Abstract
WRKY proteins play important regulatory roles in plant developmental processes such as senescence, trichome initiation and embryo morphogenesis. In strawberry, only FaWRKY1 (Fragaria × ananassa) has been characterized, leaving numerous WRKY genes to be identified and their function characterized. The publication of the draft genome sequence of the strawberry genome allowed us to conduct a genome-wide search for WRKY proteins in Fragaria vesca, and to compare the identified proteins with their homologs in model plants. Fifty-nine FvWRKY genes were identified and annotated from the F. vesca genome. Detailed analysis, including gene classification, annotation, phylogenetic evaluation, conserved motif determination and expression profiling, based on RNA-seq data, were performed on all members of the family. Additionally, the expression patterns of the WRKY genes in different fruit developmental stages were further investigated using qRT-PCR, to provide a foundation for further comparative genomics and functional studies of this important class of transcriptional regulators in strawberry.
Introduction
Members of the WRKY class of transcription factors, which are ubiquitous among higher plants, exhibit sequence-specific DNA-binding and are capable of activating or repressing the transcription of downstream target genes [1]. Proteins in this superfamily contain either one or two highly conserved signature domains of approximately 60 amino acid residues, including the conserved WRKYGQK sequence followed by a zinc finger structure in the C-terminal region [2]. Studies have also shown that the conserved WRKY domain can have slightly longer sequences, such as WRKYGKK and WEKYGQK [3], or that it can be replaced by WKKY, WKRY, WSKY, WIKY, WRIC, WRMC, WRRY or WVKY. The conservation of the WRKY domain is mirrored by a remarkable conservation of its cognate binding site, the W box (TTGACC/T) [4–6]. The WRKY domain facilitates binding of the protein to the W box or the SURE (sugar-responsive cis-element) element in the promoter regions of target genes [7, 8]. As described by Eulgem et al. (2000), WRKY proteins can be divided into three major groups based on both the number of WRKY domains and the specific features of their zinc-finger-like motif: WRKY proteins with two WRKY domains containing a C2H2 zinc-finger motif belong to group I, whereas most proteins with one WRKY domain containing a C2H2 zinc-finger motif belong to group II, which can be further divided into five subgroups (IIa, IIb, IIc, IId and IIe). Generally, the same type of finger motif is characteristic for group I and group II members (C-X4-5-C-X22-23-H-X1-H,). Group III consists of a small number of genes characterized by a single WRKY domain with a C2HC zinc-finger motif.
Since the first cDNA encoding a WRKY protein, SPF1, was cloned from sweet potato (Ipomoea batatas) [9], numerous members of the family have been characterized from several plant species, and they have been found to be involved in various physiological processes under normal growth conditions and under various biotic and abiotic stresses [10–20]. There are at least 72 WRKY family members in Arabidopsis thaliana and at least 109 in rice (Orza sativa). Furthermore, 55 WRKY genes have been identified in the cucumber (Cucumis sativus) genome and 59 putative grapevine (Vitis vinefera) WRKY transcription factors were also identified following a search of various genomic and proteomic grapevine databases [21,22]. Many of the A. thaliana WRKY proteins appear to be involved in regulating the balance between salicylic acid (SA)- and jasmonic acid (JA)-dependent defense pathways. One example is AtWRKY70, a common regulatory component of SA- and JA-dependent defense signaling, which mediates the cross-talk between these antagonistic pathways and is a positive regulator of R-gene mediated resistance and systemic defense responses [23,24]. Microarray analyses have revealed that expression of some of the A. thaliana WRKY transcripts is strongly regulated by various abiotic stresses, such as salinity, drought and cold [25,26]. Moreover, abiotic stresses (salinity, drought and cold) and phytohormone treatments were reported to result in changes in the transcript levels of 54 rice WRKY genes [20]. Finally, WRKY proteins are also known to play important regulatory roles in developmental processes such as senescence, trichome initiation and embryo morphogenesis [27–30].
While the WRKY family has been well studied in model experimental plants, such as A. thaliana and rice, less is known about their function and regulation in other species, including those of agronomic or horticultural value. In this study we used as our experimental system F. vesca, the wild strawberry, which is diploid, unlike cultivated strawberry (Fragaria × ananassa) is octoploid. F. vesca therefore has a relatively small (~240 Mb) genome, which has been sequenced, as well as a short life cycle (3.5 to 4 months) and a facile transformation system. These characteristics have resulted in a substantial increase in the number of physiological and molecular studies of this species. In cultivated strawberry, only FaWRKY1 has been characterized to date and it has been shown to be involved in mediating defense responses to the fungus Colletotrichum acutatum. Specifically, the expression of FaWRKY1 is up-regulated in strawberry following C. acutatum infection, treatments with elicitors, and wounding [31]. However, to our knowledge, there are no reported studies of WRKY genes from F. vesca and nothing is known of their potential association with fruit development and ripening. This is of particular interest given that strawberry is not only an economically important cultivated fruit crop, but also a model system for studies of these processes.
In this current study we identified a total of 59 FvWRKY genes from the recently reported F. vesca ‘Hawaii 4’ genome sequence [32]. We report here the classification, annotation and phylogenetic evaluation of these genes, together with an assessment of conserved motifs and the results of expression profiling of members of the WRKY gene family, based on RNA-seq data. The expression patterns of WRKY genes in different fruit developmental stages were further investigated using quantitative real-time reverse transcription PCR (qRT-PCR). Our results provide a foundation for further comparative genomics and functional studies of this class of transcriptional regulators in strawberry.
Materials and Methods
Identification of Putative F. vesca WRKY Genes
To generate a comprehensive list of F. vesca WRKY genes, annotated strawberry protein sequences were downloaded from the public databases, F. vesca BioView Gene Model Database (https://strawberry.plantandfood.co.nz/) and Genome Database for Rosaceae (http://www.rosaceae.org/species/fragaria/fragaria_vesca/genome_v2.0.a1). The Arabidopsis WRKY gene family database was obtained from TAIR (The Arabidopsis Information Resource, http://www.arabidopsis.org) and used for comparative analysis.
Gene Structure Construction, Phylogenetic Analysis and Classification of the F. vesca WRKY Family
All identified F. vesca WRKY (FvWRKY) genes were classified into different groups based on the AtWRKY classification scheme, and the alignment of FvWRKY and AtWRKY DNA-binding domains was performed using Clustal X 2.147 [33] with default settings. The phylogenetic trees were created using MEGA 5.0 [34] and the neighbor-joining method. Bootstrap values were calculated for 1,000 iterations.
Analysis of the FvWRKY Exon-Intron Structures and Chromosomal Location
The exon-intron organization of the FvWRKY genes and their location on strawberry chromosomes were determined based on information available at the National Centre for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov).
Plant Material and Fruit Pre-Treatments
F.Vesca (‘Hawaii 4’) were cultivated in growth chamber at 22±1°C in a 13/11 h dark/light photoperiod. Fruit samples were harvested at 18, 24, 30, 36 and 42 DAF (days after flowering). At each developmental stage, ten representative fruits were sampled, snap-frozen in liquid nitrogen and kept at -80°C until further use. For different pre-treatment experiments, two stages (18 and 36 DAF) were selected for sucrose and hormone treatments. The fruits were cut in half longitudinally, and half was used for processing while the other half was used as a control. The hormones used for treatments were indole-3-acetic acid (IAA) and abscisic acid (ABA), each at a concentration of 100 μM and the sucrose concentration used was 50 μM. All experiments were performed at 25°C.
RNA Extraction, RNA-Seq Based Expression Analysis and Real-Time PCR Analysis
Total RNA was extracted strawberry from ruit harvested at 18, 24, 30, 36 and 42 DAF using the Plant RNA Kit (Omega) according to the manufacturer’s instructions. And Total RNA was reverse transcribed into cDNA by the Invitrogen reverse transcription kit (SuperScript III Reverse Transcriptase). The RNA was subjected to RNA-seq analysis using an Illumina Genome Analyzer at Beijing Novogene Corporation. Real-time PCR was performed using a Light Cycler® 96 SW1.1 Real Time PCR System (Roche), with SYBR-Green (Takara, Dalian, China). The primer sequences used (S1 Table) were designed based on WRKY gene sequences using the Beacon designer software. These sequences were subsequently verified using the BLAST tool at NCBI and a dissociation curve was also analyzed after the PCR reaction to confirm their specificity. Each reaction was carried out in a 10 μL volume, consisting of 5 μL SYBR, 3.5 μL ddH2O, 1 μL diluted template (1 μL of the generated first-strand cDNA diluted by 9 μL ddH2O) and 0.25 μL of each of two gene specific primers. The following program was used for RT-PCR: 95°C for 10 min followed by 40 cycles at 95°C for 20 s, 54°C for 20 s, 72°C for 20 s.
Results
Identification and Annotation of the Strawberry WRKY Family and Its Chromosomal Distribution
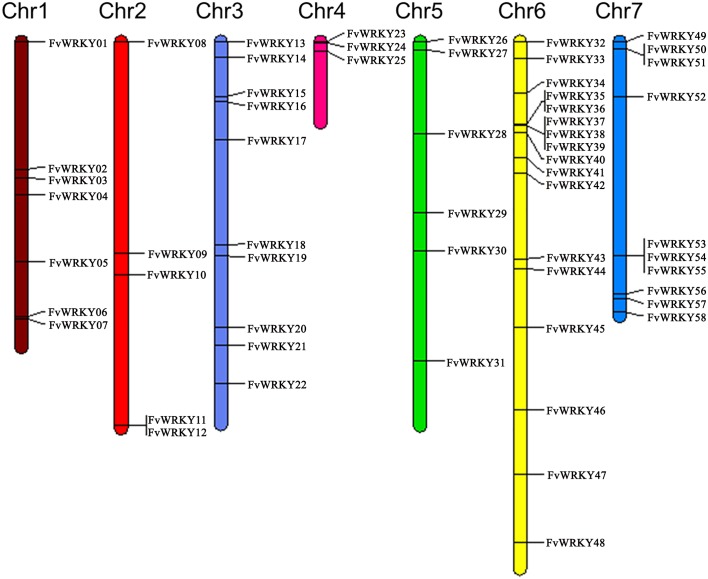
A total of 59 full length gene sequences were identified in the strawberry genome as putative members of the WRKY superfamily All the 59 gene sequences were obtained from the F. vesca Whole Genome v2.0.a1 Assembly & Annotation (https://www.rosaceae.org/species/fragaria_vesca/genome_v2.0.a1). All 58 identified WRKY genes were mapped to the respective chromosomes and were renamed from FvWRKY1 to FvWRKY58, based on their order on the chromosomes, from chromosomes 1 to 7 (Fig 1) [35]. The parameters used to characterize each of the predicted FvWRKY proteins are listed in Table 1, and included the deduced protein length, molecular weight, isoelectric point, aliphatic index and grand average of hydropathicity. The deduced length of the FvWRKY proteins ranged from 155 (FvWRKY45) to 1,348 amino acids (FvWRKY55), while the pI values ranged from 4.92 (FvWRKY31) to 9.97 (FvWRKY29), which suggests that different FvWRKY proteins might operate in different microenvironments.
Fig 1. Chromosomal distribution of the Fragaria vesca L. WRKY gene family.
Chromosome size is indicated by relative length. The putative WRKY genes from FvWKRY1 to FvWRKY58 were renamed based on their placement on the chromosomes. Only one gene, FvWRKY59, was not assigned to any chromosome.
Table 1. WRKY genes identified in Fragaria vesca.
| Number | Proposed | Gene ID | Chr | ORF (aa) | MW (kDa) | pI | Ai | GRAVY | Instability index (II) | WRKYGQK | Domain pattern | Group | Zinc finger |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | |||||||||||||
| 1 | FvWRKY01 | 101313240 | 1 | 467 | 50.9959 | 6.68 | 66 | -0.733 | 58.94 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| 2 | FvWRKY02 | 101302596 | 1 | 600 | 64.5571 | 5.2 | 61.07 | -0.713 | 46.27 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 3 | FvWRKY03 | 101310739 | 1 | 319 | 34.7101 | 9.61 | 62.41 | -0.584 | 52.08 | 1 | C-X5-C-X23-H-X-H | IId | C2H2 |
| 4 | FvWRKY04 | 101299520 | 1 | 243 | 27.8051 | 9.08 | 60.95 | -0.807 | 54.19 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 5 | FvWRKY05 | 101295368 | 1 | 425 | 46.6975 | 6.79 | 69.39 | -0.574 | 51.93 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 6 | FvWRKY06 | 101307337 | 1 | 317 | 35.3775 | 6.66 | 54.76 | -0.747 | 66.56 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 7 | FvWRKY07 | 101311038 | 1 | 205 | 22.9693 | 6.15 | 47.56 | -0.83 | 42.61 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 8 | FvWRKY08 | 101300971 | 2 | 348 | 38.908 | 9.71 | 67.82 | -0.749 | 56.22 | 1 | C-X5-C-X23-H-X-H | IId | C2H2 |
| 9 | FvWRKY09 | 101295677 | 2 | 334 | 36.1088 | 9.54 | 58.14 | -0.488 | 52.56 | 1 | C-X5-C-X23-H-X-H | IId | C2H2 |
| 10 | FvWRKY10 | 101302035 | 2 | 342 | 37.0599 | 9.51 | 62.78 | -0.57 | 51.89 | 1 | C-X5-C-X23-H-X-H | IId | C2H2 |
| 11 | FvWRKY11 | 101297319 | 2 | 382 | 42.3033 | 6.27 | 62.57 | -0.656 | 54.58 | 1 | C-X5-C-X23-H-X-H | IIa | C2H2 |
| 12 | FvWRKY12 | 101297610 | 2 | 333 | 36.8124 | 8.89 | 67.69 | -0.655 | 47.47 | 1 | C-X5-C-X23-H-X-H | IIa | C2H2 |
| 13 | FvWRKY13 | 101300421 | 3 | 571 | 61.544 | 7.15 | 58.37 | -0.742 | 49.19 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 14 | FvWRKY14 | 101291408 | 3 | 505 | 55.3735 | 6.4 | 65.94 | -0.917 | 41.08 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 15 | FvWRKY15 | 101311736 | 3 | 355 | 39.182 | 7 | 45.58 | -0.943 | 58.42 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 16 | FvWRKY16 | 101292382 | 3 | 497 | 53.6208 | 5.92 | 52.86 | -0.74 | 51.9 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 17 | FvWRKY17 | 101314070 | 3 | 277 | 30.4295 | 5.19 | 46.86 | -0.789 | 65.15 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 18 | FvWRKY18 | 101291609 | 3 | 547 | 59.5604 | 6.22 | 67.62 | -0.59 | 40.27 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 19 | FvWRKY19 | 101307469 | 3 | 573 | 62.3768 | 6.73 | 54.42 | -0.847 | 57.7 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 20 | FvWRKY20 | 101305151 | 3 | 627 | 68.6763 | 7.68 | 42.33 | -1.046 | 49.75 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 21 | FvWRKY21 | 101313293 | 3 | 734 | 79.1599 | 5.87 | 51.42 | -0.788 | 51.55 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 22 | FvWRKY22 | 101301660 | 3 | 378 | 41487.2 | 5.67 | 45.95 | -0.93 | 58.88 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 23 | FvWRKY23 | 101292010 | 4 | 313 | 34.1893 | 6.45 | 46.04 | -0.915 | 65.87 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 24 | FvWRKY24 | 101295510 | 4 | 499 | 54.7651 | 7.69 | 43.77 | -1.029 | 57.49 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 25 | FvWRKY25 | 101309152 | 4 | 615 | 67.7268 | 6.67 | 58.28 | -0.812 | 46.46 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 26 | FvWRKY26 | 101293392 | 5 | 309 | 35.2252 | 5.14 | 58.71 | -0.777 | 61.02 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 27 | FvWRKY27 | 101310412 | 5 | 355 | 40.0883 | 4.96 | 57.15 | -0.755 | 63.99 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 28 | FvWRKY28 | 101314661 | 5 | 672 | 72.805 | 6.95 | 54.96 | -0.834 | 55.83 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 29 | FvWRKY29 | 101291727 | 5 | 301 | 32.8982 | 9.97 | 57.04 | -0.674 | 53.72 | 1 | C-X5-C-X23-H-X-H | IId | C2H2 |
| 30 | FvWRKY30 | 101294370 | 5 | 520 | 57.9652 | 5.02 | 57.13 | -0.981 | 60.31 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 31 | FvWRKY31 | 101296670 | 5 | 282 | 32.3806 | 4.92 | 53.19 | -1.126 | 69.82 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 32 | FvWRKY32 | 101307803 | 6 | 319 | 35.7679 | 9.58 | 70.56 | -0.641 | 55.3 | 1 | C-X5-C-X23-H-X-H | IId | C2H2 |
| 33 | FvWRKY33 | 101307611 | 6 | 727 | 80.232 | 5.88 | 61.49 | -0.753 | 50.05 | 2 | C-X4-C-X23-H-X-H | I | C2H2 |
| 34 | FvWRKY34 | 101296583 | 6 | 491 | 53.1033 | 7.56 | 64.81 | -0.588 | 56.05 | 1 | C-X5-C-X23-H-X-H | IIb | C2H2 |
| 35 | FvWRKY35 | 101295066 | 6 | 320 | 35.7606 | 5.39 | 73.78 | -0.553 | 53.99 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 36 | FvWRKY36 | 101296502 | 6 | 297 | 33.5437 | 7.17 | 69.6 | -0.564 | 51.69 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 37 | FvWRKY37 | 101297362 | 6 | 231 | 25.9552 | 9.32 | 67.1 | -0.659 | 49.84 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 38 | FvWRKY38 | 101297067 | 6 | 344 | 38.7567 | 5.41 | 61.77 | -0.729 | 48.9 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 39 | FvWRKY39 | 101297653 | 6 | 339 | 38.2152 | 5.64 | 58.38 | -0.761 | 46.04 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 40 | FvWRKY40 | 101311683 | 6 | 517 | 57.4636 | 7.63 | 43.58 | -1.016 | 62.51 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 41 | FvWRKY41 | 101301793 | 6 | 296 | 33.308 | 5.44 | 59.26 | -0.795 | 58.73 | 1 | C-X4-C-X23-H-X-H | NG | C2H2 |
| 42 | FvWRKY42 | 101296019 | 6 | 478 | 52.1898 | 9.14 | 55.46 | -0.889 | 49.8 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 43 | FvWRKY43 | 101301515 | 6 | 268 | 29.9031 | 5.12 | 59.25 | -0.797 | 53.14 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 44 | FvWRKY44 | 101301598 | 6 | 326 | 36.0794 | 8.8 | 64.02 | -0.789 | 51.99 | 1 | C-X5-C-X23-H-X-H | IIa | C2H2 |
| 45 | FvWRKY45 | 101307027 | 6 | 155 | 17.7763 | 5.13 | 43.29 | -1.139 | 49.85 | WRKYGKK | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 46 | FvWRKY46 | 101308483 | 6 | 368 | 41.6158 | 6.46 | 53.83 | -0.982 | 62.25 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 47 | FvWRKY47 | 101305864 | 6 | 519 | 56652.8 | 8.57 | 58.65 | -0.84 | 64.22 | 2 | C-X4-C-X22-H-X-H | I | C2H2 |
| C-X4-C-X23-H-X-H | |||||||||||||
| 48 | FvWRKY48 | 101293115 | 6 | 213 | 23.5498 | 8.76 | 45.87 | -0.982 | 49.77 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 49 | FvWRKY49 | 101295163 | 7 | 353 | 39.2606 | 5.23 | 56.01 | -0.684 | 49.55 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 50 | FvWRKY50 | 101307333 | 7 | 226 | 25.3266 | 9.16 | 67.35 | -0.781 | 38.1 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 51 | FvWRKY51 | 101304921 | 7 | 345 | 37.7218 | 8.5 | 51.45 | -0.71 | 49.42 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 52 | FvWRKY52 | 101312284 | 7 | 254 | 28.6893 | 7.76 | 44.84 | -1.021 | 52.76 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
| 53 | FvWRKY53 | 101315104 | 7 | 379 | 42.4161 | 6.63 | 54.64 | -0.731 | 51.64 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 54 | FvWRKY54 | 101306674 | 7 | 340 | 37.927 | 6.43 | 53.91 | -0.835 | 49.12 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 55 | FvWRKY55 | 101307732 | 7 | 1348 | 152.8182 | 6.25 | 91.05 | -0.27 | 42.6 | 1 | C-X7-C-X23-H-X-H(C) | NG | C2HC |
| 56 | FvWRKY56 | 101295568 | 7 | 372 | 40.9578 | 6.64 | 66.88 | -0.591 | 53 | 1 | C-X7-C-X23-H-X-H(C) | III | C2HC |
| 57 | FvWRKY57 | 101309009 | 7 | 1333 | 151.6718 | 6.81 | 89.56 | -0.28 | 47.5 | 1 | C-X7-C-X23-H-X-H(C) | NG | C2HC |
| 58 | FvWRKY58 | 101304426 | 7 | 430 | 46.3448 | 5.12 | 53.77 | -0.7 | 62.01 | 1 | C-X5-C-X23-H-X-H | IIe | C2H2 |
| 59 | FvWRKY59 | 101290922 | Un | 190 | 21.6712 | 9.45 | 48.74 | -0.878 | 41.59 | 1 | C-X4-C-X23-H-X-H | IIc | C2H2 |
Abbreviations: Ai, aliphatic index; Chr, chromosome number; GRAVY, grand average of hydropathicity; MW, molecular weight; NG, no chromosomal group identified; ORF, open reading frame; pI, isoelectric point; RDM, random chromosome; UN, unknown chromosome.
Using TblastN, 58 out of the 59 FvWRKY genes could be mapped to F. vesca chromosomes, indicating an unevenly comprehensive distribution of FvWRKYs within strawberry genome (Fig 1). One WRKY gene (101290922) could not be conclusively mapped to any chromosome and was renamed FvWRKY59. Chromosome 6, contained the largest number (17) of FvWRKY genes, followed by chromosome 3 and 7, each of which had ten FvWRKY genes. Seven were mapped to chromosome 1 and three to chromosome 4. Chromosomes 2 and 5 had five and six FvWRKY genes, respectively. According to the definition of a gene cluster provided by Holub [36], twelve FvWRKY genes were present in five clusters, of which two were on each of chromosomes 6 and 7, and one cluster was on chromosome 2 (Fig 1).
FvWRKY Exon and Intron Organization
To investigate the structural evolution of the F. vesca WRKY family, the exon-intron patterns were analyzed. The number of introns ranged from one to twenty (Fig 2), and a large number of genes contained two introns, while eight genes contained three introns and nine contained four introns. The FvWRKY44 and FvWRKY57 genes contained seven introns, FvWRKY2 and FvWRKY13 contained eight introns and FvWRKY26 and FvWRKY35 contained nine introns. FvWRKY43 and FvWRKY55 contained ten and twenty introns, respectively. Two genes contained only one intron and FvWRKY50 had no introns.
Fig 2. Structures of the FvWRKY genes.
Gene names are indicated on the left. Exons, represented by black or red boxes, were drawn to scale. Dashed lines connecting two exons represent an intron. Intron phases 0, 1 and 2 are indicated by numbers 0, 1 and 2, respectively. WRKY domains in the corresponding proteins are marked in red.
The phylogenetic relationship of the FvWRKY proteins was examined by multiple sequence alignment of their WRKY domains, which were highly conserved (S1 Fig). The intron located in the conserved WRKY domain could be classified as either a R-type intron or a V-type intron. R-type introns are spliced prior to an arginine residue (R), while V-type introns are spliced before a valine residue, located six amino acids after the second cysteine residue of the zinc finger C2H2 motif (S1 Fig). The V-type intron was observed in the WRKY domains of genes belonging to groups IIa and IIb, whereas the R-type intron was widely distributed amongst all other FvWRKY groups (S1 Fig).
Phylogenetic Analysis, Classification and Motif Analysis of the F. vesca WRKY Gene Family
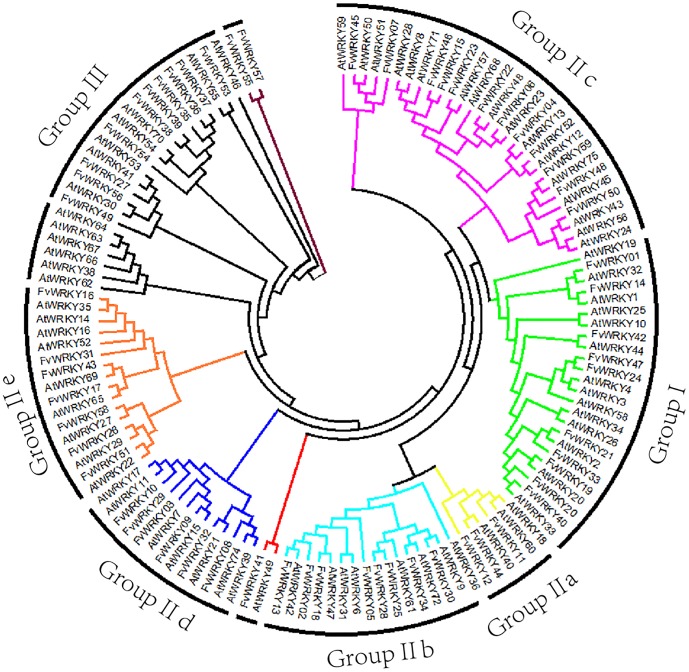
The distribution of structural domains can provide important insights into the evolution and relationship between highly divergent sequences [36]. Sequence comparisons, as well as phylogenetic and structural analyses showed that the A. thaliana WRKY domains could be classified into four large groups, termed groups I, II, III and IV [2]. Ten of the FvWRKY proteins belong to Group I, members of which contain two complete WRKY domains and a C2H2-type zinc finger motif (C-X4-C-X22-23-H-X1-H). Ten FvWRKY proteins, each with a single WRKY domain, were assigned to Group III, which is characterized by a C-X4-C-X23-H-X1-C C2HC zinc-finger structure. Finally, thirty-four FvWRKY proteins, possessing a single WRKY domain, were assigned to Group II, members of which differ from Group III FvWRKY proteins based on their C2H2-type zinc-finger structure (C-X5-C-X23-H-X1-H). Group II could be further subdivided into five distinct subgroups with 3, 8, 12, 6 and 7 members, respectively, based on their primary amino-acid sequence (Group IIa, IIb, IIc, IId and IIe). FvWRKY41 exhibited sequence divergence in the unique WRKY domain and so was not classified into any group. FvWRKY55 and FvWRKY57 were classified into Group IV, since they contained only one WRKY domain and a C2HC zinc-finger motif, however, they were distinct from other Group III members (Fig 3).
Fig 3. Phylogenetic tree based on WRKY domain sequences from Fragaria vesca and Arabidopsis thaliana.
The phylogenetic tree was created using the MEGA 5.0 software. Reliability of the predicted tree was tested using bootstrapping with 1,000 replicates. Numbers at the nodes indicate how often the group to the right appeared amongst the bootstrap replicates. Branch lines of subtrees are colored indicating different WRKY subgroups.
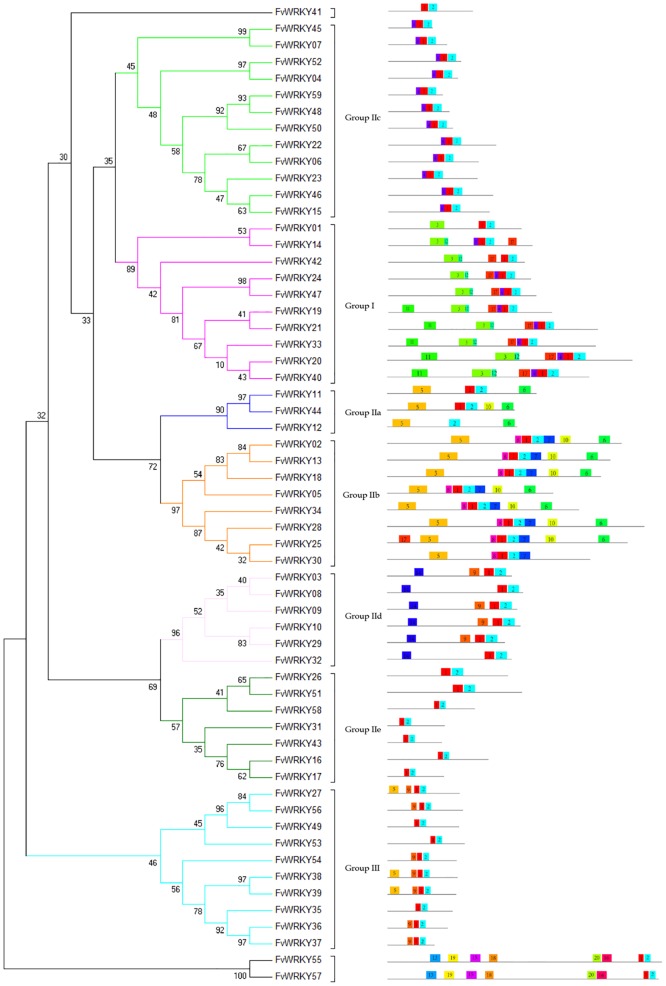
Conserved motifs other than the WRKY domain were detected by visual inspection (S1 Table). Four members (FvWRKY3, 9, 10 and 29) of Group IId were found to contain the HARF (RTGHARFRR [A/G]P) motif, whose function has not been clearly determined, and a putative leucine zipper structure was detected in FvWRKY11 (Group IIa). The LxxLL co-activator and LxLxLx repressor motifs were found in eight (FvWRKY3, 31, 46, 49, 50, 54, 55 and 57) and thirteen (FvWRKY11, 12, 25, 27, 28, 30, 32, 33, 34, 35, 50, 55 and 57) proteins, respectively, while FvWRKY50, 55 and 57 were observed to have both active repressor and co-activator motifs (Fig 4).
Fig 4. Phylogenetic tree based on the deduced FvWRKY domains and their associated motifs.
The phylogenetic tree was constructed using the MEGA 5 software. The reliability of the predicted tree was tested using bootstrapping with 1,000 replicates. Numbers at the nodes indicate how often the group to the right appeared amongst the bootstrap replicates. Subtree branch lines are colored indicating different WRKY subgroups. The motif composition related to each FvWRKY protein is displayed on the right-hand side. The motifs, numbered 1–20, are displayed in different colored boxes. The sequence information for each motif is provided in Table 2.
As shown in Table 2, twenty other distinct motifs were identified by the Multiple Expectation Maximization for Motif Elicitation online tool (http://meme-suite.org/tools/meme). The numbers of FvWRKY genes in each subgroup were compared with the corresponding numbers in other plant species, i.e., A. thaliana [37], rice [38], grapevine [22] and cucumber [21] (Table 3). They were found to be generally similar to the distribution in grapevine, although an clear expansion was detected in Group III.
Table 2. Analysis and distribution of conserved motifs in Fragaria vesca (strawberry) FvWRKY proteins.
| Motif | E-value | Width | Best possible match |
|---|---|---|---|
| 1 | 5.0e-1205 | 25 | DGY[RA]WRKYGQK[PV][IV]K[GN][SN]P[YH]PR[SG]YY[RK]C |
| 2 | 3.4e-994 | 29 | QGCP[VA][RK]K[QH]V[EQ]R[SA]S[ED]DPS[IM][LVF][IV]TTYEGEH[NT][HC] |
| 3 | 5.6e-314 | 50 | DGYNWRKYGQKQVKGSE[YFN]PRSYY[KR]CT[HF]PNCP[VT]KKKVERS[LH][DE]G[QH]ITEI[IV]YK |
| 4 | 1.6e-113 | 15 | PR[FV][AV][FV][QM]T[KRT]SE[VI]D[IH]L[DE] |
| 5 | 4.8e-090 | 46 | ELES[LAV]Q[AE]E[LM][QS][RE][VMG][RN]EE[NA][QK][RKQ]L[RK][GK][MV]LEQ[MTV]T[KE][SDN]Y[NEQ]AL[QE][TM][HKQ][LF]L[DE][VIL][LMR][QT][NQK][EQ]Q[LQ] |
| 6 | 1.9e-064 | 29 | [LTK][VI][ESAQ][ATQ][AM][TA][AKS][AS][IL]T[AKS]DP[NKT]F[TQ][AST][AV]LAAA[IL][ST]S[IS][IMV]G[NGT] |
| 7 | 1.1e-061 | 27 | PLP[PM][AS]A[TM][AS]MASTTSAAA[SA]MLLSGS[MS][STP]S |
| 8 | 7.6e-038 | 15 | [KR]ARVSVRAR[CS][ED][AST]P[MT][IML] |
| 9 | 6.5e-034 | 26 | KSRE[TS][FS][SK][DG]R[CR][HG][CS][SY]K[KR]RKT[SR][HV][KS][WR]TK[DR]V |
| 10 | 5.2e-026 | 25 | [CF][GS][SF][SY][MV][AP]T[IL]S[AT]S[AS][PS]FPT[IV]TLDLT[RS]SP |
| 11 | 3.2e-025 | 41 | [YCHS][LFS][TAY][IL][PV][PN][GA][LFIM][ST]P[TA][ADEST][LF]L[DES]SP[VM][FLM]L[SAP][NS][SM][NKLQ][AIT][LEQ]PSPTTG[SAKT]F[PALS][AFMNS][QLPV][AIKPS][FMNP][NDGV][WGHY] |
| 12 | 9.6e-019 | 13 | G[EQ]H[ND]H[PA]KPQ[PS][NT][RK][RP] |
| 13 | 2.0e-018 | 50 | CLG[EV]D[DG]VR[GV][IV]G[IV][CY]GMPGIGKTTIARAVY[DE]EI[TV]CQFEHYCFL[DE]NVKDGFKN |
| 14 | 4.2e-016 | 23 | [TA]Dx[AT]VSKFKKV[IV]SLLNR[GT][RG][HT][AG][HR][AF] |
| 15 | 7.1e-016 | 50 | WEDELEKIK[EK]IPH[LM]EIQ[GV]VLRTS[LY][DN]GL[DE]P[LS]QKDIFLDIACFFRGM[DE]KGYV |
| 16 | 8.3e-015 | 48 | A[IL]C[HY]C[ST][FL]KGNHG[EL]Y[EK]F[NS]F[QT]LLDWGF[RS]T[DN]R[FI]L[EQ]SDHMFL[AG]YVPWS[EQ]YR[CF] |
| 17 | 1.4e-010 | 29 | [DE][ED][GD][DE][DE][DES]E[PS][ENR][SA]KR[RW][KNR][IKM][ED][GANV]x[AN][SEI]E[VM][AS][AI][PAS][GHT][REKS][ART][IV] |
| 18 | 1.8e-010 | 41 | WEI[IV]RQQS[IV]K[DE]PGKRSRLW[IV]YEDV[AD][HR]V[FL]TQN[MT]ATDAVE[CG]IM |
| 19 | 4.0e-010 | 49 | KKVLLVLDDVE[NT]FAQIEALLGKQ[CH]SFGGGSRII[IV]TTRDIQSLSGV[NQ][AE]RY |
| 20 | 5.5e-010 | 41 | WFN[HN]QC[KR]G[FS]SV[NT]V[KQ]LP[PQ]NWFD[DN][EK]FLGFA[IL]C[AV]VS[DN]FKG[AP]HND |
Table 3. Size of the WRKY groups and sub-groups in different plant species.
| Group II | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group I | IIa | IIb | IIc | IId | IIe | GroupIII | NG | Total | |
| Strawberry | 10 | 3 | 8 | 12 | 6 | 7 | 10 | 3 | 59 |
| Arabidopsis | 16 | 3 | 8 | 17 | 7 | 8 | 14 | 1 | 74 |
| Rice | 15 | 4 | 8 | 15 | 7 | 11 | 36 | 96 | |
| Grapevine | 12 | 3 | 8 | 15 | 6 | 7 | 6 | 2 | 59 |
| Cucumber | 10 | 4 | 4 | 16 | 8 | 7 | 6 | 55 | |
Strawberry, Fragaria vesca. Arabidopsis, Arabidopsis thaliana. Rice, Oryza sativa. Grapevine, Vitis vinifera, Cucumber, Cucumis sativus, NG, no group identified.
FvWRKY Expression Profiles in Different Organs and Fruit Developmental Stages
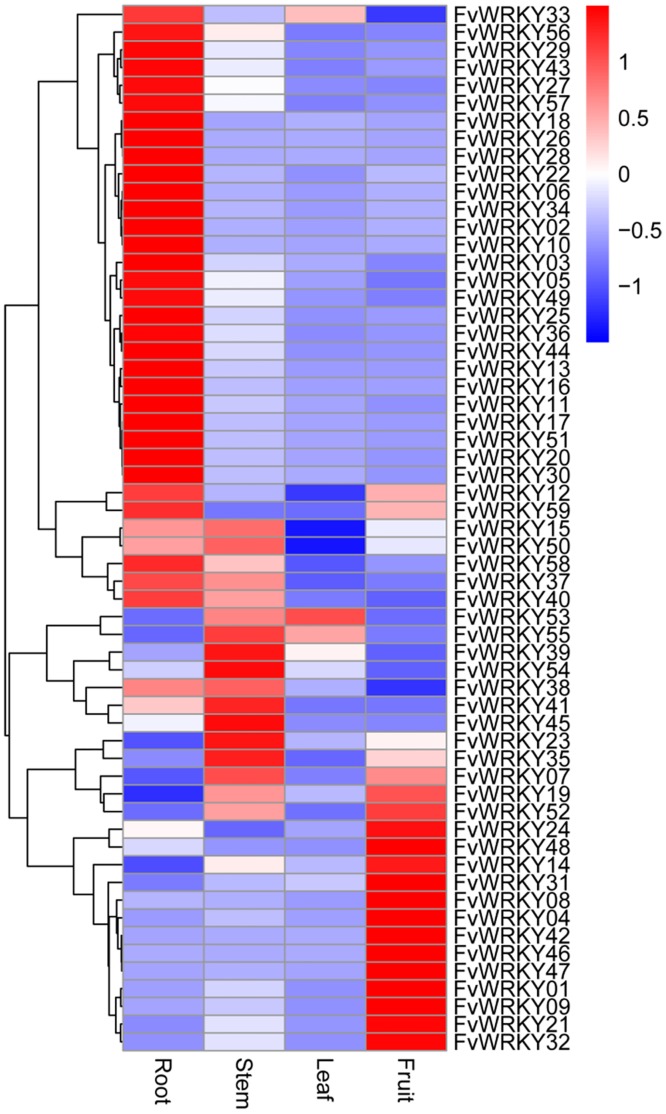
The expression patterns of all the FvWRKY genes were analyzed roots, stems, leaves and fruits of F. vesca ‘Hawaii 4’ grown under normal conditions. Among the 59 predicted genes, 52 genes were expressed in at least one of the four tissues (Fig 5). Expression of the other seven genes was not detected by real-time RT-PCR; however, it may be that they are expressed in other organs, only expressed in response to specific biotic or abiotic stresses, or are pseudogenes.
Fig 5. qRT-PCR validation of FvWRKY expression in different tissues.
Red and blue boxes indicate high and low expression levels, respectively, for each gene.
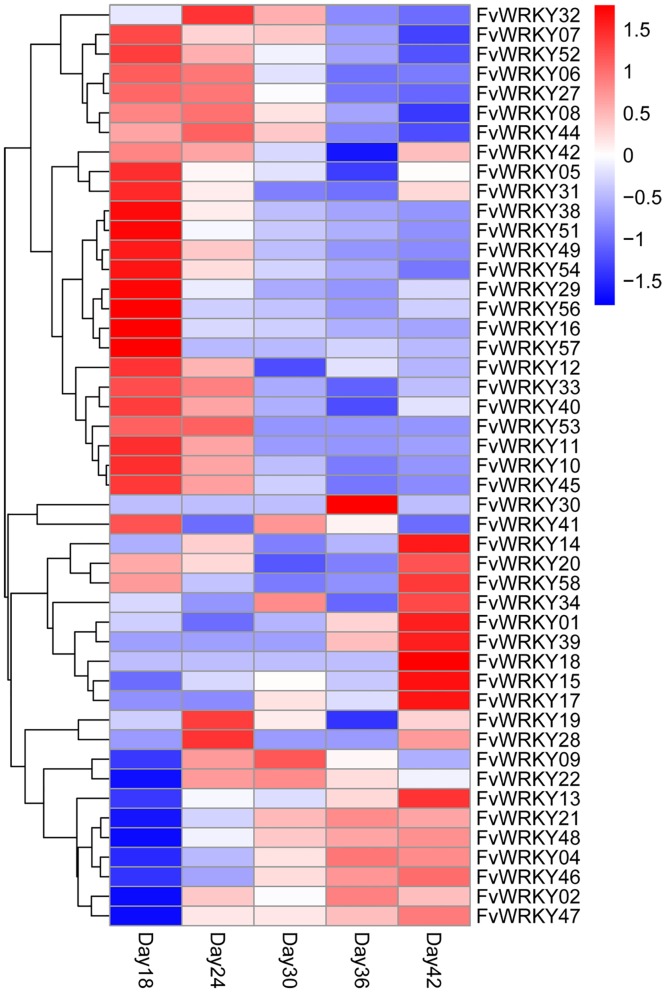
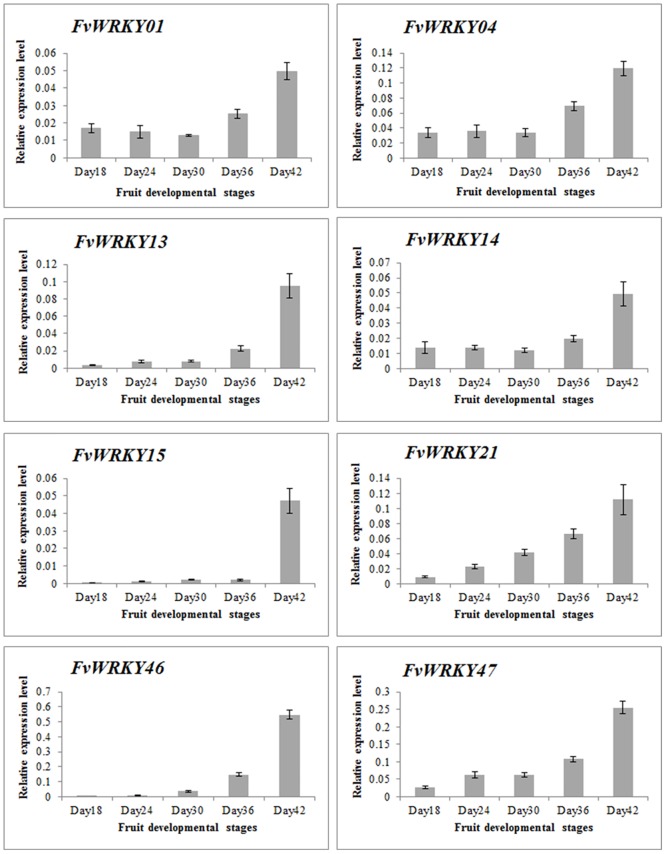
We next evaluated the expression of the FvWRKY genes during fruit development and ripening, based on RNA-seq data. Of the 59 predicted genes, 46 showed expression in fruits (Fig 6), and the expression of FvWRKY1, FvWRKY4, FvWRKY13, FvWRKY14, FvWRKY15, FvWRKY21, FvWRKY46 and FvWRKY47 was up-regulated during fruit development and ripening (Fig 7). The greatest increase in expression was observed for FvWRKY46 and FvWRKY47 at 42 DAF.
Fig 6. RNA-seq data showing FvWRKY gene expression during fruit development and ripening.
Red and blue boxes indicate high and low expression levels, respectively, for each gene. Day18, 18 days after flowering; Day24, 24 days after flowering; Day30, 30 days after flowering; Day36, 36 days after flowering; Day42, 42 days after flowering.
Fig 7. qRT-PCR validation of FvWRKY expression during fruit development and ripening.
Red and blue boxes indicate high and low expression levels, respectively, for each gene. Day18, 18 days after flowering; Day24, 24 days after flowering; Day30, 30 days after flowering; Day36, 36 days after flowering; Day42, 42 days after flowering.
Exogenous ABA, IAA and Sucrose Induce Accumulation of FvWRKY Transcripts in Fruits
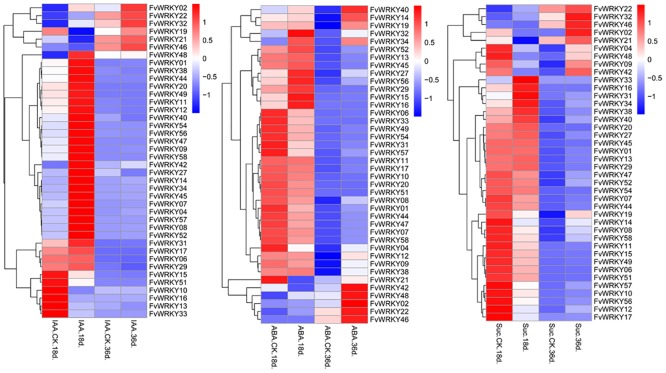
We used qRT-PCR analysis to examine the expression of the FvWRKY genes in 18 DAF and 36 DAF fruits in response to exogenous IAA, ABA and sucrose. After IAA treatment, most WRKY genes were expressed in the early stages of fruit development. Thirty-four genes were detected in 18 DAF fruit and the expression of WRKY02, 19, 21, 22, 32 and 48 was induced in 36 DAF fruits (Fig 8). Addition of ABA, which is known to regulate fruit development and ripening [39], resulted in similar expression patterns as those resulting from the sucrose treatment (Fig 8). The expression levels of most of the WRKY genes was reduced by the sucrose treatment and expression was only be detected in 18 DAF fruits, while only ten WRKY genes were expressed in 36 DAF fruits and were also induced by sucrose (Fig 8).
Fig 8. qRT-PCR expression analysis of FvWRKY genes in 18 days after flowering (DAF) and 36 DAF fruits treated with IAA, ABA or sucrose.
Red and blue boxes indicate high and low expression levels, respectively, for each gene. DAF, days after flowering; IAA.CK.18d., controlled trials of 18 DAF fruits treated with IAA (100 μM); IAA.18d., 18 DAF fruits treated with IAA (100 μM); IAA.CK.36d. controlled trials of 36 DAF fruits treated with IAA (100 μM);IAA.36d., 36 DAF fruits treated with IAA (100 μM); ABA.CK.18d., controlled trials of 18 DAF fruits treated with ABA (100 μM); ABA.18d., 18 DAF fruits treated with ABA (100 μM); ABA.CK.36d. controlled trials of 36 DAF fruits treated with ABA (100 μM);IAA.36d., 36 DAF fruits treated with ABA (100 μM); Suc.CK.18d., controlled trials of 18 DAF fruits treated with sucrose (50 μM); Suc.18d., 18 DAF fruits treated with sucrose (50 μM); Suc.CK.36d. controlled trials of 36 DAF fruits treated with sucrose (50 μM); Suc.36d., 36 DAF fruits treated with sucrose (50 μM).
Discussion
Organization of the F. vesca WRKY Gene Family
The size of the F. vesca WRKY gene family (59; genome size 240 Mb) is smaller than those of A. thaliana (72, genome size 125 Mb) and rice (96; genome size 480 Mb). When the number of WRKY genes in the different subgroups was further compared with those of A. thaliana, rice, grapevine, cucumber and F. vesca (Table 3), we found that the numbers of WRKY genes in Group I, Group IIc and III showed the greatest diferrences between F. vesca and A. thaliana and rice, but that there were similar percentages of genes in Group I of F. vesca, cucumber and grapevine. It has been reported that 80% of the rice WRKY gene loci are located in duplicated regions and that gene duplication events have lead to the generation of new WRKY genes [20]. Compared with cucumber (genome size ~367 Mb) [21] and grapevine (genome size ~400 Mb) [22], the size of the WRKY family in F. vesca is similar (55 and 59 WRKY genes in cucumber and grapevine, respectively); however F. vesca contain fewer genes in Group IIc and more in Group III.
The structure of the phylogenetic tree based on an alignment of the WRKY domains of F. vesca and A. thaliana indicated that the 59 FvWRKY proteins can be divided into three major groups (I, II and III) as previously described for other plant species [2] together with one smaller group (IV). Members within the same group or subgroup within group II shared a similar gene structure (intron/exon organization), length and amino-acid motif composition, indicating their close evolutionary relationship.
Group I WRKY proteins, with two WRKY domains, constitute approximately 20% of the entire FvWRKY family, which s comparable in size to those of cucumber and grapevine, but smaller than those of A. thaliana and rice (Table 3). A significant expansion exists in poplar (Populus tremula), where Group I contains approximately 50% of the entire PtWRKY family. The subgroups of Group II in F. vesca, A. thaliana, rice and grapevine are similar in size, except that there is a reduction in Group IIc in F. vesca. In F. vesca, the members of Group IIa and IIb are characterized by the presence of motifs 5 and 6, whereas Group IId contains the HARF sequence (motif 14), as described in A. thaliana [2]. Group III proteins with the C-X7-C-X23-H-X1-H domain pattern, were found to be smaller than in A. thaliana and even more so than in rice, but larger than in grapevine and cucumber. In the phylogenetic tree, group III was most closely related to the large subgroup IIe+d. A search of the Plant Transcription Factor Database showed that the earliest known evolutionary occurrence of group III genes was in the lycophyte, Selaginella moellendorffii. There were also no evidence of any sequenced plant species that contain only members of group I and III but some species have only members of group I and II, such as the moss, Physcomitrella patens [5]. We therefore propose that group III may have evolved from group II, as exemplified by group IIe of strawberry. Gene orthology can provide a starting point for genetic studies that aim to define the function of a candidate gene. In the F. vesca genome, both the FvWRKY55 and FvWRKY57 genes contain the domain pattern that is typical of group III, although it has a motif that is not present in other group III members, so we classified these two WRKY genes into a small group IV (Fig 4). The biological functions of group IV WRKY genes remain to remains to be determined.
Gene duplication and divergence events have been suggested to be the main contributors to evolutionary momentum. The current gene duplication analysis indicated that 12 of 59 FvWRKY genes were associated with either tandem or segmental duplication events and three pairs of the WRKY genes (FvWRKY11/FvWRKY12, FvWRKY35/FvWRKY36 and FvWRKY38/ FvWRKY39) appeared to have undergone tandem duplication (Fig 4). Our data suggest a low frequency of tandem and a high frequency of segmental duplication events exists amongst the FvWRKY genes, which is consistent with results from A. thaliana. It has been noted that segmental duplication may more often be retained due to sub-functionalization, without increasing the likelihood of gene rearrangement.
FvWRKY Genes Involved in Growth and Development
We evaluated the expression patterns of all of the predicted coding members of the FvWRKY family in different strawberry organs at various developmental stages. The data revealed that many of the genes could be grouped together based on their abundant expression in specific organs, possibly reflecting their involvement in a common metabolic and/or developmental process. Indeed, increasing numbers of studies suggest a role for WRKY genes in developmental processes, including seed and trichome development, dormancy and germination, and senescence [7, 30, 40–45]. The expression analysis revealed that FvWRKY4, 46 and 48 of Group IIc were highly expressed in fruits and that their expression increased during fruit development, suggesting a putative role in the regulation of fruit development and ripening. Involvement of WRKY genes within same group in the same developmental processes has been reported previously in both grapevine and A. thaliana [40, 44, 45]. In A. thaliana, AtWRKY6 (Group IIb), has been shown to be strongly upregulated during leaf senescence and to target a senescence-specific receptor-like kinase (SIRK/FRK1) [46]. Other A. thaliana WRKY genes are also upregulated in the leaf transcriptome during senescence, and AtWRKY53 and AtWRKY70 are known to regulate leaf senescence [47–49]. In this current study, the expression of only FvWRKY53 and FvWRKY55 (Group III) was detected in leaves, while most genes were expressed in roots and fruits.
Addition of Exogenous ABA, IAA and Sucrose Induces FvWRKY Expression
We investigated the interaction between hormone and metabolite signaling pathways and FvWRKY gene expression patterns in fruit. Almost all the FvWRKY genes were expressed in fruit that had been treated with exogenous ABA and IAA. In early stages of fruit development, most FvWRKY genes were induced by the IAA treatment, but only a few by ABA. Sucrose treatment was seen to inhibit the expression of most FvWRKY genes in fruit. Only the expression of FvWRKY16, FvWRKY31, FvWRKY33 and FvWRKY34 were induced by sucrose in the early stages of fruit development, while the expression of FvWRKY2, FvWRKY21, FvWRKY22, FvWRKY32 and FvWRKY46 was induced by sucrose in later fruit developmental stages. We observed that several FvWRKY genes that were induced by ABA, IAA and sucrose showed the same expression patterns in the late fruit development stage, notably FvWRKY2, FvWRKY22 and FvWRKY46. This suggests that these three FvWRKY may share the ABA, IAA and sucrose signaling network.
Conclusions
In the present study, identified total of 59 F. vesca WRKY genes and characterized their expression profiles in different organs and fruit developmental stages. Among the 59 predicted genes, 52 genes were expressed in at least one of four organs. qRT-PCR analyses were used to assess the expression of FvWRKY genes in 18 DAF and 36 DAF fruits in response to exogenous IAA, ABA and sucrose treatments. This analysis suggested that some FvWRKY genes may operate in the same ABA, IAA and sucrose signaling network. Our results also indicate that positive selection may have driven the functional divergence of duplicated genes during the expansion of group III WRKY genes. Based on the results presented here, we propose that a subset of WRKY proteins contribute to the regulation of strawberry fruit development and ripening.
Supporting Information
(XLS)
(TIF)
This file lists the primer sequences used for real-time quantitative RT-PCR validation of the RNA-seq data.
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No. 31370323), Beijing Natural Science Foundation and Scientific Research Key Program of Beijing Municipal Commission of Education (Project No. KZ20130020018) and the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (Project No. CIT&TCD201404098).
References
- 1.Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7: 491–498. [DOI] [PubMed] [Google Scholar]
- 2.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci.2000; 5: 199–206. [DOI] [PubMed] [Google Scholar]
- 3.Rose CA, Liu Y, Shen QXJ. The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol.2007;49 (6): 827–842. [Google Scholar]
- 4.Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci.2005;10(2):71–78. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YJ, Wang LJ. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol.2005; 5:1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rushton Deena L., Tripathi Prateek, Rabara Roel C., Lin Jun, Ringler Patricia, Boken Ashley K., et al. (2012) WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol J.2012; 10 (1):2–11. 10.1111/j.1467-7652.2011.00634.x [DOI] [PubMed] [Google Scholar]
- 7.Rushton PJ, Macdonald H, Hutty AK, Lazarus CM, Hooley R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of a-Amy2 genes. Plant Mol Biol.1995;29: 691–702. [DOI] [PubMed] [Google Scholar]
- 8.Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C.A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugarresponsive elements of the iso1 promoter. Plant Cell.2003; 15: 2076–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 59 upstream regions of genes coding for sporamin and b-amylase from sweet potato. Mol Gen Genet. 1994; 244:563–571. [DOI] [PubMed] [Google Scholar]
- 10.Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J.1996; 15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 11.Dellagi A, Heilbronn J, Avrova A, Montesano M, Palva ET, Stewart HE, et al. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol Plant-Microbe Interact.2000; 13: 1092–1101. [DOI] [PubMed] [Google Scholar]
- 12.Huang T, Duman JG. Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Mol Biol. 2002;48:339–350. [DOI] [PubMed] [Google Scholar]
- 13.Pnueli L, Hallak HE, Rozenberg M, Cohen M, Goloubinoff P,Kaplan A. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J.2002; 31:319–330. [DOI] [PubMed] [Google Scholar]
- 14.Mantri NL, Ford R, Coram TE, Pang EC. Transcriptional profiling of chickpea genes differentially regulated in response to highsalinity, cold and drought. BMC Genomics.2007; 8:303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato N, Dubouzet E, Kokabu Y, Yoshida S, Taniguchi Y, Dubouzet JG, et al. Identification of a WRKY protein as a transcriptional regulator benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol.2007; 48:8–18. [DOI] [PubMed] [Google Scholar]
- 16.Marchive C, Mzid R, Deluc L, Barrieu F, Pirrello J, Gauthier A.Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J Exp Bot.2007; 58:1999–2010. [DOI] [PubMed] [Google Scholar]
- 17.Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stress in transgenic Arabidopsis plants. Plant Biotechnol J.2008; 6: 486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- 18.Liu JJ, Ekramoddoullah AK. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome.2009;52:77–88. 10.1139/G08-106 [DOI] [PubMed] [Google Scholar]
- 19.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research.2005; 12:9–26. [DOI] [PubMed] [Google Scholar]
- 20.Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol.2008; 49:865–879. 10.1093/pcp/pcn061 [DOI] [PubMed] [Google Scholar]
- 21.Ling Jian, Jiang Weijie, Zhang Ying, Yu Hongjun, Mao Zhenchuan, Gu Xingfang, et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics.2011; 12:471 10.1186/1471-2164-12-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Min, Vannozzi Alessandro, Wang Gang, Liang Ying-Hai, Tornielli Giovanni Battista, Zenoni Sara, et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Horticulture Research.2014;1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2: e123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoth C, Ringler J, Dangl JL, Eulgem T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact.2007;20: 120–128. [DOI] [PubMed] [Google Scholar]
- 25.Karam BS, Rhonda CF, Luis OS. Transcription factors in plant defense and stress response. Curr Opin Plant Biol.2002;5:430–436. [DOI] [PubMed] [Google Scholar]
- 26.Motoaki S, Mari NJ, Ishida TN, Miki F, Youko O, Asako K, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J.2002;31:279–292. [DOI] [PubMed] [Google Scholar]
- 27.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J.2007; 50:347–363. [DOI] [PubMed] [Google Scholar]
- 28.Johnson SC, Kolevski B, Smyth DR.Transparent testa glabra2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002; 14:1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagace M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta.2004; 219:185–189. [DOI] [PubMed] [Google Scholar]
- 30.Robatzek S, Somssich IE.Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev.2002;16:1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Encinas-Villarejo S, Maldonado AM, Amil-Ruiz F, de los Santos B, Romero F, Pliego-Alfaro F, et al. Evidence for a positive regulatory role of strawberry (Fragaria x ananassa) Fa WRKY1 and Arabidopsis AtWRKY75 proteins in resistance. J Exp Bot.2009; 60(11):3043–3065. 10.1093/jxb/erp152 [DOI] [PubMed] [Google Scholar]
- 32.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, et al. The genome of woodland strawberry (Fragaria vesca). Nat Genet.2011;43:109–116. 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics.2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol.2011; 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M. Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol.2012; 12:130 10.1186/1471-2229-12-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holub EB.The aims race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet.2001; 2:516–527. [DOI] [PubMed] [Google Scholar]
- 37.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res.2005; 12(1): 9–26. [DOI] [PubMed] [Google Scholar]
- 38.Xie Z, Zhang ZL, Zou X. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol.2005;137:176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia Hai-Feng, Chai Ye-Mao, Li Chun-Li, Lu Dong, Luo Jing-Jing, Qin Ling, et al. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol.2011; 157:188–199. 10.1104/pp.111.177311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robatzek S, Somssich IE.Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev.2002;16:1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jing SJ, Zhou X, Song Y, Yu DQ. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul.2009;58:181–190. [Google Scholar]
- 42.Jiang W, Yu D.Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol.2009; 9: 96 10.1186/1471-2229-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A.MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA.2005;102(48):17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo S, Zhang Z, Tong T.Cloning and characterization of cellular senescence associated genes in human fibroblasts by suppression subtractive hybridization. Exp Cell Res.2004; 298: 465–472. [DOI] [PubMed] [Google Scholar]
- 45.Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004, 55:853–867. [DOI] [PubMed] [Google Scholar]
- 46.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J.2001; 28:123–133. [DOI] [PubMed] [Google Scholar]
- 47.Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009;58: 333–346. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Mei, Yuan Bing, Leng Ping.The role of ABA in triggering the ethylene biosynthesis and ripening of tomato fruit. J Exp Bot.2009; 60 (6): 1579–1588. 10.1093/jxb/erp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulker B, Shahid Mukhtar M, Somssich IE.The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta.2007;226:125–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(TIF)
This file lists the primer sequences used for real-time quantitative RT-PCR validation of the RNA-seq data.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.