ABSTRACT
Spermatogonial stem cells (SSCs), which are unipotent stem cells in the testes that give rise to sperm, can be converted into germline-derived pluripotent stem (gPS) by self-induction. The androgenetic imprinting pattern of SSCs is maintained even after their reprogramming into gPS cells. In this study, we used an in vitro neural differentiation model to investigate whether the imprinting patterns are maintained or altered during differentiation. The androgenetic patterns of H19, Snrpn, and Mest were maintained even after differentiation of gPS cells into NSCs (gPS-NSCs), whereas the fully unmethylated status of Ndn in SSCs was altered to somatic patterns in gPS cells and gPS-NSCs. Thus, our study demonstrates epigenetic alteration of genomic imprinting during the induction of pluripotency in SSCs and neural differentiation, suggesting that gPS-NSCs can be a useful model to study the roles of imprinted genes in brain development and human neurodevelopmental disorders.
KEYWORDS: Androgenetic imprinting, germline-derived pluripotent stem cells, in vitro model, neural stem cells, spermatogonial stem cells
Introduction
Genomic imprinting is a molecular epigenetic mechanism for controlling imprinted gene expression. Genomic imprinting is involved in development, including fetal and placental growth, cell proliferation, and adult behavior by stage-specific DNA methylation changes of imprinted genes. Imprinted genes, which have parental-specific methylation of CpG-rich domains established during gametogenesis, exhibit paternal or maternal monoallelic expression.1,2 Abnormal expression of imprinted genes causes genetic diseases including cancers and neurological disorders, such as Prader-Willi and Angelman syndromes,3 and Beckwith-Wiedemann syndrome.4
Spermatogonial stem cells (SSCs; also called germline stem cells) are precursor cells that give rise to sperm; SSCs can be converted into germline-derived pluripotent stem (gPS) cells by self-induction under defined culture conditions.5,6 gPS cells have the capacity to differentiate into 3 germ layers and exhibit androgenetic imprinting patterns of H19 and Igf2r like in SSCs.5 The androgenetic imprinting pattern in gPS cells is particularly interesting as it suggests that gPS cells can represent a unique model system to study the role of imprinted genes in development and the contribution of imprinted genes to various diseases. In particular, studies of in vitro differentiation of gPS cells can provide insights into the contribution of paternally imprinted genes to the development of specialized organs. In the present study, we assessed whether the androgenetic state affects the in vitro neural differentiation potential of gPS cells and whether paternal imprinting is maintained or altered when gPS cells differentiate into neural cell lineages.
Results and discussion
Derivation of NSCs from gPS cells
To determine the in vitro differentiation potential of gPS cells into neural stem cells (NSCs), embryoid body (EB) differentiation methods were applied to gPS cells (Fig. 1A). Four to 7 d after EB differentiation, neural differentiation was observed through the neurite formation from EBs and outgrowth of bipolar-shaped cells. One month later, outgrowing NSCs were mechanically isolated and transferred into new dishes for homogenous culture. Morphology of NSCs derived from gPS cells (gPS-NSCs) was similar to that of embryonic stem cell (ESC)-derived NSCs (ES-NSCs), which were derived using the same protocol, and to that of fetal forebrain–derived NSCs (FB-NSCs), which were derived from forebrain of mouse fetus at embryonic day 13.5 by digestion (Fig. 1B). gPS-NSCs appeared to be bipolar and showed lattice growth typical of NSCs.7 As shown in Fig. 1C, gene expression analysis by RT-PCR showed that gPS-NSCs expressed the NSC-specific marker genes, Sox2, Nestin, Pax6, Glast, Fabp7, Mash1, and Olig2, similar to the two other NSC types. In addition, global gene expression profile of gPS-NSCs gene array analysis was similar to those of FB-NSCs and ES-NSCs (Fig. 1D and 1E). Immunocytochemistry analysis confirmed that gPS-NSCs uniformly expressed the NSC-specific marker proteins Sox2 and Nestin. gPS-NSCs were multipotent, because they were able to differentiate into neurons, astrocytes, and oligodendrocytes, which was confirmed by the expression of class III β-tubulin (Tuj1) and the glial markers glial fibrillary acidic protein (GFAP) and O4, respectively (Fig. 1F). gPS-NSCs proliferated at a slower rate than FB-NSCs and ES-NSCs (Fig. S1). However, self-renewability and differentiation potential of gPS-NSCs were similar to those of FB-NSCs and ES-NSCs. These results indicate that gPS cells have neural differentiation ability and differentiate into NSCs that exhibit molecular and cellular characteristics of normal NSCs and can be further differentiated into neuronal and glial cells despite their uniparental origin.
Figure 1.
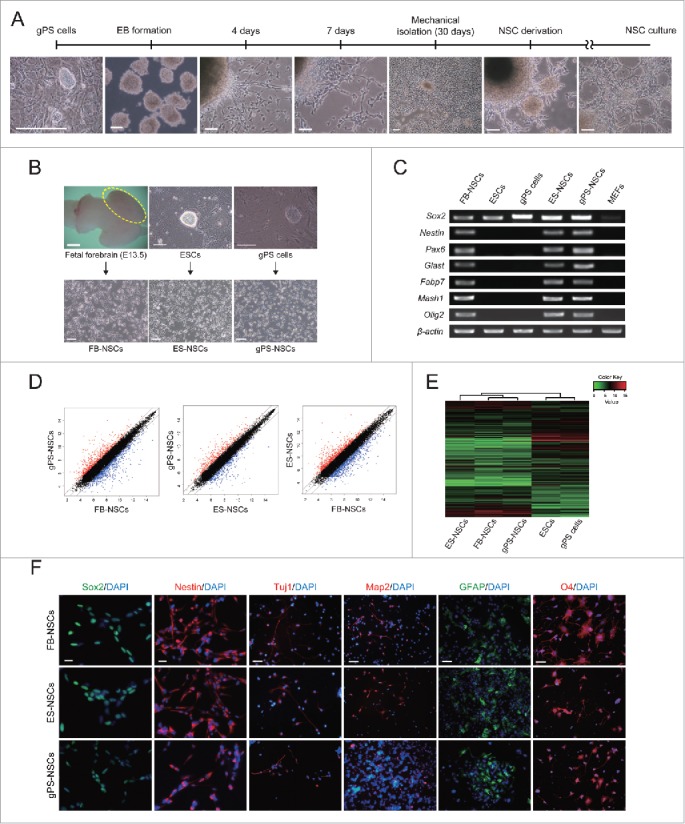
Establishment and characterization of neural stem cells derived from gPS cells. (A) Establishment of NSCs from gPS cells. Scale bars = 50 μm. (B) Phase contrast images of mouse fetal brain, FB-NSCs, ESCs, ES-NSCs, gPS cells, and gPS-NSCs. Scale bars = 100 μm. (C) Expression of NSC-specific genes in the derived NSC lines. (D) Pairwise scatter plots comparing the global gene expression patterns between gPS-NSCs, FB-NSCs and ES-NSCs. (E) Heat map clustering of the global gene expression profiles of ES-NSCs, FB-NSCs, gPS-NSCs, ESCs, and gPS cells. (F) Immunocytochemistry of derived NSC cells using the markers for NSCs (Sox2 and Nestin), neurons (Tuj1), astrocytes (GFAP), and oligodendrocytes (O4). Scale bar = 20 μm.
Alteration of DNA methylation patterns in imprinted genes during reprogramming and redifferentiation
Using bisulfite sequencing and pyrosequencing analysis, we assessed whether the DNA methylation pattern of imprinted genes was maintained or altered during reprogramming of SSCs into gPS cells and redifferentiation into gPS-NSCs (Fig. 2A and 2B). We analyzed the DNA methylation status of multiple CpG sites in the paternally imprinted gene H19 and maternally imprinted genes, Snrpn, Mest, and Ndn. As expected, differentially methylated regions (DMRs) of all 4 genes had a somatic imprinting pattern in FB-NSCs, which are similar to previously reported results by Kim et al., whereas aberrant hypermethylation of Mest (but not other imprinted genes) was detected in ESCs and ES-NSCs (Fig. S2).8 Mest hypermethylation has been previously described in ESCs.9 The aberrant methylation of imprinted genes like Mest in ESCs and ES-NSCs can be readily detected in human ESCs.10 Since adult SSCs acquire paternal methylation imprints, they had a fully methylated paternal imprinted gene, H19, and fully unmethylated maternal imprinted genes, Snrpn, Mest, and Ndn. We further investigated changes in the methylation status of imprinted genes during induction of pluripotency and redifferentiation into NSCs. The androgenetic status of H19, Snrpn, and Mest was maintained, whereas a gain of methylation of Ndn was detected in gPS cells and gPS-NSCs, indicating that the imprinting pattern can be altered in some imprinted genes by reprogramming (Fig. 2A and 2B). When the methylation levels were compared in the three NSC types, the difference was noticeable in H19, Snrpn, and Mest, while the percentage of methylated CpG sites in Ndn of gPS-NSCs was almost identical to that in other NSCs (Fig. S3).
Figure 2.
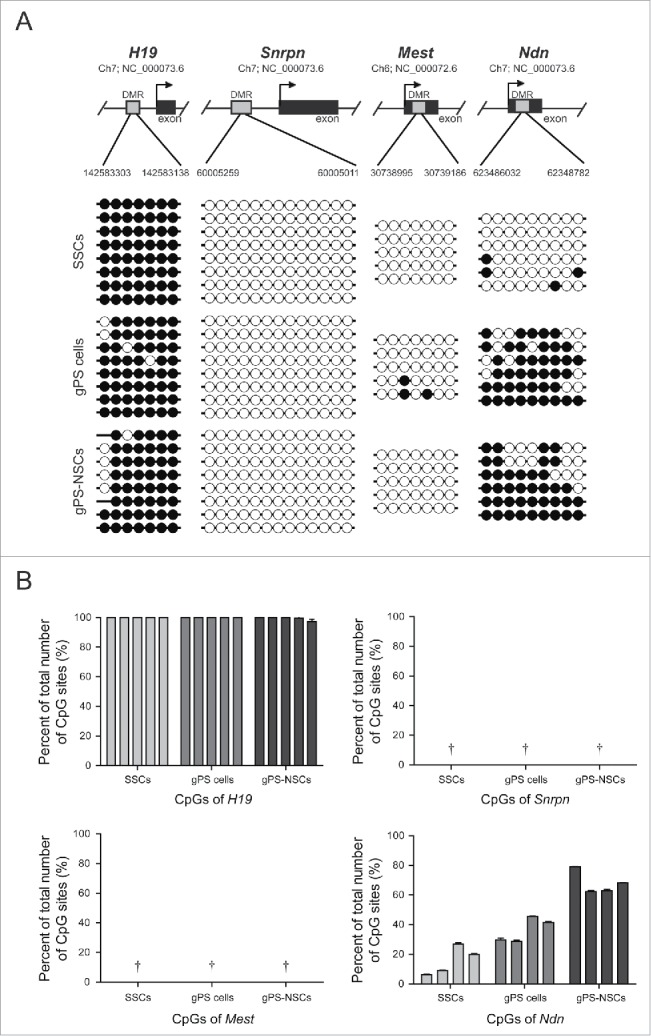
Analysis of DNA methylation. (A) Bisulfite DNA sequencing of H19, Snrpn, Mest, and Ndn in SSCs, gPS cells, and gPS-NSCs. Each line represents a single clone. (B) DNA methylation analysis and pyrosequencing using PyroMark Q96. The graphs show the methylation percentages of cytosine in the target regions of H19, Snrpn, Mest, and Ndn. Means ± s.e.m. are shown for three independent experiments († = the numbers were not detectable because of 0%).
After reprogramming of SSCs into gPS cells, de novo DNA methylation was observed in the maternally imprinted gene Ndn. Alteration of the uniparental imprinting status in Ndn was previously reported by Kim et al. (2013), who found a loss of DNA methylation during induction of pluripotency in parthenogenetic NSCs. Ndn plays an important role in proliferation and apoptosis of NSCs and in neuronal development and terminally differentiated neurons.11-13 Embryo development requires proper expression of both maternally and paternally imprinted genes. Although uniparental embryos are not capable of full-term development due to defects in genomic imprinting, androgenetic ESCs and gPS cells can generate normal chimeras.5,14 Using blastocyst injection of androgenetic ESCs, Dinger et al. demonstrated their contribution to the development of chimeric fetal brains. This suggests that proper expression of imprinted genes, such as Ndn, which has a role in neurogenesis, is associated with a transition from the androgenetic imprinting pattern to somatic-like patterns during reprogramming of SSCs into gPS cells. Interestingly, although Ndn and Snrpn were located on same locus and controlled the expression by the same imprinting center, the imprinting pattern of Snrpn was maintained androgenetic imprints while the pattern of Ndn was changed into somatic patterns after reprogramming and redifferentiation. As reported by Hanel et al., DNA methylation of Ndn was especially tissue-specific rather than other imprinted genes. The completely unmethylated pattern seen in sperm becomes partially methylated during development, especially in brain, and this change of imprinting patterns related the gene expression is regulated by binding of transcription factors.12
In contrast to the somatic imprinting patterns of H19, Snrpn, and Mest in FB-NSCs, uniparental SSCs, gPS cells, and gPS-NSCs showed androgenetic imprinting of these genes. Unlike the imprinting pattern of Ndn, those of H19, Snrpn, and Mest were not altered during reprogramming of SSCs into gPS cells and differentiation of gPS cells into NSCs. We found that the status of maintenance and changing of methylation patterns in 4 genes was sustained even after further differentiation into neuron and astrocyte (Fig. 3). The summary data of methylation analyses for four imprinted genes is shown in the Table S1. These results suggest that if epigenetic changes do not occur during reprogramming, the imprinting patterns can be maintained even during differentiation in neuronal development. This notion is also supported by Dinger et al., who found that the androgenetic imprinting pattern of Snrpn and Igf2r in androgenetic ESCs was maintained after differentiation into neural progenitor cells. Taken together, our results suggest that during reprogramming and redifferentiation, some imprinted loci retained an androgenetic imprinting pattern, while others did not.
Figure 3.
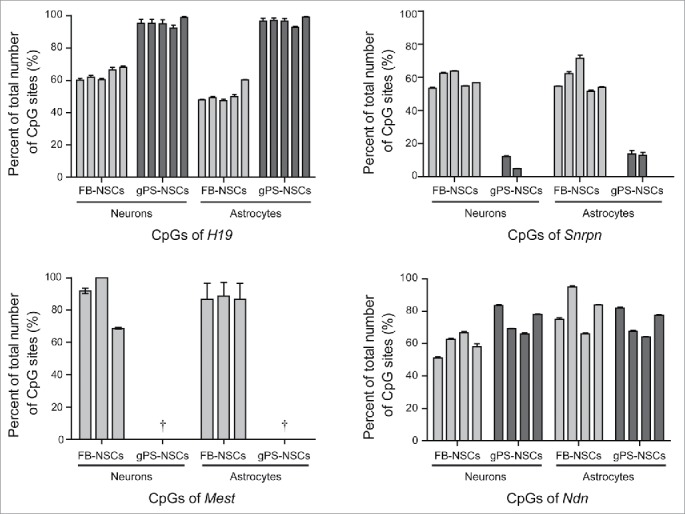
Pyrosequencing analysis of H19, Snrpn, Mest and Ndn in neurons and astrocytes differentiated from gPS-NSCs and FB-NSCs. Cytosine methylation percentages in the target regions are shown. Means ± s.e.m. are shown for three independent experiments († = the numbers were not detectable because of 0%).
Determination of imprinted gene expression in gPS-derived neural stem cells and their neural progeny
To examine whether the methylation status correlates with the expression levels of maternally expressed genes (Ube3a and Igf2r) and paternally expressed genes (Snrpn, Mest, Ndn, Impact, and Igf2), we analyzed their expression in three different NSC types by real-time PCR. Consistent with the results of gene array analysis, we found that the expression levels of Ube3a, Snrpn, and Mest were higher in gPS-NSCs than in FB-NSCs and ES-NSCs, whereas that of Igf2r was lower (Fig. 4). The result of methylation analyses, bisulfite sequencing and pyrosequencing, of the DMRs of the imprinted genes Snrpn and Mest correlated with their expression levels in NSCs. However, there was no detectable expression of H19 in all three NSCs. This is in line with the results of a previous study that H19 expression is not always correlated with DMR methylation.15 The similar expression level of Igf2 in all three NSCs is able to support that Igf2 is under tissue-specific control. Methylated paternal allele of Igf2 has observed in adult tissue included brain and leads to correlate with expression. In fetal brain, however, the paternal allele of Igf2 is predominantly unmethylated like maternal allele and showed rare expression.16 While the functional significance of allelic methylation pattern in paternal allele is not clear at present, this epigenetic modification is able to considered one of the needed mechanism during brain development.
Figure 4.

Expression analysis of imprinted genes in NSC lines, and in neurons and astrocytes differentiated from each NSC line. mRNA levels of maternally and paternally expressed imprinted genes. The expression levels of ES-NSCs, gPS-NSCs and differentiated cells from them were compared with those in FB-NSCs and differentiated cells from FB-NSCs; expression in FB-NSCs and differentiated cells from FB-NSCs were set to 1 for other samples. Means ± s.e.m. are shown for three independent experiments.
Recently, numerous lines of evidence have emerged indicating that imprinted gene expression can occur in a cell type–specific manner in the brain.17,18 A number of imprinted genes have distinct expression patterns and functions within neuronal subtypes.19 Thus, we analyzed gene expression levels in gPS-NSCs differentiated into neurons and astrocytes to determine whether the imprinting status in androgenetic cells correlates with the expression levels of the imprinted genes and affects their cell type–specific expression patterns (Fig. 4). Ube3a expression was higher in gPS-NSCs and astrocytes differentiated from gPS-NSCs than in the equivalent biparental cells, FB-NSCs and ES-NSCs (Fig. 4), although Ube3a is a maternally expressed gene in the brain.20 Therefore, the parental-specific expression Ube3a, unlike that of Igf2r, appears to be disrupted. However, it has been reported that Ube3a expression is cell type–specific in the brain and that Ube3a is expressed from both parental alleles in neural progenitor cells and glial cells, which can explain the expression of Ube3a in gPS-NSCs and astrocytes differentiated from gPS-NSCs.21 Nonetheless, it is still unclear why Ube3a expression in androgenetic cells was higher than in biparental cells.
Although it is cell type–dependent, the expression of the paternally expressed genes Snrpn, Mest, Ndn, Impact, and Igf2 tends to be parental-specific in gPS-NSCs. High androgenetic expression of these genes, especially of Impact in gPS-NSCs and astrocytes differentiated from gPS-NSCs, Igf2 in neurons derived from gPS-NSCs, and Ube3a in gPS-NSCs and astrocytes derived from gPS-NSCs suggests that androgenetic cells in the brain of the chimeric mouse by androgenetic ESCs result in abnormal contribution of certain areas of the chimeric brain.22 All of those results for imprinted gene expressions are summarized in Table S2.
In this study, we showed that gPS cells reprogrammed from SSCs by self-induction of pluripotency can be differentiated into NSCs and further differentiated into neurons and astrocytes, and that androgenetic genomic imprinting was mostly maintained during reprogramming and neural differentiation in the imprinted genes analyzed. The androgenetic imprinting status of gPS-NSCs did not limit their differentiation into neuronal and glial cells. We also found that androgenetic parental-specific expression was cell type–specific, which resulted in high expression of paternally imprinted genes in gPS-NSCs and their differentiated cells compared with biparental cells, which are known to be involved in brain development. This finding can explain the abnormal contribution of androgenetic cells observed in the chimeric brain by androgenetic ESCs reported by Keverne et al. and behavioral retardation in imprinted gene–related human brain diseases. Overall, our results suggest that an in vitro model of gPS cell differentiation can be a useful tool for investigation of the roles of the imprinted genes in mammalian brain development and human neuronal diseases associated with imprinting.
Materials and methods
ESC and gPS cell culture
ESCs and gPS cells were cultured as described in a previous study.5
Neural stem cell derivation from gPS cells, ESCs, and mouse fetal forebrain
FB-NSCs were derived from the embryonic day 13.5 fetal forebrain [C57BL6/N mice (KoaTech)].23 For derivation of NSCs from gPS cells and ESCs, after EB differentiation of 20–30 d in NSC medium, neural clumps containing NSCs were mechanically isolated and plated on gelatin-coated cell culture dishes for expansion of NSCs. NSC medium and defined culture conditions used in this study have been previously described.23
Differentiation of NSCs into neuronal and glial cells
NSCs (5 × 104 cells) were seeded into 4 wells of a poly-l-ornithine/laminin–coated 6-well plate in neuronal differentiation medium. After 4 d, neuron maturation was initiated by changing the medium to a medium without fibroblast growth factor 2 (Peprotech, 100-18b). For differentiation into astrocytes, the cells were cultured for 4 d in the medium containing 10% heat-inactivated fetal bovine serum (WelGene, S001-01). For differentiation into oligodendrocytes, the cells were cultured for 6 d in oligodendrocyte differentiation medium. The protocol and detailed information of medium have been previously described in our report.23
Immunocytochemistry
Cells on glass coverslips were fixed in 4% paraformaldehyde for 15 min at room temperature and washed with Dulbecco's phosphate buffered saline (DPBS; Hyclone Laboratories, SH30378). Cells were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, X100) in DPBS for 10 min at room temperature and then blocked with 2% of diluted bovine serum albumin (Sigma-Aldrich, A8412) in DPBS. Cells were rinsed and incubated in primary antibody solution overnight at 4°C. After washing in DPBS, cells were incubated in secondary antibody for 1 h at room temperature. Antibodies and their dilutions are listed in Table S3.
RT-PCR and quantitative RT-PCR
We used an RNeasy Kit (Qiagen, 74104) to extract total RNA following the supplier's instructions. Total RNA (1 μg) was reverse-transcribed into cDNA using an Omniscript RT Kit (Qiagen, 205110) following the manufacturer's protocol. MEFs were used as a negative control to assess the expression of NSC-specific markers on NSCs. All RT-PCR reactions used Ex Taq Polymerase (TaKaRa, RR001) and were performed for 30 cycles for all markers except for Nestin, for which 27 cycles were used. The primers used to amplify cDNA samples are listed in Table S4. Imprinted gene expression levels were evaluated by quantitative RT-PCR using a 7500 Real-Time PCR System (Applied Biosystems) and SYBR Green (Thermo Scientific, K0221). Primer sequences are listed in Table S4.
Genomic DNA isolation and bisulfite treatment
Genomic DNA was isolated using a G-spin Total DNA Extraction Kit (iNtRON, 17045). Genomic DNA (1 μg) was modified using an EpiTect Bisulfite Kit (Qiagen, 59104) according to the manufacturer's instructions.
DNA methylation sequencing analysis
The modified DNA was amplified by nested PCR with primers listed in Table S4. All reactions used Ex Taq Polymerase. PCR reactions were performed by preheating the mixtures at 94°C for 2 min followed by 40 cycles of 94°C for 30 s, 50–60°C for 30 s, and 72°C for 30 s. The amplified products were purified using a QIAquick Gel Extraction Kit (Qiagen, 28704) and subcloned into the TA cloning vector (pCR 2.1; Invitrogen, K2020). Individual clones were sequenced using an M13 forward primer. The data were visualized and aligned using QUMA (Quantification tool for Methylation Analysis; http://quma.cdb.riken.jp/).
Pyrosequencing analysis
The modified DNA was amplified by PCR with primer sets that included biotinylated reverse primers. For Mest, commercial primer sets (Qiagen, 978746) were used. The H19 and Snrpn reactions were performed in a PCR mixture (total volume 25 μl) containing 0.5 mM oligonucleotides and 12.5 μl of HotStarTaq Master Mix (Qiagen, 203603). Initial denaturation was performed at 95°C for 15 min and was followed by 45 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s, and extension at 72°C for 5 min. The Ndn reaction was performed with Ex Taq Polymerase as follows: denaturation at 94°C for 4 min 30 s, followed by 45 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 30 s, and extension at 72°C 7 min. Biotinylated PCR products were purified using streptavidin-Sepharose beads (GE Healthcare, 17-5113-01) and sequenced using the PyroMark Gold Q96 reagent (Qiagen, 972804) with the sequencing primers listed in Table S4. All samples were analyzed in triplicate.
Global gene expression and microarray analysis
Total RNA was prepared using Qiagen RNeasy columns according to the manufacturer's instructions. Hybridization to Illumina WG-6 BeadChips was conducted using standard Illumina protocols. Raw expression data was normalized using quantile normalization implemented in the limma package in R/Bioconductor. Hierarchical clustering with complete linkage was augmented by a heatmap generated using the gplots package in R. A linear regression model was applied to the normalized data (log2 scale) to assess the linear correlation between two samples.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (grant number HI12C0337 (A120392)) and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (grant number 2014007212 and 2014R1A2A2A01007212).
References
- 1.Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol 1999; 154:635-47; PMID:10079240; http://dx.doi.org/ 10.1016/S0002-9440(10)65309-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genetics 2001; 2:21-32; PMID:11253064; http://dx.doi.org/ 10.1038/35047554 [DOI] [PubMed] [Google Scholar]
- 3.Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Medical Genetics 2000; 97:136-46; PMID:11180221; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 4.Brown KW, Villar AJ, Bickmore W, Clayton-Smith J, Catchpoole D, Maher ER, Reik W. Imprinting mutation in the Beckwith-Wiedemann syndrome leads to biallelic IGF2 expression through an H19-independent pathway. Hum Mol Genetics 1996; 5:2027-32; PMID:8968759; http://dx.doi.org/ 10.1093/hmg/5.12.2027 [DOI] [PubMed] [Google Scholar]
- 5.Ko K, Tapia N, Wu G, Kim JB, Bravo MJ, Sasse P, Glaser T, Ruau D, Han DW, Greber B, et al.. Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell 2009; 5:87-96; PMID:19570517; http://dx.doi.org/ 10.1016/j.stem.2009.05.025 [DOI] [PubMed] [Google Scholar]
- 6.Ko K, Arauzo-Bravo MJ, Kim J, Stehling M, Scholer HR. Conversion of adult mouse unipotent germline stem cells into pluripotent stem cells. Nat Protoc 2010; 5:921-8; PMID:20431537; http://dx.doi.org/ 10.1038/nprot.2010.44 [DOI] [PubMed] [Google Scholar]
- 7.Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al.. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 2005; 3:e283; PMID:16086633; http://dx.doi.org/ 10.1371/journal.pbio.0030283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MJ, Choi HW, Jang HJ, Chung HM, Arauzo-Bravo MJ, Scholer HR, Do JT. Conversion of genomic imprinting by reprogramming and redifferentiation. J Cell Sci 2013; 126:2516-24; PMID:23525019; http://dx.doi.org/ 10.1242/jcs.122754 [DOI] [PubMed] [Google Scholar]
- 9.Chang G, Liu S, Wang F, Zhang Y, Kou Z, Chen D, Gao S. Differential methylation status of imprinted genes in nuclear transfer derived ES (NT-ES) cells. Genomics 2009; 93:112-9; PMID:18948186; http://dx.doi.org/ 10.1016/j.ygeno.2008.09.011 [DOI] [PubMed] [Google Scholar]
- 10.Kim KP, Thurston A, Mummery C, Ward-van Oostwaard D, Priddle H, Allegrucci C, Denning C, Young L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res 2007; 17:1731-42; PMID:17989250; http://dx.doi.org/ 10.1101/gr.6609207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z, Fujiwara K, Minamide R, Hasegawa K, Yoshikawa K. Necdin controls proliferation and apoptosis of embryonic neural stem cells in an oxygen tension-dependent manner. J Neurosci 2013; 33:10362-73; PMID:23785149; http://dx.doi.org/ 10.1523/JNEUROSCI.5682-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanel ML, Wevrick R. Establishment and maintenance of DNA methylation patterns in mouse Ndn: implications for maintenance of imprinting in target genes of the imprinting center. Mol Cell Biol 2001; 21:2384-92; PMID:11259587; http://dx.doi.org/ 10.1128/MCB.21.7.2384-2392.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang FW, Han ZB, Deng CY, He HJ, Wu Q. Conservation of genomic imprinting at the NDN, MAGEL2 and MEST loci in pigs. Genes Genetic Sys 2012; 87:53-8; PMID:22531794; http://dx.doi.org/ 10.1266/ggs.87.53 [DOI] [PubMed] [Google Scholar]
- 14.Dinger TC, Eckardt S, Choi SW, Camarero G, Kurosaka S, Hornich V, McLaughlin KJ, Muller AM. Androgenetic embryonic stem cells form neural progenitor cells in vivo and in vitro. Stem Cells 2008; 26:1474-83; PMID:18369101; http://dx.doi.org/ 10.1634/stemcells.2007-0877 [DOI] [PubMed] [Google Scholar]
- 15.Li C, Chen Z, Liu Z, Huang J, Zhang W, Zhou L, Keefe DL, Liu L. Correlation of expression and methylation of imprinted genes with pluripotency of parthenogenetic embryonic stem cells. Hum Mol Genetics 2009; 18:2177-87; PMID:19324901; http://dx.doi.org/ 10.1093/hmg/ddp150 [DOI] [PubMed] [Google Scholar]
- 16.Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 1994; 120:2933-43; PMID:7607083 [DOI] [PubMed] [Google Scholar]
- 17.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 1991; 64:849-59; PMID:1997210; http://dx.doi.org/ 10.1016/0092-8674(91)90513-X [DOI] [PubMed] [Google Scholar]
- 18.Pedone PV, Cosma MP, Ungaro P, Colantuoni V, Bruni CB, Zarrilli R, Riccio A. Parental imprinting of rat insulin-like growth factor II gene promoters is coordinately regulated. J Biol Chem 1994; 269:23970-5; PMID:7929045 [PubMed] [Google Scholar]
- 19.Plasschaert RN, Bartolomei MS. Genomic imprinting in development, growth, behavior and stem cells. Development 2014; 141:1805-13; PMID:24757003; http://dx.doi.org/ 10.1242/dev.101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genetics 1997; 17:75-8; PMID:9288101; http://dx.doi.org/ 10.1038/ng0997-75 [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genetics 2003; 12:837-47; PMID:12668607; http://dx.doi.org/ 10.1093/hmg/ddg106 [DOI] [PubMed] [Google Scholar]
- 22.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res 1996; 92:91-100; PMID:8861727; http://dx.doi.org/ 10.1016/0165-3806(95)00209-X [DOI] [PubMed] [Google Scholar]
- 23.Lee S, Lee HJ, Hwang HS, Ko K, Han DW, Ko K. Optimization of Matrigel-based culture for expansion of neural stem cells. Anim Cells Syst 2015; 19:175-80; http://dx.doi.org/ 10.1080/19768354.2015.1035750 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


