ABSTRACT
Many epigenetic association studies have attempted to identify DNA methylation markers in blood that are able to mirror those in target tissues. Although some have suggested potential utility of surrogate epigenetic markers in blood, few studies have collected data to directly compare DNA methylation across tissues from the same individuals. Here, epigenomic data were collected from adipose tissue and blood in 143 subjects using Illumina HumanMethylation450 BeadChip array. The top axis of epigenome-wide variation differentiates adipose tissue from blood, which is confirmed internally using cross-validation and externally with independent data from the two tissues. We identified 1,285 discordant genes and 1,961 concordant genes between blood and adipose tissue. RNA expression data of the two classes of genes show consistent patterns with those observed in DNA methylation. The discordant genes are enriched in biological functions related to immune response, leukocyte activation or differentiation, and blood coagulation. We distinguish the CpG-specific correlation from the within-subject correlation and emphasize that the magnitude of within-subject correlation does not guarantee the utility of surrogate epigenetic markers. The study reinforces the critical role of DNA methylation in regulating gene expression and cellular phenotypes across tissues, and highlights the caveats of using methylation markers in blood to mirror the corresponding profile in the target tissue.
KEYWORDS: Adipose tissue, blood, body mass index, DNA methylation, epigenomics, FASN (fatty acid synthase), HIF3A (hypoxia inducible factor 3, principal component analysis, surrogate markers
Introduction
Adipose tissue functions as a metabolic and endocrine organ that regulates dynamic storage of triglycerides and coordinates energy intake and usage.1 Blood is a mixture of numerous different specialized immune cell types, and the composition of the leukocyte population is well known to reflect particular phenotypic traits or external toxicant exposures.2 In the process of normal mammalian development, DNA methylation plays a critical role in controlling cell differentiation, such that different tissues have been shown to have distinct patterns of DNA methylation.3,4
In epigenetic epidemiology, it is often fairly straightforward to obtain blood samples in studies due to existing infrastructure to obtain those samples, and the utility of blood samples to assess many biomarkers across a large variety of disease domains. Epigenetics is inherently tissue-specific. Consequently, it is useful to know how methylation patterns are different and similar across tissues types, so that alterations in methylation in one tissue type (such as blood) can be more rigorously inferred to other tissues types (such as adipose tissue). Many studies to date have investigated methylation in single tissue types. For example, epigenome-wide association studies (EWAS) have reported that epigenetic alterations in adipose tissue are associated with metabolic process-related phenotypes such as type 2 diabetes,5,6 and may be modulated by physical activity.7,8 EWAS using blood DNA methylation showed associations of DNA methylation with immune-related conditions such as rheumatoid arthritis 9 and clinical parameters such as immunoglobulin E concentration.10 In addition, EWAS have been conducted to investigate associations of body mass index (BMI) or birth weight with DNA methylation in one tissue such as blood, and the candidate CpG sites were further examined in another tissue such as adipose tissue.11-16 Several epigenetic biomarkers associated with BMI were identified, such as DNA methylation of HIF3A.13 Some studies showed that corresponding methylation markers in peripheral blood have consistent associations as those discovered in adipose tissue. Overall, studies suggest some success of EWAS in discovering informative biomarker loci that may have potential of allowing blood to be used as a surrogate tissue for the epigenetics in adipose tissue. This calls for a more in-depth assessment of the nature of the DNA methylation profiles in different tissues from the same person. This should allow for more rigorous interpretation of surrogate tissue epigenetic methylation patterns. As epigenetic epidemiology emerges,17 the qualification and characteristics of epigenetic markers in peripheral blood need to be carefully assessed, with particular attention to the correlation with the target tissue and the phenotype of interest.
There are studies of tissue-specific DNA methylation directly comparing human brain with blood,18,19 adipose tissue with skeletal muscle,7 and blood with atrium tissue.20 In addition to pair-wise comparison between two tissues, tissue-specific methylation patterns have also been studied across multiple tissue types or primary cell lines from various tissues, including peripheral tissues, such as blood and buccal swabs, and internal tissues obtained with invasive procedures.21-25 However, despite the evidence of tissue-specific DNA methylation profiles, knowledge regarding the similarity and dissimilarity of adipose tissue and blood remains incomplete. Consequently, the question of whether DNA methylation in peripheral tissues can serve as a surrogate for the target internal tissues in relation to the phenotypic traits of interest is largely unanswered. We hypothesize that the DNA methylation profile varies across tissues and that the differentially methylated genes play important roles in tissue-specific biological functions. To investigate the hypotheses, we conduct a population-based study comparing epigenome-wide DNA methylation in adipose tissue with matched blood samples in healthy individuals.
Results
Epigenome-wide profile and tissue difference
The DNA methylation levels of 285,163 CpG sites are presented with a heatmap (Fig. 1). Hierarchical clustering well differentiated adipose tissue from blood, which strongly suggests more epigenome-wide similarity within tissue type than within a participant. In general, adipose tissue was hypomethylated compared with its blood counterpart. We further studied the epigenome-wide variation in DNA methylation with principal component analysis (PCA), which shows that the leading axis of epigenomic variation reflects the tissue type, even after adjusting for age, gender, race and BMI (P < 10−12) (Fig. 2A-B). The first three principal components explain 81.3%, 2.1%, and 0.9% of epigenome-wide variation, respectively.
Figure 1.
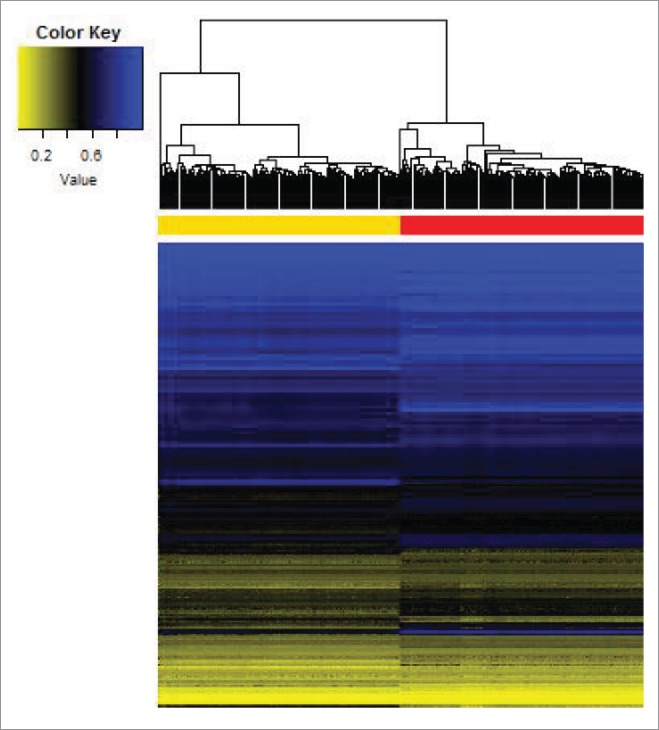
Epigenome-wide methylation level in 285,163 CpG loci characterizes tissue-specific profile across adipose tissue and blood. Hierarchical clustering is performed based on the Euclidean distance of epigenome-wide β−values, and well differentiates adipose tissue (yellow) from blood (red).
Figure 2.
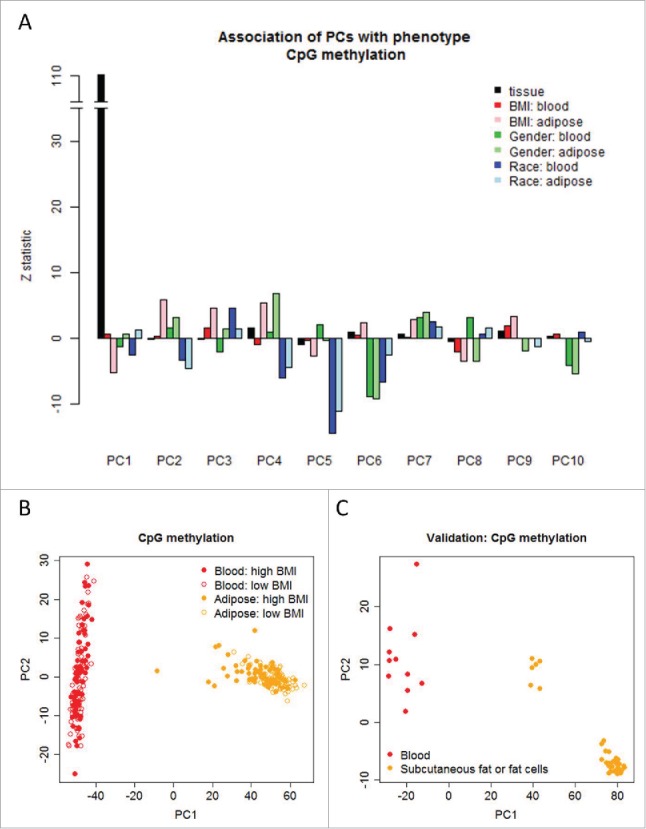
Top axes of epigenome-wide variation estimated by principal component analyses differentiate tissue type (adipose and blood), BMI, gender, and race. (a) Z-statistic characterizes the statistical association of the phenotypes with the top principal components (PCs). (b) The first PC differentiates adipose tissue from blood in the LEAP study. (c) The external validation study shows that the first PC predicted from the LEAP study differentiates adipose tissue from blood in the 47 samples archived on the GEO (GSE58622 and GSE48472).
The accurate differentiation was robust and confirmed by 4-fold cross-validation (Supplementary Fig. 1). In an effort to confirm that this is not simply attributable to a within-subject effect (e.g., it is not genetically controlled within the same individual), we sought to confirm this in another data set. The external data set 21,26 also validates the finding (Fig. 2C). Interestingly, the association of tissue type and the epigenome-wide variation was exclusively in the first principal component, but not the remaining principal components (Fig. 2A). PCA with median centering for each individual or each locus revealed similar findings (data not shown). The analyses based on average methylation of 20,073 genes are presented in Supplementary Figs. 2 and 3, which reveal similar findings compared to those using CpG methylation levels.
Figure 3.
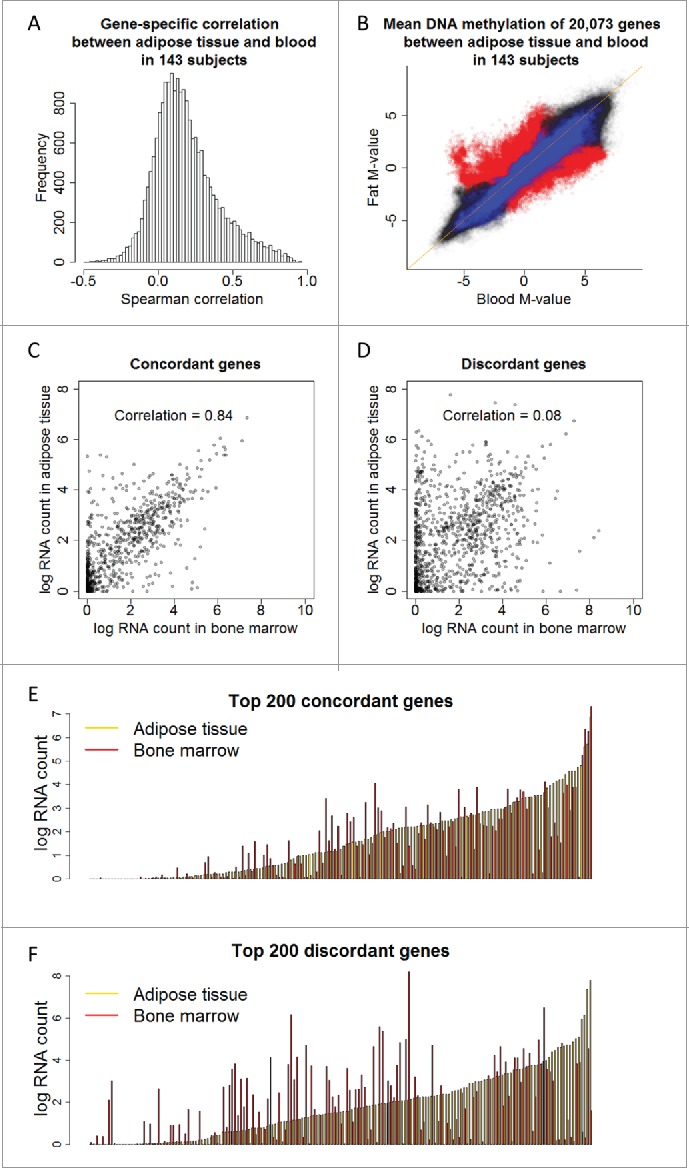
Concordant (1,966) and discordant (1,286) genes are identified using gene-specific correlation and PCA, respectively. (a) Histogram of gene-specific correlation indicates the majority of genes share low correlation across adipose tissue and blood. (b) DNA methylation M-values of 20,073 genes in 143 adipose tissues are plotted against those in the 143 matched blood samples, superimposed by the concordant genes (blue color) and discordant genes (red color). (c) RNA expression level of the concordant genes across adipose tissue and bone marrow shows high correlation, 0.84. (d) RNA expression level of the discordant genes across adipose tissue and bone marrow shows low correlation, 0.08. (e, f) RNA expression of the top 200 concordant (e) and discordant (f) genes sorted by the expression level of adipose tissue (orange).
Epigenome-wide variation, BMI, gender, and race
In order to further characterize the nature of the variation in the epigenome-wide data, we examined its association with BMI, gender, and race (Fig. 2A, Supplementary Fig. 4). The top four axes of variation were significantly associated with BMI and, notably, such association was only observed in adipose tissue and not in blood. The association with gender was observed in both blood and adipose tissue for the 6th, 7th, and 10th axes of CpG methylation variation, while the association with race was mostly observed in the 2nd, 4th, and 5th axes with consistent directionality in both tissues. The findings that the leading axes of epigenome-wide variation were associated with BMI, race, and gender were similar in analyses using gene-methylation (Supplementary Figs. 3 and 5).
Concordant and discordant genes
From the PCA of gene-average methylation, we extracted the loading of genes corresponding to the first axis. As the axis accurately differentiated adipose tissue from blood, the top 1,285 genes with largest loadings (the absolute value greater than two times the standard deviation of all loadings) were termed “discordant genes.” We calculated the gene-specific correlation between adipose tissue and blood in 143 subjects. The distribution of the correlation in 20,073 genes is shown in Fig. 3A. The majority of genes shared low correlation across the two tissue types. The 1,961 genes with a correlation greater than 0.5 across adipose tissue and blood were termed “concordant genes.” The methylation levels of all 20,073 genes (adipose tissue vs. blood) in 143 subjects are shown in Fig. 3B where concordant and discordant genes are highlighted in blue and red, respectively. The concordant genes followed closely on the diagonal line whereas the discordant genes spread over the off-diagonal region. Note the scatter plot represents an overall between-tissue correlation by collapsing all genes and subjects.
While we defined the concordant and discordant genes based on DNA methylation, we further assessed the corresponding transcriptomic pattern using previously published data.27 We compared the RNA expression of the two classes of genes in adipose tissue and bone marrow as a proxy tissue of blood. For the concordant genes, RNA expression in the two tissue types had a correlation of 0.84 (Fig. 3C and 3E), but the correlation across tissues for the discordant genes was very low, at 0.08 (Fig. 3D and 3F). In contrast, the correlation between fat tissue and thyroid was 0.71 and 0.47 for the concordant and discordant genes, respectively (Supplementary Fig. 6). Together, the analyses suggest that the concordant genes have constant gene expression pattern across tissues, but the discordant genes critical for tissue-specific biological functions have distinct epigenetic and transcription patterns. Compared with CpG-specific correlation between the adipose tissue and blood in all 285,163 sites (black bars), the CpG sites with higher correlation are enriched in concordant genes (blue bars), whereas the sites with lower correlation are enriched in discordant genes (red bars) (Supplementary Fig. 7).
Gene ontology analyses were performed to better characterize the biological features of concordant and discordant genes. The biological processes enriched in discordant genes were related to immune cell activation, lymphocyte differentiation, and blood coagulation (Table 1), and those enriched in concordant genes were related to cell adhesion and response to xenobiotic stimulus (Supplementary Table 1). The findings from GO analyses support the hypothesis that the discordant genes are critical for tissue-specific biological functions while the concordant genes are mostly responsible for maintaining basic cellular functions. While the Fisher exact test of over-representation is used to examine the enrichment of biological functions in two types of genes, we note that the P-values from such a test is based on a sampling scheme with respect to genes rather than study subjects28 and should be interpreted with caution.
Table 1.
Gene ontology enriched in discordant genes with P < 10−4.
| GOBPID* | P-Value | Odds Ratio | Expected Count | Observed Count | Size | Term |
|---|---|---|---|---|---|---|
| GO:0050900 | 3.76E-08 | 2.69 | 19.38 | 46 | 311 | leukocyte migration |
| GO:0006955 | 5.83E-08 | 1.74 | 83.68 | 132 | 1343 | immune response |
| GO:0006952 | 7.75E-08 | 1.71 | 88.98 | 138 | 1428 | defense response |
| GO:0050896 | 8.18E-08 | 1.43 | 452.50 | 531 | 7262 | response to stimulus |
| GO:0001775 | 1.64E-07 | 1.90 | 52.65 | 91 | 845 | cell activation |
| GO:0002376 | 1.68E-07 | 1.56 | 138.39 | 195 | 2221 | immune system process |
| GO:0060326 | 1.51E-06 | 2.83 | 12.84 | 32 | 206 | cell chemotaxis |
| GO:0045321 | 4.07E-06 | 1.91 | 38.82 | 68 | 623 | leukocyte activation |
| GO:0030595 | 5.18E-06 | 3.09 | 9.28 | 25 | 149 | leukocyte chemotaxis |
| GO:0050851 | 9.42E-06 | 3.14 | 8.41 | 23 | 135 | antigen receptor-mediated signaling pathway |
| GO:0097529 | 1.31E-05 | 3.28 | 7.42 | 21 | 119 | myeloid leukocyte migration |
| GO:0006954 | 1.43E-05 | 1.88 | 35.70 | 62 | 573 | inflammatory response |
| GO:0007155 | 1.55E-05 | 1.58 | 80.94 | 118 | 1299 | cell adhesion |
| GO:0022610 | 1.91E-05 | 1.57 | 81.32 | 118 | 1305 | biological adhesion |
| GO:0042129 | 2.20E-05 | 3.15 | 7.66 | 21 | 123 | regulation of T cell proliferation |
| GO:0042098 | 3.16E-05 | 2.80 | 9.66 | 24 | 155 | T cell proliferation |
| GO:0002683 | 5.17E-05 | 2.12 | 20.06 | 39 | 322 | negative regulation of immune system process |
| GO:0007166 | 5.31E-05 | 1.36 | 196.53 | 245 | 3154 | cell surface receptor signaling pathway |
| GO:0002429 | 5.70E-05 | 2.46 | 12.59 | 28 | 202 | immune response-activating cell surface receptor signaling pathway |
| GO:0070887 | 8.55E-05 | 1.40 | 141.32 | 183 | 2268 | cellular response to chemical stimulus |
| GO:0098542 | 9.54E-05 | 1.98 | 23.55 | 43 | 378 | defense response to other organism |
* GOBPID: gene ontology biological process id.
BMI and within-subject epigenome-wide correlation between adipose tissue and blood
For each subject, we calculated the correlation of DNA methylation between adipose tissue and blood, termed the within-subject correlation. As shown in Fig. 4A, the within-subject correlations between the two tissue types in all 143 subjects were all very high. BMI was significantly associated with the within-subject correlation of CpG methylation across the two tissues (P = 4.5×10−4; Fig. 4B). For the subjects with higher BMI, the DNA methylation between adipose tissue and blood was more consistent (Fig. 4C), while the epigenome-wide pattern in those with lower BMI tended to spread out with slightly lower correlation (Fig. 4D). The above analyses are based on CpG-site methylation, and the association based on average gene methylation was also highly significant (P = 6.3×10−6, Supplementary Fig. 8). Comparison of DNA methylation in adipose tissue and blood across the three BMI groups (≤25, 25–30 and ≥30) suggests that the DNA methylation profile of adipose tissue in subjects with higher BMI becomes more similar to that of blood, but not the reverse that blood DNA methylation becomes more similar to adipose tissue (Supplementary Fig. 9).
Figure 4.
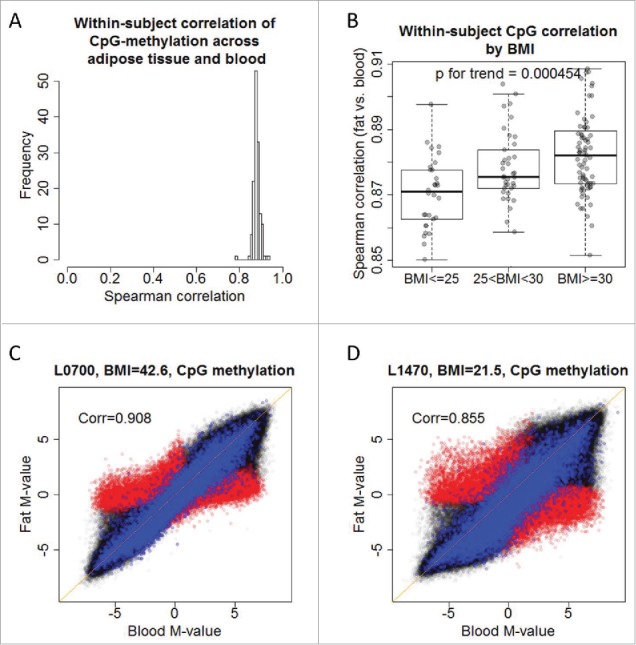
Body mass index is associated with within-subject correlation of CpG site methylation in 132 subjects. (a) DNA methylation across adipose tissue and blood shares high within-subject correlation of 285,163 CpG sites in 132 subjects. (b) Within-subject correlation of 285,163 CpG sites is associated with body mass index, P = 4.5×10−4. Subjects with higher BMI have high within-subject correlation across tissues (c,d) DNA methylation M-values of adipose tissue against those of blood in a subject with higher BMI (c) and low BMI (d). The scatter plots are superimposed by concordant (blue) and discordant (red) genes.
Blood DNA methylation as a surrogate in adipose tissue
With the available epigenome-wide data on both adipose tissue and blood from the same subjects, we evaluated whether the blood DNA methylation markers mirror those in adipose tissue. Specifically, we investigated whether the high within-subject correlation observed in Fig. 4A can be translated to qualification of blood DNA methylation as a surrogate marker for that in adipose tissue. We have introduced two different ways of examining interdependence between adipose tissue and blood: one was to focus on epigenome-wide correlation of all CpG loci or all genes within an individual, or within-subject correlation (Fig. 4A); and the other was on CpG- or gene-specific correlation across tissue types (Fig. 3A and Supplementary Fig. 7), which we argue is more relevant to most epigenetics association studies. The magnitude of CpG- or gene-specific correlation across adipose tissue and blood was not high. Only 5.2% of CpG loci and 9.8% of genes had correlation coefficients greater than 0.5. Although the magnitude of correlation was low, 19.0% CpG- and 38.9% gene-specific correlation was positively correlated and significantly different from 0 (FDR<0.05). Among these CpG loci and genes, if the modest correlation preserved across tissue types was associated with phenotypic traits of interest, then the blood DNA methylation may still serve as a surrogate marker for adipose tissue. We illustrated with the example of HIF3A in the following.
To better understand the discrepancy of high within-subject correlation and low gene-specific correlation, we illustrated with three genes, C21orf81, fatty acid synthase (FASN), and hypoxia inducible factor 3, alpha subunit (HIF3A). The three genes were selected to represent high, moderate, and low correlation of methylation across tissues. Among 20,073 genes, C21orf81 had the highest correlation (0.96) of average methylation levels between adipose tissue and blood, and HIF3A and FASN had correlation of 0.40 and −0.08, respectively. Despite their moderate to low correlation, studies have suggested that FASN is involved in regulation of body weight,29,30 and recent studies discovered DNA methylation of HIF3A is associated with body weight.12,13,15 We again looked at the scatter plot of all 20,073 genes in 143 subjects, but assessed how methylation levels of the three genes were distributed in the plot (Fig. 5A). The methylation level of C21orf81 ranged from 10 to 90% and had high correlation (0.96) between adipose tissue and blood. Methylation levels of HIF3A ranging from 45 to 57% in adipose tissue and blood had moderate correlation (0.40) across tissues. On the other hand, FASN had high methylation level (>95%), but the correlation between tissues was extremely low (−0.08). The correlation of CpG loci within these three genes revealed very similar results (Supplementary Figs. 10–12). We confirmed the previously reported findings that BMI was associated with HIF3A methylation in both adipose tissue (P = 8.9×10−5) and blood (P = 0.060) (Fig. 5D). The association of BMI with FASN was highly significant in adipose tissue (P = 1.8×10−10), but not in blood (P = 0.66) (Fig. 5B); BMI was not associated with C21orf81 methylation in either adipose tissue (P = 0.92) or blood (P = 0.91) (Fig. 5C). Similar findings were observed in CpG-specific analyses (Supplementary Figs. 13-15). The analyses suggest that the positively correlated epigenome-wide pattern does not mirror the correlation in specific genes or CpG loci, and that the qualification of being a surrogate epigenetic marker does not necessarily depend on the CpG-specific correlation and has to be evaluated case by case.
Figure 5.
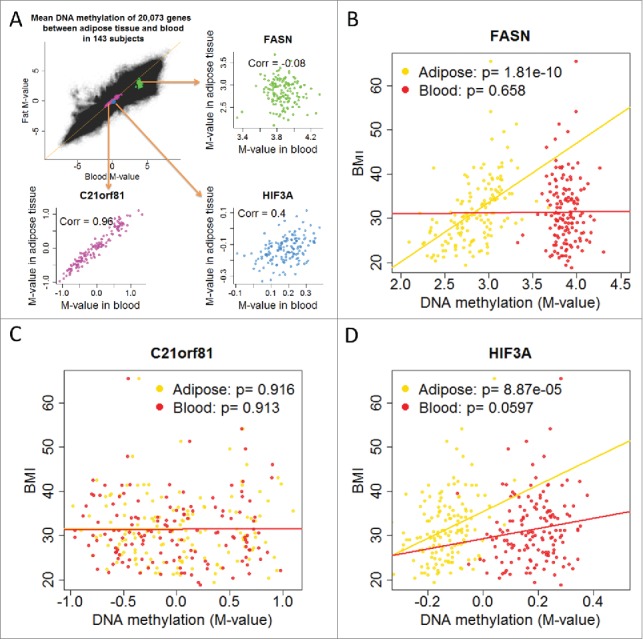
We illustrate gene-specific correlation and the association with BMI in adipose tissue and blood for three genes: HIF3A, FASN, and C21orf81. (a) DNA methylation M-values of 20,073 genes in 143 adipose tissues are plotted against those in the 143 matched blood samples, superimposed by DNA methylation levels of HIF3A, FASN, and C21orf81. The scatter plot reveals that a highly correlated epigenome-wide correlation does not necessarily suggest gene-specific correlation. (b) DNA methylation of FASN in adipose tissue is highly associated with BMI (P = 1.8×10−10), but such association does not exist in blood. (c) DNA methylation of C21orf81 in adipose tissue or blood is not associated with BMI. (d) DNA methylation of HIF3A is associated with BMI, which is highly significant in adipose tissue (P = 8.9×10−5) and marginally significant in blood (P = 0.060).
Discussion
We report, to our knowledge, the first large-scale comparison of epigenome-wide DNA methylation from adipose tissue and blood in the same people. These analyses reveal the lead axis of epigenome-wide variation is related to the difference between the two tissues and identify their concordant and discordant genes. We have interrogated the within-subject correlation and gene- or CpG-specific correlation and demonstrated the caveats and qualifications required when using blood EWAS data as surrogate markers for the target tissue. In addition to the large sample size, this study has several advantages. First, all the blood and fat samples are paired and collected from the same individuals. Second, the age of the study subjects is similar, ranging from 44 to 50 years. As age is shown to be associated with DNA methylation profile and gene expression,19,31 the narrow range minimizes its potential confounding. Third, both internal cross-validation and external validation studies were conducted and confirmed the finding that the first principal component of epigenome-wide variations differentiates adipose tissue from blood. Finally, the concordant and discordant genes identified in the study are supported by a consistent pattern in gene expression measured using RNA sequencing. The limitations of our work include the lack of information on cell decomposition and RNA profile for blood and adipose tissue, and the lack of adipose tissue from different anatomical sites. This limits our ability to conduct integrated analysis for both transcriptomics and epigenomics across a finer cellular phenotyping profile.
The discordant genes across adipose tissue and blood are enriched in biological processes related to immune response, leukocyte and lymphocyte activation or differentiation, and blood coagulation. The results support the distinct physiologic role of blood as an important discriminator in these data. Difference in DNA methylation profile across other tissues has also been studied. Distinct tissue-specific differentially methylated regions (TS-DMRs) between brain and blood were identified and the TS-DMRs were enriched near genes with biological functions related to neurodevelopment and neuronal differentiation.18 Another study found that the top axes of methylomic variations across blood and brain are related to the tissue type, anatomical regions of the brain, and age.19 In addition to comparison across non-pathological tissues, studies have also been conducted to examine the tumor and non-tumor tissues. Comparing soft-tissue sarcoma tumor or cell line and non-neoplastic fat samples, Rener et al. identified a set of CpG sites that differentiates the subphenotypes.32 A study reported a high epigenome-wide correlation (0.92) of cell-free serum DNA methylation and the matched DNA methylation in esophageal adenocarcinoma.33 An algorithm has also been developed to predict methylation profile across tissues.20 However, as shown in Fig. 5, the high epigenome-wide correlation is not directly translated to a high gene- or CpG site-specific correlation and therefore, does not necessarily guarantee clinical utility as surrogate biomarkers for the target tissue.
Since epigenomics is tissue-specific, there has been a pressing need to understand whether the DNA methylation profile in one tissue can serve as a useful surrogate for another. There are two major reasons for potentially using a surrogate: one is to investigate a biomarker in peripheral tissue that might predict a disease or a phenotypic trait. Because the target tissue for some diseases is not readily accessible, a surrogate marker in peripheral tissue such as blood enjoys many practical advantages. Many epigenome-wide association studies have adopted as a common practice that once an epigenetic biomarker for a phenotype is found in one tissue, replication using another tissue is then conducted.11-13,15 Data from the current study suggest that there may be limited utility to this approach. For example, the relationship of BMI with methylation profile in blood and adipose tissue is distinct; the DNA methylation profile of adipose tissue in subjects with higher BMI becomes more similar to that of blood. This finding is consistent with the known ability of adipose tissue in obese individuals to elaborate inflammatory cytokines, potentially mimicking similar properties of leukocytes.34 At the same time, as shown in Fig. 5, DNA methylation in HIF3A shares moderate correlation between adipose tissue and blood, and both are associated with BMI. In contrast, methylation in FASN is poorly correlated across tissues, but the DNA methylation in adipose tissue but not blood is highly associated with BMI. To serve as a biomarker, the surrogate marker has to share some correlation with the target DNA methylation, and the shared correlation has to be at least partially attributable to their association with the phenotype of interest. Since both conditions have to be satisfied, the concordant genes identified from the study do not guarantee the qualification of being a good surrogate. Moreover, the surrogate markers are phenotype-specific, e.g. a surrogate biomarker for BMI is not necessarily a marker for another trait such as asthma.
The other purpose for choosing a surrogate is to understand its biological mechanism of development. The concordant genes we identified serve this purpose. The methylation markers that are consistent across tissues reflect fundamental biological functions such as cell-cell adhesion and oxidation-reduction process. Some concordant genes may be simultaneously activated or silenced across tissues given their developmental stage.35 Although such type of surrogate markers are not necessarily related to a specific disease or phenotype, the markers may still be tissue specific: the concordant genes between blood and adipose tissue may not be identical to the ones between blood and another tissue.
It is well-acknowledged that DNA methylation profile in blood reflects the leukocyte composition,36 and algorithms were developed to account for the leukocyte mixture in the analysis.37 Adipose tissues of different anatomical origins have different gene expression.38 There is cell mixture even within adipose tissue, and genes are differentially expressed in different adipocytes 39 or within the same cell type.40 A statistical method has been proposed to adjust for latent classes of DNA methylation attributable to cell mixture.41 Hence, accounting for cellular heterogeneity within individual tissues is an important future direction for this research. It is plausible that differential developmental environments may alter the degree of heterogeneity within tissues across individuals. This difference could translate to differences in disease susceptibility or potentially even to therapeutic response in diseased tissues. DNA methylation can provide a window into the components of tissues (e.g. distinct immunophenotypes are known to have specific differentially methylated regions). This affords the unique opportunity to interrogate blood to define the profile of immune cells in any normal or disease individual using DNA methylation rather than flow cytometry, for example. Cell sorting in solid tissue or finer cellular phenotyping42 will provide better insight into the nature and importance of individual tissue heterogeneity.
The current work reinforces the fundamental nature of different DNA methylation profiles in different tissues and emphasizes the critical role of DNA methylation in regulating gene expression across tissues. We also believe this work highlights the limitations of using methylation markers in peripheral tissues such as blood to mirror the corresponding profile in the target tissue. At the same time, the cross-tissue epigenomic comparison may shed light on the developmental heterogeneity of individual tissues, providing a novel mechanism for disease susceptibility. This warrants further study, including assessing DNA methylation after tissue specific fine-phenotyping, anatomically, histologically, and temporally.
Methods
Study subjects of the LEAP project
The current study, termed the Longitudinal Effects on Aging Perinatal (LEAP) Project, was nested within the New England Family Study (NEFS),43 which comprised 17,921 offspring of pregnant women in the Collaborative Perinatal Project 44,45 from Providence, Rhode Island and Boston, Massachusetts, United States, recruited between 1959 and 1974. In the LEAP study, four hundred Providence-born participants were enrolled and assessed during 2010-11. Of these, 316 had adequate adipose tissue biopsy performed, 68 refused and 16 had inadequate biopsy specimens. Blood and adipose tissue DNA methylation analyses were performed on a representative sample of 143 of these 316 participants. The study protocol was approved by the institutional review boards at Brown University and Memorial Hospital of Rhode Island (#0908000028).
Collection of covariates and tissue samples
Body weight and height measures were obtained using a calibrated stadiometer by trained personnel, and converted into body mass index (kg/m2). Covariates of interest, including age, race, gender, socioeconomic status, were obtained for all subjects. Due to the small sample size of non-African American minorities, race was collapsed into dichotomous categories of white and non-white. DNA was extracted from 143 adipose tissues and the matched 143 blood samples. Whole blood samples were centrifuged to obtain buffy coat, and subcutaneous adipose tissue samples were collected from the upper outer quadrant of the buttock using a 16-gauge needle and disposable syringe. DNA was extracted from adipose tissue samples or buffy coat, using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) and the Zymo Genomic DNA Clean & Concentrator Kit, according to the manufacturers' protocol.
Methylation profiling
DNA was sodium bisulfite-converted using the EZ-96 DNA Methylation-Direct and EZ DNA Methylation-Direct kits (Zymo Research, Orange, CA), according to the manufacturer's instructions. Blood and adipose tissue samples were randomly distributed across 18 BeadChips on the plates, and analyzed using the Infinium HumanMethylation450 BeadChip array (Illumina, San Diego, CA) at the Genomics Core Facility at the UCSF Institute for Human Genetics (San Francisco, CA), according to the Illumina protocols for the Infinium platform.
Background correction and dye bias correction were performed using the methylumi package in R.46 Normalization was conducted using Beta-Mixture Quantile Dilation (BMIQ) approach.47 Batch effect was adjusted with linear mixed models. For each of the CpG sites, average β−values were calculated as , where and , respectively, are the signals from the probe corresponding to the methylated and unmethylated target CpG site. Prior to analyses, probes on sex chromosomes, not on CpG sites, or CpG sites with single nucleotide polymorphisms (dbSNP entries within 10 bp of the CpG cites) were excluded, followed by a further exclusion of CpG sites with variance of M-values (the logit-transformed values from β−values) in 286 samples less than the first quartile of all variances. After filtering, there were 285,163 CpG sites included in the analyses.
External validation data of DNA methylation
To validate our finding of methylomic difference between adipose tissue and blood, we also collected data from the public repository of genomic data, Gene Expression Omnibus (GEO). The data of Dahlman et al. (GSE58622) included 30 samples of fat cells collected from 30 post-obese or never-obese women.26 The data of Slieker et al. (GSE48472) contained blood and subcutaneous fat tissue samples from six cadavers within 12 hours postmortem (mean age 65.5 years) and blood samples from five healthy volunteers (mean age 28 years).21 In total, we had 36 subcutaneous adipose tissue or fat cell samples and 11 blood samples with available epigenomic data measured by Infinium HumanMethylation450 BeadChip array.
External validation data of gene transcriptomics
Fagerberg et al. collected RNA-seq data on tissue samples from 95 human individuals representing 27 tissues in order to determine tissue specificity of all protein coding genes.27 Due to lack of blood samples in the study, we used bone marrow samples as a proxy tissue for blood. We compared the average log-transformed RNA count +1 of concordant and discordant genes (defined in the following) between bone marrow and adipose tissues to confirm the findings from our methylomic analyses, and we also compared between bone marrow and thyroid tissue as a negative control. There were 4 bone marrow samples, 3 adipose tissue samples, and 4 thyroid samples. Sample mean was calculated within each tissue type to represent the average RNA expression of the tissue.
Statistical analyses
We conducted two sets of analyses: one for methylation of 285,163 CpG loci and the other for average methylation of 20,073 genes. Two sets of analyses provided complementary information. While CpG site-based analyses better reflect the current practices in conducting EWAS, gene-based analyses avoid the multiplicity issue within a gene and serve as an appropriate approach to investigate functional annotation for genes with tissue-specific methylation profile and to perform gene ontology analyses. CpG sites from 1500 bps of a transcription start site to 3′ untranslated region of a gene were mapped to the gene. Note we did not cover all genes because we filtered CpG loci with less variability and those on sex chromosomes. Hierarchical clustering was performed based on Euclidean distance of methylation levels of 285,163 CpG sites. Epigenome-wide variations of β-values were studied using principal component analysis (PCA).48 Association of the leading principal component with the tissue type was examined using generalize estimating equations 49 assuming a constant correlation between adipose tissue and blood within the same subject and adjusting for age, gender, race, BMI, cigarette smoking and socioeconomic index. To ensure robust findings, we conducted 4-fold cross-validation for PCA. The randomly selected 214 training samples were included for PCA, and the loading of the first principal component (PC) was then used to predict the first two PCs of the remaining 72 testing samples. The internal validation process was repeated over four complementary testing sets. For external validation, we performed PCA with all 286 samples and predicted the first two PCs and the tissue type of 11 blood samples and 36 samples from subcutaneous fat or fat cells.
Within-subject correlation was calculated for all 143 subjects using Spearman correlation: the correlation of all 285,163 CpG site methylation or 20,073 average gene methylation within each subject, between adipose tissue and blood. Gene-specific correlation was calculated for all 20,073 genes with Spearman correlation: the correlation of 143 methylation values in adipose tissue and the matched 143 blood methylation values for the same gene. CpG-specific correlation was calculated similar to the gene-specific correlation. Statistical significance was evaluated with P-value and false discovery rate (FDR).50 The association of leading principal components with BMI, gender and race was investigated within the tissue type using least square estimator adjusting for covariates described above. The association of methylation M-values of FASN, HIF3A, and C21orf81 with BMI was analyzed with least square estimator adjusting for above covariates. Enrichment of gene ontology (GO) 51,52 was examined for (1) discordant genes: the genes with the absolute value of loading for the first PC greater than two times of the standard deviation, and (2) concordant genes: the genes with gene-specific correlation greater than 0.5. The reason we did not define discordant genes based on the low correlation is that the majority of genes have pretty low correlation (Fig. 3A). A scatter plot of the loading versus correlation is shown in Supplementary Fig. 16, and the absolute values of the two have significant negative correlation, −0.076 (P<2.210−6). Furthermore, with the significant discrimination by PCA (Fig. 2), the discordant genes based on the loading of the first principal component should attain better specificity. GO enrichment analysis was performed using Fisher exact test.53 Statistical analyses including data preprocessing were performed using R 3.2.0.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health [R03CA182937 to Y.T.H; R01AG048825 and RC2AG036666 to E.B.L.]. Funding for open access charge: National Institutes of Health.
References
- 1.Eriksson JW, Smith U, Waagstein F, Wysocki M, Jansson PA. Glucose turnover and adipose tissue lipolysis are insulin-resistant in healthy relatives of type 2 diabetes patients: is cellular insulin resistance a secondary phenomenon? Diabetes 1999; 48:1572-8; PMID:10426375; http://dx.doi.org/ 10.2337/diabetes.48.8.1572 [DOI] [PubMed] [Google Scholar]
- 2.Murphy K, Travers P, Walport M, Janeway C. Janeway's immunobiology. New York: Garland: Science, 2012 [Google Scholar]
- 3.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem 2012; 81:97-117; PMID:22404632; http://dx.doi.org/ 10.1146/annurev-biochem-052610-091920 [DOI] [PubMed] [Google Scholar]
- 4.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al.. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 2009; 5:e1000602; PMID:19680444; http://dx.doi.org/ 10.1371/journal.pgen.1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, Poulsen P, Ribel-Madsen R, Pedersen NL, Almgren P, et al.. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 2014; 63:2962-76; PMID:24812430; http://dx.doi.org/ 10.2337/db13-1459 [DOI] [PubMed] [Google Scholar]
- 6.Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, Fernandez AF, Friedrichsen M, Vind BF, Hojlund K, et al.. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One 2012; 7:e51302; PMID:23251491; http://dx.doi.org/ 10.1371/journal.pone.0051302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronn T, Ling C. Effect of exercise on DNA methylation and metabolism in human adipose tissue and skeletal muscle. Epigenomics 2013; 5:603-5; PMID:24283873; http://dx.doi.org/ 10.2217/epi.13.61 [DOI] [PubMed] [Google Scholar]
- 8.Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, et al.. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 2013; 9:e1003572; PMID:23825961; http://dx.doi.org/ 10.1371/journal.pgen.1003572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, et al.. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 2013; 31:142-7; PMID:23334450; http://dx.doi.org/ 10.1038/nbt.2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang L, Willis-Owen SA, Laprise C, Wong KC, Davies GA, Hudson TJ, Binia A, Hopkin JM, Yang IV, Grundberg E, et al.. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature 2015; 520:670-4; PMID:25707804; http://dx.doi.org/ 10.1038/nature14125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol 2014; 44(4):1277-87; PMID:25541553; http://dx.doi.org/ 10.1093/ije/dyu236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, Hedman AK, Sandling JK, Li LA, Irvin MR, et al.. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet 2015; 24(15):4464-79; PMID:25935004; http://dx.doi.org/ 10.1093/hmg/ddv161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, et al.. DNA methylation and body-mass index: a genome-wide analysis. Lancet 2014; 383:1990-8; PMID:24630777; http://dx.doi.org/ 10.1016/S0140-6736(13)62674-4 [DOI] [PubMed] [Google Scholar]
- 14.Engel SM, Joubert BR, Wu MC, Olshan AF, Haberg SE, Ueland PM, Nystad W, Nilsen RM, Vollset SE, Peddada SD, et al.. Neonatal genome-wide methylation patterns in relation to birth weight in the Norwegian Mother and Child Cohort. Am J Epidemiol 2014; 179:834-42; PMID:24561991; http://dx.doi.org/ 10.1093/aje/kwt433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL, Jorgensen SW, Brons C, Jansson PA, Eriksson KF, et al.. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet 2015; 24(13):3792-813; PMID:25861810; http://dx.doi.org/ 10.1093/hmg/ddv124 [DOI] [PubMed] [Google Scholar]
- 16.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, Griffiths L, Hoffman EP, Stubbs RS, Macartney-Coxson D. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol 2015; 16:8; PMID:25651499; http://dx.doi.org/ 10.1186/s13059-014-0569-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet 2013; 14:585-94; PMID:23817309; http://dx.doi.org/ 10.1038/nrg3405 [DOI] [PubMed] [Google Scholar]
- 18.Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, et al.. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol 2012; 13:R43; PMID:22703893; http://dx.doi.org/ 10.1186/gb-2012-13-6-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farre P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics Chromatin 2015; 8:19; PMID:25977707; http://dx.doi.org/ 10.1186/s13072-015-0011-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma B, Wilker EH, Willis-Owen SA, Byun HM, Wong KC, Motta V, Baccarelli AA, Schwartz J, Cookson WO, Khabbaz K, et al.. Predicting DNA methylation level across human tissues. Nucleic Acids Res 2014; 42:3515-28; PMID:24445802; http://dx.doi.org/ 10.1093/nar/gkt1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slieker RC, Bos SD, Goeman JJ, Bovee JV, Talens RP, van der Breggen R, Suchiman HE, Lameijer EW, Putter H, van den Akker EB, et al.. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 2013; 6:26; PMID:23919675; http://dx.doi.org/ 10.1186/1756-8935-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al.. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518:317-30; PMID:25693563; http://dx.doi.org/ 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 2009; 18:4808-17; PMID:19776032; http://dx.doi.org/ 10.1093/hmg/ddp445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, et al.. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 2008; 18:1518-29; PMID:18577705; http://dx.doi.org/ 10.1101/gr.077479.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al.. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 2008; 6:e22; PMID:18232738; http://dx.doi.org/ 10.1371/journal.pbio.0060022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahlman I, Sinha I, Gao H, Brodin D, Thorell A, Ryden M, Andersson DP, Henriksson J, Perfilyev A, Ling C, et al.. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int J Obes (Lond) 2015; 39(6):910-9; PMID:25783037; http://dx.doi.org/24309898 10.1038/ijo.2015.31 [DOI] [PubMed] [Google Scholar]
- 27.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al.. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014; 13:397-406; PMID:24309898; http://dx.doi.org/ 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics 2007; 23:980-7; PMID:17303618; http://dx.doi.org/ 10.1093/bioinformatics/btm051 [DOI] [PubMed] [Google Scholar]
- 29.Kovacs P, Harper I, Hanson RL, Infante AM, Bogardus C, Tataranni PA, Baier LJ. A novel missense substitution (Val1483Ile) in the fatty acid synthase gene (FAS) is associated with percentage of body fat and substrate oxidation rates in nondiabetic Pima Indians. Diabetes 2004; 53:1915-9; PMID:15220220; http://dx.doi.org/ 10.2337/diabetes.53.7.1915 [DOI] [PubMed] [Google Scholar]
- 30.Loftus TM, Jaworsky DE, Frehywot GL, Townsend CA, Ronnett GV, Lane MD, Kuhajda FP. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science 2000; 288:2379-81; PMID:10875926; http://dx.doi.org/ 10.1126/science.288.5475.2379 [DOI] [PubMed] [Google Scholar]
- 31.Glass D, Vinuela A, Davies MN, Ramasamy A, Parts L, Knowles D, Brown AA, Hedman AK, Small KS, Buil A, et al.. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol 2013; 14:R75; PMID:23889843; http://dx.doi.org/ 10.1186/gb-2013-14-7-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renner M, Wolf T, Meyer H, Hartmann W, Penzel R, Ulrich A, Lehner B, Hovestadt V, Czwan E, Egerer G, et al.. Integrative DNA methylation and gene expression analysis in high-grade soft tissue sarcomas. Genome Biol 2013; 14:r137; PMID:24345474; http://dx.doi.org/ 10.1186/gb-2013-14-12-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai R, Zhao Y, Su L, Cassidy L, Liu G, Christiani DC. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia 2012; 14:29-33; PMID:22355271; http://dx.doi.org/ 10.1593/neo.111626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lasselin J, Magne E, Beau C, Ledaguenel P, Dexpert S, Aubert A, Laye S, Capuron L. Adipose inflammation in obesity: relationship with circulating levels of inflammatory markers and association with surgery-induced weight loss. J Clin Endocrinol Metab 2014; 99:E53-61; PMID:24243638; http://dx.doi.org/ 10.1210/jc.2013-2673 [DOI] [PubMed] [Google Scholar]
- 35.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007; 447:425-32; PMID:17522676; http://dx.doi.org/ 10.1038/nature05918 [DOI] [PubMed] [Google Scholar]
- 36.Accomando WP, Wiencke JK, Houseman EA, Nelson HH, Kelsey KT. Quantitative reconstruction of leukocyte subsets using DNA methylation. Genome Biol 2014; 15:R50; PMID:24598480; http://dx.doi.org/ 10.1186/gb-2014-15-3-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13:86; PMID:22568884; http://dx.doi.org/ 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdominal Obesity Study G, von Eyben FE, Kroustrup JP, Larsen JF, Celis J. Comparison of gene expression in intra-abdominal and subcutaneous fat: a study of men with morbid obesity and nonobese men using microarray and proteomics. Ann N Y Acad Sci 2004; 1030:508-36; PMID:15659836; http://dx.doi.org/ 10.1196/annals.1329.063 [DOI] [PubMed] [Google Scholar]
- 39.Yin C, Xiao Y, Zhang W, Xu E, Liu W, Yi X, Chang M. DNA microarray analysis of genes differentially expressed in adipocyte differentiation. J Biosci 2014; 39:415-23; PMID:24845505; http://dx.doi.org/ 10.1007/s12038-014-9412-5 [DOI] [PubMed] [Google Scholar]
- 40.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res 2010; 20:1441-50; PMID:20802089; http://dx.doi.org/ 10.1101/gr.110114.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 2014; 30:1431-9; PMID:24451622; http://dx.doi.org/ 10.1093/bioinformatics/btu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majka SM, Miller HL, Helm KM, Acosta AS, Childs CR, Kong R, Klemm DJ. Analysis and isolation of adipocytes by flow cytometry. Methods Enzymol 2014; 537:281-96; PMID:24480352; http://dx.doi.org/ 10.1016/B978-0-12-411619-1.00015-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buka SL, Seidman LJ, Tsuang MT, Goldstein JM. The New England Family Study High-risk Project: neurological impairments among offspring of parents with schizophrenia and other psychoses. Am J Med Genet B Neuropsychiatr Genet 2013; 162B:653-60; PMID:24132897; http://dx.doi.org/ 10.1002/ajmg.b.32181 [DOI] [PubMed] [Google Scholar]
- 44.Hardy JB. The Johns Hopkins Collaborative Perinatal Project. Descriptive background. Johns Hopkins Med J 1971; 128:238-43; PMID:5556535 [PubMed] [Google Scholar]
- 45.Niswander KR, Gordon M. The Collaborative Perinatal Study of the National Institute of Neurological Diseases and Stroke: The women and their pregnancies. In: U. S. Department of Health E, and Welfare, ed. Washington DC, 1972 [Google Scholar]
- 46.Davis S, Du P, Bilke S, Triche TJ. M B. methylumi: Handle Illumina methylation data. R package version 2.14.0 2015 [Google Scholar]
- 47.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A β-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013; 29:189-96; PMID:23175756; http://dx.doi.org/ 10.1093/bioinformatics/bts680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rencher AC. Methods of multivariate analysis. New York: J. Wiley, 2002 [Google Scholar]
- 49.Liang KY, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika 1986; 73:13-22; http://dx.doi.org/ 10.1093/biomet/73.1.13 [DOI] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society, Series B 1995; 57:289-300; http://dx.doi.org/ 10.2307/2346101 [DOI] [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9; PMID:10802651; http://dx.doi.org/ 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gene Ontology C . Gene Ontology Consortium: going forward. Nucleic Acids Res 2015; 43:D1049-56; PMID:25428369; http://dx.doi.org/ 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisher RA. On the interpretation of chi-square from contingency tables, and the calculation of P. J Royal Statistical Society 1922; 85:87-94; http://dx.doi.org/ 10.2307/2340521 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


