Abstract
Long non-coding RNAs (lncRNAs) are a class of newly recognized DNA transcripts that have diverse biological activities. Dysregulation of lncRNAs may be involved in many pathogenic processes including cancer. Recently, we found an intergenic lncRNA, LINC00472, whose expression was correlated with breast cancer progression and patient survival. Our findings were consistent across multiple clinical datasets and supported by results from in vitro experiments. To evaluate further the role of LINC00472 in breast cancer, we used various online databases to investigate possible mechanisms that might affect LINC00472 expression in breast cancer. We also analyzed associations of LINC00472 with estrogen receptor, tumor grade, and molecular subtypes in additional online datasets generated by microarray platforms different from the one we investigated previously. We found that LINC00472 expression in breast cancer was regulated more possibly by promoter methylation than by the alteration of gene copy number. Analysis of additional datasets confirmed our previous findings of high expression of LINC00472 associated with ER-positive and low-grade tumors and favorable molecular subtypes. Finally, in nine datasets, we examined the association of LINC00472 expression with disease-free survival in patients with grade 2 tumors. Meta-analysis of the datasets showed that LINC00472 expression in breast tumors predicted the recurrence of breast cancer in patients with grade 2 tumors. In summary, our analyses confirm that LINC00472 is functionally a tumor suppressor, and that assessing its expression in breast tumors may have clinical implications in breast cancer management.
Keywords: Long non-coding RNA, LINC00472, Breast, Prognosis, Grade 2 tumor, Methylation
Introduction
As DNA sequencing technology advances, our knowledge of the human genome evolves. For example, we now classify transcription into two major categories, protein-coding and non-coding transcripts. Transcribed into messenger RNAs (mRNAs), protein-coding genes in total account for a very small percentage of transcripts (only about 2 %), whereas non-coding transcripts constitutes over 95 % of the transcriptome [1]. Among the non-coding transcripts, long non-coding RNAs (sequences longer than 200 nucleotide bases, lncRNAs) have emerged as a unique group of transcripts that have similar structures as protein-coding genes such as introns and exons, but also possess a wide range of biological functions involved in a variety of cellular activities [2–8]. Given their important roles in cell signaling and regulation, lncRNA's involvement in various diseases, especially in cancer, has been suspected and investigated [2, 3, 5–8]. However, since the functionality of lncRNAs is based on the nucleotide sequences, not peptide structures, and involves multiple molecules including proteins or other non-coding transcripts, our understanding of lncRNAs remains limited. The function and regulation of many lncRNAs and their derivatives are still unidentified or uncharacterized [9, 10].
In a previous study [11], we reported the discovery of a novel long intergenic non-coding RNA (lincRNA), LINC00472, in close link to clinical and pathologic features of breast cancer. High expression of LINC00472 was found to be associated with low tumor grade, early stage disease, estrogen or progesterone receptor positivity, and less aggressive molecular subtypes. Compared to patients with low expression, those with high expression of this lincRNA also had more favorable responses to adjuvant chemotherapy and endocrine therapy as well as survived longer. These observations have been remarkably consistent across more than a dozen clinical studies that have involved thousands of patients. Further, our in vitro experiments demonstrated that LINC00472 expression is low in breast cancer cell lines and up-regulating its expression via transfection of a LINC00472-expressing vector slows cell growth and inhibits cell migration [11].
In this article, we report our further investigation of this lincRNA in addressing three additional issues. First, we investigated which mechanism, change in gene copy number or DNA methylation, might have the potential to influence LINC00472 expression in breast cancer. Second, we were expected to further replicate our findings in microarray datasets other than the Affymetrix because our previous results mainly focused on the results from that platform. Third, since tumor grade was correlated with LINC00472 expression, and since both were associated with breast cancer survival, it would be helpful to demonstrate if LINC00472 had additional value in predicting breast cancer prognosis after eliminating the confounding effects of tumor grade. Compared to grade 1 and 3, grade 2 tumors are known to be much more heterogeneous with regard to disease prognosis. Thus, identifying additional prognostic markers for grade 2 tumors is considered necessary and valuable.
Materials and methods
Microarray-based comparative genome hybridization (aCGH)
We used the aCGH data from GEO (GSE23720) [12, 13] for copy number analysis. In the dataset, tumor DNA samples were extracted from 173 breast cancer patients, and 13 normal male DNA samples were used as reference. Genomic imbalances of the DNA samples were determined using the Agilent-014693 Human Genome CGH Microarray 244A chip. We downloaded the values obtained by circular binary segmentation (CBS) of the normalized log2 ratio Cy5/Cy3 (Cy5: label for human primary breast tumor samples; Cy3: label for the DNA pool from 13 normal male samples). Two probes (A_14_P113080 and A_14_P202474) on this Agilent chip cover the genomic region that contains the LINC00472 gene. In the same study, 193 patients had gene expression data generated by the Affymetrix Human Genome U133 Plus 2.0 Array. The Affymetrix chip has four probes (220324_at, 231136_at, 235771_at and 243974_at) mapped to different regions of the LINC00472 gene, and their values are highly correlated with one another. We used the data from probe 220324_at as we did in our previous work [11]. To investigate whether copy number variation of the LINC00472 gene contributes to its expression, we first generated a data table with both copy number and expression values of LINC00472 by matching the patients IDs, which included information from 173 patients at last. We separated these patients into low and high expression groups using the median of LINC00472 expression values as cutoff. Then we plotted the normalized copy number values (Cy5/Cy3 ratio) side by side, and calculated the Mann–Whitney U statistic between the two groups. As reference, data from the retinoblastoma 1 (RB1) gene were extracted and analyzed in the same way.
Affymetrix genome-wide human SNP array 6.0
The cBioPortal for Cancer Genomics was used to analyze raw data from a provisional study of breast invasive carcinoma in The Cancer Genome Atlas (TCGA) [14, 15]. Through May 2015, 1065 tissue samples tested both by RNA sequencing and by the Affymetrix Genome-wide Human SNP6.0 Array were available for plotting. We downloaded the expression data and copy number values of the LINC00472 and RB1 genes, and compared them using the same strategy as described above for the GSE23720 data.
Illumina HumanMethylation450 BeadChip
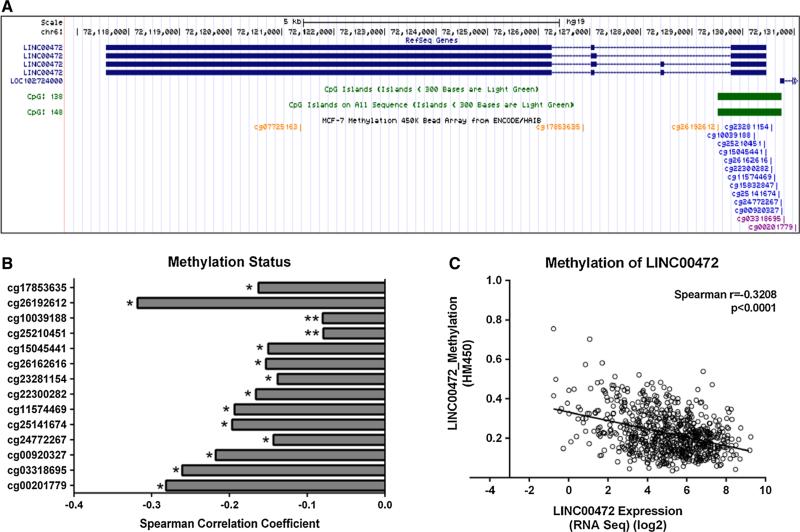
The provisional breast invasive carcinoma study from TCGA included microarray methylation data generated from the Illumina HumanMethylation450 BeadChip. This chip covers 99 % of the RefSeq genes, with an average of 17 CpG sites per gene distributed across the promoter, 5′UTR, first exon, gene body, and 3′UTR. Fifteen CpG sites are located in the LINC00472 gene (Fig. 2a), of which 14 are in the promoter and first exon regions. The cBioPortal for Cancer Genomics [14, 15] analyzes the Spearman correlation coefficient between gene expression and DNA methylation, and automatically selects the CpG site with the strongest correlation. To examine the expression-methylation correlations in detail, we downloaded the TCGA level 3 data on all the 15 CpG sites which contained normalized DNA methylation results, and performed correlation analysis with gene expression for each CpG site.
Fig. 2.
Methylation status and LINC00472 expression. a A screenshot from UCSC Genome Browser shows the CpG island around the LINC00472 promoter and probes included in the Illumina HumanMethylation450 BeadChip for measuring methylation in the CpG sites. b Bar charts demonstrate a consistent negative correlation between LINC00472 expression and methylation from all the probes. The y axis shows each probes, and x axis shows the Spearman correlation coefficient for each probe (*p ≤ 0.0001; **p < 0.05). c Scatter plot shows a negative correlation between LINC00472 expression and the methylation level around the LINC00472 promoter. Normalized DNA methylation beta values are shown in the y axis. Linear regression analysis suggests a regression line of Y = −0.02139X + 0.3321
Gene expression analysis
In our previous work on LINC00472, we only analyzed the GEO data generated from the Affymetrix Human Genome U133 plus 2.0 array or U133A array [11]. In the current study, we broadened the evaluation by analyzing four additional datasets in GEO that were based on the Agilent and Illumina platforms containing probes for LINC00472. These datasets included studies with a total of 561 breast cancer samples (Supplementary Table S1). Because different microarray platforms were used in these studies, we dichotomized the normalized LINC00472 expression data using study-specific median as cutoff to define “LINC00472_higher” (≥median) and “LINC00472_lower” (<median) for meta-analysis across the studies. Clinical and pathologic variables were also dichotomized. Associations of LINC00472 with clinical and pathologic variables were determined by odds ratios and their 95 % confidence intervals (95 % CI). Summary results, weighted by inverse-variance, were calculated based on the random-effects model, and presented in Forest plots. For datasets with survival information, Kaplan–Meier survival curves were constructed on individual studies and log-rank test was used to assess differences in survival between groups. In this survival analysis, LINC00472 expression was grouped into 3 categories based on its tertile distribution.
Analysis of grade 2 tumors
We analyzed the associations of LINC00472 expression with breast cancer survival specifically in grade 2 tumors in our study (Turin_Study), and in eight other GEO datasets that contained more than 60 patients with grade 2 tumors (Supplementary Table S2). In total, 936 patients with grade 2 tumors were included in this analysis. Kaplan–Meier survival analysis was performed on the individual studies, and LINC00472 expression levels were dichotomized based on the median in each study. Summarized results were also generated using the inverse-variance weighted random-effects model.
Statistical analysis
For data analysis, normalized LINC00472 expression intensity was analyzed as a categorical variable with low and high levels classified by median expression. Associations of LINC00472 expression with clinical and pathologic factors were determined using the Chi-square statistic. Kaplan–Meier survival curves were constructed to show survival differences according to LINC00472 expression, and the log-rank test was used for comparison. Survival outcomes considered were disease-free survival, distant relapse-free survival, relapse-free survival, and metastasis-free survival. The Mann–Whitney U statistic was used to compare differences in copy number variation. Spearman correlation coefficients were calculated for correlation analysis. Data were analyzed using the Statistical Analysis System, version 9.4 (SAS Institute Inc., Cary, NC) and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). All statistics were two-sided; p values less than 0.05 were considered significant. Review Manager (Revman Version 5.3, Copenhagen, Denmark) was used for meta-analysis.
Results
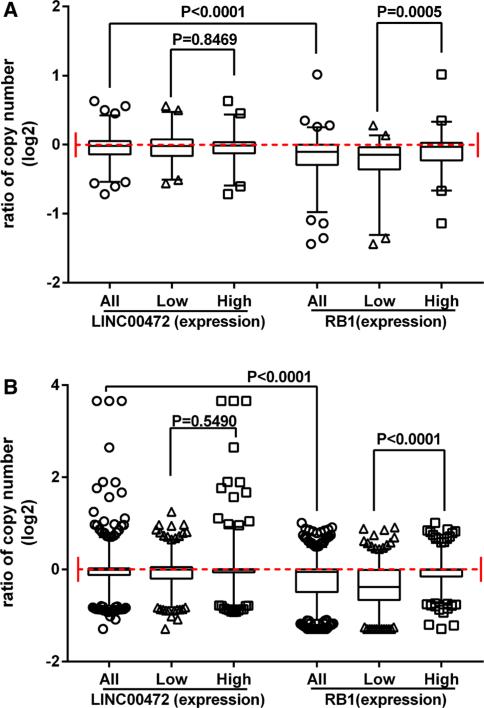
In our previous study, we found low LINC00472 expression in tumors compared to adjacent non-tumor or normal breast tissues [11], but did not know whether or not the differences were the results of copy number changes in the corresponding genomic region. To address this issue, we analyzed DNA copy number variations in relation to gene expression in two publically available datasets, one from GEO and one from TCGA. The dataset GSE23720 [12, 13] contained 173 tumor samples analyzed both by the Affymetrix gene expression microarray (Platform: GPL570) and by the Agilent CGH microarray (Platform: GPL9128). The ratio of gene copy numbers between tumor DNA and normal DNA (Cy5/Cy3) for LINC00472 distributed almost evenly around 1.0, suggesting no loss or deletion of this gene, while for the RB1 gene, which has been reported generally to be deleted in cancer tissues, most of the Cy5/Cy3 ratios were below 1.0 (Fig. 1a). Grouping the samples into high versus low LINC00472 expression showed no differences in gene copy numbers between these groups (Fig. 1a).
Fig. 1.
Copy number variation and LINC00472 expression. a Box and whiskers plot based on the dataset GSE23720 show similar distributions of copy numbers for the LINC00472 gene (left) but different distributions for the RB1 gene (right) between patients with high and low expression, correspondingly. The y axis shows the normalized signal ratio between tumor tissues (Cy5) and a pool of normal male DNA (Cy3). The whiskers cover 2.5–97.5 percentiles. p values were determined by the Mann–Whitney U test. b Box and whiskers plot based on the TCGA breast cancer study show similar distributions of copy numbers for the LINC00472 gene (left) but different distributions for the RB1 gene (right) between patients with high and low expression, correspondingly. The y axis shows the ratio of copy number values. The whiskers cover 2.5–97.5 percentiles. p values were determined by the Mann–Whitney U test
In the TCGA provisional breast cancer study, gene expression data were produced by RNA sequencing, and copy number variations were measured by the Affymetrix Genome-wide Human SNP6.0 Array. We plotted the data as we did for the GSE23720 data, and found that LINC00472 expression was not associated with copy number alteration, while many samples in this large TCGA dataset showed copy number loss or deletion in the RB1 gene (Fig. 1b). RB1 expression was positively correlated with gene copy number (Fig. 1b) as had been observed previously [16].
We next analyzed the relationship of LINC00472 expression and DNA methylation of the gene. In the TCGA provisional breast cancer study, 735 patient samples had information on gene expression by RNA Sequencing and on DNA methylation by the HumanMethylation450 chip. The Illumina HumanMethylation450 chip contains 14 methylation probes for the CpG sites in the promoter and first exon regions of the LINC00472 gene (Fig. 2a). We downloaded all the methylation data from the 14 CpG sites, and analyzed their correlations with expression of LINC00472. Our analysis showed that methylation in these CpG sites were all inversely correlated with LINC00472 expression, higher methylation, and lower expression (Fig. 2b), suggesting that the expression of this gene is regulated by promoter methylation. Across the 14 probes, the strongest correlation coefficient was −0.32 (p < 0.0001) (Fig. 2c). Further analyses of methylation with respect to disease features and patient survival revealed no significant associations between these variables (data not shown).
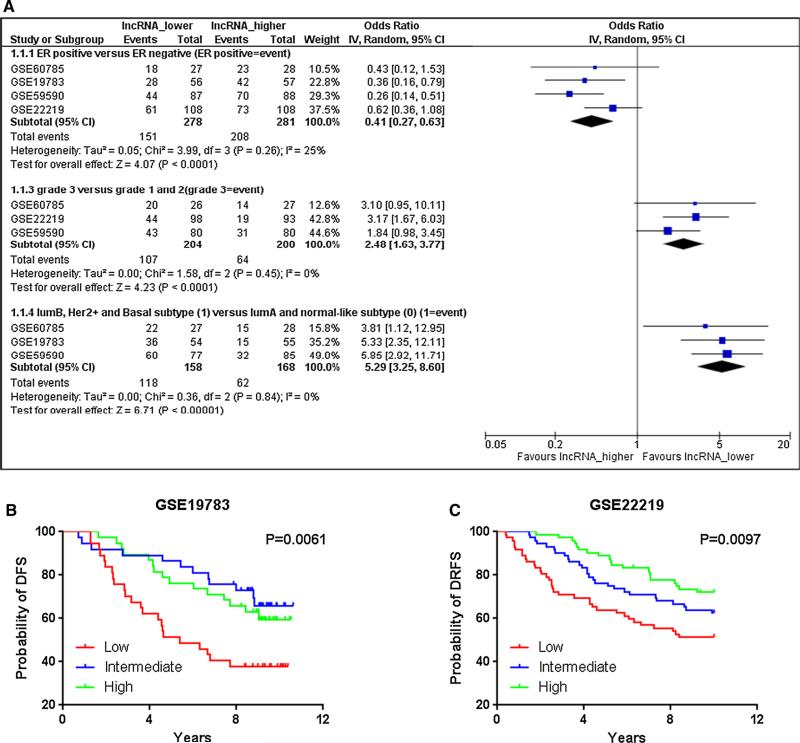
In our previous study [11], we focused exclusively on the results of the Affymetrix chip (Affymetrix Human Genome U133 plus 2.0 array and U133A array) in order to ensure that we employed consistent and reliable gene expression data for validation. In the present report, we broadened the scope of our validation by including chip results from other manufacturers. We identified four such datasets, three from the Illumina chip and one from the Agilent (Supplemental Table S1). Consistent with our previous observations, analysis of these data showed that LINC00472 expression was positively associated with estrogen receptor (ER) status, and negatively with tumor grades and aggressive molecular subtypes (Fig. 3a). Two of the datasets also had information on disease-free survival. High expression of LINC00472 was associated with favorable disease outcomes compared to low expression (p = 0.0061 and 0.0097 for GSE19783 and GSE22219, respectively) (Fig. 3b, c). These results again confirmed the findings of our previous study.
Fig. 3.
Agilent and Illumina platforms for LINC00472 expression. a A meta-analysis shows that low LINC00472 expression was associated with ER negative tumors (OR = 0.41; 95 % CI 0.27–0.63), high-grade tumors (OR = 2.48; 95 % CI 1.63–3.77), and luminal B, Her2 positive or basal-like tumors (OR = 5.29; 95 % CI 3.25–8.60). b Kaplan–Meier survival curves by low, intermediate and high LINC00472 expression in dataset GSE19783. c Kaplan–Meier survival curves by low, intermediate, and high LINC00472 expression in dataset GSE22219
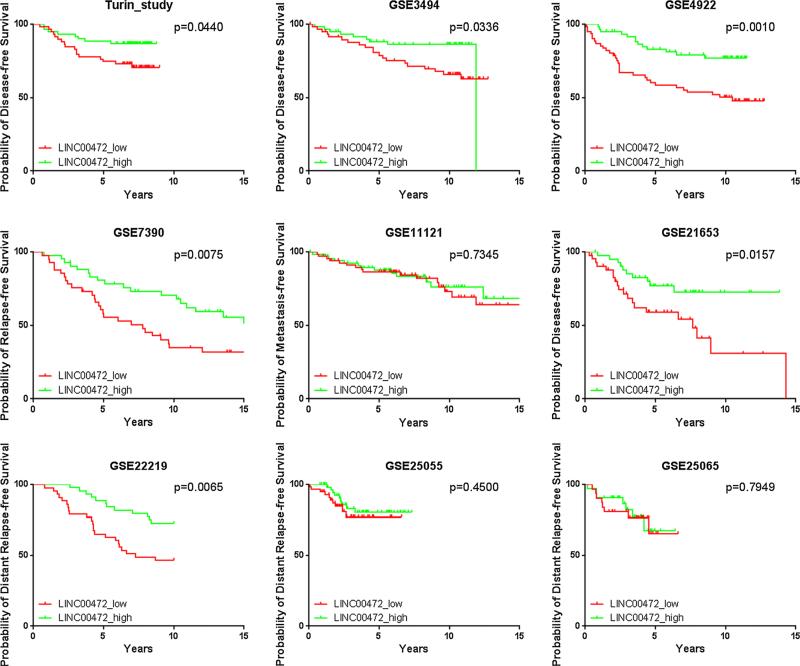
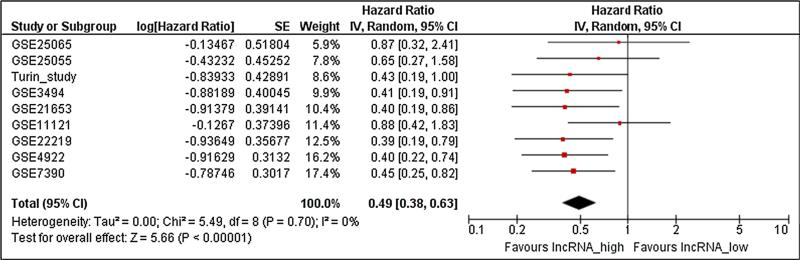
LINC00472 expression is associated with tumor grade, potentially limiting its utility for prognosis, especially in high- and low-grade tumors (grade 3 and 1) where expression is relatively homogenous [17]. To improve the accuracy of breast cancer prognosis among patients with grade 2 tumors, additional tumor features, especially molecular markers, should be considered. We therefore analyzed LINC00472 data in patients with grade 2 tumors. Nine datasets including our own had more than 60 such patients. Of the 9 studies, 6 showed high expression of LINC00472 significantly associated with favorable disease-free survival compared to low expression (Fig. 4). Meta-analysis of these studies demonstrated that patients with grade 2 breast cancer had a 50 % reduction in risk of disease relapse if their tumors expressed high levels of LINC00472 transcript (Fig. 5).
Fig. 4.
Kaplan-Meier survival curves by low and high LINC00472 expression in our study and 8 other datasets from GEO with more than 60 patients with grade 2 tumor in each dataset
Fig. 5.
Meta-analysis of associations between LINC00472 expression and disease-free survival among patients with grade 2 tumors. Summarized hazard ratio was estimated using the random-effect model and each study was weighted with its variance. High LINC00472 expression was associated with better disease-free survival (OR = 0.49; 95 % CI 0.38–0.63)
Discussion
This study further confirms that LINC00472 expression is significantly associated with breast cancer in terms of tumor grade, estrogen receptor status, and molecular subtype, and that higher expression of LINC00472 predicts better disease outcome. Our study also provides some evidence that LINC00472 expression may be regulated by DNA methylation in its promoter, whereas changes in gene copy number are not found in breast tumors and cannot account for the variation in LINC00472 expression. More importantly, levels of LINC00472 expression can be used to distinguish survival differences among breast cancer patients with grade 2 tumors. These features underscore the potential significance of LINC00472 in serving as a marker for breast cancer prognosis.
As part of our investigation, we evaluated two aspects of LINC00472 expression regulation, gene copy number, and promoter methylation, using data available online from genome-wide analysis. Data from the microarray-based comparative genome hybridization analysis and Affymetrix genome-wide human SNP genotyping array both showed no evidence of substantial deviation from standard copy number, suggesting no deletion or amplification of this gene in tumor samples. We integrated the copy number data with gene expression results, and found no differences in gene copy number between tumor samples with high versus low expression of LINC00472. These analyses indicate that expression variation of LINC00472 in breast cancer is not due to changes in gene copy number. We also compared these results with similar data for the RB1 gene which is known to have copy number loss in cancer, reinforcing the conclusion of no copy number changes in LINC00472.
The LINC00472 gene contains a CpG island in its promoter. As reported by several lncRNA profiling studies [18–20], DNA methylation in the CpG island of a lncRNA gene promoter may regulate the expression of the lncRNA gene, just like it does for coding genes. We therefore examined methylation values in the TCGA database generated from the Illumina HumanMethylation450 BeadChip, and integrated the data with gene expression results. Both our own analysis and the analysis through the cBioPortal for Cancer Genomics showed that LINC00472 expression was inversely correlated with methylation levels of the CpG sites in the promoter and first exon. Our analysis of the TCGA data also indicates that this inverse correlation exists not only in breast cancer, but in other cancer sites as well. In lung adenocarcinoma, the Spearman correlation coefficient (r) was −0.40 (p < 0.0001), in lung squamous cell carcinoma, the r was −0.30 (p < 0.0001), in uterine carcinosarcoma r was −0.30 (p < 0.0001), and in uterine corpus endometrial carcinoma r was −0.53 (p < 0.0001). Our findings suggest that promoter methylation may play a role in regulation of LINC00472 expression. Data from another GEO dataset GSE39004 [21], containing both gene expression and methylation information from 46 tumor samples, also showed a similar correlation (data not shown).
In our previous study, we used gene expression data exclusively from two microarray chips, the Affymetrix Human Genome U133 plus 2.0 and the U133A arrays. There were reports suggesting that microarray data from different platforms did not correlate well [22, 23]. We had the same impression when we compared gene expression signatures generated by different microarray platforms for breast cancer prognosis and found little overlap in genes across different signatures [24]. This phenomenon led us to think that our previous results need to be validated by other microarray platforms. In this study, we included microarray data from other manufacturers to broaden the range of data sources for validation and to rule out the possibility that our validation was limited to one type of array from a single manufacturer. We identified four datasets in GEO (Supplementary Table S1), and each contained more than 50 samples of gene expression data and clinical information that were useful for evaluation. Our meta-analysis confirmed that low LINC00472 expression was linked to breast cancer of more unfavorable prognosis.
A set of tumor samples in GEO has been analyzed both by RNA sequencing (GSE60785) and by gene expression microarray (GSE60788). The results of these analyses with regard to LINC00472 expression were highly correlated (Spearman correlation coefficient = 0.74; p < 0.0001). The associations of LINC00472 expression with ER status, tumor grade, and molecular subtype were also similar between the two platforms. The provisional breast cancer dataset in TCGA, which was used in our copy number and methylation analyses, included more than 1000 patients, but these studies were conducted relatively recently and patients in the datasets had short follow-up times. The microarray data in TCGA did not cover most long non-coding RNAs, including LINC00472, and therefore we had to use RNA sequencing data to analyze the association of LINC00472 with survival. In this analysis, patients with higher expression of LINC00472 had significantly better overall survival than patients with lower expression. Considering these methods plus the RT-qPCR that we used in our previous study [11] we conclude that the associations between LINC00472 expression and disease features are consistent in breast cancer patients regardless of the analytical methods used to measure the expression of LINC00472.
As a well-established indicator of breast cancer prognosis, tumor grade, determined on the basis of cell morphology, provides important information on the potential behaviors of malignant cells [25]. Determining tumor grade may be relatively straightforward for grade 1 or 3 breast cancers [26, 27], but not for grade 2, as reflected by the lowest degree of concordance among pathologists compared to grades 1 and grade 3 [17]. Grade 2 tumors have the most uncertainty in choice of post-surgical treatment, especially chemotherapy [17]. Several genomic tests have been developed on the basis of gene expression profiling, including Oncotype DX [28] and MammaPrint [29, 30], to assist the prediction of breast cancer prognosis for grade 2 tumors [31]. However, even for the ongoing TAILORx trial (the Trial Assigning Individualized Options for Treatment), patients with intermediate grade tumors are still randomly assigned to receive adjuvant chemotherapy or not as well as to subsequent endocrine therapy [32, 33], because risk of recurrence for these patients is uncertain. Multiple gene expression signatures have been developed with the hope that genomic-grade can predict tumor prognosis better than histologic grade [13, 34–40]. However, the gene expression signatures are comprised of distinct sets of genes with little overlap [28, 32, 36, 41–47], suggesting that substantial heterogeneity may exist and additional predictors are needed. To address this issue, we focused on the prognostic value of LINC00472 in patients with grade 2 tumors only, and found that survival in such patients was further distinguished when their LINC00472 levels were analyzed in tumor samples. Additional studies are needed to further confirm the prognostic and predictive values of LINC00472 in grade 2 tumors when confounding factors can be considered and adjusted in analysis.
Although our investigation found additional evidence in support of our finding of LINC00472 being a potential biomarker for breast cancer prognosis, more studies, especially those prospective ones where a standardized lab test is employed to measure gene expression, are still needed for further validating the results and excluding the influences of other prognostic factors or parameters. For clinical application, we also need to establish a unique cutoff for predicting prognosis, and demonstrate the sensitivity and specificity of the test. Another issue we should consider is that our findings are currently based on the analysis of fresh frozen tissues which may not be feasible or practical for application in clinic. One should test if FFPE tissue blocks can be used for testing this marker since these samples are more readily available for analysis. More research is also needed for understanding the biologic implication of LINC00472 in breast cancer.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-015-3632-8) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of Interest The authors declare that they have no conflict of interest.
References
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 4.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 6.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, Katsaros D, Loo LW, Hernandez BY, Chong C, Canuto EM, Biglia N, Lu L, Risch H, Chu WM, Yu H. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget. 2015;6(11):8579–8592. doi: 10.18632/oncotarget.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabatier R, Finetti P, Adelaide J, Guille A, Borg JP, Chaffanet M, Lane L, Birnbaum D, Bertucci F. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS One. 2011;6(11):e27656. doi: 10.1371/journal.pone.0027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bekhouche I, Finetti P, Adelaide J, Ferrari A, Tarpin C, Charafe-Jauffret E, Charpin C, Houvenaeghel G, Jacquemier J, Bidaut G, Birnbaum D, Viens P, Chaffanet M, Bertucci F. High-resolution comparative genomic hybridization of inflammatory breast cancer and identification of candidate genes. PLoS ONE. 2011;6(2):e16950. doi: 10.1371/journal.pone.0016950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Richardson AL, Schnitt SJ, Schmitt FC, Tan PH, Tse GM, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res: BCR. 2010;12(4):207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhi H, Ning S, Li X, Li Y, Wu W, Li X. A novel rean-notation strategy for dissecting DNA methylation patterns of human long intergenic non-coding RNAs in cancers. Nucleic Acids Res. 2014;42(13):8258–8270. doi: 10.1093/nar/gku575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultra-conserved regions in human cancer. Oncogene. 2010;29(48):6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465(7295):175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, Wallace TA, Issaq HJ, Zhou M, Killian JK, Stevenson HS, Karoly ED, Chan K, Samanta S, Prieto D, Hsu TY, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Investig. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvinen AK, Hautaniemi S, Edgren H, Auvinen P, Saarela J, Kallioniemi OP, Monni O. Are data from different gene expression microarray platforms comparable? Genomics. 2004;83(6):1164–1168. doi: 10.1016/j.ygeno.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Rogojina AT, Orr WE, Song BK, Geisert EE., Jr Comparing the use of Affymetrix to spotted oligonucleotide microarrays using two retinal pigment epithelium cell lines. Mol Vis. 2003;9:482–496. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Katsaros D, Shen Y, Yu H. Biological and clinical significance of MAD2 and BUB1, genes frequently appeared in the expression signatures for breast cancer prognosis. PLoS ONE. 2015;10(8):e0136246. doi: 10.1371/journal.pone.0136246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 26.Rakha EA, El-Sayed ME, Lee AH, Elston CW, Grainge MJ, Hodi Z, Blamey RW, Ellis IO. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol. 2008;26(19):3153–3158. doi: 10.1200/JCO.2007.15.5986. [DOI] [PubMed] [Google Scholar]
- 27.Dalton LW, Page DL, Dupont WD. Histologic grading of breast carcinoma. A reproducibility study. Cancer. 1994;73(11):2765–2770. doi: 10.1002/1097-0142(19940601)73:11<2765::aid-cncr2820731119>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 28.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 29.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 30.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ, Panel members Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20(8):1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010;17(4):R245–R262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 33.Sparano JA. TAILORx: trial assigning individualized options for treatment (Rx). Clin Breast Cancer. 2006;7(4):347–350. doi: 10.3816/CBC.2006.n.051. [DOI] [PubMed] [Google Scholar]
- 34.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 36.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, Klijn JG, Larsimont D, Buyse M, Bontempi G, Delorenzi M, Piccart MJ, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 37.Ma XJ, Hilsenbeck SG, Wang W, Ding L, Sgroi DC, Bender RA, Osborne CK, Allred DC, Erlander MG. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J Clin Oncol. 2006;24(28):4611–4619. doi: 10.1200/JCO.2006.06.6944. [DOI] [PubMed] [Google Scholar]
- 38.Ma XJ, Salunga R, Dahiya S, Wang W, Carney E, Durbecq V, Harris A, Goss P, Sotiriou C, Erlander M, Sgroi D. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14(9):2601–2608. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, Shioda T, Ma XJ, Sgroi DC. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13(21):6327–6334. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 41.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 42.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, Liu ET, Miller L, Nordgren H, Ploner A, Sandelin K, Shaw PM, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res BCR. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mannelqvist M, Wik E, Stefansson IM, Akslen LA. An 18-gene signature for vascular invasion is associated with aggressive features and reduced survival in breast cancer. PLoS One. 2014;9(6):e98787. doi: 10.1371/journal.pone.0098787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin ZQ, Liu JJ, Xu YC, Yu J, Ding GH, Yang F, Tang L, Liu BH, Ma Y, Xia YW, Lin XL, Wang HX. A 41-gene signature derived from breast cancer stem cells as a predictor of survival. J Exp Clin Cancer Res CR. 2014;33:49. doi: 10.1186/1756-9966-33-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X, Rodland EA, Sorlie T, Naume B, Langerod A, Frigessi A, Kristensen VN, Borresen-Dale AL, Lingjaerde OC. Combining gene signatures improves prediction of breast cancer survival. PLoS One. 2011;6(3):e17845. doi: 10.1371/journal.pone.0017845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.