Abstract
The oral streptococci are spherical Gram-positive bacteria categorized under the phylum Firmicutes which are among the most common causative agents of bacterial infective endocarditis (IE) and are also important agents in septicaemia in neutropenic patients. The Streptococcus mitis group is comprised of 13 species including some of the most common human oral colonizers such as S. mitis, S. oralis, S. sanguinis and S. gordonii as well as species such as S. tigurinus, S. oligofermentans and S. australis that have only recently been classified and are poorly understood at present. We present StreptoBase, which provides a specialized free resource focusing on the genomic analyses of oral species from the mitis group. It currently hosts 104 S. mitis group genomes including 27 novel mitis group strains that we sequenced using the high throughput Illumina HiSeq technology platform, and provides a comprehensive set of genome sequences for analyses, particularly comparative analyses and visualization of both cross-species and cross-strain characteristics of S. mitis group bacteria. StreptoBase incorporates sophisticated in-house designed bioinformatics web tools such as Pairwise Genome Comparison (PGC) tool and Pathogenomic Profiling Tool (PathoProT), which facilitate comparative pathogenomics analysis of Streptococcus strains. Examples are provided to demonstrate how StreptoBase can be employed to compare genome structure of different S. mitis group bacteria and putative virulence genes profile across multiple streptococcal strains. In conclusion, StreptoBase offers access to a range of streptococci genomic resources as well as analysis tools and will be an invaluable platform to accelerate research in streptococci. Database URL: http://streptococcus.um.edu.my.
Introduction
Streptococcus is a major genus of spherical Gram-positive bacteria which belong to the phylum Firmicutes. Streptococci are classified as alpha-hemolytic, beta-hemolytic or gamma-hemolytic according to their appearance on blood agar. Alpha-hemolysis involves the bleaching of heme iron by streptococcal hydrogen peroxide (H2O2), resulting in a greenish tinge on blood agar [1]. Alpha-hemolytic streptococci used to be known as the ‘Viridans group’ for the greenish color produced by hemolysis. However, alpha-hemolysis is not entirely consistent between different strains of individual Streptococcal species, and therefore the term ‘Viridans’ is somewhat misleading and is no longer used. These organisms are now more commonly known as the oral streptococci. Overall, the streptococci are divided into six groups, namely the Mitis, Anginosus, Salivarius, Mutans, Bovis and Pyogenic groups, using sequence analysis of the 16S rRNA gene or of a group of housekeeping genes [2–4]. In 2002, Facklam proposed a phenotypic identification scheme which included an additional new cluster called Sanguinis [5]. This cluster, containing S. sanguinis, S. gordonii and S. sinensis is sometimes included within the mitis group.
The human oral streptococci are commensals which often inhabit the gastrointestinal and genitourinary tracts, as well as the oral mucosa and tooth surfaces. In healthy individuals, streptococci can constitute more than 50% of the oral microbiota [6] and these bacteria generally possess low pathogenic potential. However, oral streptococci can invade the bloodstream, and have the potential to cause infective endocarditis (IE) or post-antineoplastic septicaemia in neutropenic patients with haematological disease. Other oral Streptococcus-associated conditions including odontofacial infections, brain abscesses and abdominal infections have also been reported [7]. Furthermore, recent work has shown that S. mitis group bacteria play a major role in exacerbating influenza infection particularly among immunocompromised individuals; Streptococcus oralis and S. mitis were found to produce neuraminidase (NA), a vital target of anti-influenza drugs. The NA activity exhibited by these oral bacteria stimulates the release of influenza virus, boosts viral M1 protein expression levels and activates the cell signaling ERK pathway, potentially enhancing viral infections [8].
The mitis group is comprised of 13 known species including S. australis, S. cristatus (formerly S. crista), S. gordonii, S. infantis, S. mitis, S. oligofermentans, S. oralis, S. parasanguinis (formerly S. parasanguis), S. peroris, S. pneumoniae, S. pseudopneumoniae, S. sanguinis (formerly S. sanguis), and the latest grouped species, S. tigurinus. Currently, the complete genome sequences of 7 species of this mitis group (S. pneumoniae, S. pseudopneumoniae, S. mitis, S. oralis, S. gordonii, S. sanguinis and S. parasanguinis) are stored on the National Center for Biotechnology Information (NCBI)’s FTP site.
Here, we present StreptoBase which provides an invaluable resources and analysis platform for research communities. Through this platform and the provided in-house designed analysis tools, users may obtain insights into the biology, phylogeny, genetic variation and virulence of particular strains or species of interest. Furthermore, we have included 27 newly sequenced, assembled and annotated genomes of novel strains from six different species of S. mitis group from our laboratory into StreptoBase. These new genomes include novel genome sequences of the recently classified species S. oligofermentans and S. tigurinus. The ultimate objective of StreptoBase is to provide a user-friendly database resource and analysis platform. Users can search, browse, visualize, download and analyze the mitis group genomes, particularly comparative whole-genome analysis on the fly using our in-house advanced bioinformatics tools, which is designed to support the expanding Streptococcus genus research community.
Materials and Methods
Datasets
Seventy-seven genome sequences of S. mitis group bacteria were downloaded from the public NCBI database. We also have included 27 novel strains/genomes of S. mitis group generated from our laboratory in a sequencing project. All 27 strains were clinical isolates from individuals with dental plaque or infective endocarditis from different geographical locations (Table 1). Of these strains, 14 strains were isolated in the United Kingdom, 10 in United States, 2 in Australia and 1 in Denmark (Table 1). S. sanguinis NCTC 7863 is also known as ATCC 10556 while S. gordonii Blackburn and Channon are designated NCTC 10231 and NCTC 7869, respectively. Additionally, a number of these S. mitis group strains including JPIIBBV4, JPIIBV3, JPIBVI, LRIIBV4, DGIIBVI and DOBICBV2 have been previously described [9]. The isolation of strain M99 was described in a study of mechanisms of platelet aggregation by oral streptococci [10]. The other two oral isolates, SK120 and SK184 have also been described by Mogens Kilian and his fellow researchers in their taxonomic study of ‘Viridans’ Streptococci conducted in 1989 [11].
Table 1. The isolation details of 27 Streptococcus strains includes isolation source, geographical area and strain author.
| Strain Name | Identified Species | Isolation source | Country | Strain Author | References |
|---|---|---|---|---|---|
| PV40 | S. gordonii | Infective endocarditis | UK | P.M. Vesey, S.D. Hogg and R.R.B. Russell, Newcastle University | |
| NCTC 7863 | S. sanguinis | Infective endocarditis | USA | White and Niven 1946 | Streptococcus sanguinis (ATCC® 10556™) |
| Blackburn | S. gordonii | Human isolate | UK | R. Hare, P.H.L.S. Colindale, London | Describe in Nobbs, A. H., et al (2007). Journal of bacteriology, 189(8), 3106–3114. |
| BVME8 | S. parasanguinis | Human oral cavity | UK | J. Manning, S.D. Hogg, Newcastle University | |
| Channon | S. gordonii | Not recorded | UK | R. Hare, Queen Charlotte’s Hospital, London | Described in Millsap, K. W. et al (1999). FEMS Immunology & Medical Microbiology, 26(1), 69–74. |
| DGIIBVI | S. tigurinus | Dental plaque | USA | M. Levine, Oklahoma University | Described in McAnally & Levine (1993) Oral Microbiol Immunol 8: 69–74 |
| DOBICBV2 | S. oligofermentans | Dental plaque | USA | M. Levine, Oklahoma University | Described in McAnally & Levine (1993) Oral Microbiol Immunol 8: 69–74 |
| FSS2 | S. gordonii | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| FSS3 | S. gordonii | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| FSS4 | S. sanguinis | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| FSS8 | S. gordonii | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| FSS9 | S. sanguinis | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| JPIIBBV4 | S. oligofermentans | Dental plaque | USA | M. Levine, Oklahoma University | Described in McAnally & Levine (1993) Oral Microbiol Immunol 8: 69–74 |
| JPIIBV3 | S. oralis | Dental plaque | USA | M. Levine, Oklahoma University | Described in McAnally & Levine (1993) Oral Microbiol Immunol 8: 69–75 |
| JPIBVI | S. tigurinus | Dental plaque | USA | M. Levine, Oklahoma University | Described in McAnally & Levine (1993) Oral Microbiol Immunol 8: 69–76 |
| LRIIBV4 | S. oligofermentans | Dental plaque | USA | M. Levine, Oklahoma University | Described in McAnally & Levine (1993) Oral Microbiol Immunol 8: 69–77 |
| M5 | S. gordonii | Dental plaque | USA | Rosan, B., University of Pennsylvania | Described in Rosan B (1973) Infect Immun 7 (2):205 |
| M99 | S. gordonii | Infective endocarditis | USA | P.M. Sullam, UCSF | Isolation described in Sullam, P.M., Valone, F.H., and Mills, J. (1987) Infect Immun 55: 1743–1750. |
| MB451 | S. sanguinis | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| MB666 | S. gordonii | Infective endocarditis | UK | S.D. Hogg, Newcastle University | |
| MW10 | S. gordonii | Not recorded | Australia | J. Manning, Sydney Dental School | |
| PJM8 | S. sanguinis | Human oral cavity | UK | J. Manning, S.D. Hogg, Newcastle University | |
| PK488 | S. gordonii | Subgingival dental plaque | USA | P. E. Kolenbrander, National Institutes of Health, MD | |
| POW10 | S. parasanguinis | Not recorded | Australia | J. Manning, Sydney Dental School | |
| SK12 | S. gordonii | Human oral cavity | Denmark | M. Kilian, Aarhus, Denmark | |
| SK120 | S. gordonii | Human oral cavity | UK | P. H. A. Sneath (provided by M. Kilian) | Described in Kilian et al (1989) INTERNATIONAL JOURNAL OF SYSTEMATIC BACTERIOLOGY, 39: 471–484. |
| SK184 | S. gordonii | Dental plaque | UK | P. Handley (provided by M. Kilian) | Described in Kilian et al (1989) INTERNATIONAL JOURNAL OF SYSTEMATIC BACTERIOLOGY, 39: 471–484. |
Briefly, the 27 S. mitis group genomes were sequenced using Next-Generation Sequencing Illumina HiSeq2000 platform. Data pre-processing was performed by a trimming approach (Phred score Q20) and assembled using CLC Genomic Workbench V6.5 (CLC BIO Inc., Aarhus, Denmark). In general, these assemblies showed high N50 values and low contig numbers, indicating high quality genome assemblies. The assembled mitis group genomes harbor an average GC content of 35% to 45% and with an average genome size of approximately 2MB (Table 2).
Table 2. The genome identity of the 27 isolated Streptococcus strains with the summary assembly results.
| Strain Name | K-mer | Contig no. | N50 (bp) | Genome Size (bp) | Identified Species | Genome coverage (%) | Genome Identity (%) | NCBI Accession numbers |
|---|---|---|---|---|---|---|---|---|
| PV 40 | 32 | 43 | 233745 | 2191051 | S. gordonii | 95 | 98 | SAMN03480623 |
| NCTC 7863 | 24 | 110 | 45631 | 3078022 | S. sanguinis | 84 | 95 | SAMN03480625 |
| Blackburn | 24 | 50 | 158790 | 2164532 | S. gordonii | 90 | 96 | SAMN03480626 |
| BVME8 | 17 | 109 | 53977 | 2122687 | S. parasanguinis | 86 | 97 | SAMN03480630 |
| Channon | 28 | 33 | 174000 | 2233600 | S. gordonii | 89 | 96 | SAMN03480628 |
| DGIIBVI | 26 | 44 | 229281 | 1885841 | S. tigurinus | 79 | 94 | SAMN03480631 |
| DOBICBV2 | 21 | 99 | 45179 | 1979216 | S. oligofermentans | 77 | 94 | SAMN03480632 |
| FSS2 | 28 | 19 | 575926 | 2185874 | S. gordonii | 92 | 98 | SAMN03481559 |
| FSS3 | 21 | 398 | 172943 | 2312061 | S. gordonii | 92 | 96 | SAMN03481560 |
| FSS4 | 28 | 63 | 389092 | 2312671 | S. sanguinis | 85 | 95 | SAMN03480635 |
| FSS8 | 25 | 41 | 286373 | 2151860 | S. gordonii | 90 | 95 | SAMN03480641 |
| FSS9 | 25 | 20 | 356680 | 2429261 | S. sanguinis | 97 | 95 | SAMN03480643 |
| JPIIBBV4 | 30 | 95 | 48467 | 1991853 | S. oligofermentans | 78 | 94 | SAMN03480680 |
| JPIIBV3 | 31 | 75 | 209178 | 1990145 | S. oralis | 79 | 94 | SAMN03480681 |
| JPIBVI | 28 | 37 | 940267 | 1792994 | S. tigurinus | 87 | 96 | SAMN03480682 |
| LRIIBV4 | 24 | 373 | 44211 | 2097683 | S. oligofermentans | 76 | 94 | SAMN03481561 |
| M5 | 28 | 67 | 145888 | 2157832 | S. gordonii | 88 | 95 | SAMN03480683 |
| M99 | 29 | 45 | 134448 | 2167061 | S. gordonii | 89 | 95 | SAMN03480687 |
| MB451 | 26 | 27 | 382788 | 2452806 | S. sanguinis | 94 | 96 | SAMN03480686 |
| MB666 | 25 | 20 | 313888 | 2308142 | S. gordonii | 90 | 96 | SAMN03480688 |
| MW10 | 28 | 27 | 247835 | 2186113 | S. gordonii | 92 | 98 | SAMN03480689 |
| PJM8 | 25 | 163 | 396031 | 2368281 | S. sanguinis | 92 | 95 | SAMN03480699 |
| PK488 | 38 | 46 | 183297 | 2262708 | S. gordonii | 91 | 96 | SAMN03480700 |
| POW10 | 14 | 117 | 30074 | 2042518 | S. parasanguinis | 77 | 96 | SAMN03480701 |
| SK12 | 25 | 28 | 235294 | 2164760 | S. gordonii | 89 | 95 | SAMN03480703 |
| SK120 | 36 | 27 | 200167 | 2145851 | S. gordonii | 90 | 96 | SAMN03480740 |
| SK184 | 26 | 53 | 210865 | 2255121 | S. gordonii | 92 | 97 | SAMN03480741 |
Genome annotation
StreptoBase currently comprises a total of 104 S. mitis group genomes (a genome collection of NCBI resources genome records plus our 27 isolated strains) from 11 species: S. australis, S. cristatus, S. gordonii, S. infantis, S. mitis, S. oligofermentans, S. oralis, S. parasanguinis, S. peroris, S. sanguinis, and S. tigurinus (Table 3).
Table 3. The species table summarizes the total number of draft and complete genomes of each S. mitis group species accordingly.
| Species | Draft Genomes | Complete Genome |
|---|---|---|
| S. australis | 1 | 0 |
| S. cristatus | 1 | 0 |
| S. gordonii | 14 | 1 |
| S. infantis | 6 | 0 |
| S. mitis | 21 | 1 |
| S. oligofermentans | 3 | 1 |
| S. oralis | 10 | 1 |
| S. parasanguinis | 8 | 2 |
| S. peroris | 1 | 0 |
| S. sanguinis | 26 | 1 |
| S. tigurinus | 6 | 0 |
To facilitate comparative analysis across different S. mitis group genomes, consistency in annotation is important. Therefore, we annotated all 104 genome sequences using the Rapid Annotation using Subsystem Technology (RAST) pipeline, which is a well-established and fully open web-based engine, supporting annotation of both complete and draft genomes[12]. The RAST pipeline enables genome identification of an array set of distinct genome components including protein-coding genes, ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), pseudogenes, gene function prediction. The RAST genome annotation works by mapping a set of genes to their corresponding subsystems as well as their metabolic reconstructions. Moreover, it predicts functional proteins assignment according to their relatedness in the subsystems of FIGfams database. Using the RAST pipeline, we predicted 213,268 Coding Sequences (CDSs), 5,140 RNAs and 4,542 tRNAs in all 104 genomes in the mitis group genomes.
To systematically predict subcellular localization of each RAST-predicted gene, we utilized the latest PSORTb subcellular localization tool (version 3.0) program [13]. PSORTb is an efficient, open-source tool which supports high precision of proteome-scale prediction coverage and refined sub-categories localization. The predicted subcellular localization sites were computationally calculated based on the values of feature variables which infer the sequences characteristics. Each of the generated values was then sorted to their respective candidate site through their estimated relativity. Besides the subcellular localization information, we also ran our in-house Perl script to estimate the GC content, hydrophobicity and molecular weight of each protein or gene.
Database structure, composition and implementation
StreptoBase was designed to provide a wide range of useful information and functionalities (Fig 1). For instance, StreptoBase provides users with some background information about S. mitis group species. Within the homepage of StreptoBase, there is a summary box which comprises the genome information stored in the database, such as number of species, strains, number of CDS, number of RNAs and number of tRNAs (Table 4), which are useful before users proceed to further genome details and downstream analyses.
Fig 1. StreptoBase structure and composition.
(A) Flowchart of functional annotation of Streptococcus genomes. (B) Diagram of the StreptoBase web server. (C) Web interface of the StreptoBase sitemap.
Table 4. StreptoBase Data Summary Table.
| Database Summary | Counts |
|---|---|
| Number of Species: | 11 |
| Number of Strains/Genomes: | 104 |
| Number of CDS: | 213,268 |
| Number of RNAs: | 5,140 |
| Number of tRNAs: | 4,542 |
Furthermore, we have compiled and gathered information from various sources on S. mitis group species, for example, news and conferences, blogs and information and recently published papers, which are available in the StreptoBase homepage. By clicking on “Browse” menu, users will see the list of 11 S. mitis group species along with their respective number of draft and complete genomes, while each “View Strains” button, enabling users to visualize all available Streptococcus genomes of any particular species, respectively. Under the “Browse Strains” page, a summarized genome description which encompasses genome size (Mbp), GC content (%) and a list of contigs, genes and rRNAs of that particular species strain are tabulated and displayed. The “Details” button allows users to access further detailed and additional data of that particular strain such as a complete list of ORFs in the genome, their corresponding functions, start and stop chromosomal positions of each ORF/gene in the “Browse ORF” page. To display all information about an ORF or gene, users can click on the “Details” button associated with the ORF. This will open the “ORF Detail” page, displaying information such as their gene type, start and stop positions, nucleotide length, amino acid sequences, functional classification, SEED subsystem (if available), direction of transcription (strand), subcellular localization, hydrophobicity (pH) as well as molecular weight (Da) will be displayed.
Streptococcus Genome Browser (SGB)
StreptoBase is equipped with a real-time and interactive Streptococcus Genome Browser (SGB), which was customised from a well-established genome browser, JBrowse [14], a fast and modern JavaScript-based genome browser which performs navigation on genome annotations and visualization of the location of genes and flanking genomic regions/genes of a selected Streptococcus strain. This interactive SGB enables users to browse genes or genomic regions with graphic-wise motion smoothly and rapidly. SGB overcomes the discontinuous transitions and provides efficient panning and zooming of a specific genomic region in each Streptococcus genome. Furthermore, users can remotely turn on or off the DNA, RNA, and CDS tracks during the navigation process, providing flexibility in customizing what to view in the SGB viewer. We have also implemented a “Search” feature in the genome browser page, allowing users to quickly search a gene by keyword or ORF ID which is not provided by JBrowse.
Real-time keyword search engine
Considering the fact that StreptoBase would host an extensive number of genes and their annotation and this information will increase periodically, the ability to rapidly search a gene in the database is crucial. To address this issue, we implemented a real-time search engine in StreptoBase using AJAX technology. This real-time search engine was designed to support asynchronous communications between web interface and MySQL database, avoiding the need to refresh the web page and allowing the visualization of search results seamlessly. The real-time search engine retrieves a list of suggested functional classifications of genes that are related to the entered keyword once a keyword is typed.
Database implementation
The web interface of StreptoBase was developed using HyperText Markup Language (HTML), HyperText Preprocessor (PHP), JavaScript, jQuery, Cascading Style Sheets (CSS) and AJAX. The StreptoBase is supported by Linux, Apache, MySQL and PHP (LAMP) architecture.
The Apache web server is equipped with Linux OS to manage the comprehensive Streptococcus genomic data housed in StreptoBase. The front end PHP framework of CodeIgniter version 2.1.3 was implemented to offer model-view-controller (MVC), dividing application data, presentation and background logic and process into three distinct modules. With this advanced feature, all Streptococcus related sources codes and biological data are arranged in a clear and organized fashion which facilitate future updating of new Streptococcus genomes into the existing database system. For Streptococcus biological data storage and management, we utilized MySQL version 14.12 in order to store the extensive Streptococcus genome information into a well-designed database schema and tables. The backend process of StreptoBase is monitored by Perl script, Python script and R script which support the efficiency and functionality of our integrated bioinformatics tools.
Additionally, users are able to download all the Streptococcus genome sequences, ORF annotation details in table format, ORF sequences, RNAs and CDSs as well as nucleotide and amino acid sequences via the “Download” menu.
Results
Database features and incorporated bioinformatics tools
The S. mitis group species are important colonizers of the oral cavity, and are occasionally associated with serious infections [15]. In addition, these organisms have recently been suggested to play important roles in the pathogenesis of influenza [8]. Therefore, the genomic study of diverse S. mitis group bacteria is essential in order to understand how these microorganisms transit from a commensal lifestyle in the mouth to subsequent pathogenesis. However, there is no existing specialized genome database available for the wide array of S. mitis group genomes for comparative genomics. While most biological genome databases only focus on the genome content and genetic variation, we have identified a need to create functional bioinformatics tools to investigate virulence determinants within genomes through comparative pathogenomics, as well as to compare the genome content and genetic variation within the S. mitis group bacteria.
Pairwise Genome Comparison (PGC) tool
We designed and customised a web-based PGC tool for S. mitis group bacteria, enabling users to select and perform pairwise comparisons between two user-selected Streptococcus genomes. A list of Streptococcus genomes is available on PGC tool of StreptoBase, allowing users to choose two Streptococcus genomes for cross strain or cross species comparison. Alternatively, users can upload their own genome sequences, either nucleotides or protein, and compare with the Streptococcus genomes in StreptoBase.
Briefly, the PGC pipeline is supported by NUCmer that is designed to align whole-genome sequences, and Circos that is a well-established tool for genome visualisation. Once users submit their jobs to our server, PGC will call NUCmer program to align user-selected genomes and in-house scripts will be used to process the genome alignment output and generate input files parsed to Circos in order to generate a circular ideogram layout of alignments. Unlike the conventional linear display of alignments, the circular layout shows the relationship between pairs of positions with karyotypes and links encoding the position, size and orientation of the related genomic elements.
Three user-defined parameters are provided in the PGC web interface including minimum percent identity (%), merge threshold (bp) and link threshold (bp). The minimum percent identity cut-off defines a homologous region (represented by links/ribbons in the Circos plot) between the two compared genomes. The merge threshold allows merging of two links/ribbons which have distance within the user-defined threshold, and the link threshold allows users to eliminate any mapped/homologous regions that have genomic size less than the user-defined cut-off. A histogram track is added in the outer ring of the circular plot to indicate the percentage of mapped regions, allowing users to quickly identify potential indels (indicated by white gaps) and mapping regions (indicated by green charts) between the two aligned genomes. The implementation of the PGC pipeline is governed using Perl scripts. This pipeline produces two types of outputs: NUCmer alignment results and the high quality Circos plot (SVG format). Users can freely download these results for publication or further analyses in the PGC result page.
The existing Microbial Genome Comparison (MGC) tool utilizes an in silico genome subtraction method to identify genetic elements specific to a group of strains [16]. While PGC tool uses genome files and NUCmer to perform pairwise genome alignment, the MGC tool uses in silico fragmented genome sequences and performs BLASTN on groups of queries. On the contrary, the VISTA Browser which is well-known for its biological application is able to perform pre-computed pairwise and multiple whole-genome alignments using both global and local alignments [17]. In contrast to circular plots and histograms that are generated by the PGC tool, the alignment results generated by VISTA Browser are displayed using VISTA track in graph plot format to show conserved regions. Additionally, the open source Java-based Artemis Comparison Tool (ACT) requires users to generate a comparison file which identifies homology regions between assembly and reference genome using programs such as BLASTN, TBLASTX or Mummer to be loaded on ACT [18]. The comparative ACT visualization is performed using Artemis components. By contrast, our PGC tool enables a single-flow process of pairwise genome alignment and instant display of the comparative alignment Circos plot.
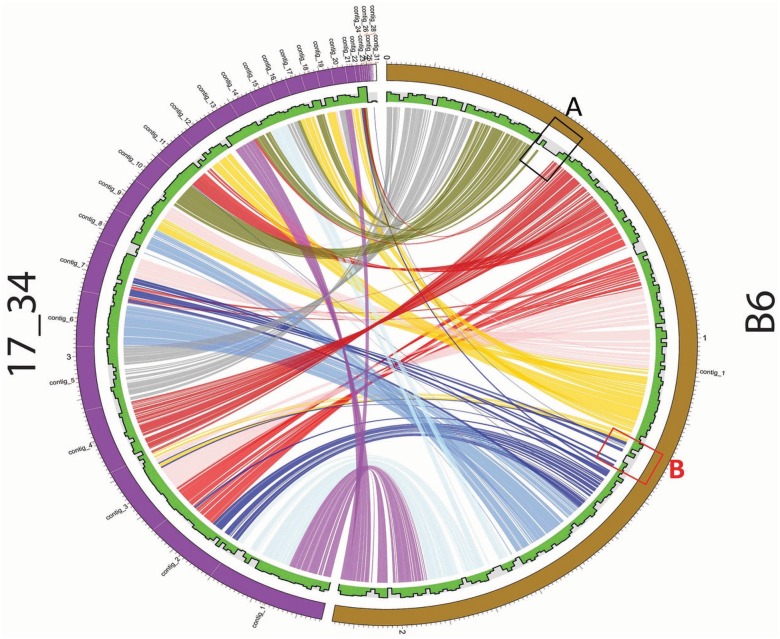
To demonstrate the utility of PGC, we compared S. mitis B6 (complete genome) and 17/34 (draft genome) as a case study in Fig 2.
Fig 2. Pairwise genome comparison between S. mitis B6 and S. mitis 17/34 using PGC tool incorporated in StreptoBase.
50% sequence identity and 50% sequence coverage were used to compare strains using the PGC tool. A and B highlight the indels of the pairwise genome comparison between S. mitis B6 and S. mitis 17/34.
The parameters were set as 80% of minimum percent, default value of 1000bp link threshold and 2000bp merge threshold. S. mitis B6 was isolated in Germany, whereas S. mitis 17/34 was isolated from the urethra of a Russian patient with urethritis. Based on the generated PGC plot, both S. mitis genomes generally shared high similarity as most of their genomic regions could be aligned (Fig 2). One of the features of PGC plot is its ability to quickly identify putative indels via visualization of the gaps in the plot chart which is supported by information displayed in the histogram track. For instance, two of the gap occurrences (Fig 2) indicate the absence of genomic regions in the S. mitis 17/34 genome. The external circular bar of the plot shows the genome size measurements which are approximately 2MB for both S. mitis genomes. Based on the gap observed in Fig 2 (indel ‘A’), the gene loss occurred close to position 400,000bp.
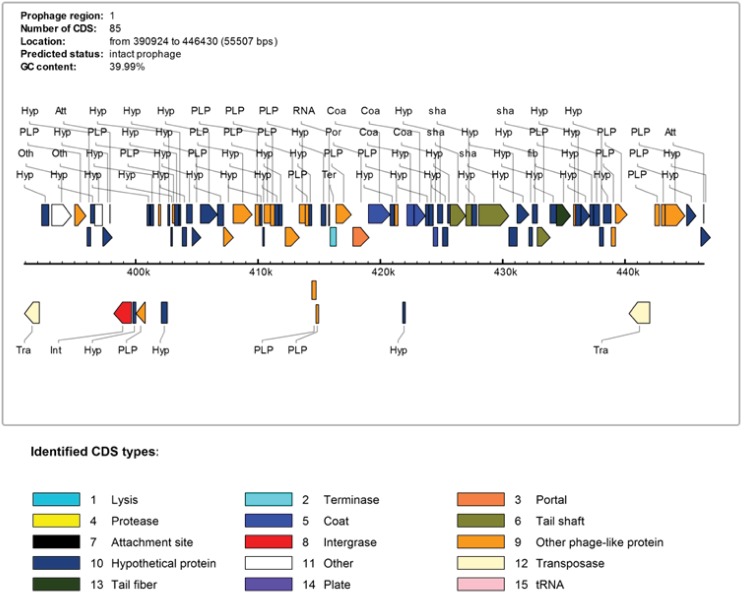
Next, we examined the genes located at indel ‘A’ in S. mitis B6 (Fig 2) by visualising this region using SGB. We identified many phage-related genes associated with this region. To further examine this region, we utilized PHAST (PHAge Search Tool) to annotate and identify prophages sequences found within S. mitis B6 genome (You Zhou et al., 2011). A 56Kb intact prophage with 82 CDSs and GC content of 39.9% was detected from 390,924bp to 446,969bp. Since S. mitis B6 is a complete genome, we can therefore imply the base pair position directly into our B6 annotation file. According to PHAST results, this intact prophage of S. mitis B6 comprised phage-associated genes including phage integrase protein, phage CI-like repressor, phage binding protein, phage portal protein, SPP1 family phage head morphogenesis protein and phage capsid proteins. Therefore, we suggest that S. mitis B6 might have recently acquired this intact prophage. The graphical display of the intact prophage with different types of phage-related genes is shown in Fig 3.
Fig 3. Intact prophage detected in S. mitis B6. This prophage has 85 predicted genes.
Based on the indel ‘B’ detected on the PGC plot in Fig 2, we have revealed a 24Kb incomplete prophage with GC content of 39.17% located at position 1356040bp to 1380128bp Interestingly, this region contains a complete atp operon regulated by the CcpA protein within this incomplete prophage of S. mitis B6 genome. The genes of the atp operon are shown in Table 5. These genes encoding ATP synthases are commonly possessed by oral streptococci for adaptation to the acidic host environment by creating a more alkaline internal system.
Table 5. The ATP synthases within the atp operon of S. mitis B6.
| Locus Tag | Gene Name | Functional annotation |
|---|---|---|
| smi_1315 | atpE | ATP synthase C chain (EC 3.6.3.14) |
| smi_1314 | atpB | ATP synthase A chain (EC 3.6.3.14) |
| smi_1313 | atpF | ATP synthase B chain (EC 3.6.3.14) |
| smi_1312 | atpH | ATP synthase delta chain (EC 3.6.3.14) |
| smi_1311 | atpA | ATP synthase alpha chain (EC 3.6.3.14) |
| smi_1310 | atpG | ATP synthase gamma chain (EC 3.6.3.14) |
| smi_1309 | atpD | ATP synthase beta chain (EC 3.6.3.14) |
| smi_1308 | atpC | ATP synthase epsilon chain (EC 3.6.3.14) |
This protective mechanism is critical especially for streptococcal acid-sensitive glycolytic enzymes [19]. Hence, it may be that the acquisition of this atp operon carried by the incomplete prophage of S. mitis B6 via horizontal gene transfer has assisted its commensal status in maintaining the optimal pH level for bioenergetics processes of S. mitis B6 cells.
Pathogenomics Profiling (PathoProT) tool
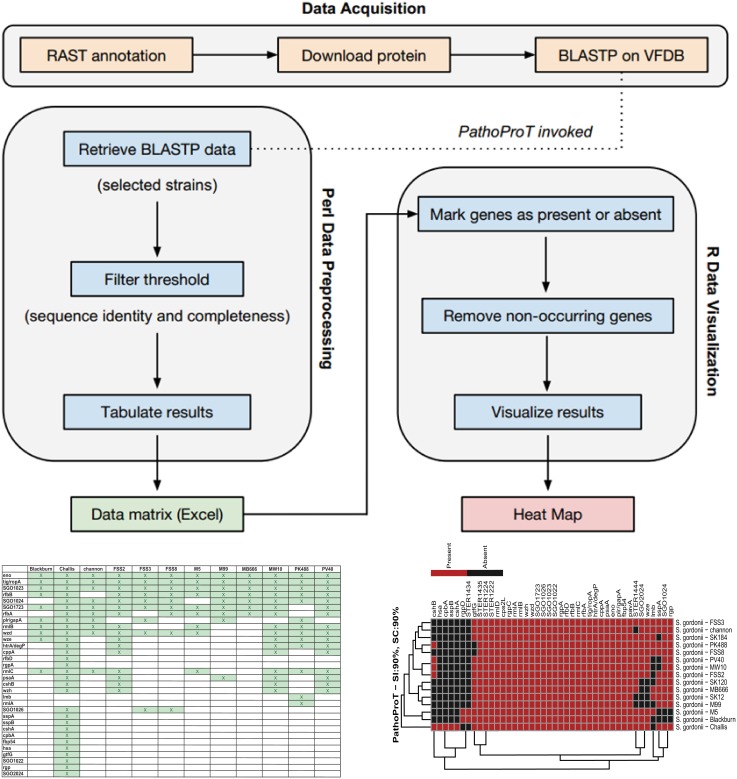
PathoProT was designed to predict virulence genes by comparing Streptococcus amino acid sequences against the Virulence Factors Database (VFDB) [20]. PathoProT utilizes the stand-alone BLAST tools downloaded from the NCBI website. VFDB (Version 2012) currently hosts a set of 19,775 experimentally verified virulence genes originating from a wide range of different bacterial species, providing a useful resource for sequence homology searches. Users can select a list of Streptococcus strains for comparative analysis and set the cut-off, for example, genome identity and completeness for the BLAST search through our provided online web form. The default parameters of PathoProT pipeline are set at 50% sequence identity and 50% sequence completeness for searching and identifying orthologous virulence genes across the selected Streptococcus genomes. However, users can apply their desired cut-offs for the homology search in order to achieve the optimal stringency levels in their analyses.
Briefly, PathoProT pipeline was mainly implemented using Perl. In-house Perl scripts will process BLAST outputs (generated by searching these query sequences against VFDB) for each RAST-predicted protein (query sequence) in the user-selected genomes and identify putative virulence based on user-defined parameters. The filtered BLAST results are consolidated and organised in a matrix table containing information of presence or absence of virulence genes (rows) and Streptococcus strain names (columns). Finally, PathoProT will pass and process this output with our in-house R scripts for hierarchical clustering (complete-linkage algorithm) and generating a heat map for visualisation. The Streptococcus strains will be sorted based on their virulence gene profiles (Fig 4) and a phylogenetic tree will be drawn, users are able to gauge the relationships among the closely-related S. mitis group species/strains as well as their corresponding virulence genes form noticeable clusters through the dendrograms. Therefore, this comparative pathogenomics analysis pipeline is able to provide excellent insight into the virulence gene profiles across different species of Streptococcus. For instance, there is no existing bioinformatics tool that serves the same functionality as PathoProT, which is to predict and allow comparison of virulence genes across different species of bacterial genomes.
Fig 4. A PathoProT flowchart.
PathoProT is mainly implemented using Perl and R scripts. The input of PathoProT would be lists of genes for the selected strains/genomes and the pipeline will generate a heat map at the end of the process.
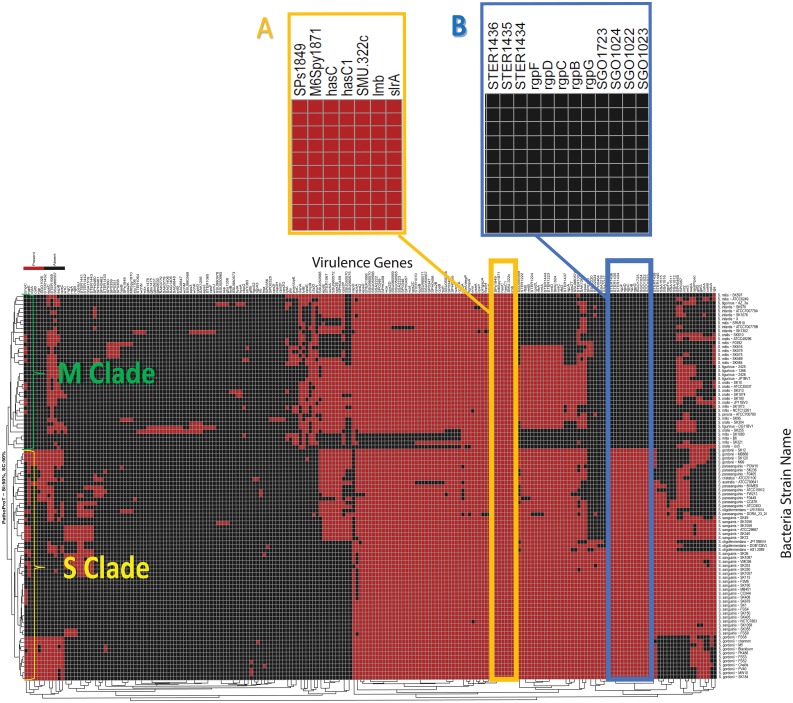
To demonstrate the features or functionalities of PathoProT, we present a comparative pathogenomics study among the S. mitis group bacteria using a threshold of 50% for both sequence identity and coverage to give an insight into their virulence gene profiles. Based on the generated PathoProT heat map, a number of putative virulence genes appear to be conserved among all the mitis group species (Fig 5). The conserved genes hasC (hasC1orSMU.322c) which encodes UTP-glucose-1-phosphate uridylyltransferase (or UDP—glucose pyrophosphorylase)(M6Spy1871) is involved in synthesis of the hyaluronic acid (HA) capsule along with two neighboring genes: hasA and hasB within the has operon.[21]. In fact, Streptococcus pneumoniae, the most pathogenic species of the S. mitis group possesses a polysaccharide capsule which contributes to bacterial pathogenesis [22]. In Streptococcus, HA is found as streptococcal capsule material in some species is an important virulence factor, camouflaging the bacteria efficiently against the recognition of host immune system [23,24] as well as protecting them against reactive oxides released by leukocytes [25]. Additionally, it is possible that HA plays a significant role in mitis group streptococcal adherence and colonization of epithelial cells, leading to bacterial resistance against phagocytosis by macrophages [26–28].
Fig 5. An informative heat map generated by PathoProT tool.
(A) A list of conserved virulence genes carried by all mitis group species and (B) The RGP synthesis related genes which can differentiate M Clade and S Clade. Presence of the virulence gene was labeled in red and absence of the virulence genes was labelled in black.
Another conserved virulence gene, slrA encodes streptococcal lipoprotein rotamase A, which is one of the major surface proteins expressed by S. pneumoniae. This gene is an important cyclophilin which modulates biological function of virulence proteins during the first stage of pneumococcal infection [29]. It is likely that the slrA gene promotes invasion of host cells and facilitates pneumococcal colonization and adherence in S. mitis group bacteria[30,31]. Furthermore, it has been reported that deficiency in slrA reduces bacterial virulence due to its impact on the adherence and internalization by epithelial and endothelial cells [29]. Likewise, the conserved lmb gene encodes a laminin-binding protein which was first identified in Streptococcus agalactiae [32]. The virtually identical adhesins were later discovered in both Streptococcus suis [33] and Streptococcus pyogenes [34,35]. The lmb adhesins have been proposed to help in bacterial pathogenesis via invasion of the damaged epithelium [36]. Overall many surface lipoproteins and adhesins that are important in virulence and pathogenic infections are highly conserved across the S. mitis group bacteria.
According to the phylogenetics tree generated on the left side of the PathoProT heat map (Fig 5), the mitis group can be clearly categorized into two clades: S Clade (S. sanguinis, S. gordonii, S. parasanguinis, S. australis, S. cristatus and S. oligofermentans) and M Clade (S. mitis, S. infantis, S. tigurinus, S. oralis and S. peroris). This phylogeny relationship of the S. mitis group species indicates the close relatedness of cross-species within M Clade and species-to-species of S Clade. Interestingly, we found the rgp genes can be used to differentiate the two different clades in the heat map. For instance, these marker genes are present in all S Clade species but absent in all the M Clade species.
The rgp genes cluster (B, C, D, F and G) is responsible for the synthesis of rhamnose-glucose polysaccharide (RGP) in Streptococcus mutans. Notably, similar genes have been found to be involved in rhamnan synthesis in Escherichia coli [37]. In fact, it has been suggested that E. coli and S. mutans share a common pathway for rhamnan synthesis based on their similarities in RGP synthesis [37]. The function of rgpB is to transfer the second rhamnose residue to a rhamnose residue on N-acetylglucosamine linked to the lipid carrier, followed by rgpF which later catalyzes the transfer of the third rhamnose residue to the second rhamnose residue of the resultant glycolipid carrier. Both rgpB and rgpF have presumably to work alternately in the elongation of the rhamnan chain. Homologous rhamnosyl transferases of rgpB and rgpF have been detected in Streptococcus thermophilus (STER1436) and Streptococcus gordonii (SGO1022). On the other hand, rgpC and rgpD genes encode the putative ABC transporters specific for RGP (homologous STER1434 in S. thermophilus and homologous SGO1024 in S. gordonii) which play role in polysaccharide export [37]. The rgpG gene (S. gordonii SGO1723 homolog) initiates the RGP synthesis by transferring N-acetylglucosamine-1-phosphate to a lipid carrier [38].
The rgp genes are also implicated in pathogenesis in several Streptococcus species. For instance, rgp plays an essential role in bacterial virulence as well as eliciting an inflammatory response in S. suis [39]. Induction of infective endocarditis by S. mutans has been reported to be triggered by rgp genes via nitric oxide release [40], platelet aggregation [41] and conferring resistance to phagocytosis by human polymorphonuclear leukocytes [42]. Therefore, S Clade S. mitis group species which produce these rhamnose rich polymers might exhibit a different pattern of pathogenesis from M Clade Streptococcus species in order to establish greater virulence and increased survival in host cells. A recent study has identified the Sanguinis group of streptococci as a common causative agent of transient bacteremia which potentially can lead to infective endocarditis. This group has also been reported to be present in a few cases of virulent septicemic infection in neutropenic patients [43].
Sequence search tools
We have incorporated two types of BLAST engines, standard BLAST and VFDB BLAST, into StreptoBase to search for the closest Streptococcus strains to the query strain. These exclusive BLAST searches are functionally based on the stand-alone BLAST tool [44] downloaded from NCBI. Both BLAST engines support three types of BLAST functions, namely, BLASTN, BLASTP and BLASTX. Users are allowed to define the genome completeness (%) and genome identity (%) on the BLAST tools submission forms. These specialized BLAST tools are aimed to facilitate users to perform similarity searches of their query sequences against Streptococcus genome sequences, gene sequences (standard BLAST) as well as against the virulence genes of VFDB (VFDB BLAST), which allows users to examine whether their genes of interest are potential virulence genes using a sequence homology approach.
Future work and conclusion
With advances in NGS technology, further Streptococcus species or strains will be sequenced and this creates an urgent need to store, browse, retrieve and analyze vast amounts of genome data and the development of specialized tools for comparative analyses of these genomes.
Here we have successfully described and demonstrated the functionalities of StreptoBase particularly our in-house designed bioinformatics pipelines for the analyses of Streptococcus genomic data.
This specialized biological database will be constantly updated in order to provide the latest genome updates and research developments associated with the Streptococcus genus, and to ensure the accuracy and usefulness of the S. mitis group species genome data and annotation. We anticipate that StreptoBase will serve as a useful resource and analysis platform particularly for comparative analyses of the S. mitis group genomes for research communities. We encourage other researchers or research groups to offer suggestions and share their annotations, opinions, and curated data with us at girg@um.edu.my.
Availability and system requirements
StreptoBase is available online at http://Streptococcus.um.edu.my. Users can download and visualize all sequences and annotations described in this paper on the StreptoBase website. Strains that have not already been deposited in the NCTC or ATCC culture collections are available on request from NSJ. This analysis platform is generally compatible with multiple type of browsers including Internet Explorer 8.x or higher, Mozilla Firefox® 10.x or higher, Safari 5.1 or higher, Chrome 18 or higher and any other equivalent browser software. This web site is best viewed at a screen resolution of 1024 × 768 pixels or higher.
Supporting Information
The genome details include genome size, number of contigs, number of ORFs, number of tRNAs, number of rRNAs, GC content as well as NCBI accession numbers of the 104 Streptococcus strains.
(XLSX)
Acknowledgments
We would like to thank all members of Genome Informatics Research Group (GIRG) in contributing to this research.
Data Availability
Data are available from StreptoBase. Accession numbers can be found within the Supporting Information files.
Funding Statement
This project was funded by University of Malaya and Ministry of Education (MOHE), Malaysia under the High Impact Research (HIR) Grant UM.C/HIR/MOHE/08 and UM Research Grant (UMRG) [Account No. UMRG: RG541-13HTM]. The funder provided support in the form of salaries for a few authors but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Barnard JP, Stinson MW (1996) The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect Immun 64: 3853–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawamura Y, Hou X-G, Sultana F, Miura H, Ezaki T (1995) Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. International journal of systematic bacteriology 45: 406–408. [DOI] [PubMed] [Google Scholar]
- 3.Bentley RW, Leigh JA, Collins MD (1991) Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. International journal of systematic bacteriology 41: 487–494. [DOI] [PubMed] [Google Scholar]
- 4.Jakubovics NS, Yassin SA, Rickard AH (2014) Community interactions of oral streptococci. Adv Appl Microbiol 87: 43–110. 10.1016/B978-0-12-800261-2.00002-5 [DOI] [PubMed] [Google Scholar]
- 5.Facklam R (2002) What happened to the streptococci: overview of taxonomic and nomenclature changes. Clinical microbiology reviews 15: 613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human Microbiome Project C (2012) Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westling K (2005) Viridans group streptococci septicaemia and endocarditis: Molecular diagnostics, antibiotic susceptibility and cinical aspects: Institutionen för medicin/Department of Medicine.
- 8.Kamio N, Imai K, Shimizu K, Cueno ME, Tamura M, et al. (2015) Neuraminidase-producing oral mitis group streptococci potentially contribute to influenza viral infection and reduction in antiviral efficacy of zanamivir. Cellular and Molecular Life Sciences 72: 357–366. 10.1007/s00018-014-1669-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAnally J, Levine M (1993) Bacteria reactive to plaque-toxin-neutralizing monoclonal antibodies are related to the severity of gingivitis at the sampled site. Oral microbiology and immunology 8: 69–74. [DOI] [PubMed] [Google Scholar]
- 10.Sullam P, Valone F, Mills J (1987) Mechanisms of platelet aggregation by viridans group streptococci. Infection and immunity 55: 1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilian M, MIKKELSEN L, HENRICHSEN J (1989) Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906). International Journal of Systematic Bacteriology 39: 471–484. [Google Scholar]
- 12.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC genomics 9: 75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nancy YY, Wagner JR, Laird MR, Melli G, Rey S, et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH (2009) JBrowse: a next-generation genome browser. Genome research 19: 1630–1638. 10.1101/gr.094607.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bancescu G, Dumitriu S, Bancescu A, Defta C, Pana M, et al. (2004) Susceptibility testing of Streptococcus mitis group isolates. Indian journal of medical research 119: 257–261. [PubMed] [Google Scholar]
- 16.Argimón S, Konganti K, Chen H, Alekseyenko AV, Brown S, et al. (2014) Comparative genomics of oral isolates of Streptococcus mutans by in silico genome subtraction does not reveal accessory DNA associated with severe early childhood caries. Infection, Genetics and Evolution 21: 269–278. 10.1016/j.meegid.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic acids research 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, et al. (2005) ACT: the Artemis comparison tool. Bioinformatics 21: 3422–3423. [DOI] [PubMed] [Google Scholar]
- 19.Lemos JA, Abranches J, Burne RA (2005) Responses of cariogenic streptococci to environmental stresses. Current issues in molecular biology 7: 95–108. [PubMed] [Google Scholar]
- 20.Chen L, Yang J, Yu J, Yao Z, Sun L, et al. (2005) VFDB: a reference database for bacterial virulence factors. Nucleic acids research 33: D325–D328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crater DL, Van de Rijn I (1995) Hyaluronic acid synthesis operon (has) expression in group A streptococci. Journal of Biological Chemistry 270: 18452–18458. [DOI] [PubMed] [Google Scholar]
- 22.Kelly T, Dillard JP, Yother J (1994) Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infection and immunity 62: 1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ (1991) Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proceedings of the National Academy of Sciences 88: 8317–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt K-H, Günther E, Courtney HS (1996) Expression of both M protein and hyaluronic acid capsule by group A streptococcal strains results in a high virulence for chicken embryos. Medical microbiology and immunology 184: 169–173. [DOI] [PubMed] [Google Scholar]
- 25.Cleary PP, Larkin A (1979) Hyaluronic acid capsule: strategy for oxygen resistance in group A streptococci. Journal of Bacteriology 140: 1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WIBAWAN IWT, Pasaribu F, Utama I, Abdulmawjood A, Laemmler C (1999) The role of hyaluronic acid capsular material of Streptococcus equi subsp. zooepidemicus in mediating adherence to HeLa cells and in resisting phagocytosis. Research in veterinary science 67: 131–135. [DOI] [PubMed] [Google Scholar]
- 27.Kim S-J, Park S-Y, Kim C-W (2006) A novel approach to the production of hyaluronic acid by Streptococcus zooepidemicus. Journal of microbiology and biotechnology 16: 1849–1855. [Google Scholar]
- 28.Chen WY, Marcellin E, Hung J, Nielsen LK (2009) Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. Journal of Biological Chemistry 284: 18007–18014. 10.1074/jbc.M109.011999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermans PW, Adrian PV, Albert C, Estevão S, Hoogenboezem T, et al. (2006) The streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonization. Journal of Biological Chemistry 281: 968–976. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez CJ, Kumar N, Lizcano A, Shivshankar P, Dunning Hotopp JC, et al. (2011) Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS One 6: e28738 10.1371/journal.pone.0028738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moscoso M, García E, López R (2006) Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. Journal of bacteriology 188: 7785–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dmitriev A, Shen A, Tkáčiková Ľ, Mikula I, Yang Y (2004) Structure of scpB-lmb intergenic region as criterion for additional classification of human and bovine group B Streptococci. Acta Veterinaria Brno 73: 215–220. [Google Scholar]
- 33.Zhang Y-M, Shao Z-Q, Wang J, Wang L, Li X, et al. (2014) Prevalent distribution and conservation of Streptococcus suis Lmb protein and its protective capacity against the Chinese highly virulent strain infection. Microbiological research 169: 395–401. 10.1016/j.micres.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 34.Terao Y, Kawabata S, Kunitomo E, Nakagawa I, Hamada S (2002) Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infection and immunity 70: 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsner A, Kreikemeyer B, Braun-Kiewnick A, Spellerberg B, Buttaro BA, et al. (2002) Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infection and immunity 70: 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, et al. (1999) Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infection and immunity 67: 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata Y, Yamashita Y, Ozaki K, Nakano Y, Koga T (2002) Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infection and immunity 70: 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita Y, Shibata Y, Nakano Y, Tsuda H, Kido N, et al. (1999) A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. Journal of bacteriology 181: 6556–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden M, Hauser H, Sanders M, Ngo TH, Cherevach I, et al. (2009) Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4: e6072 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin V, Kleschyov AL, Klein J-P, Beretz A (1997) Induction of nitric oxide production by polyosides from the cell walls of Streptococcus mutans OMZ 175, a gram-positive bacterium, in the rat aorta. Infection and immunity 65: 2074–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chia J-S, Lin Y-L, Lien H-T, Chen J-Y (2004) Platelet aggregation induced by serotype polysaccharides from Streptococcus mutans. Infection and immunity 72: 2605–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuda H, Yamashita Y, Toyoshima K, Yamaguchi N, Oho T, et al. (2000) Role of serotype-specific polysaccharide in the resistance of Streptococcus mutans to phagocytosis by human polymorphonuclear leukocytes. Infection and immunity 68: 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shelburne SA, Sahasrabhojane P, Saldana M, Yao H, Su X, et al. (2014) Streptococcus mitis strains causing severe clinical disease in cancer patients. Emerging infectious diseases 20: 762 10.3201/eid2005.130953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, et al. (2008) NCBI BLAST: a better web interface. Nucleic acids research 36: W5–W9. 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genome details include genome size, number of contigs, number of ORFs, number of tRNAs, number of rRNAs, GC content as well as NCBI accession numbers of the 104 Streptococcus strains.
(XLSX)
Data Availability Statement
Data are available from StreptoBase. Accession numbers can be found within the Supporting Information files.