Abstract
A human infection with novel avian influenza A H5N6 virus emerged in Changsha city, China in February, 2014. This is the first detected human case among all human cases identified from 2014 to early 2016. We obtained and summarized clinical, epidemiological, and virological data from this patient. Complete genome of the virus was determined and compared to other avian influenza viruses via the construction of phylogenetic trees using the neighbor-joining approach. A girl aged five and half years developed fever and mild respiratory symptoms on Feb. 16, 2014 and visited hospital on Feb. 17. Throat swab specimens were obtained from the patient and a novel reassortant avian influenza A H5N6 virus was detected. All eight viral gene segments were of avian origin. The hemagglutinin (HA) and neuraminidase (NA) gene segments were closely related to A/duck/Sichuan/NCXN11/2014(H5N1) and A/chicken/Jiangxi/12782/2014(H10N6) viruses, respectively. The six internal genes were homologous to avian influenza A (H5N2) viruses isolated in duck from Jiangxi in China. This H5N6 virus has not gained genetic mutations necessary for human infection and was suggested to be sensitive to neuraminidase inhibitors, but resistant to adamantanes. Epidemiological investigation of the exposure history of the patient found that a live poultry market could be the source place of infection and the incubation period was 2–5 days. This novel reassortant Avian influenza A(H5N6) virus could be low pathogenic in humans. The prevalence and genetic evolution of this virus should be closely monitored.
Keywords: Avian influenza, H5N6, The first detected human case, Reassortant, Sequence
1. Introduction
Avian influenza virus is a member of the Orthomyxoviridae family, Influenzavirus A genus. On the basis of the external glycoproteins hem-agglutinin (HA) and neuraminidase (NA), currently 18 HA (H1–H18) and 11 NA (N1–N11) subtypes are known. Subtypes H17N10 and H18N11 were described recently in bats (Tong et al., 2012, 2013). The avian influenza subtypes capable of infecting humans are H5N1, H5N2, H6N1, H9N2, H7N7, H7N2, H7N3, H10N7, H7N9 and H10N8 (Arzey et al., 2012; Chen et al., 2014; Cheng et al., 2011; Gao et al., 2013; Hirst et al., 2004; Koopmans et al., 2004; Ogata et al., 2008; Ostrowsky et al., 2012; To et al., 2012; Wei et al., 2013).
On Feb. 22, 2014, the National Laboratory for Influenza Surveillance at Changsha Municipal Center for Disease Control and Prevention (CDC) detected an avian influenza virus in a throat swab sample collected by a sentinel hospital of the China influenza surveillance system. The sample was tested positive for H5 but negative for N1 by the sentinel hospital. The test result was confirmed by Hunan Provincial CDC and the Chinese National Influenza Center. On Mar. 20, 2015, the virus was further confirmed to be avian-origin influenza A H5N6 by full genome sequencing conducted at Changsha Municipal CDC. The onset date of this case was earlier than all known H5N6-infected cases reported by World Health Organization (WHO) or by the National Health and Family Planning Commission (NHFPC) of the People's Republic of China, suggesting that this case from Changsha was likely the first detected human infection with the novel reassortant avian influenza A virus. Here, we report the result of a clinical investigation on this patient and the characteristics of this virus.
2. Materials and methods
2.1. Clinical and epidemiological data collection
A standardized case reporting form was used to gather the following epidemiological and clinical data: demographic characteristics; underlying medical conditions; recent exposures to pigs, poultry, or other animals; recent visits to live animal markets; clinical signs and symptoms; laboratory testing methods and results; antiviral treatment; and clinical outcomes. According to the regulations and guidelines of the NHFPC of China, data collection on this patient was part of the routine surveillance and outbreak investigation, and was therefore exempt from the oversight by institutional review board (IRB).
Close contacts, defined as individuals who had provided care to, had been living with, or had potentially been directly exposed to respiratory secretions or body fluids of the patient in 14 days before the illness onset of the patient, were identified. The IRB of Changsha CDC approved the assessment of these close contacts. Written consent was obtained from each close contact.
2.2. Viral analysis
2.2.1. RNA extraction and real-time RT-PCR
Throat swab specimens were obtained from the patient on day 2 since the illness onset (illness onset counted as day 1). Specimen collection, storage and transportation were performed according to WHO guidelines (WHO Global Influenza Surveillance Network, 2011). Real-time RT-PCR or conventional RT-PCR, or both, were used for influenza typing and subtyping by the Changsha CDC, Hunan provincial CDC, and the Chinese National Influenza Center (CNIC). The sample was con-firmed to contain M and HA genes of influenza A virus with subtype H5, but the NA gene could not be subtyped. Throat swab specimens were also obtained from the patient on days 7, 17, 24, and 27 since illness onset and showed similar results.
The virus was inactivated by heating at 65 °C for 30 min prior to RNA extraction. RNA was extracted from the throat-swab sample using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. Primers and probes specific to seasonal influenza viruses (H1, H3, or B), avian influenza viruses (H5, H7, H9) and neuraminidase (N1, N2, N9) were used to sequence the virus. Sequences for comparison were provided by the CINC and have been published elsewhere (World Health Organization, 2014).
On day 8 since illness onset of the patient (Feb 23, 2014), five environmental samples were collected and stored in viral transport medium, including one sample of poultry drinking-water and one sample of water used for cleaning slaughtered poultry from the epidemiologically linked live poultry trading site (Fig. S1), and two swabs of poultry cages and one sample of sewage water from the epidemiologically linked restaurant. These samples were tested by real-time RT-PCR or conventional RT-PCR or both for influenza typing and subtyping by the Changsha CDC, using primers and probes specific to H5, H7, H9, N1, N2, and N9 provided by the CNIC.
2.2.2. Genome sequencing and phylogenetic analysis
A total of 8 universal primer sets were used to amplify the full genome of all influenza A viruses for sequencing (Hoffmann et al., 2001), with the use of SuperScript® III One-Step RT-PCR System with Platinum® Taq RT-PCR (Life Technologies Corporation, USA). PCR products were sequenced by Life Technologies biotechnology (Shanghai, China) CO. Full genome sequences of the virus from the patient were deposited in the Influenza Virus Database of the National Center for Biotechnology Information (accession number KR063684-91).
The sequence homology of each gene of the virus was analyzed with the online Basic Local Alignment Search Tool (BLAST). The ClustalW Multiple alignments were constructed using the BioEdit Sequence Alignment Editor (USA, Borland). The phylogenetic trees for the HA and NA genes were constructed using the neighbor-joining method provided by MEGA 5.2 software, coupled with the Tamura-Nei model for nucleotide substitution. One thousand bootstrap data sets were generated to evaluate the reliability of the phylogenetic tree. The molecular characteristics of each gene of the virus were summarized.
3. Results
This patient was a five-year-old kindergarten student living with her father. Looking for employment, the father migrated with his daughter from Chenzhou to Changsha, Hunan province, on Feb. 5, 2014. From Feb. 5 to Feb. 10, the child stayed at home as her father worked outside. During the four days following Feb. 10, the girl went out with her cousin and was exposed to a live poultry trading site for about 5 min a day.
Fig. S1 shows the path of the patient's activity outside her house during the four days. Every morning, she and her cousin went out for breakfast nearby, bypassing a restaurant with live chicken and wild birds in cages. After breakfast, they shopped for snacks in a supermarket across
the street. Then they went to shop for vegetables at a vegetables trading site next to a live poultry trading site with live chicken and ducks for sale. About 5 min later, they returned home. The girl stayed home on Feb. 15, 2014. On Feb. 16, 2014, she developed influenza-like illness (ILI) including sore throat and fever (38.0 °C). Her father bought her some herb medicine (Table 1), but her situation got no better. On Feb. 17, 2014, her father brought her to the outpatient care of Changsha Central Hospital which is a sentinel hospital for influenza surveillance. Physical examination found a body temperature of 38.9 °C and a red pharynx. Due to the presentation of ILI symptoms, a throat swab sample was collected to be tested for influenza virus. In addition, a blood sample was drawn for routine tests and found that the number of white blood cells was 23.12 × 109/L, twice the upper bound of the normal range, the proportion of neutrophils was higher than the normal range, and the proportion of lymphocytes was lower than the normal range. These test results suggested a probable bacterial infection. A diagnosis of “suppurative tonsillitis” was made by the outpatient physician, and a combination of antibiotics, febrifuge and herb medicine was prescribed for treatment (Table 1). The patient was not hospitalized and returned home in the evening.
Table 1.
Clinical characteristics, treatment, and outcome of the patient infected with the avian influenza A(H5N6) virus in Changsha, China in 2014.
| Signs and treatment | Starting day | Stopping day | Details |
|---|---|---|---|
| Fever | 1 | 2 | 38.0–38.9 °C |
| Sore throat | 1 | 3 | |
| Chills | 3 | 3 | |
| Other complications | None | ||
| Bacterial co-infection | Probable (white blood cell count was twice the upper bound of normal range) | ||
| Antibiotic treatment | 2 | 4 | Amoxicillin and Clavulanate Potassium (two 25 mg/kg doses given orally) |
| Febrifuge treatment | 2 | 4 | Ibuprofen suspension (four 5 mg/kg doses given orally) |
| Traditional Chinese medicine | 1 | 11 | Kangbingdu oral liquid, Pudilan anti-inflammatory oral liquid, Andrographolide |
| Antiviral treatment | 12 | 18 | Tamiflu (75 mg twice a day) |
The fever and sore throat lasted for 3 days, and the patient felt chilly after that. Throat swab samples were collected on day 7 and day 11 respectively (Fig. S2), which were tested positive for H5 but negative for N1 by Changsha Municipal CDC using Real-time PCR. The results were confirmed by Hunan Provincial CDC and China CDC. In an attempt to reduce the viral activity, antivirals (Oseltamivir) were prescribed on day 11 and taken for seven days (75 mg twice a day). Tests on the blood sample drawn on day 2 showed increased levels of white blood cells, neutrophilic granulocytes, monocytes, proportion of neutrophils, and proportion of monocytes. The number of lymphocytes was normal but proportion of lymphocytes was lower than the normal range (Table S1).
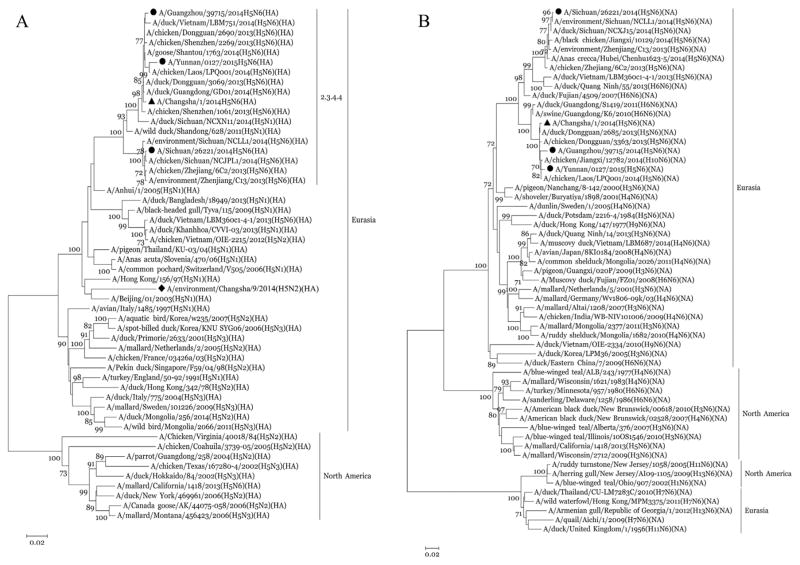
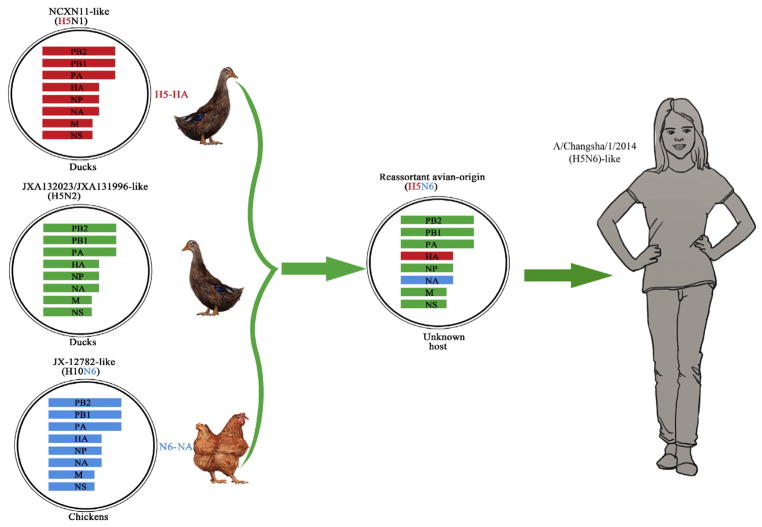
Real-time RT-PCR and full genome sequencing found the throat swabs obtained on day 7 and day 11 to be positive for avian influenza A H5N6 virus [The virus designated as A/Changsha/1/2014(H5N6)] and negative for influenza viruses H1, H3, B, H5N1, H7N9, and H9N2. Phylogenetic analyses revealed that the HA gene of A/Changsha/1/2014(H5N6) was closely related to A/duck/Sichuan/NCXN11/2014(H5N1) virus (98% identity), and the neuraminidase (NA) gene was closely related to A/chicken/Jiangxi/12782/2014(H10N6) virus (99% identity) (Table 2, Fig. 1). The RNA polymerase basic subunit (PB) 1 protein, PB2 protein and nucleocapsid protein (NP) genes were closely related to A/duck/Jiangxi/JXA132023/2013(H5N2) virus (99% identity). The sequences of the remaining viral genes were closely related (99% identity) to A/duck/Jiangxi/JXA131996/2013(H5N2) virus, which was isolated in poultry in Jiangxi province, China (Table 2). Phylogenetic analysis demonstrated that the new isolate in this study, A/Changsha/1/2014(H5N6), is a novel triple reassortant H5N6 virus, similar to A/Sichuan/26221/2014 and A/Guangzhou/39715/2014, the two isolates from human H5N6 cases reported in Sichuan and Guangdong provinces in 2014, but different from A/Yunnan/0127/2015, the isolate from a human H5N6 case in Yunnan province in 2015 which is a complex reassortant virus. The A/Changsha/1/2014 (H5N6) virus might have acquired its HA gene from A/duck/Sichuan/NCXN11/2014(H5N1)-like viruses, NA gene from A/chicken/Jiangxi/12782/2014(H10N6)-like viruses and the six internal genes from viruses similar to the avian influenza A(H5N2) viruses in ducks of Jiangxi province, China (Table 2, Fig. 2). Each gene was of an avian origin. The A/Chang-sha/1/2014(H5N6) isolate shared moderate to high levels of nucleotide identity in the eight gene segments with the other three isolates from human H5N6 cases in China: PB2:84.2 ~ 98.9%, PB1: 88.8 ~ 99.1%, PA: 88.0 ~ 99.0%, HA: 95.4 ~ 99.1%, NP: 92.8 ~ 99.3%, NA: 89.6 ~ 99.8%, M: 90.0 ~ 99.7%, and NS: 84.2 ~ 98.7%. In particular, A/Changsha/1/2014(H5N6) was closely related (98.5 ~ 99.7% identity) to A/Guang-zhou/39,715/2014 (Table 3), which was isolated from a 59-year-old male who developed symptoms on Dec. 4, 2014 in Guangzhou, Guang-dong province in southern China (Yang et al., 2015). Guangdong province is located to the south of Hunan province. It is also clear that A/Changsha/1/2014(H5N6) and the other three isolates from human H5N6 cases belong to the Eurasian lineage of avian influenza viruses, and their HA genes belong to clade 2.3.4.4 H5 viruses reported recently (Fig. 1) (Smith et al., 2015). A/Changsha/1/2014 (H5N6) was clustered with A/Guangzhou/39715/2014(H5N6) and the avian strains from Laos and Vietnam in the phylogenetic trees for both HA (Fig. 1A) and NA (Fig. 1B), suggesting these viruses likely had a common source.
Table 2.
Nucleotide identity of A/Changsha/1/2014 (H5N6) virus genes. The date of this BLAST search was Apr 10, 2015.
| Viral genea | Closest influenza virus relative | Nucleotide identity (%) |
|---|---|---|
| PB2 | A/duck/Jiangxi/JXA132023/2013(H5N2) | 99.6 |
| PB1 | A/duck/Jiangxi/JXA132023/2013(H5N2) | 99.3 |
| PA | A/duck/Jiangxi/JXA131996/2013(H5N2) | 99.5 |
| HA | A/duck/Sichuan/NCXN11/2014(H5N1) | 98.0 |
| NP | A/duck/Jiangxi/JXA132023/2013(H5N2) | 99.7 |
| NA | A/chicken/Jiangxi/12782/2014(H10N6) | 99.2 |
| M | A/duck/Jiangxi/JXA131996/2013(H5N2) | 99.7 |
| NS | A/duck/Jiangxi/JXA131996/2013(H5N2) | 99.0 |
PB2: RNA polymerase basic subunit 2; PB1: RNA polymerase basic subunit 1; PA: RNA polymerase acidic subunit; HA: haemagglutinin; NP: nucleoprotein; NA: neuraminidase; M: matrix gene; NS: non-structural gene.
Fig. 1.
Phylogenetic analysis of the HA (panel A) and NA(panel B) genes of the novel influenza A(H5N6) virus isolated from a patient in Changsha, China in 2014. The sequence of the H5N6 virus in our study is marked by ▲, the sequence of the H5N2 virus in our study is marked by ◆, and the sequences of H5N6 viruses isolated from the other three H5N6 cases in China are marked by ●.
Fig. 2.
Possible source of the genes of the avian influenza A(H5N6) virus isolated from a patient in Changsha, China in 2014.
Table 3.
Nucleotide identity levels between the human isolate A/Changsha/1/2014 (H5N6) and the other three human isolates of influenza A(H5N6) viruses in China during 2014–2015.
| Strain | Nucleotide identity, %
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | MP | NS | |
| A/Sichuan/26221/2014(H5N6) | 97.9 | 97.8 | 98.4 | 95.5 | 98.2 | 89.6 | 98.3 | 98.3 |
| A/Guangzhou/39715/2014(H5N6) | 98.9 | 99.1 | 99.0 | 99.1 | 99.3 | 98.5 | 99.7 | 98.7 |
| A/Yunnan/0127/2014(H5N6) | 84.2 | 88.8 | 88.0 | 95.4 | 92.8 | 98.8 | 90.0 | 84.2 |
To identify key mutation sites, the amino acid sequence of A/Chang-sha/1/2014 (H5N6) was compared to those of the H5N6, H5N1, H7N9, H10N8 and H9N2 viruses reported in the influenza virus database. There are six basic amino acids (R, K) at the protein cleavage site (amino acids 338–346) of the A/Changsha/1/2014 (H5N6) HA protein, similar to a human H5N6 isolate (A/Guangzhou/39715/2014),a duck H5N6 isolate (A/duck/Dongguan/3069/2013) and a human H5N1 isolate (A/Hubei/1/2010) but different from those in human H7N9, H10N8 and H9N2 isolates which are known to be low pathogenic in poultry (Table 4), indicating that A/Changsha/1/2014 (H5N6) might be highly pathogenic in poultry (Senne et al., 1996). Substitutions Q226L and G228S at the 210-loop in the HA protein that may facilitate human infection (Srinivasan et al., 2013) were not found in A/Chang-sha/1/2014 (H5N6), A/Guangzhou/39,715/2014 (H5N6), A/duck/Dongguan/3069/2013(H5N6), or A/Hubei/1/2010(H5N1) viruses (Table 4). The resistance-conferring mutation, R294K in NA, did not occur in A/Changsha/1/2014(H5N6), indicating possible sensitivity of this virus to neuraminidase inhibitors (McKimm-Breschkin et al., 1998). However, the substitution S31N was noted in the M2 gene, indicating possible drug resistance of this virus to adamantanes (Table 4) (Hay et al., 1985; Pinto et al., 1992). A deletion of A/Changsha/1/2014(H5N6) virus was observed in the stalk region of NA residue 69 to 73 (Table 4), which was also found in A/Guangzhou/39715/2014 (H5N6), A/duck/Dongguan/3069/2013(H5N6), A/Hubei/1/2010(H5N1) and A/Changsha/2/2013(H7N9) but not in A/Jiangxi/IPB13/2013(H10N8) and A/Lengshuitan/11197/2013(H9N2) (Table 4). Other mutations in the A/Changsha/1/2014(H5N6) were also observed, such as N30D and T215A in M1, L89V in PB2, and P42S in the NS1 (Table 4). E627K, another mutation in PB2 facilitating adaption to mammalian hosts (Hatta et al., 2001), appeared in two human H5N6 isolates (A/Guangzhou/39715/2014, A/Yunnan/0127/2015) and a human H10N8 isolate A/Jiangxi/IPB13/2013, but was absent in A/Changsha/1/2014 (H5N6), a duck H5N6 isolate (A/duck/Dongguan/3069/2013) and other virus in Table 4.
Table 4.
Molecular analysis of important amino acids in the HA, NA, PB2, PB1, PB1-F2, NS1, M1 and M2 proteins of 7 avian influenza virus strain.
| Viral protein | Sites | Amino acid position | Virus 1c | Virus 2d | Virus 3e | Virus 4f | Virus 5g | Virus 6h | Virus 7i | Function |
|---|---|---|---|---|---|---|---|---|---|---|
| HA | Cleavage site | 338–346 | RERRRKRGL | RERRRKRGL | RERRRKRGL | RERRRKRGL | -EIPK GRGL | -ELIQGRGL | –SRSSRGL | Indicating pathogenic effects in poultry (Senne et al., 1996) |
| Q226L | 226/238a | Q | Q | Q | Q | L | Q | L | Q226L,G228S increases binding affinity for a-2,6-linked sialic acid receptor (Srinivasan et al., 2013) | |
| G228S | 228/240a | G | G | G | G | G | G | G | ||
| NA | R294K | 294/290b | R | R | R | R | R | R | R | R294K reduces susceptibility to oseltamivir and zanamivir (McKimm-Breschkin et al., 1998) |
| 69–73 | Deletion | Deletion | Deletion | Deletion | Deletion | No | No | Deletion of amino acids 69–73: increased virulence in mice (McKimm-Breschkin et al., 1998) | ||
| PB2 | L89V | 89 | V | V | V | V | V | V | V | Enhanced polymerase activity and increased virulence in mice (Gao et al., 2013) |
| E627K | 627 | E | K | E | E | E | K | E | Mammalian host adaptation (Hatta et al., 2001) | |
| D701N | 701 | D | D | D | D | D | D | D | Enhanced transmission in guinea pigs (Chen et al., 2014) | |
| PB1 | H99Y | 99 | H | H | H | H | H | H | H | H5 virus transmissible among ferrets (Gao et al., 2013) |
| I368V | 368 | I | I | I | I | V | V | V | ||
| PB1-F2 | Full length | Nonfunctional PB1-F2 protein due to mutation | Nonfunctional PB1-F2 protein due to mutation | Nonfunctional PB1-F2 protein due to mutation | 90 aa | 52 aa | 52 aa | 90 aa | Increased virulence in mice (Gao et al., 2013) | |
| NS1 | P42S | 42 | S | S | S | S | S | S | S | Increased virulence in mice (Jiao et al., 2008) |
| 218–230 | No | No | No | No | Deletion | Deletion | Deletion | Lack of PDZ domain binding motif:decreased virulence in mice (Jackson et al., 2008) | ||
| M1 | N30D | 30 | D | D | D | D | D | D | D | Increased virulence in mice (Fan et al., 2009) |
| T215A | 215 | A | A | A | A | A | A | A | ||
| M2 | S31N | 31 | N | S | S | S | N | N | N | Reduced susceptibility to amantadine and rimantadine (Hay et al., 1985; Pinto et al., 1992) |
Substitutions of particular concern are shown in bold.
Amino acid sites of H3/H5N6.
Amino acid sites of N9/N6.
A/Changsha/1/2014(H5N6).
A/Guangzhou/39715/2014(H5N6).
A/duck/Dongguan/3069/2013(H5N6).
A/Hubei/1/2010(H5N1).
A/Changsha/2/2013(H7N9).
A/Jiangxi/IPB13/2013(H10N8).
A/Lengshuitan/11197/2013(H9N2).
H5 and N2 genes were detected in the poultry drinking-water sample from the live poultry trading site which the patient had visited, and the result was confirmed by sequencing. This H5 gene belongs to the Eurasian lineage, but is not closely related to the one isolated from the patient (Fig. 1A).
4. Discussion
A novel avian influenza A H5N6 virus [A/Changsha/1/2014(H5N6)] was isolated in China from a patient with fever and mild respiratory symptoms who recovered after a few days, implying the presence of mild cases or even asymptomatic carriers of avian influenza H5N6 in human. Three human infections with avian influenza A H5N6 virus in China have been previously reported — one in Sichuan province on May 7, 2014, one in Guangdong province on Dec 23, 2014, and one in Yunnan province on February 9, 2015, all presenting severe pneumonia (Pan et al., 2016; Yang et al., 2015; World Health Organization, 2015). However, the human case infected with influenza A/Changsha/1/2014(H5N6) virus reported in this study is the earliest among all known cases.
Our phylogenetic analysis showed that the A/Changsha/1/2014(H5N6) and A/Guangzhou/39715/2014 isolates are both novel triple reassortant H5N6 virus, and they likely shared a common source. A/Sichuan/26221/2014(H5N6) is also a triple reassortant virus but with somewhat different sources of genes as compared to A/Changsha/1/2014(H5N6): HA gene belonging to Clade 2.3.4.4 H5, internal genes belonging to Clade 2.3.2.1 H5, and NA gene closely related to H6N6 avian virus (Pan et al., 2016). A/Yunnan/0127/2015(H5N6) (GenBank: KT245143–50) is a complex reassortant that might have acquired its HA gene from A/chicken/Tonghai/802/2014(H5N1)-like viruses, NA gene from A/chicken/Jiangxi/12782/2014(H10N6)-like viruses, and the six internal genes from viruses similar to the H9N2, H7N9 and H10N8 viruses of poultry origin, China. These results indicated the diversity in the sources of the gene segments of the reassortant H5N6 virus in China during 2014–2015.
All eight viral gene segments of A/Changsha/1/2014 (H5N6) were of avian origin. Previous and our studies confirmed that the H5N6 virus can be transmitted from poultry to humans. Live poultry markets have been suggested to be an important source of human infection with avian influenza A H5N1, H7N9 and H10N8 viruses (Shi et al., 2013; Wan et al., 2011; Zhang et al., 2014). The field investigation in our study to establish the source of the infection did not reach a clear conclusion. The patient had visited a live poultry market multiple times a few days before illness onset and could have acquired the infection during one of those visits, indicating an incubation period of 2–5 days, similar to other influenza viruses. We did not find avian influenza A H5N6 in the environmental samples collected form the live bird market that the patient had visited.
With amino acids at the protein cleavage site similar to other H5 viruses, the HA protein of human H5N6 isolate A/Changsha/1/2014(H5N6) virus might be highly pathogenic in poultry (Senne et al., 1996). Based on the amino acid sequence of A/Changsha/1/2014(H5N6), it is most probably sensitive to neuraminidase inhibitors, but resistant to adamantanes. In addition, A/Changsha/1/2014(H5N6) encoded a deletion in the stalk region of NA, and other mutations were observed, such as N30D and T215A in M1, L89V in PB2, and P42S in the NS1. These substitutions were associated with increased virulence in mice (Fan et al., 2009; Jiao et al., 2008). In this study, the patient infected with A/Changsha/1/2014(H5N6) virus did not present severe symptoms such as pneumonia. The young age of the patient might have played a role in the mildness of the symptoms, as the other three known H5N6-infected cases were all adults (Pan et al., 2016; Yang et al., 2015; World Health Organization, 2015). Another possible contributor to the mildness is the lack of mutation E627K in PB2 protein. This mutation was found to promote viral adaption to mammalian cells (Hatta et al., 2001) and was seen in two human H5N6 isolates (A/Guangzhou/39715/2014, A/Yunnan/0127/2015) and a human H10N8 isolate A/Jiangxi/IPB13/2013, all of which were associated with severe respiratory symptoms and fatality in human cases (Chen et al., 2014; Pan et al., 2016).
Some probable person-to-person transmission of avian influenza A(H5N1) (Wang et al., 2008) and A(H7N9) (Qi et al., 2013) viruses has been reported. In this study, medical observation and laboratory testing of close contacts of the patient showed no evidence of infection. No secondary infections were reported for the other three cases either. Mutations Q226L and G228S (in the H3 numbering system) of the HA protein of avian influenza H2, H3 and H7 subtypes were shown to be associated with increased affinity of HA to the glycan-receptors in the lower respiratory tracts of humans (Liu et al., 2013; Srinivasan et al., 2013; van Riel et al., 2006; Yang et al., 2010; Zhang et al., 2013), although these mutations not necessarily facilitate the binding of H5 (Zhang et al., 2013). We did not find these mutations in A/Changsha/1/2014 (H5N6) HA protein. These epidemiological and genetic observations suggest that avian influenza A(H5N6) virus has not yet gained transmissibility from person to person (Liu et al., 2013; Srinivasan et al., 2013; van Riel et al., 2006; Yang et al., 2010; Zhang et al., 2013).
Two surveillance methods are employed by the China influenza surveillance system — viral culture and nucleic acid screening. Viral culture used to be the only method for detecting influenza viruses. However, from January to March, 2014, the nucleic acid screening was added for screening all the samples collected from the routine surveillance, in response to the epidemic of human infections with avian influenza A H7N9 virus in China. The H5N6 case we reported here was first detected through the China influenza surveillance system using the nucleic acid screening, suggesting that this method is sensitive and efficient in discovering novel influenza viruses.
Our study is limited in the following aspects. First, we were not able to isolate the virus from the patient's throat swab specimens for further research. Second, this or similar H5N6 viruses were not found in the live poultry market to which the patient was exposed, and therefore the source of this human infection cannot be pinpointed.
Despite the lack of efficient poultry-to-human and human-to-human transmission of influenza A(H5N6) viruses at current stage, the recent outbreaks of this virus in poultry in Asia (Wong et al., 2015; Wu et al., 2015) and the four human cases in a one-year period (Pan et al., 2016; Yang et al., 2015; World Health Organization, 2015) warrant close monitoring of the viral evolution and timely detection of new human cases. The possibility of reassortment of H5N6 with other avian influenza strains, e.g., H7N9 and H10N8, into a new highly pathogenic strain of pandemic potential should not be ruled out. In addition to strengthening surveillance, we recommend resources be allocated to initiate development of vaccines targeting avian influenza A(H5N6) for both poultry and human.
Acknowledgments
This study was supported by the Hunan Provincial Health Medicine Research Project (B2015-153, to Rusheng Zhang), Yang Yang was supported by the MIDAS grant from NIH/NIGMS (U54 GM111274).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.meegid.2016.03.010.
Contributor Information
Tianmu Chen, Email: 13698665@qq.com.
Faming Chen, Email: 351985738@qq.com.
Shi-Ping Wang, Email: spwang@126.com.
References
- Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng YM, Iannello P, Barr I, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. Influenza Virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Yuan H, Gao RB, Zhang JX, Wang DY, Xiong Y, Fan GY, Yang F, Li XD, Zhou JF, Zou SM, Yang L, Chen T, Dong LB, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang SW, Zhang YP, Li H, Gong T, Shi Y, Ni XS, Li JX, Zhou J, Fan JY, Wu JW, Zhou XF, Hu MH, Wan JG, Yang WZ, Li DX, Wu GZ, Feng ZJ, Gao GF, Wang Y, Jin Q, Liu MB, Shu YL. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Cheng VCC, Chan JFW, Wen X, Wu WL, Que TL, Chen H, Chan KH, Yuen KY. Infection of immunocompromised patients by avian H9N2 influenza A virus. J Infect. 2011;62:394–399. doi: 10.1016/j.jinf.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Fan SF, Deng GH, Song JS, Tian GB, Suo YB, Jiang YP, Guan YT, Bu ZG, Kawaola Y, Chen HL. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology. 2009;384:28–32. doi: 10.1016/j.virol.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Gao RB, Cao B, Hu YW, Feng ZJ, Wang DY, Hu WF, Chen J, Jie ZJ, Qiu HB, Xu K, Xu XW, Lu HZ, Zhu WF, Gao ZC, Xiang NJ, Shen YZ, He ZB, Gu Y, Zhang ZY, Yang Y, Zhao X, Zhou L, Li XD, Zou SM, Zhang Y, Li XY, Yang L, Guo JF, Dong J, Li Q, Dong LB, Zhu Y, Bai T, Wang SW, Hao P, Yang WZ, Zhang YP, Han J, Yu HJ, Li DX, Gao GF, Wu GZ, Wang Y, Yuan ZH, Shu YL. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular-basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M, Astell CR, Griffith M, Coughlin SM, Moksa M, Zeng T, Smailus DE, Holt RA, Jones S, Marra MA, Petric M, Krajden M, Lawrence D, Mak A, Chow R, Skowronski DM, Tweed SA, Goh S, Brunham RC, Robinson J, Bowes V, Sojonky K, Byrne SK, Li Y, Kobasa D, Booth T, Paetzel M. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis. 2004;10:2192–2195. doi: 10.3201/eid1012.040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105:4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao PR, Tian GB, Li YB, Deng GH, Jiang YP, Liu C, Liu WL, Bu ZG, Kawaoka Y, Chen HL. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Liu D, Shi WF, Shi Y, Wang DY, Xiao HX, Li W, Bi YH, Wu Y, Li XB, Yan JH, Liu WJ, Zhao GP, Yang WZ, Wang Y, Ma JC, Shu YL, Lei FM, Gao GF. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, McDonald M, Colman PM, Hart GJ, Bethell RC, Varghese JN. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–2462. doi: 10.1128/jvi.72.3.2456-2462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Yamazaki Y, Okabe N, Nakamura Y, Tashiro M, Nagata N, Itamura S, Yasui Y, Nakashima K, Doi M, Izumi Y, Fujieda T, Yamato S, Kawada Y. Human H5N2 avian influenza infection in Japan and the factors associated with high H5N2-neutralizing antibody titer. J Epidemiol. 2008;18:160–166. doi: 10.2188/jea.JE2007446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowsky B, Huang A, Terry W, Anton D, Brunagel B, Traynor L, Abid S, Johnson G, Kacica M, Katz J, Edwards L, Lindstrom S, Klimov A, Uyeki TM. Low pathogenic avian influenza A (H7N2) virus infection in immunocompromised adult, New York, USA, 2003. Emerg Infect Dis. 2012;18:1128–1131. doi: 10.3201/eid1807.111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Gao RB, Lv Q, Huang SH, Zhou ZH, Yang L, Li XD, Zhao X, Zou XH, Tong WB, Mao SL, Zou SM, Bo H, Zhu XP, Liu L, Yuan H, Zhang MH, Wang DQ, Li ZM, Zhao W, Ma ML, Li YQ, Li TS, Yang HP, Xu JN, Zhou LJ, Zhou XY, Tang W, Song Y, Chen T, Bai T, Zhou JF, Wang DY, Wu GZ, Li DX, Feng ZJ, Gao GF, Wang Y, He SS, Shu YL. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect. 2016;72:52–59. doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. Influenza-virus m2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ Br Med J. 2013;347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senne DA, Panigrahy B, Kawaoka Y, Pearson JE, Suss J, Lipkind M, Kida H, Webster RG. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: Amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 1996;40:425–437. [PubMed] [Google Scholar]
- Shi JZ, Deng GH, Liu PH, Zhou JP, Guan LZ, Li WH, Li XY, Guo J, Wang GJ, Fan J, Wang JL, Li YY, Jiang YP, Liu LL, Tian GB, Li CJ, Chen HL. Isolation and characterization of H7N9 viruses from live poultry markets—implication of the source of current H7N9 infection in humans. Chin Sci Bull. 2013;58:1857–1863. [Google Scholar]
- Smith GJ, Donis RO World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5 Evolution Working Group. Nomenclature updates resulting from the evolution of avian influenza A (H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013–2014. Influenza Other Respir Viruses. 2015;9:271–276. doi: 10.1111/irv.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLoS One. 2013;8:e49597. doi: 10.1371/journal.pone.0049597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KKW, Ng KHL, Que TL, Chan JMC, Tsang KY, Tsang AKL, Chen HL, Yuen KY. Avian influenza A H5N1 virus: a continuous threat to humans. Emerg Microbes Infect. 2012;1:e25. doi: 10.1038/emi.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SX, Li Y, Rivailler P, Conrardy C, Castillo DAA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu XY, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SX, Zhu XY, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen XF, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu WL, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RAM, Osterhaus A, Kuiken T. H5N1 virus attachment to lower respiratory tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- Wan XF, Dong LB, Lan Y, Long LP, Xu CL, Zou SM, Li Z, Wen LY, Cai ZP, Wang W, Li XD, Yuan F, Sui HT, Zhang Y, Dong J, Sun SH, Gao Y, Wang M, Bai T, Yang L, Li DX, Yang WZ, Yu HJ, Wang SW, Feng ZJ, Wang Y, Guo YJ, Webby RJ, Shu YL. Indications that live poultry markets are a major source of human H5N1 influenza virus infection in China. J Virol. 2011;85:13432–13438. doi: 10.1128/JVI.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Feng Z, Shu Y, Yu H, Zhou L, Zu RQ, Huai Y, Dong J, Bao CJ, Wen LY, Yang P, Zhao W, Dong LB, Zhou MH, Liao QH, Yang HT, Wang M, Lu XJ, Shi ZY, Wang W, Gu L, Zhu FC, Li Q, Yin WD, Yang WZ, Li DX, Uyeki TM, Wang Y. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet. 2008;371:1427–1434. doi: 10.1016/S0140-6736(08)60493-6. [DOI] [PubMed] [Google Scholar]
- Wei SH, Yang JR, Wu HS, Chang MC, Lin JS, Lin CY, Liu YL, Lo YC, Yang CH, Chuang JH, Lin MC, Chung WC, Liao CH, Lee MS, Huang WT, Chen PJ, Liu MT, Chang FY. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med. 2013;1:771–778. doi: 10.1016/S2213-2600(13)70221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Global Influenza surveillance Network. Mannual for the laboratory diagnosis and virological surveillance of influenza. 2011 http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf.
- Wong FYK, Phommachanh P, Kalpravidh W, Chanthavisouk C, Gilbert J, Bingham J, Davies KR, Cooke J, Eagles D, Phiphakhavong S, Shan SH, Stevens V, Williams DT, Bounma P, Khambounheuang B, Morrissy C, Douangngeun B, Morzaria S. Reassortant highly pathogenic influenza A(H5N6) virus in Laos. Emerg Infect Dis. 2015;21:511–516. doi: 10.3201/eid2103.141488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. WHO information for molecular diagnosis of influenza virus-update. 2014 (cited 5 June 2015) http://who.int/infuenza/gisrs_laboratory/molecular_diagnosis_influenza_virus_humans_update_201403.pdf?ua=1.
- World Health Organization. WHO: human infection with avian influenza A(H5N6) virus—China. 2015 (cited 31Mar 2015, On 9 February 2015) http://flu.org.cn/en/news-18190.html.
- Wu HB, Lu RF, Peng XR, Xu LH, Cheng LF, Lu XY, Jin CZ, Xie TS, Yao HP, Wu NP. Novel reassortant highly pathogenic H5N6 avian influenza viruses in poultry in China. Infect Genet Evol. 2015;31:64–67. doi: 10.1016/j.meegid.2015.01.019. [DOI] [PubMed] [Google Scholar]
- Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Mok CKP, Peiris JSM, Zhong NS. Human Infection with a novel avian influenza A(H5N6) virus. N Engl J Med. 2015;373:487–489. doi: 10.1056/NEJMc1502983. [DOI] [PubMed] [Google Scholar]
- Zhang W, Shi Y, Lu XS, Shu YL, Qi JX, Gao GF. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science. 2013;340:1463–1467. doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- Zhang T, Bi YH, Tian HA, Li XW, Liu D, Wu Y, Jin T, Wang Y, Chen QJ, Chen Z, Chang JY, Gao GF, Xu B. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis. 2014;20:2076–2079. doi: 10.3201/eid2012.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]