Summary
Cell-autonomous defense mechanisms are potent strategies that protect individual cells against intracellular pathogens. The Rab-family GTPase Rab32 was previously shown to restrict the intracellular human pathogen Salmonella Typhi, but its potential broader role in antimicrobial defense remains unknown. We show that Rab32 represents a general cell-autonomous, antimicrobial defense that is counteracted by two Salmonella effectors. Mice lacking Rab-32 or its nucleotide exchange factor BLOC-3 are permissive to S. Typhi infection and exhibit increased susceptibility to S. Typhimurium. S. Typhimurium counters this defense pathway by delivering two type III secretion effectors, SopD2, a Rab32 GAP, and GtgE, a specific Rab32 protease. An S. Typhimurium mutant strain lacking these two effectors exhibits markedly reduced virulence, which is fully restored in BLOC-3-deficient mice. These results demonstrate that a cell-autonomous, Rab32-dependent host defense pathway plays a central role in the defense against vacuolar pathogens and describe a mechanism evolved by a bacterial pathogen to counter it.
Graphical abstract

Introduction
Microbial pathogens encounter a variety of host innate immune mechanisms that restrict their survival and growth. Pathogens are most often sensed by the host through pattern-recognition receptors, which can detect microbial products or microbial-induced tissue injury (Broz and Monack, 2013; Dixit and Kagan, 2013; Medzhitov, 2007). Pathogen recognition is then followed by host defense responses that, when successful, limit pathogen replication (Yang et al., 2013; Jarczak et al., 2013; Nairz et al., 2014; Nish and Medzhitov, 2011; Voehringer, 2013). In turn, and as part of the everlasting evolutionary “arms race,” pathogens have evolved mechanisms to avoid their detection or to survive the defense responses (Diacovich and Gorvel, 2010; Reddick and Alto, 2014). Although much is known about host-sensing mechanisms to detect microbial pathogens, much less is known about host-effector mechanisms to control microbial infections or the microbial virulence factors that may have specifically evolved to counter them. Greater emphasis has been placed on the role of immune cells in pathogen defense. However, the first, and evolutionary the oldest, line of defense against microbial infection is composed of cell-intrinsic or cell-autonomous mechanisms that operate in most cells of the body (Deretic, 2011; Randow et al., 2013). These mechanisms, which tend to be conserved across phyla, provide protection to individual cells in the body and, in metazoan, synergize with the immune system to confer whole-body protection against pathogens. Some of these cell-autonomous mechanisms have adapted to operate in a rather specific manner. For example, members of the tripartite motif (TRIM) protein family, such as TRIM5α (Rajsbaum et al., 2014) and the single-stranded DNA cytosine deaminase APOBEC3 (Harris and Dudley, 2015), have evolved to restrict the replication of retroviruses. Others, however, have the capacity to restrict the growth a broad range of pathogens. For example, a family of interferon-inducible GTPases has been found to restrict the growth of a variety of intracellular microbial pathogens (Hunn et al., 2011; Kim et al., 2012).
The bacterial pathogen Salmonella enterica comprises many serovars that can infect a large and diverse number of vertebrate species (Grassl and Finlay, 2008; Ohl and Miller, 2001). While some serovars such as Salmonella enterica Typhimurium (S. Typhimurium) can infect a broad range of hosts, others are extremely host specific. Salmonella enterica Typhi (S. Typhi), for example, can only infect humans, where it causes typhoid fever, a systemic disease that results in ∼200,000 deaths worldwide, mostly children in developing countries (Crump and Mintz, 2010; Parry et al., 2002; Raffatellu et al., 2008). We recently discovered that expression of a single gene from S. Typhimurium in S. Typhi allows this bacterium to replicate in cells and tissues of a nonpermissive host (Spanò and Galán, 2012). This gene, gtgE, encodes for a protease that targets the Rab-family GTPase Rab32 (Spanò and Galán, 2012; Spanò et al., 2011). In specialized cells, these GTPases are known to orchestrate the maturation and assembly of lysosome-related organelles such as melanosomes and T cell granules (Dell'Angelica, 2004; Luzio et al., 2014; Raposo and Marks, 2007; Wasmeier et al., 2006). However, the potential role of these GTPases in other cells has not been investigated. Removal of Rab32 or its exchange factor, the biogenesis of lysosome-related organelle complex 3 (BLOC-3) (Gerondopoulos et al., 2012), allowed the replication of human-specific serovar S. Typhi in mouse macrophages (Spanò and Galán, 2012). We report here that the Rab32-dependent, cell-autonomous pathway restricts the replication of both S. Typhi and S. Typhimurium in mouse tissues. We also show that S. Typhimurium counters this host defense pathway by delivering SopD2, a type III secretion effector protein with GAP activity toward Rab32 that works in conjunction with GtgE. We find that an S. Typhimurium mutant strain lacking these two effectors exhibits a drastic reduction in mouse virulence, which is fully reversed in a mouse lacking BLOC-3. These results demonstrate that a Rab32-dependent defense pathway plays a central role in the control of the replication of an intracellular pathogen. Furthermore, our results describe a remarkable adaptation evolved by broad-host Salmonella to counter this cell-autonomous pathogen-restriction pathway.
Results
A Rab32/BLOC-3-Dependent Pathway Restricts the Replication of Human-Adapted and Broad-Host Salmonellae in Mouse Tissues
The observation that removal of Rab32 or its exchange factor BLOC-3 results in increased survival of S. Typhi in mouse macrophages suggested the possibility that this pathway may constitute a general mechanism of pathogen restriction. We therefore tested the susceptibility of Rab32- or BLOC-3-deficient mice to S. Typhimurium or S. Typhi infection. We found that both mouse strains exhibited a marked increase in susceptibility to S. Typhimurium. Intraperitoneal infection of either of these mouse strains with S. Typhimurium resulted in significantly higher bacterial loads in systemic tissues compared to wild-type control animals (Figures 1A and 1B). Furthermore, BLOC-3-deficient mice could be productively and persistently infected with the human-specific serovar S. Typhi (Figure 1C). While wild-type animals cleared the S. Typhi infection, a significant number of CFUs (colony-forming units) were recovered from systemic tissues of BLOC-3-deficient mice (Figure 1C). We have previously shown that heterologous expression of the S. Typhimurium type III secretion effector GtgE in S. Typhi allows its replication in nonpermissive cells. We also showed that this effector exerts this function by proteolytically targeting Rab32. We therefore reasoned that GtgE must allow S. Typhimurium to overcome the Rab32 pathogen-restriction mechanism and that its absence should result in a significant virulence phenotype. However, we found that deletion of gtgE from S. Typhimurium or expression of its catalytic mutant (GtgEH151A) resulted in a very modest phenotype in mouse virulence (Figure 1D). Taken together, these results indicate that the Rab32/BLOC-3 pathway is important for the control of Salmonella infections and that S. Typhimurium must target this pathway with redundant mechanisms.
Figure 1. The Rab32-BLOC-3-Dependent Pathogen-Restriction Pathway Limits the Growth of S. Typhimurium and S. Typhi in a Mouse Model of Infection.
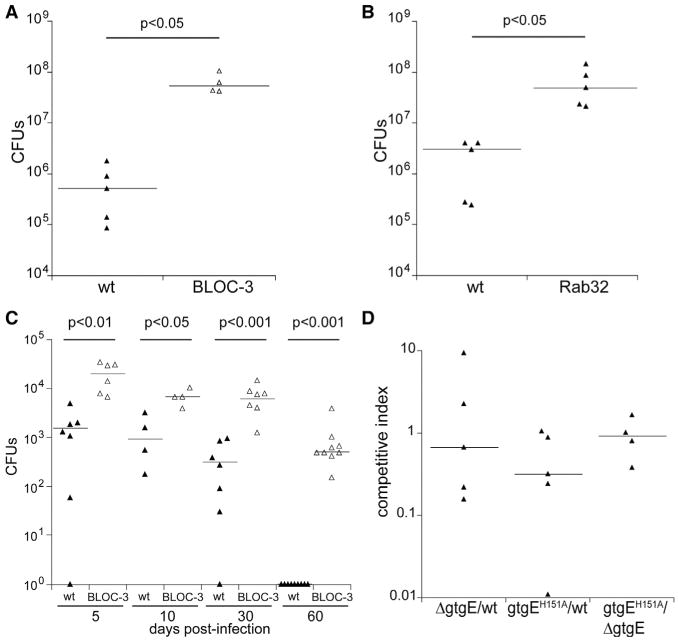
(A and B) C57BL/6 (WT), C57B6.C3-Pde6brd1 Hps4le/J (BLOC-3 deficient), or Rab32-deficient mice were intraperitoneally infected with 102 CFU of wild-type S. Typhimurium, and 5 days after infection the levels of the CFU in the spleens were enumerated. Each triangle represents the bacterial load for an individual animal, and horizontal bars indicate medians of the CFU. The p values of the differences in bacterial loads between the indicated animals determined by the Wilcoxon-Mann-Whitney test are shown.
(C) C57BL/6 (WT) or C57B6.C3-Pde6brd1 Hps4le/J (BLOC-3 deficient) mice were infected intraperitoneally with 105 CFU of wild-type S. Typhi, and bacterial loads in the spleens were enumerated at the indicated days after infection. Each triangle represents the bacterial load for an individual animal, and horizontal bars indicate the medians of the CFU. The p values of the difference sin bacterial loads between the indicated animals determined by the Wilcoxon-Mann-Whitney test are shown.
(D) C57BL/6 mice were intraperitoneally infected with equal numbers (103 CFU each) of the wild-type, ΔgtgE, or gtgEE151A (encoding a GtgE catalytic mutant) strains (in the indicated combinations), and 5 days after infection, the levels of the different strains in the spleen of infected mice were enumerated. Each triangle represents the competitive index for the indicated strains in an individual mouse, and the horizontal bars are the medians of the competitive indices.
The S. Typhimurium Effector Proteins SopD2 and GtgE Prevent the Recruitment of Rab32 to the S. Typhimurium-Containing Vacuole
We have previously observed that Rab32 is efficiently recruited to the S. Typhi-containing vacuoles, but not to the S. Typhimurium-containing vacuoles (SCVs) (Spanò and Galán, 2012). We have also shown that an S. Typhi strain engineered to express the S. Typhimurium type III secretion effector protein GtgE does not recruit Rab32 to its vacuole (Spanò and Galán, 2012). However, we observed that only a small proportion (<10%) of the vacuoles containing the S. Typhimurium ΔgtgE (strain carrying a deletion within gtgE) mutant strain colocalized with Rab32 (Figures 2A, 2B, and S1, available online). This observation, coupled with the mild virulence phenotype associated with the absence of gtgE (see Figure 1D), suggested the presence of an additional S. Typhimurium effector protein(s) that may target the Rab32/BLOC-3 host defense pathway by preventing the recruitment of Rab32 to the SCV. We investigated this possibility by examining the presence of Rab32 in vacuoles containing an S. Typhimurium mutant lacking several of its type III secretion effector proteins in addition to GtgE (Bruno et al., 2009). Since this mutant lacks the effectors required for bacterial internalization, we reintroduced a gene encoding SopB, which by itself can mediate S. Typhimurium internalization into cultured epithelial cells (Zhou et al., 2001). We found that the majority of the vacuoles containing this effector-deficient strain were decorated with Rab32, a proportion similar to that observed in S. Typhi-containing vacuoles (Figures 2A, 2B, and S1). These results suggested that one or more of the effectors missing from this effector-deficient S. Typhimurium strain must be involved in preventing the recruitment of Rab32 to the SCV. Among the effector genes absent from the S. Typhimurium multieffector mutant strain are slrP (Tsolis et al., 1999), sopA (Wood et al., 2000), and sopD2 (Brumell et al., 2003; Schroeder et al., 2010), which are absent or pseudogenes in S. Typhi (Parkhill et al., 2001). Given the observation that S. Typhi is unable to prevent the recruitment of Rab32 to its vacuole, we reasoned that in addition to gtgE, one of the other effectors absent from S. Typhi, must be responsible for preventing the recruitment of this Rab GTPase to the SCV. We tested this hypothesis by specifically removing each one of these effectors from an S. Typhimurium ΔgtgE mutant strain and testing the resulting strains for their ability to recruit Rab32 to their vacuoles. We observed limited colocalization of Rab32 with the SCV in cells infected with a ΔgtgE/ΔslrP/ΔsopA S. Typhimurium mutant strain (Figures 2A, 2B, and S1), indicating that SlrP and SopA are not involved in preventing the recruitment of Rab32 to the SCV. In contrast, we found that the majority of the SCVs of cells infected with the S. Typhimurium ΔgtgE/ΔsopΔ2 mutant were decorated with Rab32 (Figures 2A, 2B, and S1). Absence of gtgE and sopD2 did not result in a destabilization of the SCV since most bacteria were enclosed within a Rab32-containing vacuole. Furthermore, expression of SopD2 in S. Typhi effectively prevented the recruitment of Rab32 to the S. Typhi-containing vacuole, indicating that, similar to GtgE, this effector by itself is sufficient to prevent the recruitment of Rab32 to the SCV (Figures 2A, 2B, and S1). We observed minimal recruitment of Rab32 to the SCV in cells infected with the S. Typhimurium ΔsopD2 strain (Figures 2A, 2B, and S1), indicating that SopD2 works in a functionally redundant manner with GtgE. Taken together, these results show that SopD2 and GtgE work together to prevent the recruitment of Rab32 to the SCV.
Figure 2. The S. Typhimurium Effector Proteins SopD2 and GtgE Prevent the Recruitment of Rab32 to the S. Typhimurium-Containing Vacuole.
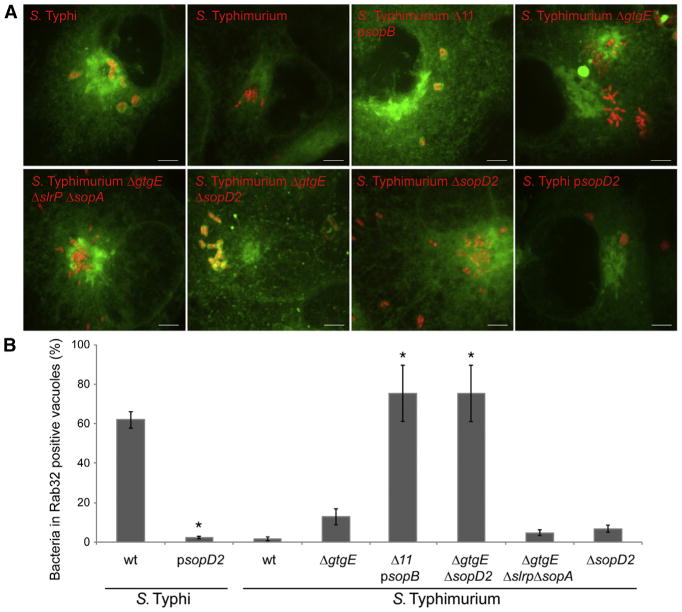
(A and B) COS-1 cells expressing CFP-Rab32 (green) were infected with the indicated strains of S. Typhi or S. Typhimurium expressing mCherry (red), and 3 hr after infection, the infected cells were visualized by fluorescence microscopy. Images show maximum-intensity projections of confocal z stacks (scale bars, 5 μm) (A). The mean ± SEM of the percentage of Salmonella-containing vacuoles that were positive for Rab32 in three independent experiments are shown (B). At least 100 bacteria were counted in each experimental condition. *p < 0.01 (Student's t test) for the difference with the values obtained in cells infected with wild-type S. Typhi or S. Typhimurium. The strain indicated as S. Typhimurium Δ11 psopB lacks 11 type III secretion effector proteins and carries a plasmid encoding sopB (see Experimental Procedures and text for details). See also Figure S1.
SopD2 Is GAP for Rab32
We investigated the mechanisms by which SopD2 prevents the localization of Rab32 to the SCV. We compared the levels of Rab32 in cells infected with S. Typhimurium ΔgtgE or ΔgtgE sopD2 mutant strains. We found no differences in the levels of Rab32 in cells infected with either of these strains (Figure S2), indicating that, unlike GtgE, SopD2 does not alter the levels of Rab32 in infected cells. We then sought to identify potential SopD2-interacting host proteins. We infected cultured mouse macrophages with an S. Typhimurium ΔsopD2 strain expressing plasmid-borne, FLAG-epitope-tagged SopD2 or equally tagged, unrelated type III secretion system (T3SS) effector proteins as specificity controls. Interacting proteins were then identified by immunoaffinity purification and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. These analyses identified the highly related Rab GTPases Rab8A, Rab8B, and Rab10 as potential SopD2-specific interacting proteins in two independent experiments (Table S1). The interactions between SopD2 and Rab8A, Rab8B, or Rab10 were subsequently verified in transient cotransfection experiments with epitope-tagged versions of these proteins (Figure S3).
Small GTP-binding proteins including Rab GTPases exert their regulatory function by alternating between GTP (active)- or GDP (inactive)-bound configurations (Lee et al., 2009; Stenmark, 2009). The transition between these two states is regulated by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP leading to activation, and GTPase-activating proteins (GAPs), which stimulate nucleotide hydrolysis (Barr and Lambright, 2010). Consequently, GEFs and GAPs often display higher affinity for the GDP- and GTP-bound forms of their cognate GTPases, respectively. We tested the interaction of SopD2 with the GDP- or GTP-bound forms of Rab8A, Rab8B, and Rab10 using the GTP- and GDP-locked mutants of these GTPases in transient coexpression experiments. We found that SopD2 preferentially bound to the GTP-locked forms of Rab8A, Rab8B, and Rab10 (Figure 3A), thus suggesting the possibility that this effector maybe a GAP for these GTPases. We therefore examined the ability of SopD2 to stimulate the intrinsic GTPase activity of Rab8A and Rab10 in vitro using purified proteins. We found that SopD2 exhibited robust GAP activity toward both of these GTPases, although it did not show measurable GAP activity toward the Rho-family GTPase Rac1 (Figure 3B).
Figure 3. SopD2 Is a GAP for Rab-Family GTPases.
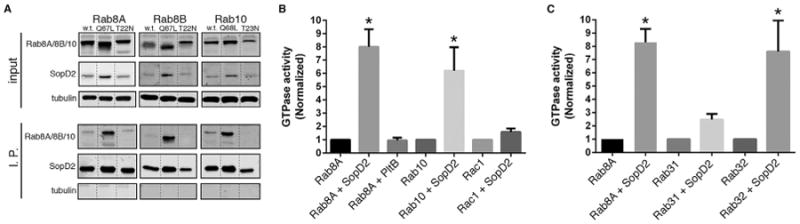
(A) HeLa cells were transiently cotransfected with plasmids expressing FLAG-epitope-tagged SopD2 along with plasmids expressing GFP-tagged wild-type (WT), dominant-negative (T22N or T23N), or constitutively active (Q67L or Q68L) forms of Rab8A, Rab8B, or Rab10. Sixteen hours after transfection, cell lysates were analyzed by immunoprecipitation with anti-FLAG and western immunoblotting with anti-GFP, FLAG, and tubulin (as loading control) antibodies. Dotted lines indicate places where the experimentally relevant lanes were spliced together (all lanes originate from a single gel).
(B and C) The indicated purified GTPases were incubated alone or in the presence of purified SopD2 or the irrelevant protein PltB (as a negative control). The intrinsic GTPase activity of the different GTPases was then measured as indicated in Experimental Procedures. Values represent the ratio of GTPase activities observed in the presence or absence of SopD2 (or the indicated control protein) and are the mean ± SD of three independent determinations. *p < 0.001 (Student's t test) relative to incubation in the absence of SopD2. See also Figures S2–S4 and Table S1.
These results suggested that through its GAP activity, SopD2 disrupts membrane trafficking pathways controlled by Rab8 and/or Rab10 that may be required for the delivery of Rab32 and Rab32-dependent cargo to the SCV. To investigate this hypothesis, we examined the effect of disrupting the pathways controlled by these GTPases on the recruitment of Rab32 to the SCV. Since the activities of these highly related Rab GTPases have been shown to be often redundant (Sato et al., 2014), we sought to disrupt the pathway(s) they controlled by expressing mutant forms of Rab8A, Rab8B, and Rab10. Although expression of constitutive active GTP-locked mutants of these GTPase reduced the presence of Rab32 on the SCV, expression of GDP-locked, dominant-negative mutants did not alter the ability of S. Typhimurium to recruit Rab32 to the SCV (Figure S4). These results suggested that these GTPases might not be the relevant targets for the SopD2 activity associated with its ability to block the delivery of Rab32 to the SCV. GAPs for Rab GTPases often display activity toward multiple members of this protein family (Albert and Gallwitz, 1999; Barr and Lambright, 2010; Fukuda, 2011). In fact, our analysis of SopD2-interacting proteins also detected interactions with other Rab family GTPases (Table S1), although inconsistently, suggesting the possibility that this effector may be able to engage other Rab GTPases. Given the importance of Rab32 in Salmonella restriction (Spanò and Galán, 2012), we specifically tested the GAP activity of SopD2 toward this GTPase. We found that SopD2 is a potent GAP for Rab32. In contrast, SopD2 did not display significant GAP activity toward Rab31 (Figure 3C), indicating that despite its ability to target several Rab GTPases, there is still specificity in its enzymatic activity. These results indicate that SopD2 interferes with the Rab32-dependent pathogen-restriction pathway by targeting Rab32 itself.
SopD2 Catalytic Activity Mimics Eukaryotic GAPs
Most GAP proteins exert their function by inserting a critical arginine residue into the active site of their cognate GTPases, thus stabilizing the transition state (Lee et al., 2009). We therefore inspected the SopD2 sequence for the presence of potential catalytic arginine residue and constructed mutants in which candidate arginine residues were replaced by alanine (Figure S5). The resulting mutants were purified, and their potential GAP activity was examined in vitro. Purified proteins retained a chromatographic behavior indistinguishable from wild-type, indicating that the mutations introduced did not cause gross conformational changes in these GTPases (Figure 4A). Deletion of the first 27 amino acids (containing R19) or changing R45 or R65 to alanine did not affect the ability of SopD2 to catalyze nucleotide exchange on Rab8A (Figure 4B). In contrast, changing R315 to alanine completely abolished the GAP activity of SopD2 toward all Rab GTPases tested, including Rab32 (Figures 4B and 4C). Furthermore, the SopD2R315A mutant was unable to complement the ability of an S. Typhimurium Δgt ΔsopD2 strain to prevent the recruitment of Rab32 to the SCV (Figures 4D and S6), even though the mutant protein was translocated into cells in a manner indistinguishable from wild-type (Figure S7). These results implicate a critical arginine in the catalytic activity of SopD2 and indicate that this effector protein carries out its function in a manner similar to other eukaryotic GAP proteins. The crystal structure of SopD2 (D'Costa et al., 2015) does not show significant structural similarity to any known eukaryotic GAPs. Similar to eukaryotic GAPs for small GTPases, however, the catalytic arginine is on an extended loop (arginine “finger loop”) at the C-terminal end of SopD2 (Figure 4E). Therefore, SopD2 has evolved to mimic the fundamental aspects of the catalytic mechanism of host GAPs but within a unique structural context. This observation is reminiscent of other bacterially encoded effector proteins that have evolved to mimic eukaryotic cell function through convergent evolution (Stebbins and Galán, 2001).
Figure 4. SopD2 Catalytic Activity Mimics Eukaryotic GAPs.
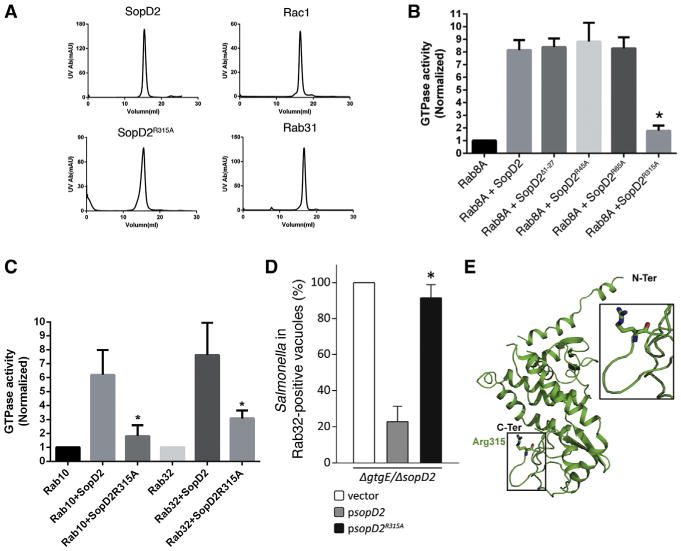
(A) Size-exclusion chromatography profiles of purified proteins used in these experiments.
(B and C) The indicated purified Rab GTPases were incubated with purified SopD2 or the indicated mutants, and the intrinsic GTPase activity of the different GTPases was then measured as indicated in Experimental Procedures. Values represent the ratio of GTPase activities observed in the presence or absence of the different SopD2 preparations and are the mean ± DS of three independent determinations. *p < 0.001 (Student's t test) relative to incubation with wild-type SopD2.
(D) HeLa cells expressing YFP-Rab32 were infected with S. Typhimurium ΔgtgE ΔsopD2 strains carrying a plasmid encoding sopD2, sopD2R315A, or the plasmid vector alone, and 4 hr after infection the infected cells were fixed, stained with an antibody against S. Typhimurium LPS, and visualized by fluorescence microscopy. Values represent the mean ± SD of the percentage of Salmonella-containing vacuoles that are positive for Rab32 and have been normalized according to the values observed in cells infected with the S. Typhimurium ΔgtgE ΔsopD2 strain carrying the vector control, which was considered to be 100% (the actual value for the control was 59% ± 3%). The values were obtained from three independent experiments in which at least 100 cells for each experimental condition were examined. *p < 0.01 (Student's t test) for the difference with the values obtained in cells infected with S. Typhimurium ΔgtgE ΔsopD2 complemented with a wild-type copy of SopD2.
(E) Crystal structure of SopD2 depicting the location of the catalytic arginine. The visualization of the atomic structure (PDB No. 5CQ9) was carried using PyMol. See also Figures S5–S7.
SopD2 and GtgE Redundantly Target the Rab32/BLOC-3 Pathogen-Restriction Pathway
Our results show that to block the delivery of Rab32 to the SCV in cultured cells, S. Typhimurium requires the functionally redundant activities of the T3SS effector proteins SopD2 and GtgE. We therefore examined the relative contribution of these effectors to S. Typhimurium virulence in a mouse model of infection. Mice infected with a ΔsopD2 S. Typhimurium mutant had lower bacterial loads in systemic tissues than those infected with wild-type (Figure 5A). While this phenotype was more pronounced than the phenotype observed for ΔgtgE mutant strain (Figure 5B), the effect of removing sopD2 was still relatively modest. Removal of both gtgE and sopD2, however, resulted in a drastic virulence reduction (Figures 5A and 5B). Mice infected with the ΔgtgE ΔsopD2 double mutant exhibited CFU levels almost five orders of magnitude lower in systemic sites than those in mice infected with wild-type S. Typhimurium (Figure 5A). Furthermore, mice that received a dose of the S. Typhimurium ΔgtgE ΔsopD2 double mutant equivalent to ∼1,000 LD50 for the wild-type strain did not succumb to infection (Figure 5B). The virulence defect was fully reversed by expressing a plasmid-borne wild-type copy of sopD2 (Figures 5C and 5D), but not by a plasmid expressing the catalytic mutant sopD2R315A (Figure 5D). The virulence defect of the double-mutant strain was comparable to the phenotype observed in an S. Typhimurium ΔspiA mutant strain lacking a functional T3SS encoded within its pathogenicity island 2 (Figures 5A and 5E), which is one of the most virulence-attenuating mutations reported in S. Typhimurium (Hensel et al., 1995; Ochman et al., 1996). These results indicate that SopD2 and GtgE work in a functionally redundant manner to confer virulence to S. Typhimurium.
Figure 5. Functional Redundancy of SopD2 and GtgE in S. Typhimurium Virulence.
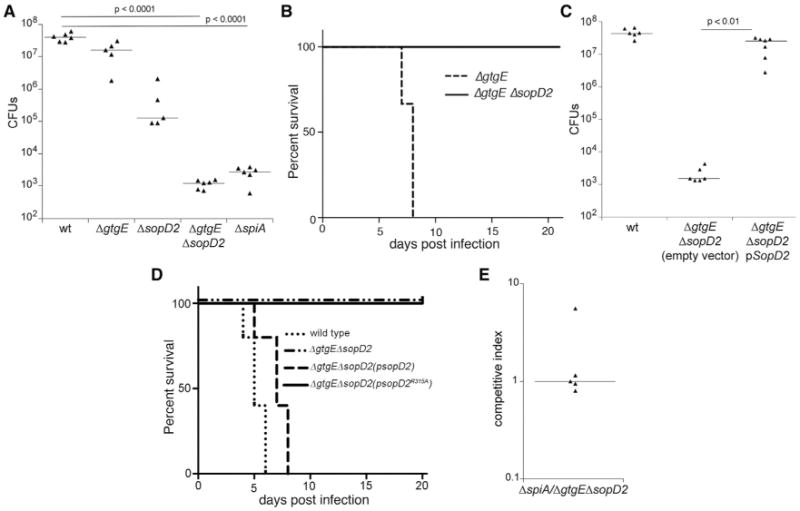
(A) C57BL/6 mice were intraperitoneally infected with 103 CFU of the indicated S. Typhimurium strains, and 5 days after infection, the levels of the different strains in the spleen of the infected mice were enumerated. Each triangle represents the bacterial load for an individual animal, and horizontal bars indicate the geometric median of the CFU. The significant p values of the differences in bacterial loads between the indicated strains determined by the Wilcoxon-Mann-Whitney test are shown.
(B) Survival of C57BL/6 mice orally infected with 107 CFU of the S. Typhimurium ΔgtgE (n = 3) or ΔgtgE ΔsopD2 (n = 4) mutant strains.
(C) C57BL/6 mice were intraperitoneally infected with 103 CFU of the indicated strains of S. Typhimurium, and 5 days after infection, the levels of the different strains in the spleen of infected mice were enumerated. The significant p values of the differences in bacterial loads between the indicated conditions determined by the Wilcoxon-Mann-Whitney test are shown.
(D) Survival of C57BL/6 mice orally infected with 107 CFU of S. Typhimurium ΔgtgE ΔsopD2 mutant strain carrying a plasmid encoding wild-type SopD2 or its catalytic mutant SopD2R315A (n = 5 for each strain).
(E) C57BL/6 mice were intraperitoneally infected with equal numbers (103 CFU each) of the S. Typhimurium ΔspiA and ΔsopD ΔgtgE mutant strains, and 5 days after infection, the levels of the different strains in the spleen of infected mice were enumerated. Each triangle represents the competitive index for the indicated strains in an individual mouse, and the horizontal bars are the medians of the competitive indices.
We then examined whether the activity of GtgE and SopD2 converge into the same host restriction pathway. We reasoned that if this were the case, the drastic virulence defect of the ΔsopD2/ΔgtgE double-mutant strain should be reversed in a mouse where the Rab32/BLOC-3-dependent pathway is not operational. Consistent with this hypothesis, the virulence defect of the ΔsopD2/ΔgtgE double mutant was fully reversed in a BLOC-3-deficient mouse (Figure 6). Similar numbers of CFUs in systemic tissues were recovered from BLOC-3−/− mice infected with the S. Typhimurium ΔsopD2 ΔgtgE mutant or with the wild-type strain. Taken together, these results indicate that S. Typhimurium targets the Rab32/BLOC-3-dependent pathogen-restriction pathway with the functionally redundant activities of two T3SS effector proteins, SopD2 and GtgE. In addition, these findings demonstrate that the Rab32/BLOC-3 cell-autonomous pathway plays a central role in host defense against an intracellular pathogen.
Figure 6. SopD2 and GtgE Redundantly Target the Rab32/BLOC-3 Pathogen-Restriction Pathway.
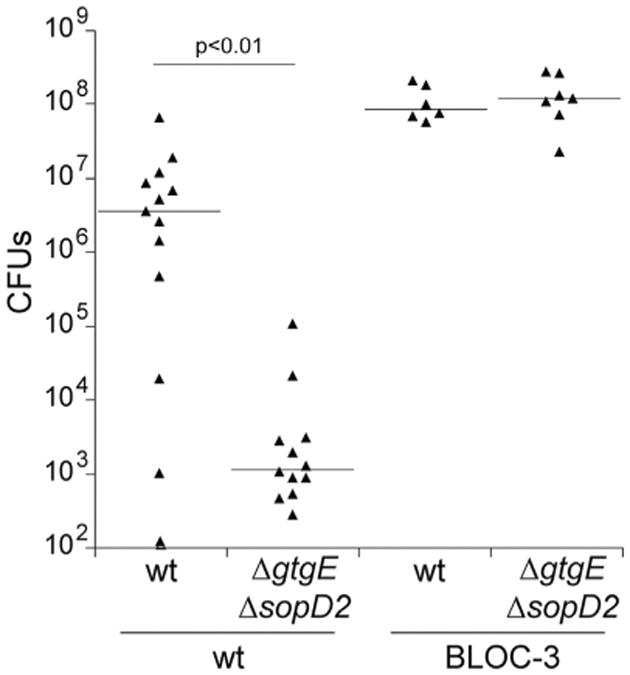
C57BL/6- or BLOC-3-deficient mice were intraperitoneally infected with 102 CFU of the indicated strains of S. Typhimurium, and 5 days after infection, the levels of the different strains in the spleen of infected mice were enumerated. Each triangle represents the bacterial load for an individual animal, and horizontal bars indicate medians of the CFU. The significant p values of the differences in bacterial loads between the indicated conditions determined by the Wilcoxon-Mann-Whitney test are shown.
Discussion
Many microbial pathogens occupy membrane-bound compartments within infected cells, which often shield them from detection by cytoplasmic sensor or protect them from cytoplasmic antimicrobial factors (Creasey and Isberg, 2014). Pathogens that enter the endocytic network, however, must subvert membrane traffic to avoid transport to degradative intracellular compartments. Many pathogenic bacteria accomplished this subversion with effector proteins delivered by specialized machines, which can directly modulate membrane traffic (Asrat et al., 2014; Alix et al., 2011). As a result, intracellular bacterial pathogens often build highly specialized intracellular niches that can sustain their replication. This is a formidable task, as cells are endowed with powerful cell-autonomous mechanisms of self-defense that allow them to detect, kill, or severely limit the growth of vacuolar pathogens. For example, a number of pattern-recognition receptors have evolved to be positioned within the endocytic network and are thus well poised for vacuolar pathogen detection (Sanjuan et al., 2009). Furthermore, effector mechanisms such as a family of interferon-inducible GTPases are endowed with the ability to target microbial pathogen-containing vacuoles and orchestrate a defense response (Hunn et al., 2011; Kim et al., 2012).
We have previously described a Rab32-dependent pathway that restricts the replication of the human-adapted S. Typhi in macrophages of mice, a nonpermissive species (Spanò and Galán, 2012). We have shown here that S. Typhi can replicate in mice defective for Rab32 or its GEF BLOC-3, and that these mice are more susceptible to infection with the broad-host S. Typhimurium. These observations demonstrate that this pathway plays an essential role in restricting the replication of these intracellular pathogens. Consistent with a direct role in controlling bacterial intracellular replication, Rab32 is efficiently recruited to the Salmonella-containing vacuole presumably delivering an antimicrobial factor. Although the specific nature of the antimicrobial factor that limits Salmonella replication is unknown, it is well established that Rab32, in conjunction with other Rab GTPases and multiprotein assemblies known as BLOC-1, BLOC-2, and BLOC-3, coordinates the delivery of specific cargo to lysosome-related compartments such as melanosomes, T cells, and platelet-dense granules (Bultema et al., 2012; Dell'Angelica, 2004; Luzio et al., 2014; Raposo and Marks, 2007). This cargo includes antimicrobial compounds as well as enzymes required for the synthesis of melanin and its phenyl oxidase intermediates, many of which have potent antimicrobial activity (Aspengren et al., 2009; Benado et al., 2009; Bultema et al., 2012; Dell'Angelica, 2004; Denat et al., 2014). In insects, this pathway has been shown to play a central role in the defense response against microbial pathogens (Jiang et al., 2010; Lu et al., 2014). Our results indicate that this general pathway is also operational in vertebrates and constitutes a cell-autonomous mechanism to control intracellular pathogen replication.
We have also described here a mechanism that the intracellular bacterial pathogen S. Typhimurium has evolved to block this Rab32-dependent host defense pathway. This mechanism involves the delivery of two type III secretion effector proteins that in a functionally redundant manner but utilizing different biochemical mechanisms are able to neutralize the Rab32-dependent delivery of antimicrobial compounds to the SCV. One of the effector proteins is a protease, GtgE, which can specifically target and cleave Rab32 (Spanò and Galán, 2012; Spanò et al., 2011). The other effector protein is SopD2, which can directly inactivate Rab32 working as GAP for this GTPase. An S. Typhimurium strain lacking these two effector proteins exhibited a drastic virulence-attenuation phenotype demonstrating the potential effectiveness of this pathogen-restriction pathway. The virulence phenotype was completely reversed in a mouse in which this pathway was absent by virtue of a mutation in BLOC-3, the nucleotide exchange factor for Rab32.
Both gtgE and sopD2 are either absent (gtgE) or a pseudo-gene (sopD2) in the human-adapted S. Typhi (Parkhill et al., 2001). The loss of the ability to neutralize the Rab32-dependent pathogen-restriction mechanism is most likely central to the evolutionary process that eventually led to the loss of the ability of S. Typhi to explore other hosts. The loss of these effectors from S. Typhi would suggest the lack of evolutionary pressure to keep them within the human host, which in turn would imply that this restriction pathway is not operational in humans. However, the high degree of conservation of the components of this pathogen-restriction pathway suggests otherwise. Rather, our results suggest that in humans, S. Typhi must occupy a niche(s) different than in mice and that in this niche(s) this pathway is not operational.
Microbial pathogens often use virulence factors that mimic the activity of eukaryotic proteins as a powerful means to modulate eukaryotic cell function. That mimicry can sometimes be evident in the high degree of primary amino acid sequence or structural similarities of the virulence factors with the eukaryotic protein they mimic. However, mimics in bacterial pathogens have also emerged by convergent evolution, and in this case, they do not display overt structural or amino acid sequence similarity with the protein they mimic (Orchard and Alto, 2012; Stebbins and Galán, 2000, 2001). SopD2 does not exhibit structural or amino acid sequence similarity to known eukaryotic GAPs. However, to exert its activity it uses the same “arginine-finger” mechanism utilized by eukaryotic GAPs and a similar scaffold to present this critical residue to its target GTPase. This is therefore an example of convergently evolved mimicry to eukaryotic-protein function.
We found that, at least in vitro, the GAP activity of SopD2 can be directed to Rab GTPases other than Rab32, such as Rab8A, Rab8B, and Rab10. The ability to target several Rab GTPases in vitro has been previously observed for other eukaryotic GAPs. However, it is unclear whether the GAP activity of SopD2 toward other GTPases contributes to its ability to block the Rab32-dependent pathogen-restriction pathway by preventing Rab32 recruitment to the SCV. Our inability to block Rab32 recruitment by interfering with Rab8A, Rab8B, or Rab10 function suggests that these Rab GTPases may not be a relevant target of SopD2 in the context of this specific function. However, it is possible that by targeting these GTPases, SopD2 may influence other unknown aspects of Salmonella biology. In fact, it has been recently reported that SopD2 can interfere with Rab7 function by poorly understood mechanisms that involve a domain located within its first 150 amino acids (D'Costa et al., 2015). However, the catalytic site responsible for the GAP activity described here is located at the C terminus, and therefore the function of SopD2 described here is unrelated to the previously reported activity.
Our results demonstrate the importance of a cell-autonomous, Rab32-dependent defense mechanism against Salmonella. However, a recent genome-association study has identified a polymorphism within Rab32 that correlates with increased susceptibility to Mycobacterium leprae (Zhang et al., 2011), suggesting that this pathway may also be important for the control of other intracellular vacuolar pathogens. We have also described a remarkable mechanism evolved by Salmonella to counter this pathway by redundantly targeting Rab32 with two type III secretion effector proteins with different biochemical activities (Figure 7). These findings constitute a remarkable example of the evolutionary arms race between a pathogen and its host and may provide the basis for the development of therapeutic strategies to combat infectious diseases by vacuolar pathogens.
Figure 7. Model for the Rab32/BLOC-3-Dependent, Cell-Autonomous Defense Pathway and the Mechanisms Evolved by the Broad-Host S. Typhimuriumto Counter It.
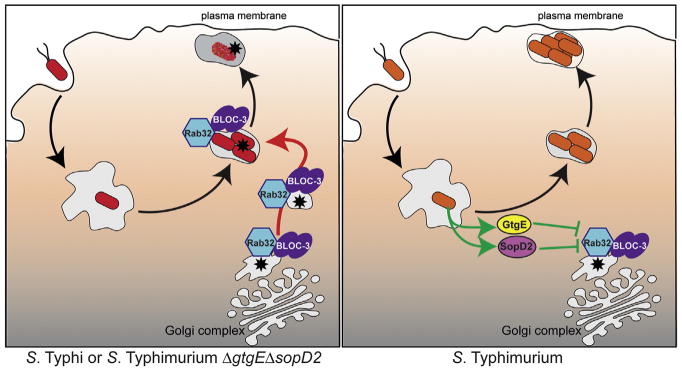
Asterisk represents an antimicrobial factor delivered to the Salmonella-containing vacuole.
Experimental Procedures
Bacterial Strains and Plasmids
The wild-type Salmonella enterica serovar Typhi strain ISP2825 (Galán and Curtiss, 1991) and the wild-type Salmonella enterica serovar Typhimurium strain SL1344 (Hoiseth and Stocker, 1981) have been described previously. All the S. Typhimurium deletion strains were constructed by standard recombinant DNA and allelic exchange procedures as previously described (Kaniga et al., 1994) and are listed in Table S1. All the plasmids used in this study were constructed using standard recombinant DNA techniques and are listed in Table S2.
Tissue Culture Infection Experiments
Overnight cultures of the different Salmonella strains were diluted 1/20 in LB (Lennox) broth containing 0.3 M NaCl and grown until they reached an OD600 of 0.9. Cells were infected with the different strains of S. Typhi and S. Typhimurium in Hank's balanced salt solution (HBSS) at a multiplicity of infections of 30 and 5, respectively, to obtain similar numbers of intracellular bacteria per cell. One hour postinfection, cells were washed three times with HBSS and incubated in DMEM supplemented with 100 μg/ml gentamicin for 30 min to kill extracellular bacteria. Cells were then washed, and fresh DMEM containing 5 μg/ml gentamicin was added to avoid cycles of reinfection. Type III secretion-mediated protein translocation into cultured Henel-407 cells was carried out as previously described (Collazo and Galán, 1997).
Animal Infection Experiments
All animal experiments were conducted according to protocols approved by Yale University's Institutional Animal Care and Use Committee. B6.C3-Pde6brd1 Hps4le/J mouse strain (BLOC-3 deficient), which carries a spontaneous nonsense mutation in the light ear gene encoding for Hps4, was purchased from Jackson Laboratory. Rab32−/− mouse embryos were obtained from the NIH-supported Deltagen and Lexicon Knockout Mouse Resource, and animals were derived at the Yale University Genome Editing Center. Overnight cultures of the different Salmonella strains were diluted 1/20 in LB containing 0.3 M NaCl and grown until they reached an OD600 of 0.9. Eight- to twelve-week-old C57BL/6-, BLOC-3-, or Rab32-deficient mice were infected intraperitoneally or orally (by stomach gavage) with different S. Typhi or S. Typhimurium strains at the doses indicated in the figure legends. To enumerate bacterial loads in the spleens, mice were sacrificed 4–5 days after infection, organs were homogenized in 3 ml PBS containing 0.05% sodium deoxycholate, and dilutions were plated on LB plates to determine CFU.
For competition experiments, 8- to 12-week-old C57BL/6 mice were infected intraperitoneally with a mixture containing 1:1 of the competing strains. For the competition experiment reported in Figure 1A, an S. Typhimurium strain carrying a kanamycin-resistance cassette within a gene that does not affect virulence was used as wild-type. This strain was competed to ΔgtgE or GtgEH151A, which have been previously described (Spanò et al., 2011). Inocula were plated on LB plates or kanamycin-selective plates to measure the number of CFUs of the competing strains. To enumerate bacterial loads in the spleens, mice were sacrificed 4–5 days after infection, organs were homogenized in 3 ml PBS containing 0.05% sodium deoxycholate, and dilutions were plated on LB or LB-containing kanamycin plates. The competitive index was calculated as the ratio of CFU of strain A over strain B recovered from the spleens, normalized by the ratio of the CFU of strain A over strain B in the inoculum.
Viral Transduction
Mouse Rab32 N-terminally fused to cyan fluorescent protein (CFP) was expressed through viral transduction using a LZRS-based retroviral vector. Pseudotyped virus was produced by cotransfecting 4 μg pLZRS-CFP-Rab32, 4 μg pVSVG, and 4 μg pGag/Pol plasmids in a 10 cm dish of HEK293 cells using 30 μl of Fugene 6 Transfection Reagent from Roche. HEK293 culture supernatant was collected 48 hr after transfection and used 1:10 to transduce COS-1. Transduced cells were infected with Salmonella strains 40 hr after transduction.
Imaging
Live-cell imaging was performed at 37°C in a temperature-, humidity-, and CO2-controlled live chamber (Pathology Devices). COS-1 or HeLa cells (as indicated) were plated on 35 mm glass-bottom MatTek dishes previously coated with 0.2 mg/ml fibronectin, transduced with virus expressing CFP-Rab32, and 40 hr later infected with the indicated S. Typhi or S. Typhimurium strains. Cells were imaged 3 hr postinfection using a 60 × oil objective (numerical aperture, 1.4) of an Improvision spinning disc confocal microscope equipped with a Nikon TE2000. Quantifications are averages of three experiments, in which at least 100 intracellular bacteria were counted in each condition.
Rab32 Levels in S. Typhimurium-Infected Cells
For the analysis of Rab32 levels after bacterial infection, COS-1 cells were transduced with virus expressing CFP-Rab32, and 40 hr later they were infected with different Salmonella strains. Two and a half hours after infection, cells were lysed in SDS-PAGE loading buffer. Western-blot analysis was performed using Odyssey infrared imaging system (LI-COR Biosciences). A rabbit polyclonal anti-GFP (Invitrogen) was used at 1:10,000 dilution.
Coaffinity Purification and Liquid Chromatography-Tandem Mass Spectrometry Analysis
Raw 264.7 cells (2 × 108 in each condition) were infected with S. Typhimurium ΔsopD2 bearing a plasmidexpressing 3×FLAG-tagged SopD2 (pSB4829) (sample 1), with S. Typhimurium ΔpipA bearing a plasmid expressing 3×FLAG-tagged PipA (sample 2), or with S. Typhimurium ΔSopA bearing a plasmid expressing 3×FLAG-tagged SopA (sample 3) as irrelevant effector controls. Five hours postinfection, cells were scraped, washed with PBS, and lysed at 4°C in 10 mM HEPES, 150 mM NaCl, 1 mM EDTA, and 0.2% Triton X-100 (pH 7.4) (lysis buffer) in the presence of a protease inhibitor cocktail. The following steps were all performed at 4°C. After centrifugation at 5,000 × g for 10 min, the supernatants were incubated with 50 μl protein A Sepharose for 1 hr and centrifuged again at 5,000 × g. The precleared supernatants were incubated with 15 μl anti-FLAG M2 Affinity Gel beads (Sigma) O/N at 4°C. At the end of the incubation, beads were washed three times with lysis buffer and once with TBS and incubated with 300 μl 0.1 M glycine (pH 7.2) for 5 min. After centrifugation, the supernatants were collected, and the acid elution was repeated twice. All elutions were pooled together and precipitated with 1% TCA and 0.05% Na deoxycholate. Precipitates were washed twice in acetone, dried and resuspended in SDS-loading buffer, and run on a 10% SDS-PAGE. After Coomassie staining, lanes were sliced and subjected to standard in-gel digestion as described previously (Liu et al., 2012). Briefly, the protein samples were treated with DTT to reduce disulfide bonds and then alkylated with iodoacetamide (IAM). Trypsin digestion was allowed to occur overnight. Resulting peptides were extracted from the gel matrix and re-suspended in aqueous buffer before final LC-MS/MS analysis. Nanoflow reverse-phase LC separation was carried out on a Proxeon EASY-nLC system (Thermo Scientific). The capillary column (75 μm × 150 mm, PICOFRIT, New Objective) was packed in house. A methanol slurry containing 5 μm and 100 Å Magic C18AQ silica-based particles (Microm BioResources Inc.) was forced to run through an empty capillary (with afrit in the end) using a pressurized device. The LC mobile phase comprised of solvent A (97% H2O, 3% acetonitrile [ACN], and 0.1% formic acid [FA]) and solvent B (100% ACN and 0.1% FA). The nano-LC separation was performed with the following gradient: B was increased from 7% to 35% in 40 min, raised to 90% in 3 min, and kept therefor 10 min before going back to 100% A for column equilibration. At the moment when peptides were eluted from the capillary column, they were electrosprayed directly onto a linear ion trap mass spectrometer (LTQ Velos, ThermoElectron) for MS and MS/MS analysis. A data-dependent mode was enabled for peptide fragmentations with one full MS scan followed by collision-induced dissociation (CID) of the ten most intense peptide ions. Dynamic exclusion was enabled to preclude repeated analyses of the same precursor ion. MS/MS scans were processed and searched using MASCOT (Matrix Science Ltd.). The resulting peptide and protein assignments were filtered to keep only those identifications with scores above extensive homology. Proteins identified in sample 1 and absent in sample 2 and sample 3 were considered significant.
GAP Assay
Purified recombinant proteins Rac1, Rab8A, Rab10, Rab31, and Rab32 were prepared following standard procedures as previously described (Stebbins and Galán, 2000). Measurement of the GAP activities of the different preparations was carried out using a GRPase-GAP assay kit (Innova Biosciences). Briefly, purified GTPases and SopD2 and its mutants (0.2 nmol each) were incubated in 200 μl of GTPase reaction buffer from the GTPase assay kit for 3 hr. Reactions were terminated by the addition of 50 μl gold mix and stabilized by the addition of 20 μl stabilizer 2 min later. After 30 min, the enzymatic GTP hydrolysis activity was obtained at an absorbance of 635 nm using a microplate reader (Molecular Devices).
Supplementary Material
Highlights.
A Rab32-dependent mechanism of defense restricts the replication of Salmonella
Salmonella counters this host defense pathway with the effectors SopD2 and GtgE
SopD2 is a GAP and GtgE a protease, which both target Rab32
A mutant lacking these effectors exhibits drastic virulence reduction
Acknowledgments
We thank members of the J.E.G. laboratory for careful review of this manuscript. We thank Jana Kamanova and Hui Sun for technical advice and reagents. X.G. was supported in part by a James Hudson Brown-Alexander Brown Coxe Postdoctoral Fellowship. This work was supported by National Institute of Allergy and Infectious Diseases grants AI055472 and AI079022 (to J.E.G.).
Footnotes
Author Contributions: S.S. conducted experiments shown in Figures 1, 2, 5, 6, S1, and S2 and Table S1; X.G. conducted experiments shown in Figures 3 and 4; S.H. contributed to experiments shown in Figures 3, 4, S3, S4, and S6; M.L.-T. contributed to experiments shown in Figure 5 and Table S1; J.E.G was involved in the design, interpretation, and supervision of this study; J.E.G. wrote the paper with input from S.S. and comments from all authors.
Supplemental Information: Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2016.01.004.
References
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem. 1999;274:33186–33189. doi: 10.1074/jbc.274.47.33186. [DOI] [PubMed] [Google Scholar]
- Alix E, Mukherjee S, Roy CR. Subversion of membrane transport pathways by vacuolar pathogens. J Cell Biol. 2011;195:943–952. doi: 10.1083/jcb.201105019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspengren S, Hedberg D, Sköld HN, Wallin M. New insights into melanosome transport in vertebrate pigment cells. Int Rev Cell Mol Biol. 2009;272:245–302. doi: 10.1016/S1937-6448(08)01606-7. [DOI] [PubMed] [Google Scholar]
- Asrat S, de Jesús DA, Hempstead AD, Ramabhadran V, Isberg RR. Bacterial pathogen manipulation of host membrane trafficking. Annu Rev Cell Dev Biol. 2014;30:79–109. doi: 10.1146/annurev-cellbio-100913-013439. [DOI] [PubMed] [Google Scholar]
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benado A, Nasagi-Atiya Y, Sagi-Eisenberg R. Protein trafficking in immune cells. Immunobiology. 2009;214:507–525. doi: 10.1016/j.imbio.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13:551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- Brumell JH, Kujat-Choy S, Brown NF, Vallance BA, Knodler LA, Finlay BB. SopD2 is a novel type III secreted effector of Salmonella typhimurium that targets late endocytic compartments upon delivery into host cells. Traffic. 2003;4:36–48. doi: 10.1034/j.1600-0854.2003.40106.x. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galán JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema JJ, Ambrosio AL, Burek CL, Di Pietro SM. BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J Biol Chem. 2012;287:19550–19563. doi: 10.1074/jbc.M112.351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo CM, Galán JE. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- Creasey EA, Isberg RR. Maintenance of vacuole integrity by bacterial pathogens. Curr Opin Microbiol. 2014;17:46–52. doi: 10.1016/j.mib.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JA, Mintz ED. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa VM, Braun V, Landekic M, Shi R, Proteau A, McDonald L, Cygler M, Grinstein S, Brumell JH. Salmonella disrupts host endocytic trafficking by SopD2-mediated inhibition of Rab7. Cell Rep. 2015;12:1508–1518. doi: 10.1016/j.celrep.2015.07.063. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Denat L, Kadekaro AL, Marrot L, Leachman SA, Abdel-Malek ZA. Melanocytes as instigators and victims of oxidative stress. J Invest Dermatol. 2014;134:1512–1518. doi: 10.1038/jid.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol. 2010;8:117–128. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol. 2013;117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- Galán JE, Curtiss R., 3rd Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–2139. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassl GA, Finlay BB. Pathogenesis of enteric Salmonella infections. Curr Opin Gastroenterol. 2008;24:22–26. doi: 10.1097/MOG.0b013e3282f21388. [DOI] [PubMed] [Google Scholar]
- Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Hunn JP, Feng CG, Sher A, Howard JC. The immunity-related GTPases in mammals: a fast-evolving cell-autonomous resistance system against intracellular pathogens. Mamm Genome. 2011;22:43–54. doi: 10.1007/s00335-010-9293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarczak J, Kościuczuk EM, Lisowski P, Strzałkowska N, Jóźwik A, Horbańczuk J, Krzyżewski J, Zwierzchowski L, Bagnicka E. Defensins: natural component of human innate immunity. Hum Immunol. 2013;74:1069–1079. doi: 10.1016/j.humimm.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Bossio JC, Galán JE. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Mishra A, Lambright DG. Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic. 2009;10:1377–1389. doi: 10.1111/j.1600-0854.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao B, Novik V, Galán JE. Quantitative Proteomics of Intracellular Campylobacter jejuni Reveals Metabolic Reprogramming. PLoS Pathog. 2012;8:e1002562. doi: 10.1371/journal.ppat.1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Zhang Q, Zhang J, Yang B, Wu K, Xie W, Luan YX, Ling E. Insect prophenoloxidase: the view beyond immunity. Front Physiol. 2014;5:252. doi: 10.3389/fphys.2014.00252. http://dx.doi.org/10.3389/fphys.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Hackmann Y, Dieckmann NM, Griffiths GM. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb Perspect Biol. 2014;6:a016840. doi: 10.1101/cshperspect.a016840. http://dx.doi.org/10.011101/cshperspect.a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Nairz M, Haschka D, Demetz E, Weiss G. Iron at the interface of immunity and infection. Front Pharmacol. 2014;5:152. doi: 10.3389/fphar.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nish S, Medzhitov R. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity. 2011;34:629–636. doi: 10.1016/j.immuni.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001;52:259–274. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- Orchard RC, Alto NM. Mimicking GEFs: a common theme for bacterial pathogens. Cell Microbiol. 2012;14:10–18. doi: 10.1111/j.1462-5822.2011.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, Wilson RP, Winter SE, Bäumler AJ. Clinical pathogenesis of typhoid fever. J Infect Dev Ctries. 2008;2:260–266. doi: 10.3855/jidc.219. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R, García-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddick LE, Alto NM. Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol Cell. 2014;54:321–328. doi: 10.1016/j.molcel.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev. 2009;227:203–220. doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- Sato T, Iwano T, Kunii M, Matsuda S, Mizuguchi R, Jung Y, Hagiwara H, Yoshihara Y, Yuzaki M, Harada R, Harada A. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J Cell Sci. 2014;127:422–431. doi: 10.1242/jcs.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder N, Henry T, de Chastellier C, Zhao W, Guilhon AA, Gorvel JP, Méresse S. The virulence protein SopD2 regulates membrane dynamics of Salmonella-containing vacuoles. PLoS Pathog. 2010;6:e1001002. doi: 10.1371/journal.ppat.1001002. http://dx.doi.org/10.1371/journal.ppat.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Galán JE. A Rab32-dependent pathway contributes to Salmonella typhi host restriction. Science. 2012;338:960–963. doi: 10.1126/science.1229224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Liu X, Galán JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci USA. 2011;108:18418–18423. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galán JE. Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Galán JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Townsend SM, Miao EA, Miller SI, Ficht TA, Adams LG, Bäumler AJ. Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D. Protective and pathological roles of mast cells and basophils. Nat Rev Immunol. 2013;13:362–375. doi: 10.1038/nri3427. [DOI] [PubMed] [Google Scholar]
- Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, Seabra MC. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175:271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MW, Jones MA, Watson PR, Siber AM, McCormick BA, Hedges S, Rosqvist R, Wallis TS, Galyov EE. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell Microbiol. 2000;2:293–303. doi: 10.1046/j.1462-5822.2000.00054.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Bazhin AV, Werner J, Karakhanova S. Reactive oxygen species in the immune system. Int Rev Immunol. 2013;32:249–270. doi: 10.3109/08830185.2012.755176. [DOI] [PubMed] [Google Scholar]
- Zhang F, Liu H, Chen S, Low H, Sun L, Cui Y, Chu T, Li Y, Fu X, Yu Y, et al. Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nat Genet. 2011;43:1247–1251. doi: 10.1038/ng.973. [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen LM, Hernandez L, Shears SB, Galán JE. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol. 2001;39:248–259. doi: 10.1046/j.1365-2958.2001.02230.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


