Abstract
Identifying the genes required for environmental sex determination is important for understanding the evolution of diverse sex determination mechanisms in animals. Orthologs of Drosophila orphan receptor Fushi tarazu factor-1 (Ftz-F1) are known to function in genetic sex determination. In contrast, their roles in environmental sex determination remain unknown. In this study, we have cloned and characterized the Ftz-F1 ortholog in the branchiopod crustacean Daphnia magna, which produces males in response to environmental stimuli. Similar to that observed in Drosophila, D. magna Ftz-F1 (DapmaFtz-F1) produces two splicing variants, αFtz-F1 and βFtz-F1, which encode 699 and 777 amino acids, respectively. Both isoforms share a DNA-binding domain, a ligand-binding domain, and an AF-2 activation domain and differ only at the A/B domain. The phylogenetic position and genomic structure of DapmaFtz-F1 suggested that this gene has diverged from an ancestral gene common to branchiopod crustacean and insect Ftz-F1 genes. qRT-PCR showed that at the one cell and gastrulation stages, both DapmaFtz-F1 isoforms are two-fold more abundant in males than in females. In addition, in later stages, their sexual dimorphic expressions were maintained in spite of reduced expression. Time-lapse imaging of DapmaFtz-F1 RNAi embryos was performed in H2B-GFP expressing transgenic Daphnia, demonstrating that development of the RNAi embryos slowed down after the gastrulation stage and stopped at 30–48 h after ovulation. DapmaFtz-F1 shows high homology to insect Ftz-F1 orthologs based on its amino acid sequence and exon-intron organization. The sexually dimorphic expression of DapmaFtz-F1 suggests that it plays a role in environmental sex determination of D. magna.
Introduction
Sex determination is a fundamental biological process that governs the development of sexual characteristics, including the sexual differentiation of gonads, and affects the sexually dimorphic behavior, physiology, and morphology. The mechanism can be broadly categorized into two groups according to their primary causal factors: genetic sex determination (GSD) and environmental sex determination (ESD) [1–3]. In GSD, the sex-specific developmental pathway is resulted from the genetic segregation of genes, usually positioned on sex chromosomes. The ESD, however, relies on the environmental cues such as temperature, photoperiod, nutrition, and population density to induce molecular cascades for activation of alternate sex-determining genes [4, 5]. Currently, while GSD mechanism is well reported, the molecular basis of ESD has not yet been clarified. Analyzing the function of genes involved in ESD and unraveling the sex-determining pathways is crucial to understanding the origin and evolution of sex-determining pathways.
The water flea Daphnia magna, a crustacean living in freshwater ponds, undergoes switching of its reproductive strategy between asexual and sexual reproduction, depending on the environmental conditions [6]. Healthy D. magna produce female offspring by parthenogenesis or asexual cycle. Alternatively, in response to environmental stimuli such as insufficient food, short photoperiod and/or increased population density, it produces males, which allows for the fertilization of haploid eggs by sexual reproduction to produce resting eggs that can survive in harsh conditions [7,8]. The environmentally dependent production of males is a key process in the life cycle of Daphnia and leads to increased genetic diversity and fitness to overcome adverse conditions necessary for survival [9].
For male production in D. magna ESD, juvenile hormone (JH) and the DM domain gene DapmaDsx1 are currently known to be essential. JH stimulates germ cells at the late stage of oogenesis leading to the development of males from ovulated eggs [10–13]. In response to the JH signal, DapmaDsx1 is expressed and maintained to regulate development of male traits during embryogenesis [14], suggesting that JH-dependent DapmaDsx1 activation is necessary for the environmentally dependent production of males. However, genes that mediate JH signaling and DapmaDsx1 activation remain unknown.
Fushi tarazu factor-1 (Ftz-F1) is a member of the orphan nuclear receptor family involved in the genetic regulation of various developmental processes and was first identified in Drosophila melanogaster [15]. Subsequently, Ftz-F1 orthologs have been isolated from a wide range of animals and have several different names including steroidogenic factor-1 (Sf-1) [16], adrenal-4-binding protein (Ad4BP) [17] and nuclear hormone receptor-25 (nhr-25) [18]. The vertebrate Ftz-F1 orthologs, Sf-1 genes, are strongly linked to steroid biosynthesis and sex-determination pathways. In mammals, Sf-1 genes are expressed in steroidogenic tissues, play a key role in regulating steroidogenesis, and are involved in the testis-determining pathway during genetic sex determination [19,20]. Recent studies found that Drosophila Ftz-F1 participates in JH signaling by interacting with Methoprene-tolerant (MET), a hormone receptor protein that directs JH-mediated gene activation [21,22]. Therefore, it is reasonable to hypothesize that the Ftz-F1 ortholog may be a factor that mediates JH signaling and environmental sex determination in Daphnia.
In this study, we identified a D. magna Ftz-F1 ortholog (DapmaFtz-F1) that produces two splicing variants, αFtz-F1 and βFtz-F1, both of which exhibited sexual dimorphism in their expression during embryogenesis. Our findings suggest that DapmaFtz-F1 is possibly required for male production in D. magna ESD.
Materials and Methods
Daphnia Strain and Culture Conditions
The Daphnia magna strain (NIES clone) was obtained from the National Institute for Environmental Studies (NIES; Tsukuba, Japan) and maintained as previously described [23] using ADaM [24] as the culture medium. Transgenic D. magna that exhibits ubiquitous and constitutive expression of GFP under the control of D. magna elongation factor 1 α-1 (EF1α-1) gene promoter [25] was used for microinjection experiments. To obtain male embryos, adult daphnids (2–3 weeks old) were treated with 1 μg/L of synthetic juvenile hormone analog, Fenoxycarb (Wako Pure Chemical; Osaka, Japan) [12]. Then, the ovulated eggs were collected and used for subsequent experiments.
Cloning of the D. magna Ftz-F1 Gene
Male and female daphnids were collected separately and briefly washed. Homogenization was performed with beads using a Micro Smash machine MS-100 (TOMY; Tokyo, Japan) in the presence of Sepasol-RNA I reagent (Nacalai Tesque Inc.; Kyoto, Japan). Total RNA was isolated according to the manufacturer’s protocol, which was followed by phenol/chloroform extractions. The purified total RNA was converted to first strand cDNA with SuperScript III Reverse Transcriptase (Invitrogen; Carlsbad, CA, USA), utilizing random primers (Invitrogen) according to the manufacturer’s recommended protocol. DapmaFtz-F1 cDNA fragments that code for the DNA-binding domain (DBD) and the ligand-binding domain (LBD) were obtained from female cDNA by PCR with AmpliTaq DNA polymerase (Applied Biosystems; Foster City, CA, USA) using degenerate primers that were designed based on the conserved amino acid sequences of the DBD (5′-GAAGAACTGTGTCCNGTBTGYGG-3′) and the LBD (5′-ARTTTCATYTGRTCGTCAACCTT-3′). Amplified DNA fragments were cloned into a pGEM-T Easy Vector System (Promega Corp.; Madison, WI, USA) and sequenced.
Full-length cDNAs were completed by 5′ and 3′ rapid amplification of cDNA ends (RACE) with a GeneRacer Kit (Invitrogen) and a SMARTer RACE cDNA Amplification Kit (Clontech Laboratories Inc.; Mount View, WI, USA), respectively. The primer sequences for the RACE experiments were as follows: 5′-RACE gene specific primer (5′-TCCTCCGCCGGACGGGTGATTATTTG-3′); 5′-RACE nested primer (5′-TTCGCGGATCAATGGCGGAACTTTAGC-3′); 3′-RACE gene specific primer (5′-ATTCTCCGTCCGGCAGCAGCGTCTAC-3′); and 3′-RACE nested primer (5′-TCCACTTGCCGCATCACTCGGCTATC-3′). These amplification products were purified from an agarose gel after electrophoresis and were cloned into a TOPO vector, using a TOPO cloning kit (Invitrogen) for sequencing. The sequencing reaction was performed using a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA) and the DNA sequences were analyzed using the BLAST program.
Phylogenetic Analysis of the D. magna Ftz-F1 Gene
Amino acid sequences of Ftz-F1 family genes were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/) as shown Table 1, and the whole amino acid sequences of each protein were used to construct the phylogenetic tree. Multiple sequence alignments, based on the amino acid sequences, were constructed using the Clustal W [26] in MEGA version 6.06 [27]. The following settings were used for the analysis: pairwise alignment parameters: gap opening penalty = 6.00, gap extension penalty = 0.21, and identity protein weight; matrix multiple alignment parameters: gap opening penalty = 10.00, gap extension penalty = 0.24, delay divergent cut-off = 30%, and gap separation distance = 4. The phylogenetic reconstruction was performed using the p-distance algorithm and the neighbor-joining method implemented in MEGA.
Table 1. Accession numbers of Ftz-F1 ortholog genes used in this study.
| Common name | Scientific name | Gene name (definition in NCBI) | Accession no. |
|---|---|---|---|
| Water flea | Daphnia magna | Ftz-F1 | This work (LC105700, LC105701) |
| Water flea | Daphnia pulex | Ftz-F1 | EFX77612.1 |
| German cockroach | Blattella germanica | Ftz-F1 | CAQ57670.1 |
| Red flour beetle | Tribolium castaneum | Ftz-F1 | EFA01263.1 |
| Yellow fever mosquito | Aedes aegypti | Ftz-F1 | AAF82307.1 |
| Silkworm | Bombyx mori | Ftz-F1 | NP_001037528.2 |
| Shrimp | Metapenaeus ensis | Ftz-F1 | AAD41899.1 |
| Fruit fly | Drosophila melanogaster | Ftz-F1 | AAA28542.1 |
| Mouse | Mus musculus | Sf-1 | AAB28338.1 |
| Zebrafish | Danio rerio | Nr5a2 | NP_571538.1 |
| Medaka | Oryzias latipes | Ftz-F1 | BAA32394.1 |
| Roundworm | Caenorhabditis elegans | Nhr-25 | CAA91028.1 |
Temporal Expression Analysis by Quantitative Real-Time PCR
Male and female embryos were collected at 0, 6, 12, 18, 24, 30, 48, and 72 h after oviposition. These time points correspond to several embryonic stages described in [25,28,29]. To have three biological replicates, the collected embryos at each stage were divided into three groups. Each group was subjected to total RNA purification as described above. The number of embryos and amounts of purified total RNAs in each group were shown in S1 Table. cDNAs were synthesized using 1 μg of total RNAs as mentioned above. Of each cDNA pool, 1/120 volume was used as a template for qRT-PCR, providing us an equation (1) for calculation of number of embryos subjected to qRT-PCR.
| (1) |
PCR was performed using a SYBR GreenER qPCR SuperMix Universal Kit (Invitrogen) with Mx3005P Real-Time PCR System (Agilent Technologies; CA, USA). In the presence of the appropriate primer pairs, real-time PCR amplifications were performed in triplicate at the following conditions: 2 min at 50°C and 10 min 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Gel electrophoresis and dissociation curve analyses were performed to confirm the correct amplicon size and the absence of non-specific bands. Copy number of DapmaFtz-F1 mRNAs was measured by the quantification method, which relates the PCR signal to the input copy number by using a calibration curve obtained by a dilution series of plasmid that contains sequences corresponding to each primer set. Finally, copy number obtained by qRT-PCR was divided by N, resulting in copy number of transcripts in one embryo. The oligonucleotide sequences for qRT-PCR are shown in Fig 1B.
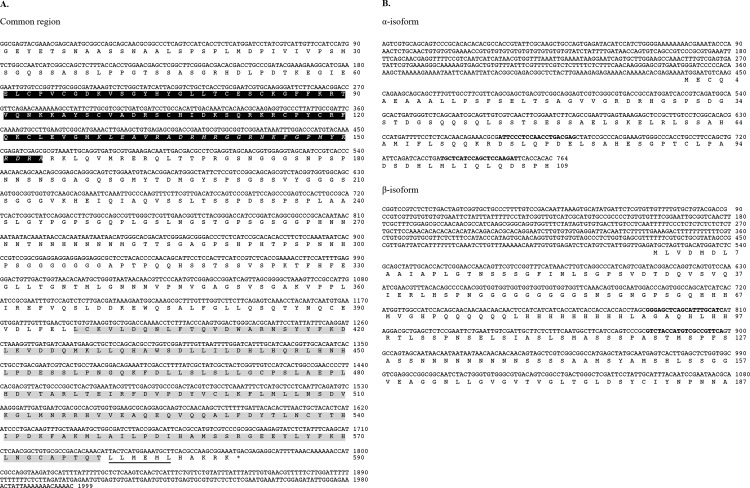
Fig 1. Nucleotide and deduced amino acid sequences of D. magna Ftz-F1.
(A) The DapmaFtz-F1 common region. Black and grey shaded amino acids indicate the putative DNA-binding domain (DBD) and ligand-binding domain (LBD) respectively, based on the alignment of amino acid sequences of signature domains. The Ftz-F1 box (italicized) and the activation factor-2 (AF-2) core (underlined) motifs are indicated. (B) Nucleotide sequence of DapmaFtz-F1 isoform-specific regions. Deduced amino acid sequences starting from the first methionine for each isoform are also indicated. Locations of the primers used in qRT-PCR are emboldened. Numbers on the right indicate the nucleotide and amino acid positions.
Gene Function Analysis by RNA Interference
RNA-mediated interference with 100 μM of Ftz-F1_699 siRNA (5′-CCAGUCUCUGACGAUAA-3′) and Ftz-F1 918 siRNA (5′-GCACACACCUUCUCCAAAU-3′) were employed to knock down DapmaFtz-F1 function in vivo by the method of microinjection into Daphnia eggs [30]. Eggs were obtained from a D. magna transgenic line at 2–3 weeks of ages, directly after the ovulation and placed in ice-cold M4 media that contained 80 mM sucrose. The injection solutions contained the specified siRNAs, mixed with 0.02 μM Alexa Fluor 568 dye (LifeTechnologies Inc.; Grand Island, NY, USA) as a marker to check whether an appropriate volume of solution was injected. The injected eggs were incubated in a 96-well plate at 23°C. A random sequence (5′-GGUUAAGCCGCCUCACAUTT-3′) that did not affect Daphnia embryogenesis [31] was utilized as a control siRNA (Control_416 siRNA). The phenotypes of injected embryos were carefully observed by time-lapse imaging from 3 h to 30 h after ovulation by fluorescence microscopy. At 24 h after injection, total RNAs were isolated from two embryos injected with Control_416 or Ftz-F1_918 siRNA in three replicates and were converted to cDNAs as described above. RT-qPCR analysis was performed with the same protocol mentioned above except that two primers, the FTZ-F1-realtime-5 (5′-CGCACACCTTCTCCAAATAA-3′) and FTZ-F1-realtime-3 (5′-TTACCAGTCAACAGTCCCTCAAAA-3′) were used to amplify the common region of Ftz-F1 gene.
Results and Discussion
Characterization of cDNAs Encoding D. magna Ftz-F1
To examine the existence of the Ftz-F1 ortholog in D. magna, we designed degenerate primers for amplification of the DapmaFtz-F1 cDNA fragment that codes for the DBD and the LBD regions (Fig 1A). After cloning and sequencing the amplified DNA fragments, a BLAST analysis revealed that the putative amino acid sequence shows high homology to Dr. melanogaster nuclear hormone receptor Ftz-F1. Therefore, we designated this gene as DapmaFtz-F1 (i.e., D. magna Ftz-F1 gene).
To identify full-length DapmaFtz-F1 cDNA, 5′ and 3′ RACE reactions were performed using cDNAs of male and female adults. The sequences were assembled into two different isoforms, αFtz-F1 and βFtz-F1, which are composed of 2,763 and 3,078 nucleotides, respectively. The open reading frames (ORFs) for αFtz-F1 and βFtz-F1 encode 699 and 777 amino acid residues respectively. They differed at the 5′ UTR and 5′ region of the ORF (Fig 1B). No sex-specific transcript was found from the RACE experiments.
Features of D. magna Ftz-F1 Proteins
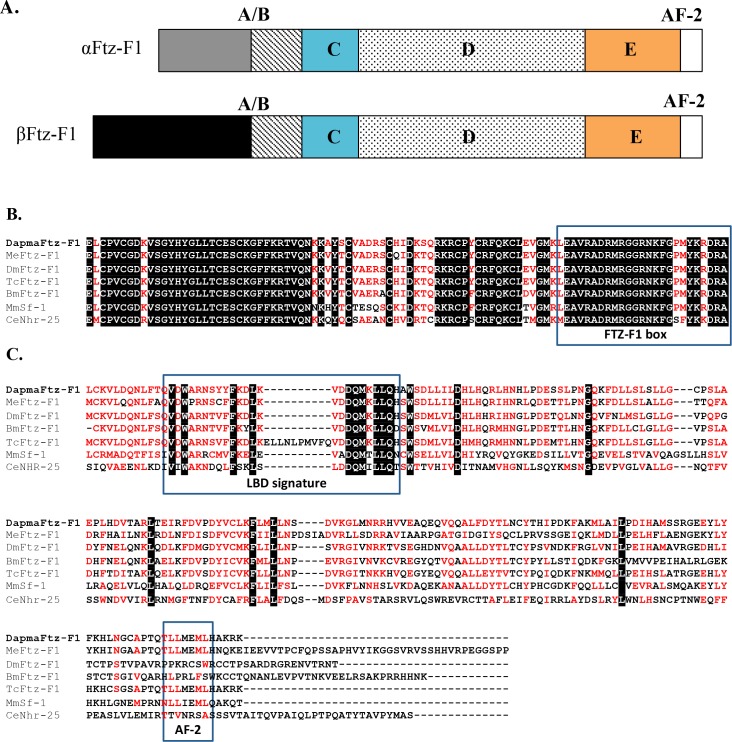
We compared amino acid sequences of DapmaFtz-F1 proteins with those of Ftz-F1 orthologs from Drosophila and various animals. The multiple alignment revealed that DapmaFtz-F1 proteins were predicted to have the typical structure of a nuclear receptor, which consists of an A/B region, a conserved zinc finger DBD at a DNA sequence recognition C region, a hinge D region, and lastly, an LBD follow by an activation function-2 (AF-2) at the E region (Fig 2A). Both αFtz-F1 and βFtz-F1 proteins consist of identical 590 amino acid sequences, which include the DBD (94 aa) and the LBD (182 aa) regions (Fig 1A). They have a different amino acid sequence at the A/B region, where αFtz-F1 and βFtz-F1 have 109 aa and 187 aa, respectively (Fig 1B). No conserved motif among Ftz-F1 orthologs was found in the A/B region of the α- or β-isoform. The DBD region and the LBD regions of DapmaFtz-F1 were aligned with other Ftz-F1 orthologs, namely Metapenaeus ensis Ftz-F1, Dr. melanogaster Ftz-F1, Bombyx mori Ftz-F1, Tribolium castaneum Ftz-F1, Mus musculus Sf-1, and Caenorhabditis elegans nhr-25 (Fig 2B and 2C). In the C region, a sequence named the Ftz-F1 box, adjacent to the zinc-finger motif [32], was conserved (Figs 1A and 2B). This DNA sequence recognition region is the most conserved region of the amino acid sequence. In the E region, the LBD signature and AF-2 motif that is required for ligand binding [32–34] was also found (Figs 1A and 2C). Both regions of DapmaFtz-F1 were the most homologous to those of insect orthologs.
Fig 2. Evolutionary conserved domains of D. magna Ftz-F1.
(A) Schematic diagram of the Ftz-F1 regions that are divided into A/B, C, D and E regions. (B) Alignment of the C region and the Ftz-F1 box (boxed), and (C) alignment of the E region showing the LBD signature domain (boxed) and AF-2 motif (boxed). Identical amino acids are shaded in black whereas amino acids with similar characteristics are colored in red. MeFtz-F1 is the Me. ensis (shrimp) protein; DmFtz-F1 is the Dr. melanogaster (fruit fly) protein; BmFtz-F1 is the B. mori (silkworm) protein; and TcFtz-F1 is the T. castaneum (beetle) protein. MmSf-1 is the steroidogenic factor-1 of Mu. musculus (mouse); and CeNhr-25 is the nuclear hormone receptor-25 of C. elegans (roundworm).
Phylogenetic Analysis of DapmaFtz-F1
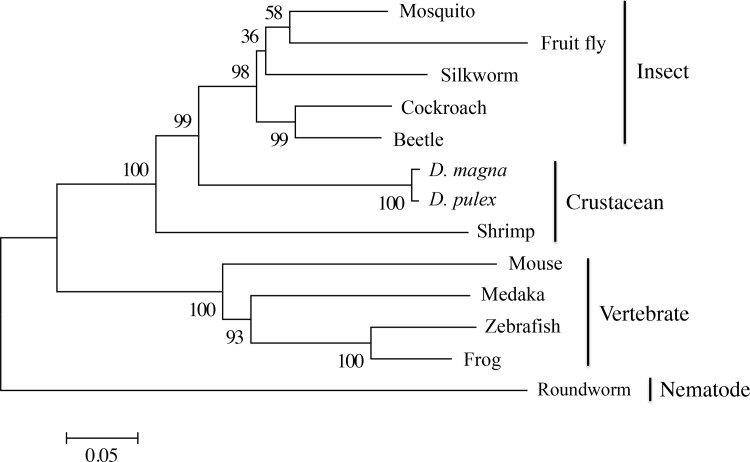
To analyze the evolutionary relationship of DapmaFtz-F1 further, a phylogenetic tree of DapmaFtz-F1 with 12 other Ftz-F1 related genes was constructed by the neighbor-joining method, using whole amino acid sequences (Fig 3). The topology of the phylogenetic relationship between Ftz-F1 orthologs was in good agreement with the taxonomic relationship between insects and crustaceans. Compared to the Ftz-F1 ortholog of a shrimp belonging to malacostracan crustaceans, the branchiopod crustacean Daphnia Ftz-F1 was more closely related to insect Ftz-F1 orthologs. Our result supports the hypothesis that insects originated from branchiopod crustaceans [35].
Fig 3. Phylogenetic tree of the amino acid sequences of the DBD and LBD Ftz-F1 nuclear hormone receptor subfamily.
The percentages of the replicate tree in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The bar indicates branch length and corresponds to the mean number of the differences (P<0.05) per residue along each branch. Evolutionary distances were computed using the p-distance method.
Genomic Organization of the DapmaFtz-F1 Gene
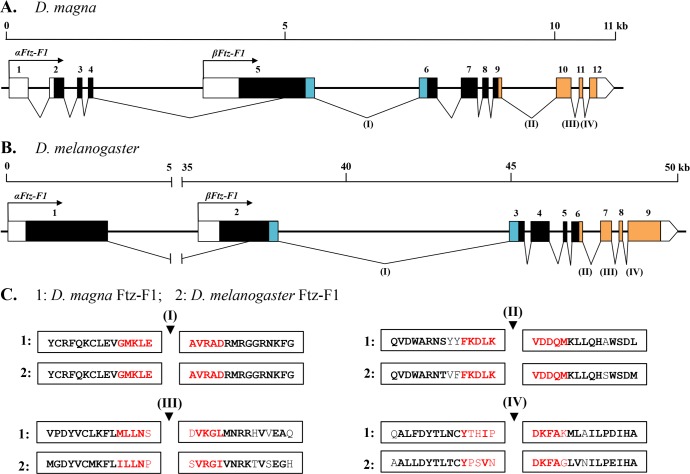
Next, we mapped the DapmaFtz-F1 transcripts to the genomic sequences and examined the exon-intron structure. The genomic structure of the DapmaFtz-F1 gene is composed of 12 exons, spread over ~11 kb of genomic DNA as illustrated in Fig 4A. All exon-intron junctions possessed the consensus “GT-AG” nucleotides at their 5′ and 3′ splicing sites. αFtz-F1 contains all 12 exons except a partial deletion of exon 5, whereas the βFtz-F1 lacks exons 1–4. The region encoding the DBD is within two exons (i.e., exon 5 and 6) that are separated by a large intron of 2,006 bp, and the LBD region is located in four exons, which are exons 9–12.
Fig 4.
Genomic structural organization of (A) the D. magna Ftz-F1 gene and (B) the Dr. melanogaster Ftz-F1 gene. The numbered boxes are exons, and the intervening lines are introns. Colored boxes indicate coding regions; blue represents DBD, whereas orange represents LBD. Empty boxes indicate untranslated regions. Each isoform possesses a unique coding sequence at the 5′end, with black arrows indicating the transcription start site. Scale bars are provided at the top of each diagram for the size in kilobases. Numbers I, II, III, and IV indicate the location of intron splice sites that are conserved between D. magna and Dr. melanogaster. (C) Putative conserved splice sites mapped to the conserved domain of Ftz-F1 from (1) D. magna and (2) Dr. melanogaster. The amino acid sequences shown are from DBD (Number I) and LBD (Numbers II, III, and IV). Red amino acids indicate 10 residues (five residues for pre- and post-introns, respectively) around the intron position assigned as the splice site, whereas further homology up and downstream of the intron is represented in black. Bold amino acid residues are residues shared between two species. Black triangles indicate the location of the intron within the splice site.
The exon arrangement is similar to the structural organization of Dr. melanogaster Ftz-F1 (Fig 4B) [36,37]. The common region between αFtz-F1 and βFtz-F1 is composed of eight exons and seven introns in both D. magna and Dr. melanogaster. Importantly, four of the seven intron positions are conserved between the two species (I, II, III, and IV in Fig 4C). The first position (I) is at the end of the C region just upstream of the Ftz-F1 box within the DBD. The other three intron positions (II, III, and IV) are located at the E region or LBD. Taken together with the result of the phylogenetic analysis, we speculate that DapmaFtz-F1 has diverged from an ancestral gene common to branchiopod crustacean and insect Ftz-F1 genes.
DapmaFtz-F1 mRNA Expression during Embryogenesis
Because the genes related to sex determination and differentiation are known to show sex-specific differences in the abundance of transcripts [14], we next investigated the sexual differences of DapmaFtz-F1 expression at various embryonic stages using a quantitative real-time PCR assay. Isoform-specific amplification was achieved by designing primers at the 5′ end of each coding region of αFtz-F1 and βFtz-F1 transcripts, as shown in Fig 1B. Adults were exposed to the JH agonist Fenoxycarb 9 h before oviposition, a critical stage for sex determination [13], inducing the ovulated eggs to develop as males. The eggs that developed as females were collected from unexposed mothers. The result of qRT-PCR analyses is presented in S1 File and Fig 5.
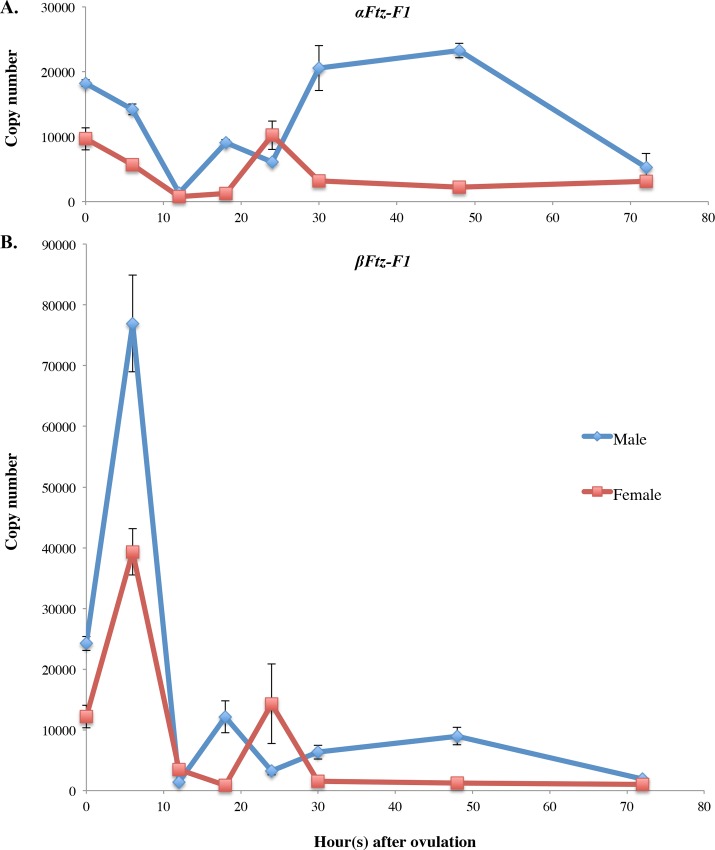
Fig 5. Temporal expression profiles of the DapmaFtz-F1 gene in embryonic developmental stages of D. magna.
(A) DapmaFtz-f1 gene expression levels of the α-isoform and (B) the β-isoform in one embryo of males (blue line) and females (red line). Embryonic development was staged at 0 h (single cell egg), 6 h (late gastrulation stage), 12 h (cephalic-appendage developing stage), 18 h (early thoracic appendage-developing stage), 24 h (after hatching embryo), 30 h (middle carapace-developing stage), 48 h (further developed thoracic appendages and antennae embryo), and 72 h (juvenile Daphnia) after oviposition. Results are shown as copy numbers of transcript per egg. The copy numbers were measured from three independent qPCR amplifications and error bars represent standard error values across samples.
Just after oviposition (0 h), αFtz-F1 expression in males was almost two-fold higher than in females. Between 0 and 12 h, αFt-zF1 expression gradually decreased in both males and females. In the later embryonic stages (post 12 h) αFtz-F1 was absent in females, except for a detectable peak at 24 h; in males it was expressed at each time point but was depleted by 72 h (Fig 5A), when the embryos become juveniles and swim out from their mother’s brood chamber. The temporal change in βFtz-F1 expression was more prominent, especially during early embryogenesis (Fig 5B). At 0 h, the quantity of βFtz-F1 mRNA was also two-fold higher in males. This isoform was activated three-fold at 6 h during the gastrulation stage and then dropped at 12 h in both males and females. During middle and late embryogenesis, the β isoform exhibited sexually dimorphic expression; however, its expression level was lower than that of the α isoform.
As summarized in Table 2, the DapmaFtz-F1 transcripts are dominantly expressed in males at most embryonic stages, except at 24 h for the αFtz-F1 isoform, and at 12 h and 24 h for the βFtz-F1 isoform, suggesting that DapmaFtz-F1 may play a role in regulating male trait development during embryogenesis.
Table 2. Sexual differences in αFtz-F1 and βFtz-F1 expression in D. magna during embryogenesis.
Ftz-F1 expression was normalized using Ribosomal L32 expression as a reference gene. The fold difference was obtained by normalizing male expression to female expression.
| Time after ovulation (h) | αFtz-F1 expression | αFtz-F1 expression | ||
|---|---|---|---|---|
| Fold difference | Std. dev. | Fold difference | Std. dev. | |
| 0 | 1.88 | ± 0.09 | 1.98 | ± 0.17 |
| 6 | 2.52 | ± 0.25 | 1.96 | ± 0.35 |
| 12 | 1.74 | ± 0.31 | 0.41 | ± 0.05 |
| 18 | 7.32 | ± 0.72 | 14.49 | ± 5.40 |
| 24 | 0.60 | ± 0.06 | 0.22 | ± 0.07 |
| 30 | 6.49 | ± 1.90 | 4.12 | ± 1.30 |
| 48 | 10.33 | ± 0.87 | 7.06 | ± 1.95 |
| 72 | 1.68 | ± 1.21 | 1.91 | ± 0.34 |
Std. dev. = standard deviation.
Time-Lapse Imaging of DapmaFtz-F1 RNAi Embryos
To examine the roles of the DapmaFtz-F1 gene, DapmaFtz-F1 expression was knocked down using the RNAi method [30] in male and female embryos. We used transgenic Daphnia, carrying the GFP gene fused to histone H2B gene [25], because the nuclear stain with the H2B-GFP protein enhances the visualization of cell dynamics in live embryos. Embryonic development was recorded by time-lapse imaging from 3 h to 30 h after ovulation. The development of knockdown embryos was similar to the control egg (non-injected) until the gastrulation stage (6 h). Subsequently, GFP intensity started to weaken and the development of RNAi embryos slowed down and did not develop normally (S1 Movie). All of the Ftz-F1 siRNA-injected embryos failed to hatch, but their development slowly continued up to the eye developmental stage, because we observed that some of the injected embryos developed eye pigment after a 48 h incubation (Table 3). There was no difference in the RNAi phenotype between male and female embryos that were observed in this experiment.
Table 3. Summary of the siRNA microinjection experiment for phenotype observation.
Excluding the Control_416 siRNA-injected embryos, all Ftz-F1 siRNA-injected embryos developed abnormally, including failure to shed the outer egg membrane and slow development compared to that observed with normal eggs.
| siRNA (100 μM) | Sex of egg | Number of injected eggs | Survived for 24 h | Developed eye pigment by 48 h |
|---|---|---|---|---|
| Control_416 | Female | 9 | 7 | 7 |
| Control_416 | Male | 12 | 10 (*6) | 4 |
| Ftz-F1_699 | Female | 11 | 7 | 4 |
| Ftz-F1_918 | Female | 10 | 8 | 8 |
| Ftz-F1_918 | Male | 19 | 12 (*6) | 6 |
* The embryos were subjected to total RNA isolation for qRT-PCR analysis.
To confirm that the DapmaFtz-F1 expression has been successfully knocked down during RNAi, total RNA from 24 h siRNA injected-male embryos was isolated, and the DapmaFtz-F1 expression level was measured. The qRT-PCR analysis (S2 File) showed that 90 ± 5% of expression was suppressed when compared to the control, indicating that RNAi effectively occurred in the siRNA-injected embryos. In short, the results suggested that DapmaFtz-F1 was essential for embryonic development of D. magna.
Although embryonic lethality prevented us from analyzing sex-specific functions of DapmaFtz-F1, its expression pattern suggested three potential roles in Daphnia ESD. First, in one cell embryos at 0 h after ovulation, male embryos exhibited two-fold higher expression of both isoforms compared to that observed in females (Table 2), suggesting that JH-dependent activation of DapmaFtz-F1 occurred during oogenesis, and the synthesized transcripts were deposited into eggs as maternal RNAs. This may imply that DapmaFtz-F1 may be a direct target of JH, and these maternal transcripts may function in activating DapmaDsx1 during male development. Consistent with this assumption, we found several candidates of Ftz-F1- binding site in the DapmaDsx1 promoter (S3 File). Second, during the gastrulation stage, βFtz-F1 was transiently activated and highly expressed in males (Fig 5). Importantly, this isoform was more abundant compared to the αFtz-F1 transcript, suggesting that βFtz-F1 may have a dominant role in sex determination at this stage. Third, in the later stages of embryogenesis, the expression of the αFtz-F1 transcript was observed to be dominant, which may suggest that αFtz-F1 is responsible for male trait development. In previous studies, the Me. ensis Ftz-F1 was also detected in the testis [34] and Ftz-F1α of Xenopus laevis was discovered in the developing gonads and testis [38]. In addition, the homolog of Ftz-F1 in mammalians, Sf-1, is a critical regulator of normal development of the hypothalamic-pituitary-gonadal axis during reproduction and sexual differentiation [20]. These findings indicated that the role of Ftz-F1 orthologs in sexual development is evolutionarily conserved among species. To understand these possible functions of DapmaFtz-F1, more sophisticated methods for analyzing gene functions, such as tissue-specific and inducible knockdown methods, would be required in the future.
Conclusion
In this study, we identified the Ftz-F1 gene from D. magna and showed that DapmaFtz-F1 is very closely related to insect Ftz-F1 orthologs. Additionally, our study revealed that DapmaFtz-F1 expression during embryogenesis is sexually dimorphic for both isoform variants. We speculate that DapmaFtz-F1 may have sex-specific functions in environmental sex determination of D. magna.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
The development of three siRNA-injected embryos with one non-injected embryo as control (top) was recorded from 3–30 h after ovulation.
(AVI)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files. All sequence files are available from the DDBJ database (http://www.ddbj.nig.ac.jp/index-e.html) (accession numbers LC105700, LC105701).
Funding Statement
This work was supported by JSPS KAKENHI Grant Number 26840105 (to YK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bull J. Sex determining mechanisms: an evolutionary perspective. Experientia. 1985; 1285–1296. [DOI] [PubMed] [Google Scholar]
- 2.Marin I, Baker B. The Evolutionary Dynamics of Sex Determination. Science. 1998;281: 1990–1994. [DOI] [PubMed] [Google Scholar]
- 3.Zarkower D. Establishing sexual dimorphism: conservation amidst diversity? Nat Rev Genet. 2001;2: 175–185. [DOI] [PubMed] [Google Scholar]
- 4.Korpelainen H. Sex ratios and condition required for environmental sex determination animals. Biol Rev. Blackwell Publishing Ltd; 1990;65: 147–184. [DOI] [PubMed] [Google Scholar]
- 5.Crews D, Bull JJ. Mode and tempo in environmental sex determination in vertebrates. Semin Cell Dev Biol. 2009;20: 251–255. 10.1016/j.semcdb.2009.02.004 [DOI] [PubMed] [Google Scholar]
- 6.Hebert PDN. The population biology of Daphnia (Crustacea, Daphnidae). Biol Rev. Blackwell Publishing Ltd; 1978;53: 387–426. [Google Scholar]
- 7.Hobk A, Larsson P. Sex determination of Daphnia magna. Ecology. 1990;71: 2255–2268. [Google Scholar]
- 8.Kleiven OT, Larsson. P, Hobk A. Sexual reproduction in Daphnia magna requires three stimuli. Oikos. 1992;65: 197–206. [Google Scholar]
- 9.Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281: 1986–1990. [PubMed] [Google Scholar]
- 10.Oda S, Tatarazako N, Watanabe H, Morita M, Iguchi T. Production of male neonates in Daphnia magna (Cladocera, Crustacea) exposed to juvenile hormones and their analogs. Chemosphere. 2005;61: 1168–1174. [DOI] [PubMed] [Google Scholar]
- 11.Olmstead AW, Leblanc GA. Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J Exp Zool. 2002;293: 736–739. [DOI] [PubMed] [Google Scholar]
- 12.Tatarazako N, Oda S, Watanabe H, Morita M, Iguchi T. Juvenile hormone agonists affect the occurrence of male Daphnia. Chemosphere. 2003;53: 827–833. [DOI] [PubMed] [Google Scholar]
- 13.Kato Y, Kobayashi K, Oda S, Tatarazako N, Watanabe H, Iguchi T. Sequence divergence and expression of a transformer gene in the branchiopod crustacean, Daphnia magna. Genomics. Elsevier Inc.; 2010;95: 160–165. 10.1016/j.ygeno.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 14.Kato Y, Kobayashi K, Watanabe H, Iguchi T. Environmental Sex Determination in the Branchiopod Crustacean Daphnia magna: Deep Conservation of a Doublesex Gene in the Sex-Determining Pathway. PLoS Genet. 2011;7: e1001345 10.1371/journal.pgen.1001345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda H, Sonoda S, Brown JL, Scott MP, Wu C. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 1990;4: 624–35. [DOI] [PubMed] [Google Scholar]
- 16.Lala DS, Rice D a, Parker KL. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol. 1992;6: 1249–1258. [DOI] [PubMed] [Google Scholar]
- 17.Honda S-I, Morohashi K-I, Nomura M, Takeya H, Kitajimaf M, Omura T. Ad4BP Regulating Steroidogenic P-450 Gene Is a Member of Steroid Hormone Receptor Superfamily. J Biol Chem. 1993;268: 7494–7502. [PubMed] [Google Scholar]
- 18.Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans Ortholog of ftz-f1, Is Required for Epidermal and Somatic Gonad Development. Dev Biol. 2000;221: 259–272. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda Y, Lala DS, Luo X, Kim E, Moisan MP, Parker KL. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol Endocrinol. 1993;7: 852–860. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda Y, Shen WH, Ingraham H a, Parker KL. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol Endocrinol. 1994;8: 654–662. [DOI] [PubMed] [Google Scholar]
- 21.Dubrovsky EB, Dubrovskaya V a., Bernardo T, Otte V, DiFilippo R, Bryan H. The Drosophila FTZ-F1 nuclear receptor mediates juvenile hormone activation of E75A gene expression through an intracellular pathway. J Biol Chem. 2011;286: 33689–33700. 10.1074/jbc.M111.273458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardo T, Dubrovsky EB. The Drosophila Juvenile Hormone Receptor Candidates Methoprene-tolerant (MET) and Germ Cell-expressed (GCE) Utilize a Conserved LI XX L Motif to Bind the FTZ-F1 Nuclear. 2012;287: 7821–7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato Y, Kobayashi K, Oda S, Colbourn JK, Tatarazako N, Watanabe H, et al. Molecular cloning and sexually dimorphic expression of DM-domain genes in Daphnia magna. Genomics. 2008;91: 94–101. [DOI] [PubMed] [Google Scholar]
- 24.Kluttgen B, Dulmer U, Engels M, Ratte H. ADaM, an artificial freshwater for the culture of zooplankton. Water Res. 1994;28: 743–746. [Google Scholar]
- 25.Kato Y, Matsuura T, Watanabe H. Genomic integration and germline transmission of plasmid injected into crustacean Daphnia magna eggs. PLoS One. 2012;7: e45318 10.1371/journal.pone.0045318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sagawa K, Yamagata H, Shiga Y. Exploring embryonic germ line development in the water flea, Daphnia magna, by zinc-finger-containing VASA as a marker. Gene Expr Patterns. 2005;5: 669–678. [DOI] [PubMed] [Google Scholar]
- 29.Mittmann B, Ungerer P, Klann M, Stollewerk A, Wolff C. Development and staging of the water flea Daphnia magna (Straus, 1820; Cladocera, Daphniidae) based on morphological landmarks. Evodevo. 2014;5: 12 10.1186/2041-9139-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato Y, Shiga Y, Kobayashi K, Tokishita SI, Yamagata H, Iguchi T, et al. Development of an RNA interference method in the cladoceran crustacean Daphnia magna. Dev Genes Evol. 2011;220: 337–345. 10.1007/s00427-011-0353-9 [DOI] [PubMed] [Google Scholar]
- 31.Asada M, Kato Y, Matsuura T, Watanabe H. Early Embryonic Expression of a Putative Ecdysteroid-Phosphate Phosphatase in the Water Flea, Daphnia magna (Cladocera: Daphniidae). J Insect Sci. 2014;14: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yussa M, Löhr U, Su K, Pick L. The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mech Dev. 2001;107: 39–53. [DOI] [PubMed] [Google Scholar]
- 33.Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, et al. A canonical structure for the ligand-binding domain of nuclear receptor. Nat Struct Biol. 1996;3: 87–94. [DOI] [PubMed] [Google Scholar]
- 34.Chan SM, Chan KM. Characterization of the shrimp eyestalk cDNA encoding a novel fushi tarazu-factor 1 (FTZ-F1). FEBS Lett. 1999;454: 109–114. [DOI] [PubMed] [Google Scholar]
- 35.Glenner H, Thomsen PF, Hebsgaard MB, Sorensen M V., Willerslev E. EVOLUTION: The Origin of Insects. Science. 2006;314: 1883–1884. [DOI] [PubMed] [Google Scholar]
- 36.Lavorgna G, Ueda H, Clos J, Wu C. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of the fushi tarazu. Science. 1991;252: 848–851. [DOI] [PubMed] [Google Scholar]
- 37.Yamada M, Murata T, Hirose S, Lavorgna G, Suzuki E, Ueda H. Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development. 2000;127: 5083–5092. [DOI] [PubMed] [Google Scholar]
- 38.Takase M, Nakajima T, Nakamura M. FTZ-F1alpha is expressed in the developing gonad of frogs. Biochim Biophys Acta. 2000;1494: 195–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
The development of three siRNA-injected embryos with one non-injected embryo as control (top) was recorded from 3–30 h after ovulation.
(AVI)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All sequence files are available from the DDBJ database (http://www.ddbj.nig.ac.jp/index-e.html) (accession numbers LC105700, LC105701).