Chalbos D., Haegeman G., Vanden Berghe W. (2009). Hyperactivated NF-kappaB and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells Mol Cell Biol 29 5488–5504 10.1128/MCB.01657-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor C. M., Murphy E. A. (2012). A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny J Virol 86 9854–9865 10.1128/JVI.01278-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A., Hayden M. S., Ghosh S. (2011). Crosstalk in NF-κB signaling pathways Nat Immunol 12 695–708 10.1038/ni.2065 . [DOI] [PubMed] [Google Scholar]
- Pillebout E., Weitzman J. B., Burtin M., Martino C., Federici P., Yaniv M., Friedlander G., Terzi F. (2003). JunD protects against chronic kidney disease by regulating paracrine mitogens J Clin Invest 112 843–852 10.1172/JCI200317647 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz J. L., Baltimore D. (2002). Two pathways to NF-kappaB Mol Cell 10 693–695 10.1016/S1097-2765(02)00697-4 . [DOI] [PubMed] [Google Scholar]
- Portela D., Patel R., Larson-Keller J. J., Ilstrup D. M., Wiesner R. H., Steers J. L., Krom R. A., Paya C. V. (1995). OKT3 treatment for allograft rejection is a risk factor for cytomegalovirus disease in liver transplantation J Infect Dis 171 1014–1018 10.1093/infdis/171.4.1014 . [DOI] [PubMed] [Google Scholar]
- Prösch S., Staak K., Stein J., Liebenthal C., Stamminger T., Volk H.-D., Krüger D. H. (1995). Stimulation of the human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNFalpha is mediated via induction of NF-kappaB Virology 208 197–206 10.1006/viro.1995.1143 . [DOI] [PubMed] [Google Scholar]
- Quezada S. A., Jarvinen L. Z., Lind E. F., Noelle R. J. (2004). CD40/CD154 interactions at the interface of tolerance and immunity Annu Rev Immunol 22 307–328 10.1146/annurev.immunol.22.012703.104533 . [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi V. R., Nazarian A. A., Li C. C., Gore S. L., Sridharan R., Imbalzano A. N., Smale S. T. (2006). Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response Genes Dev 20 282–296 10.1101/gad.1383206 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi V. R., Braas D., Bhatt D. M., Cheng C. S., Hong C., Doty K. R., Black J. C., Hoffmann A., Carey M., Smale S. T. (2009). A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling Cell 138 114–128 10.1016/j.cell.2009.04.020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable R. R., Rivero A., Rodriguez A., Wilson J., Daniels J., Jenkins G., Larson T., Hellinger W. C., Spivey J. R., Paya C. V. (2001). Allograft rejection predicts the occurrence of late-onset cytomegalovirus (CMV) disease among CMV-mismatched solid organ transplant patients receiving prophylaxis with oral ganciclovir J Infect Dis 184 1461–1464 10.1086/324516 . [DOI] [PubMed] [Google Scholar]
- Razonable R. R., Humar A., AST Infectious Diseases Community of Practice (2013). Cytomegalovirus in solid organ transplantation Am J Transplant 13 (Suppl 4), 93–106 10.1111/ajt.12103 . [DOI] [PubMed] [Google Scholar]
- Reeves M. B., Compton T. (2011). Inhibition of inflammatory interleukin-6 activity via extracellular signal-regulated kinase-mitogen-activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells J Virol 85 12750–12758 10.1128/JVI.05878-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M., Sinclair J. (2013). Epigenetic regulation of human cytomegalovirus gene expression: impact on latency and reactivation. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention, pp. 330–346. Edited by Reddehase M. J. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Reeves M. B., MacAry P. A., Lehner P. J., Sissons J. G., Sinclair J. H. (2005). Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers Proc Natl Acad Sci U S A 102 4140–4145 10.1073/pnas.0408994102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke P., Fietze E., Döcke W. D., Kern F., Ewert R., Volk H. D. (1994a). Late acute rejection in long-term renal allograft recipients. Diagnostic and predictive value of circulating activated T cells Transplantation 58 35–41 10.1097/00007890-199407000-00007 . [DOI] [PubMed] [Google Scholar]
- Reinke P., Fietze E., Ode-Hakim S., Prösch S., Lippert J., Ewert R., Volk H. D. (1994b). Late-acute renal allograft rejection and symptomless cytomegalovirus infection Lancet 344 1737–1738 10.1016/S0140-6736(94)92887-8 . [DOI] [PubMed] [Google Scholar]
- Ruland J. (2011). Return to homeostasis: downregulation of NF-κB responses Nat Immunol 12 709–714 10.1038/ni.2055 . [DOI] [PubMed] [Google Scholar]
- Sasaki T., Kojima H., Kishimoto R., Ikeda A., Kunimoto H., Nakajima K. (2006). Spatiotemporal regulation of c-Fos by ERK5 and the E3 ubiquitin ligase UBR1, and its biological role Mol Cell 24 63–75 10.1016/j.molcel.2006.08.005 . [DOI] [PubMed] [Google Scholar]
- Seckert C. K., Griebl M., Buttner J., Freitag K., Lemmermann N., Hummel M., Liu X.-F., Abecassis M., Angulo A., other authors (2013). Immune surveillance of cytomegalovirus latency and reactivation in murine models: link to ‘memory inflation’. In Cytomegaloviruses: From Molecular Pathogenesis to Therapy, pp. 374–416. Edited by Reddehase M. J. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Shaulian E., Karin M. (2002). AP-1 as a regulator of cell life and death Nat Cell Biol 4 E131–E136 10.1038/ncb0502-e131 . [DOI] [PubMed] [Google Scholar]
- Simon C. O., Seckert C. K., Dreis D., Reddehase M. J., Grzimek N. K. (2005). Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs J Virol 79 326–340 10.1128/JVI.79.1.326-340.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart D. E., Vincent K. J., Arthur M. J., Eickelberg O., Castellazzi M., Mann J., Mann D. A. (2001). JunD regulates transcription of the tissue inhibitor of metalloproteinases-1 and interleukin-6 genes in activated hepatic stellate cells J Biol Chem 276 24414–24421 10.1074/jbc.M101840200 . [DOI] [PubMed] [Google Scholar]
- Söderberg-Nauclér C., Fish K. N., Nelson J. A. (1997). Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors Cell 91 119–126 10.1016/S0092-8674(01)80014-3 . [DOI] [PubMed] [Google Scholar]
- Stein J., Volk H. D., Liebenthal C., Krüger D. H., Prösch S. (1993). Tumour necrosis factor alpha stimulates the activity of the human cytomegalovirus major immediate early enhancer/promoter in immature monocytic cells J Gen Virol 74 2333–2338 10.1099/0022-1317-74-11-2333 . [DOI] [PubMed] [Google Scholar]
- Stenberg R. M., Thomsen D. R., Stinski M. F. (1991). Structural analysis of the major immediate early gene of human cytomegalovirus J Virol 49 190–984 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teferedegne B., Green M. R., Guo Z., Boss J. M. (2006). Mechanism of action of a distal NF-kappaB-dependent enhancer Mol Cell Biol 26 5759–5770 10.1128/MCB.00271-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y. (2005). JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress Oncogene 24 7567–7578 10.1038/sj.onc.1208901 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viedt C., Dechend R., Fei J., Hänsch G. M., Kreuzer J., Orth S. R. (2002). MCP-1 induces inflammatory activation of human tubular epithelial cells: involvement of the transcription factors, nuclear factor-kappaB and activating protein-1 J Am Soc Nephrol 13 1534–1547 10.1097/01.ASN.0000015609.31253.7F . [DOI] [PubMed] [Google Scholar]
- Weber A., Wasiliew P., Kracht M. (2010). Interleukin-1 (IL-1) pathway Sci Signal 3 cm1 . [DOI] [PubMed] [Google Scholar]
- Wolter S., Doerrie A., Weber A., Schneider H., Hoffmann E., von der Ohe J., Bakiri L., Wagner E. F., Resch K., Kracht M. (2008). c-Jun controls histone modifications, NF-kappaB recruitment, and RNA polymerase II function to activate the ccl2 gene Mol Cell Biol 28 4407–4423 10.1128/MCB.00535-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullaert A., Heyninck K., Beyaert R. (2006). Mechanisms of crosstalk between TNF-induced NF-kappaB and JNK activation in hepatocytes Biochem Pharmacol 72 1090–1101 10.1016/j.bcp.2006.07.003 . [DOI] [PubMed] [Google Scholar]
- Zerbini L. F., Wang Y., Cho J. Y., Libermann T. A. (2003). Constitutive activation of nuclear factor kappaB p50/p65 and Fra-1 and JunD is essential for deregulated interleukin 6 expression in prostate cancer Cancer Res 63 2206–2215 . [PubMed] [Google Scholar]
- Zhang Z., Schlachta C., Duff J., Stiller C., Grant D., Zhong R. (1995). Improved techniques for kidney transplantation in mice Microsurgery 16 103–109 10.1002/micr.1920160212 . [DOI] [PubMed] [Google Scholar]
- Zhang Z., Kim S. J., Varghese T., Thomas G., Hummel M., Abecassis M. (2008). TNF receptor independent activation of the cytomegalovirus major immediate early enhancer in response to transplantation Transplantation 85 1039–1045 10.1097/TP.0b013e318168449c . [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li Z., Yan S., Wang X., Abecassis M. (2009). TNF-alpha signaling is not required for in vivo transcriptional reactivation of latent murine cytomegalovirus Transplantation 88 640–645 10.1097/TP.0b013e3181b242a7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data