ABSTRACT
The RAB class of small GTPases includes the major regulators of intracellular communication, which are involved in vesicle generation through fusion and fission, and vesicular trafficking. RAB proteins also play an imperative role in neuronal maintenance and survival. Recent studies in the field of neurodegeneration have also highlighted the process of autophagy as being essential for neuronal maintenance. Here we review the emerging roles of RAB proteins in regulating macroautophagy and its impact in the context of neurodegenerative diseases.
KEYWORDS: autophagy, endocytosis, GTPase, intracellular trafficking, neurodegeneration, neuronal maintenance, RAB proteins, stress response
Intracellular trafficking is essential for neurons—a specialized type of cell that has long axonal processes that extend up to a few meters long—for maintaining polarity, stimulus dependent receptor uptake and degradation, and for axonal transport of organelles. RAB (Ras-genes from rat brain) GTPases are critical regulators that provide identity to vesicles and aid in the formation, movement and fusion of their respective membranes. RAB proteins thus play an indispensable role in neuronal processes such as NTF/neurotrophin-mediated signaling1 and thus consequently in neurite outgrowth.2 Another such active molecular process regulated by RAB proteins is macroautophagy (hereafter autophagy) a cellular catabolic process essential for neuronal maintenance. Defects in this pathway are strongly associated with several neurodegeneration disorders.3 The process of autophagy involves the formation of a double-layered membrane that engulfs the cellular contents to be degraded, followed by the fusion of the double-layered vesicle with a lysosome for cargo degradation. Classically, autophagy was viewed as being induced upon starvation, but recent literature highlights its importance as a housekeeping mechanism for clearing damaged organelles and potentially toxic protein aggregates. Thus, its requirement in cells of neuronal origin becomes important, and recent discoveries have shed light on various regulators, including a few on the role of a number of RAB proteins involved in multiple steps of autophagy.4 A number of studies also suggest the importance of RAB proteins in maintenance of healthy neurons, and how an overdose of RAB might reverse the phenotype inflicted by accumulation of misfolded proteins.5 Figure 1 provides a schematic view on the proposed role of the core autophagic machinery and its regulation by RAB proteins. In this article, we attempt to discuss isolated studies that ascribe aberration of this RAB-mediated autophagic regulation on the pathobiology of neurodegenerative diseases.
Figure 1.
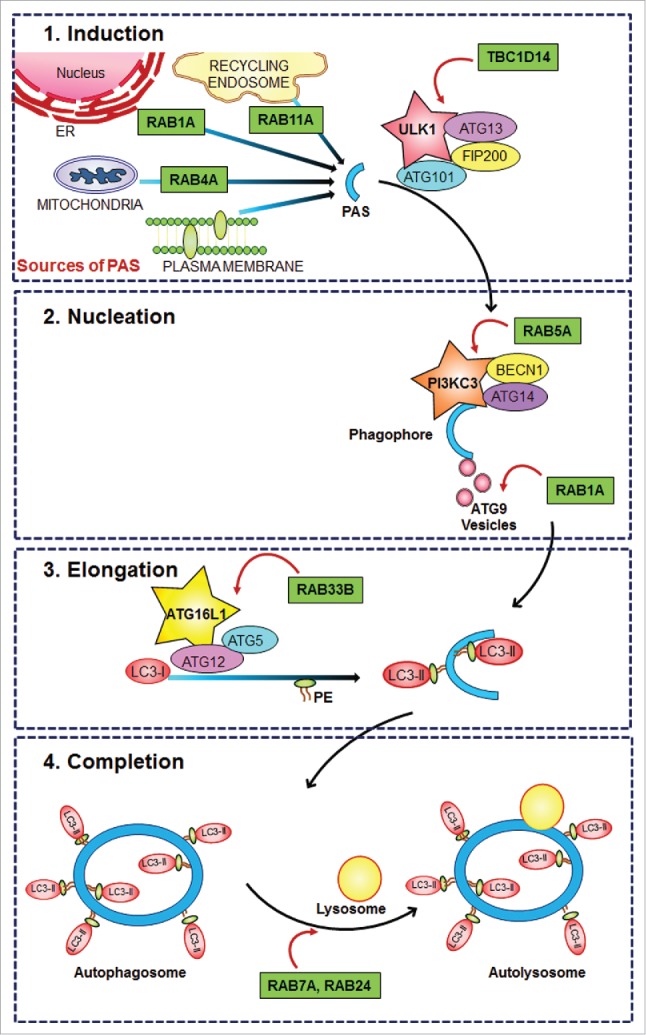
Core autophagic machinery and its regulation by RAB proteins. Schematic diagram showing the role of the core autophagic machinery and its regulation by RAB proteins. 1. Induction: The initial phagophore assembly site (PAS) could be contributed to by any of the multiple organelles, each requiring a set of proteins regulated by RAB activity. The signal is most often under the control of ULK1, the activity of which can also be regulated by RAB accessory proteins. 2. Nucleation: At the PAS, binds the initial set of proteins that help in membrane elongation/expansion, mediated by activated lipid (PtdIns3P), to which bind proteins such as PIK3C3/VPS34, that is a direct effector of activated RAB5. 3. Elongation: The ubiquitin-like conjugation machinery of ATG12–ATG5–ATG16L1 is also under the control of an ER resident RAB, RAB33B. 4. Completion: The fusion of the final double-layered autophagosome with the acidic lysosome for degradation of cargo is also under the control of activity of the late endosome-specific RAB7A and RAB24. Protein names in star-shaped boxes represent the proteins that are directly regulated by the respective RAB proteins.
RAB-regulated autophagy in neuronal survival
An early report suggested the requirement of RAB24 for survival of injured neurons.6 Both Rab24 and Map1lc3b/Lc3 gene transcripts were upregulated, and the 2 proteins colocalized in injured motor neurons as well as under proteotoxic stress induced by proteasomal blockade. The authors postulated RAB24-mediated autophagy to be required for nerve regeneration.6 A similar, activity-dependent relocalization of RAB24 was observed on pharmacological induction of autophagy, although in CHO cells.7 Recent studies have elegantly shown the role of RAB24 in autophagic clearance.8 Through the use of electron microscopy, it was suggested that it is not during the formation of the autophagosome that RAB24 plays a role. In fact, an early study had hinted at its role in maturation of autophagosomes as they reported the presence of RAB24 on late endocytic compartments.9 Overexpression of mutant HTT (huntingtin) protein with an expanded polyQ repeat in a defective RAB24 background leads to the accumulation of polyglutamine aggregates in HeLa cells.8 Thus, it would be safe to conclude that RAB24 not only plays a role in basal autophagy, but also helps in clearing the protein load through autophagy, thereby highlighting its importance in post-mitotic cell types, such as neurons. Supporting this finding was the genetic study of canine heredity ataxia that ascribed mutations in RAB24 to be a cause of the disease.10 The authors also described Purkinje neurons harboring a number of uncleared late autophagic vesicles.10 Thus, RAB24-mediated autophagy induction might protect neurons against cell death.
Abnormal RABs in neurodegenerative diseases
The importance of RABs in neuronal survival and maintenance became obvious with the identification of mutations in the RAB7A gene, which were found to be associated with the Charcot-Marie-Tooth disease type 2B, an axonal neuropathy.11 Cultured Purkinje neurons require the activity of RAB7A for efficient clearance of autophagosomes on trophic factor depletion.12 Specifically, RAB7A facilitates the crosstalk between endosomes and autophagosomes. An example of this crosstalk is the compensatory clearance of cargo by the other pathway if one is blocked. In the retina of Drosophila, for example, upregulation of RAB7A protein induced on blocking autophagy leads to compensatory degradation of ninaE/rhodopsin through endo-lysosomal degradation, thus preventing retinal degeneration.13 The significance of increased levels of RAB4A, RAB5A, RAB7A and RAB27A gene transcripts seen in brains of patients with Alzheimer disease (AD), or mild cognitive impairment is yet to be understood.14 Upregulation of RAB4A in AD, might be a consequence of AD-associated mitochondrial damage, as RAB4A is required for mitochondrial clearance in HeLa cells.15 RAB5A protein levels are also upregulated in mice models of Lafora disease, a neurodegenerative disorder associated with polyglucosan inclusions.16 Such a compensatory upregulation of RAB in pathologies such as Lafora disease could also be speculated. In brains of patients with dementia with Lewy bodies, large RAB7A-positive endosomes are seen, as well as increases in the protein level.17 The upregulation of RAB7A in such conditions could possibly represent a failed attempt for neuronal survival since MTOR (mechanistic target of rapamycin [serine/threonine kinase])-dependent autophagy induction (using a rapamycin analog) in a murine model of tauopathy also leads to an increase in RAB7A levels.18 A list of RAB proteins having a role in autophagy and mutated or misexpressed in neurodegenerative diseases is provided inTable 1.
Table 1.
List of neurodegenerative disorders with abnormal activities for RAB GTPase.
| Alzheimer disease (AD) and mild | RAB4A: Required for mitochondrial autophagy.14 |
| RAB5A: Regulates activity of PIK3C3-BECN1 complex.14 | |
| cognitive impairment (MCI) | RAB7A: Required for autophagosome-lysosome fusion.14 |
| RAB27A: Transcripts were upregulated in brains of AD and MCI patients.14 | |
| RAB4 and RAB6A are upregulated in the triple-transgenic mouse model of AD.33 | |
| Amyotrophic lateral sclerosis (ALS) | RAB1A: Involved in endoplasmic reticulum-mediated autophagosome formation, and accumulated as inclusions in neurons of sporadic ALS patients.31 |
| RAB1A: Overexpression rescued inclusion formation in mouse model of ALS.32 | |
| Charcot-Marie-Tooth type 2B (CMT2B) | RAB7A: Mutations resulting in its enhanced activity are a causative factor for familial CMT2B.11 |
| Dementia with Lewy bodies (DLB) | RAB7A: Levels are increased in brains of patients with DLB.17 |
| Huntington disease (HD) | RAB11A: Involved in recycling endosome-mediated autophagosome formation, interacts with HTT and is inactive in knock-in mouse model of HD.34 |
| Lafora disease (LD) | RAB5A: Required for autophagosome formation, is upregulated in mouse model of LD.16 |
Note. MCI, mild cognitive impairment.
RAB regulators and their role in neurodegeneration
Functional studies on disease-associated proteins have led to the identification of novel regulators of RAB proteins and their involvement in autophagy. For example, wild-type HTT is required for movement of RAB7A-positive vesicles through neurons of a Drosophila model of Huntington disease.19 Any perturbation of HTT activity might thus impact RAB7A-mediated autophagy, and could be a cause of buildup of uncleared autophagosomes in Huntington disease. Another instance where a disease-associated protein regulates autophagy through RAB activity is that of LRRK1 (leucine-rich repeat kinase 1) involved in Parkinson disease (PD). Upon autophagic induction, LRRK1 mediates RAB7A GTPase activity through the regulation of TBC1D2—a RAB7A GTPase activating protein—and a loss in its activity leads to accumulation of autolysosomes and undegraded cargo.20 PARK2/Parkin, mutations in which are also associated with PD, also regulates the activity of the RAB-GTPase activating proteins TBC1D15 and TBC1D17, which are required for routing of damaged mitochondria to autophagosomes through RAB7.21 These proteins thus have an impact on the hydrolytic activity of RAB GTPases and thus might have a functional outcome on autophagy.
Crosstalk of ATG and RAB proteins
The biogenesis of the autophagosome requires the formation of double-membrane vesicles. Yet the aqueous intracellular environment would not favor de novo synthesis of a hydrophobic membrane. Therefore, the autophagosomal membrane precursor, the phagophore, is thought to originate from an intracellular organelle or a vesicle, onto which ATG proteins bind and induce membrane elongation/expansion. The organelle that contributes to the phagophore membrane in mammalian cells has been an enigma for a number of years. Recent discoveries, however, have established the contribution of almost all intracellular organelles. The different membrane sources reported for the biogenesis of the phagophore include the plasma membrane, recycling endosomes, endoplasmic reticulum, mitochondria and Golgi body.22 One can assume synaptic vesicles to be such a membrane donor as they employ RAB26-dependent autophagy elongation molecules: ATG16L1 and MAP1LC3B.23 In this regard, it is also interesting to note that early endosomal RAB5A-induced autophagy can lead to clearance of HTT aggregates, albeit in cellular and fly models of Huntington disease.24 The interaction between BECN1/Beclin 1, RAB5A, and PIK3C3/VPS34 is required for both the processes of autophagy as well as endocytosis.25 Based on this evidence we propose the partitioning of RAB5A into the autophagic elongation and early endosomal compartments, and channeling into each depending upon the specific cellular requirement. Thus, the RAB5A-decorated enlarged endosomes observed in models and patients of Alzheimer disease could be a consequence of decreased autophagy; in addition leading to enhanced generation of amyloid beta.26
RAB proteins rescue neurodegeneration by inducing autophagy
The possibility that RAB proteins could serve as stress response molecules comes from a study that demonstrated enhanced extracellular secretion of SNCA/alpha-synuclein through a RAB11A-dependent mechanism following an autophagy block.27 These examples do not come as a surprise, as overexpression of RAB proteins alleviates neurodegeneration in multiple models. For example overexpression of RAB11A in Drosophila models of Parkinson and Huntington diseases leads to a decrease of SNCA aggregates, reduced neuronal death and synaptic dysfunction.28,29 Since RAB11A plays a positive role in autophagic maturation,30 one could speculate that the alleviation of symptom is an outcome of induced autophagy.
Development of disease could be a consequence of RAB1A misfolding and inactivation as was seen in a model of amyotrophic lateral sclerosis.32 Mutant FUS (FUS RNA binding protein) is responsible for one of the familial forms of amyotrophic lateral sclerosis, and impairs autophagy. The impaired autophagic phenotype is reversed by overexpression of RAB1A in a mutant FUS background,34 thus underscoring the nexus between RABs, autophagy and neurodegeneration. Further studies on RAB proteins and RAB regulators in neurodegenerative disorders could offer insight into the complex process of autophagy and its role in neurodegeneration.
Abbreviations
- AD
Alzheimer disease
- ATG
autophagy related
- DLB
dementia with lewy bodies
- GTPase
guanosine-5′-triphosphatase
- HD
Huntington disease
- HTT
Huntingtin
- LRRK1
leucine-rich repeat kinase 1
- PAS
phagophore assembly site
- PD
Parkinson disease
- RAB
Ras related protein in brain
- TBC
Tre-2, Bub2 and Cdc16 domain containing protein
- ULK1
unc-51 like autophagy activating kinase 1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank our colleague Ms. Neha Vithani for her help with the figure.
Funding
Work on autophagy in our laboratory is supported by a sponsored research grant from the Science and Engineering Research Board, Department of Science & Technology, Govt. of India (to SG).
References
- [1].Bucci C, Alifano P, Cogli L. The role of RAB proteins in `neuronal cells and in the trafficking of neurotrophin receptors. Membranes 2014; 4:642–77; PMID:25295627; http://dx.doi.org/ 10.3390/membranes4040642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Villarroel-Campos D, Gastaldi L, Conde C, Caceres A, Gonzalez-Billault C. RAB-mediated trafficking role in neurite formation. J Neurochem 2014; 129:240–8; PMID:24517494; http://dx.doi.org/ 10.1111/jnc.12676 [DOI] [PubMed] [Google Scholar]
- [3].Kiriyama Y, Nochi H. The Function of Autophagy in Neurodegenerative Diseases. Int J Mol Sci 2015; 16:26797–812; PMID:26569220; http://dx.doi.org/ 10.3390/ijms161125990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ao X, Zou L, Wu Y. Regulation of autophagy by the RAB GTPase network. Cell Death Differ 2014; 21:348–58; PMID:24440914; http://dx.doi.org/ 10.1038/cdd.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].D'Adamo P, Masetti M, Bianchi V, More L, Mignogna ML, Giannandrea M, Gatti S. RAB GTPases and RAB-interacting proteins and their role in the control of cognitive functions. Neurosci Bio Behav Rev 2014; 46 Pt 2:302–14 [DOI] [PubMed] [Google Scholar]
- [6].Egami Y, Kiryu-Seo S, Yoshimori T, Kiyama H. Induced expressions of RAB24 GTPase and LC3 in nerve-injured motor neurons. Biochem Biophys Res Commun 2005; 337:1206–13; PMID:16236257; http://dx.doi.org/ 10.1016/j.bbrc.2005.09.171 [DOI] [PubMed] [Google Scholar]
- [7].Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-RAB24. Traffic 2002; 3:472–82; PMID:12047555; http://dx.doi.org/ 10.1034/j.1600-0854.2002.30704.x [DOI] [PubMed] [Google Scholar]
- [8].Yla-Anttila P, Mikkonen E, Happonen KE, Holland P, Ueno T, Simonsen A, Eskelinen EL. RAB24 facilitates clearance of autophagic compartments during basal conditions. Autophagy 2015; 11:1833–48; PMID:26325487; http://dx.doi.org/ 10.1080/15548627.2015.1086522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Olkkonen VM, Dupree P, Killisch I, Lutcke A, Zerial M, Simons K. Molecular cloning and subcellular localization of three GTP-binding proteins of the RAB subfamily. J Cell Sci 1993; 106(Pt 4):1249–61; PMID:8126105 [DOI] [PubMed] [Google Scholar]
- [10].Agler C, Nielsen DM, Urkasemsin G, Singleton A, Tonomura N, Sigurdsson S, Tang R, Linder K, Arepalli S, Hernandez D, et al.. Canine hereditary ataxia in old english sheepdogs and gordon setters is associated with a defect in the autophagy gene encoding RAB24. PLoS Genet 2014; 10:e100391; http://dx.doi.org/ 10.1371/journal.pgen.1003991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bucci C, De Luca M. Molecular basis of Charcot-Marie-Tooth type 2B disease. Biochem Soc Trans 2012; 40:1368–72; PMID:23176482; http://dx.doi.org/ 10.1042/BST20120197 [DOI] [PubMed] [Google Scholar]
- [12].Bains M, Zaegel V, Mize-Berge J, Heidenreich KA. IGF-I stimulates RAB7-RILP interaction during neuronal autophagy. Neurosci Lett 2011; 488:112–7; PMID:20849920; http://dx.doi.org/ 10.1016/j.neulet.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Midorikawa R, Yamamoto-Hino M, Awano W, Hinohara Y, Suzuki E, Ueda R, Goto S. Autophagy-dependent rhodopsin degradation prevents retinal degeneration in Drosophila. J Neurosci 2010; 30:10703–19; PMID:20702701; http://dx.doi.org/ 10.1523/JNEUROSCI.2061-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ginsberg SD, Mufson EJ, Alldred MJ, Counts SE, Wuu J, Nixon RA, Che S. Upregulation of select RAB GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer's disease. J Chem Neuroanat 2011; 42:102–10; PMID:21669283; http://dx.doi.org/ 10.1016/j.jchemneu.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Talaber G, Miklossy G, Oaks Z, Liu Y, Tooze SA, Chudakov DM, Banki K, Perl A. HRES-1/RAB4 promotes the formation of LC3(+) autophagosomes and the accumulation of mitochondria during autophagy. PloS One 2014; 9:e84392; PMID:24404161; http://dx.doi.org/ 10.1371/journal.pone.0084392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Puri R, Suzuki T, Yamakawa K, Ganesh S. Dysfunctions in endosomal-lysosomal and autophagy pathways underlie neuropathology in a mouse model for Lafora disease. Hum Mol Genet 2012; 21:175–84; PMID:21965301; http://dx.doi.org/ 10.1093/hmg/ddr452 [DOI] [PubMed] [Google Scholar]
- [17].Higashi S, Moore DJ, Minegishi M, Kasanuki K, Fujishiro H, Kabuta T, Togo T, Katsuse O, Uchikado H, Furukawa Y, et al.. Localization of MAP1-LC3 in vulneRABle neurons and Lewy bodies in brains of patients with dementia with Lewy bodies. J Neuro Pathol Exp Neurol 2011; 70:264–80; PMID:21412173; http://dx.doi.org/ 10.1097/NEN.0b013e318211c86a [DOI] [PubMed] [Google Scholar]
- [18].Frederick C, Ando K, Leroy K, Heraud C, Suain V, Buee L, Brion JP. Rapamycin ester analog CCI-779/Temsirolimus alleviates tau pathology and improves motor deficit in mutant tau transgenic mice. J Alzheimers Dis 2015; 44:1145–56; PMID:25408212 [DOI] [PubMed] [Google Scholar]
- [19].White JA 2nd, Anderson E, Zimmerman K, Zheng KH, Rouhani R, Gunawardena S. Huntingtin differentially regulates the axonal transport of a sub-set of RAB-containing vesicles in vivo. Hum Mol Genet 2015; 24:7182–95; PMID:26450517; http://dx.doi.org/ 10.1093/hmg/ddv415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Toyofuku T, Morimoto K, Sasawatari S, Kumanogoh A. Leucine-Rich Repeat Kinase 1 Regulates Autophagy through Turning On TBC1D2-Dependent RAB7 Inactivation. Mol Cell Biol 2015; 35:3044–58; PMID:26100023; http://dx.doi.org/ 10.1128/MCB.00085-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial RAB GAPs govern autophagosome biogenesis during mitophagy. ELife 2014; 3:e01612; PMID:24569479; http://dx.doi.org/ 10.7554/eLife.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep 2011; 3:25; PMID:22162728; http://dx.doi.org/ 10.3410/B3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Binotti B, Pavlos NJ, Riedel D, Wenzel D, Vorbruggen G, Schalk AM, Kuhnel K, Boyken J, Erck C, Martens H, et al.. The GTPase RAB26 links synaptic vesicles to the autophagy pathway. Elife 2015; 4:e05597; PMID:25643395; http://dx.doi.org/ 10.7554/eLife.05597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. RAB5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649–60; PMID:18430781; http://dx.doi.org/ 10.1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McKnight NC, Zhong Y, Wold MS, Gong S, Phillips GR, Dou Z, Zhao Y, Heintz N, Zong WX, Yue Z. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet 2014; 10:e1004626; PMID:25275521; http://dx.doi.org/ 10.1371/journal.pgen.1004626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cataldo AM, Mathews PM, Boiteau AB, Hassinger LC, Peterhoff CM, Jiang Y, Mullaney K, Neve RL, Gruenberg J, Nixon RA. Down Syndrome Fibroblast Model of Alzheimer-Related Endosome Pathology: Accelerated Endocytosis Promotes Late Endocytic Defects. Am J Pathol 2008; 173:370–84; PMID:18535180; http://dx.doi.org/ 10.2353/ajpath.2008.071053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Poehler AM, Xiang W, Spitzer P, May VE, Meixner H, Rockenstein E, Chutna O, Outeiro TF, Winkler J, Masliah E, et al.. Autophagy modulates SNCA/α-synuclein release, thereby generating a hostile microenvironment. Autophagy 2014; 10:2171–92; PMID:25484190; http://dx.doi.org/ 10.4161/auto.36436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Breda C, Nugent ML, Estranero JG, Kyriacou CP, Outeiro TF, Steinert JR, Giorgini F. RAB11 modulates α-synuclein-mediated defects in synaptic transmission and behaviour. Hum Mol Genet 2015; 24:1077–91; PMID:25305083; http://dx.doi.org/ 10.1093/hmg/ddu521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Steinert JR, Campesan S, Richards P, Kyriacou CP, Forsythe ID, Giorgini F. RAB11 rescues synaptic dysfunction and behavioural deficits in a Drosophila model of Huntington's disease. Hum Mol Genet 2012; 21:2912–22; PMID:22466800; http://dx.doi.org/ 10.1093/hmg/dds117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Szatmari Z, Kis V, Lippai M, Hegedus K, Farago T, Lorincz P, Tanaka T, Juhasz G, Sass M. RAB11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Mol Biol Cell 2014; 25:522–31; PMID:24356450; http://dx.doi.org/ 10.1091/mbc.E13-10-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Soo KY, Halloran M, Sundaramoorthy V, Parakh S, Toth RP, Southam KA, McLean CA, Lock P, King A, Farg MA, et al.. RAB1-dependent ER-Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS. Acta Neuropathol 2015; 130:679–97; PMID:26298469; http://dx.doi.org/ 10.1007/s00401-015-1468-2 [DOI] [PubMed] [Google Scholar]
- [32].Soo KY, Sultana J, King AE, Atkinson RAK, Warraich ST, Sundaramoorthy V, Blair I, Farg MA, Atkin JD. ALS-associated mutant FUS inhibits macroautophagy which is restored by overexpression of RAB1. Cell Death Discov 2015; 1:15030; http://dx.doi.org/ 10.1038/cddiscovery.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Soejima N, Ohyagi Y, Nakamura N, Himeno E, Iinuma KM, Sakae N, Yamasaki R, Tabira T, Murakami K, Irie K, et al.. Intracellular accumulation of toxic turn amyloid-β is associated with endoplasmic reticulum stress in Alzheimer's disease. Curr Alzheimer Res 2013; 10:11–20; PMID:22950910 [PubMed] [Google Scholar]
- [34].Li X, Sapp E, Chase K, Comer-Tierney LA, Masso N, Alexander J, Reeves P, Kegel KB, Valencia A, Esteves M, et al.. Disruption of RAB11 activity in a knock-in mouse model of Huntington's Disease. Neurobiol Dis 2009; 36:374–83; http://dx.doi.org/ 10.1016/j.nbd.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]


