A novel and affordable fragment library was assembled and applied to the model protease endothiapepsin in a crystallographic fragment-screening experiment.
Keywords: structural biology, medicinal chemistry, fragment screening, X-ray crystallography, protein–ligand structures
Abstract
Crystallographic screening of the binding of small organic compounds (termed fragments) to proteins is increasingly important for medicinal chemistry-oriented drug discovery. To enable such experiments in a widespread manner, an affordable 96-compound library has been assembled for fragment screening in both academia and industry. The library is selected from already existing protein–ligand structures and is characterized by a broad ligand diversity, including buffer ingredients, carbohydrates, nucleotides, amino acids, peptide-like fragments and various drug-like organic compounds. When applied to the model protease endothiapepsin in a crystallographic screening experiment, a hit rate of nearly 10% was obtained. In comparison to other fragment libraries and considering that no pre-screening was performed, this hit rate is remarkably high. This demonstrates the general suitability of the selected compounds for an initial fragment-screening campaign. The library composition, experimental considerations and time requirements for a complete crystallographic fragment-screening campaign are discussed as well as the nine fully refined obtained endothiapepsin–fragment structures. While most of the fragments bind close to the catalytic centre of endothiapepsin in poses that have been observed previously, two fragments address new sites on the protein surface. ITC measurements show that the fragments bind to endothiapepsin with millimolar affinity.
1. Introduction
Fragments are small organic compounds with molecular weights (MWs) in the range from 150 to 300 Da (Rees et al., 2004 ▸) which can be used to map the functional surface of a protein. In contrast to larger molecules (MW = 300–500 Da), which are typically studied as candidates in high-throughput screening (HTS) experiments, fragments are mostly weak-binding entities with affinities in the high-micromolar to millimolar range (Rees et al., 2004 ▸; Hajduk & Greer, 2007 ▸). In contrast to the larger molecules used in HTS, they have the advantage that a relatively large chemical space can be explored using a comparatively small selection of compounds (Hajduk & Greer, 2007 ▸; Fink & Reymond, 2007 ▸). Furthermore, the ligand efficiency, defined as the contribution of binding free energy per non-H atom of the fragment (Hopkins et al., 2004 ▸), is generally higher than that of larger molecules. This makes fragments much more amenable to subsequent optimization into potent drug-like molecules through a manageable number of synthesis steps. Within the last decade, fragment screening has evolved from a mere concept into a widely applied technique in drug development (Schulz & Hubbard, 2009 ▸). Many pharmaceutical companies nowadays apply fragment screening in their drug-discovery projects (Baker, 2013 ▸). In contrast to the widespread use of fragment screening in industry, academic research has been rather slow in taking up fragment-screening experiments as a tool to elucidate protein function. One explanation for this seeming reluctance may be the nonavailability of suitable (and affordable) substance libraries and the experimental effort associated with a fragment-screening campaign. In order to help remedy this situation, we assembled a small, versatile and affordable fragment library of 96 compounds, which was designed for particular use in crystallographic fragment-screening experiments. The resulting library was validated against the aspartic protease endothiapepsin (EP). EP is a monomer consisting of 330 amino acids (molecular weight 34 kDa) with an active site that is defined by two aspartic acids (Asp35 and Asp219) and one catalytic water molecule. EP represents a model system for pepsin-like aspartic proteases, which play a role in several important diseases. EP-related pharmacologically relevant proteases include plasmepsins, a target for malaria, β-secretase, which is involved in Alzheimer’s disease, and the protease renin, a nephritic enzyme involved in hypertension.
2. Materials and methods
2.1. Library assembly and composition
The assembly of the presented library started with an examination of the PDBeChem database (http://www.ebi.ac.uk/pdbe-srv/pdbechem/; Dimitropoulos et al., 2006 ▸). The version of PDBeChem from May 2012 contained 14 149 protein–ligand structures. By selecting ligands which contained between three and ten C atoms, up to five N atoms and between one and ten O atoms, the number of candidate compounds was reduced to about 2000. From these 2000, about 100 compounds were selected manually. The criteria were chemical diversity but also practical aspects such as price, compound availability and compound stability. In addition to these selections, some compounds not matching these criteria (e.g. NAD+) were selected because of their widespread occurrence and their importance in protein-function elucidation. The list of all selected compounds is given in Supplementary Table S1. Most of the compounds were purchased from Sigma–Aldrich Co. LLC, Taufkirchen, Germany, except for compounds 1, 2, 6, 7 and 48, which were purchased from Merck KGaA, Darmstadt, Germany, and compound 3, which was purchased from Biomol GmbH, Hamburg, Germany. In all cases, the highest purity grade available was acquired. All compounds were then used as purchased without any further purification. Subsequently, 1 M stock solutions of the compounds in DMSO (Roth GmbH & Co. KG, Karlsruhe, Germany) were prepared. The DMSO used was of p.a. grade (≥99.8% purity). In cases where the compounds were not sufficiently soluble to allow the preparation of a 1 M stock solution, a saturated solution was used.
2.2. Preparation and crystallization
EP was prepared and crystallized following the protocol described by Köster et al. (2011 ▸). Monoclinic crystals were grown in 400 nl sitting drops that consisted of 50% reservoir solution [0.1 M sodium acetate pH 4.6, 0.1 M ammonium acetate pH 4.6, 28%(w/v) PEG 4000] and 50% protein solution (4.8 mg ml−1 protein in 0.1 M sodium acetate pH 4.6). Crystallization experiments were carried out in 96-well plates (CrystalQuick Lp) using a Gryphon robot (Art Robbins Instruments).
2.3. Soaking of crystals
The soaking procedure was designed such that it included cryoprotection following Köster et al. (2011 ▸). Only one fragment was soaked per crystal. For the soaking experiment, 96 soaking buffers consisting of 7.5 µl reservoir solution, 2.5 µl glycerol and 1 µl fragment stock solution were prepared. At least two crystals per fragment were incubated in 2 µl soaking buffer for 48 h, harvested with a loop and flash-cooled in liquid nitrogen.
2.4. Data collection and processing
Diffraction data sets were collected on beamline BL14.1 at the BESSY II storage ring (Mueller et al., 2012 ▸, 2015 ▸) using a Pilatus 6M detector (Dectris). In order to save time, no data-collection strategy was calculated, and a standard rotation range of 200° was collected from each crystal. All data sets were processed using XDS (Kabsch, 2010 ▸) through the automated GUI and expert system XDSAPP (Krug et al., 2012 ▸; Sparta et al., 2016 ▸). All relevant data-collection and processing parameters are shown in Table 1 ▸.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outer shell.
| Data set | EP–29 | EP–30 | EP–39 | EP–42 | EP–50 | EP–51 | EP–53 | EP–91 | EP–97 |
|---|---|---|---|---|---|---|---|---|---|
| X-ray source and beamline | BL14.1, BESSY II | ||||||||
| Wavelength (Å) | 0.9184 | ||||||||
| Detector | Pilatus 6M | ||||||||
| No. of crystals | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Crystal-to-detector distance (mm) | 180.8 | 180.8 | 180.8 | 180.8 | 180.8 | 180.8 | 180.8 | 168.0 | 180.8 |
| Rotation range per image (°) | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Total rotation range (°) | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
| Resolution range (Å) | 42.7–1.10 (1.17–1.10) | 42.7–1.31 (1.39–1.31) | 42.7–1.17 (1.24–1.17) | 42.8–1.35 (1.43–1.35) | 42.6–1.25 (1.33–1.25) | 42.8–1.27 (1.35–1.27) | 42.7–1.44 (1.52–1.44) | 41.1–1.28 (1.36–1.28) | 42.7–1.03 (1.09–1.03) |
| Space group | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 |
| Unit-cell parameters | |||||||||
| a (Å) | 45.05 | 45.22 | 45.23 | 45.32 | 45.18 | 45.30 | 45.21 | 45.27 | 45.22 |
| b (Å) | 72.90 | 73.27 | 72.99 | 72.91 | 73.52 | 72.94 | 73.11 | 73.13 | 72.94 |
| c (Å) | 52.10 | 52.79 | 52.53 | 52.65 | 53.10 | 52.51 | 52.58 | 52.70 | 52.66 |
| α (°) | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| β (°) | 108.73 | 109.08 | 109.17 | 109.23 | 109.44 | 109.14 | 109.25 | 109.31 | 109.2 |
| γ (°) | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| Mosaicity (°) | 0.083 | 0.118 | 0.070 | 0.237 | 0.175 | 0.133 | 0.084 | 0.083 | 0.057 |
| Total No. of reflections | 472468 | 290171 | 402395 | 261794 | 336405 | 317317 | 226321 | 310118 | 548947 |
| Unique reflections | 125106 | 77955 | 107230 | 70867 | 90188 | 84800 | 58437 | 82413 | 148808 |
| Multiplicity | 3.78 | 3.72 | 3.75 | 3.69 | 3.73 | 3.74 | 3.87 | 3.76 | 3.69 |
| Completeness (%) | 96.8 (92.4) | 99.4 (97.9) | 98.7 (96.8) | 99.0 (95.2) | 99.6 (98.9) | 99.6 (99.3) | 97.7 (95.7) | 99.7 (99.3) | 93.5 (74.2) |
| Mean I/σ(I) | 13.7 (2.7) | 10.0 (2.2) | 12.1 (2.4) | 18.7 (2.3) | 16.0 (2.3) | 14.2 (2.2) | 12.2 (2.0) | 12.7 (1.9) | 13.6 (2.1) |
| R r.i.m./R meas (%) | 6.2 (53.8) | 8.6 (65.6) | 6.6 (57.2) | 6.4 (64.8) | 6.1 (65.4) | 6.7 (65.7) | 8.7 (72.7) | 7.2 (68.7) | 5.3 (57.6) |
| Wilson B factor (Å2) | 12.5 | 17.6 | 14.5 | 17.1 | 16.1 | 16.4 | 19.3 | 16.8 | 12.2 |
| ISa (Diederichs, 2010 ▸) | 41.8 | 14.5 | 31.2 | 77.7 | 53.4 | 47.2 | 40.7 | 45.3 | 25.8 |
2.5. Structure refinement and fragment-binding analysis
Directly following data processing with XDSAPP, an automated refinement procedure was invoked using PHENIX (Adams et al., 2010 ▸). The starting model was based on PDB entry 3t6i (Köster, 2012 ▸), from which all ligands and water molecules in the active site were removed. Electron-density maps and coordinate files of each experiment obtained from the automated refinement were inspected manually to judge the presence or absence of the expected fragment from the difference electron density. Subsequently, the identified hits were subjected to several rounds of alternating model building in Coot (Emsley et al., 2010 ▸) and refinement in PHENIX (Adams et al., 2010 ▸). For all fragments, occupancy refinement was carried out. All relevant structure-refinement and validation statistics are shown in Table 2 ▸.
Table 2. Structure-refinement and validation statistics.
| Data set | EP–29 | EP–30 | EP–39 | EP–42 | EP–50 | EP–51 | EP–53 | EP–91 | EP–97 |
|---|---|---|---|---|---|---|---|---|---|
| Refinement program | PHENIX v.1.9-1692 | ||||||||
| Resolution limits (Å) | 42.7–1.10 | 39.4–1.31 | 42.7–1.17 | 34.7–1.35 | 39.6–1.25 | 42.8–1.27 | 42.7–1.44 | 34.8–1.28 | 42.7–1.03 |
| Completeness (%) | 96.9 | 99.5 | 98.8 | 99.1 | 99.7 | 99.8 | 97.9 | 99.8 | 93.6 |
| Data cutoff [F/σ(F)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. of reflections | |||||||||
| Working set | 118847 | 74055 | 101857 | 67321 | 85676 | 80559 | 54387 | 80309 | 141365 |
| Test set | 6255 | 3898 | 5360 | 3543 | 4510 | 4240 | 2862 | 2099 | 7440 |
| R work (%) | 11.45 | 12.75 | 11.46 | 11.68 | 11.27 | 11.58 | 12.01 | 13.80 | 10.99 |
| R free (%) | 12.97 | 15.83 | 13.87 | 14.16 | 14.02 | 14.49 | 15.91 | 16.40 | 12.55 |
| DPI† (Å) | 0.029 | 0.055 | 0.034 | 0.057 | 0.041 | 0.046 | 0.080 | 0.029 | 0.023 |
| No. of atoms | |||||||||
| Total | 2876 | 2763 | 2860 | 2856 | 2838 | 2887 | 2851 | 2889 | 2950 |
| Protein | 2470 | 2459 | 2472 | 2468 | 2464 | 2458 | 2459 | 2473 | 2488 |
| Fragment | 13 | 12 | 10 | 8 | 12 | 28 | 12 | 16 | 12 |
| Other ligands | 64 | 66 | 74 | 54 | 54 | 48 | 50 | 92 | 54 |
| Waters | 329 | 226 | 304 | 326 | 308 | 353 | 330 | 308 | 396 |
| R.m.s. deviations | |||||||||
| Bonds (Å) | 0.007 | 0.007 | 0.007 | 0.009 | 0.010 | 0.007 | 0.008 | 0.006 | 0.006 |
| Angles (°) | 1.3 | 1.2 | 1.2 | 1.3 | 1.4 | 1.2 | 1.3 | 1.2 | 1.3 |
| Average B factors (Å2) | |||||||||
| Protein | 11.8 | 13.9 | 12.6 | 12.8 | 13.3 | 12.8 | 14.8 | 13.0 | 12.2 |
| Fragment | 21.4 | 13.3 | 23.4 | 25.1 | 16.8 | 34.2 | 20.8 | 21.1 | 16.0 |
| Other ligands | 39.8 | 37.7 | 35.1 | 34.4 | 34.4 | 34.5 | 34.9 | 40.5 | 31.2 |
| Waters | 28.2 | 29.3 | 28.9 | 27.9 | 31.7 | 28.7 | 30.1 | 28.4 | 29.0 |
| Ramachandran plot | |||||||||
| Outliers (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.06 | 0.0 |
| Allowed (%) | 1.4 | 1.7 | 1.4 | 1.7 | 1.7 | 1.7 | 2.0 | 1.4 | 1.7 |
| Favoured (%) | 98.6 | 98.3 | 98.6 | 98.3 | 98.3 | 98.3 | 98.0 | 98.6 | 98.3 |
| PDB code | 4y38 | 4y3j | 4ze6 | 4y3y | 4y48 | 4y4d | 4y4g | 4zea | 4y4j |
DPI was calculated using the Online_DPI server (Kumar et al., 2015 ▸).
2.6. Structure comparison
All superimpositions of the complex structures were performed in Coot (Emsley et al., 2010 ▸) using either the SSM or the LSQKAB superimpose functionality or in PyMOL (v.1.2r3pre; Schrödinger) using the ALIGN module.
2.7. Isothermal titration calorimetry (ITC)
The fragments which were found to bind to EP in the crystallographic experiments were subsequently analyzed by displacement ITC using a MicroCal ITC200 system (GE Healthcare) at 25°C. The sample cell was filled with 300 µl of purified EP (Köster, 2012 ▸) at a concentration of 50 µM in 0.1 M sodium acetate buffer pH 4.6, 3%(v/v) DMSO and 2 mM fragment. The same solution lacking any fragment was used to determine the thermodynamic profile of the reference ligand SAP114 (molecule 9; Kuhnert et al., 2015 ▸). This ligand was present at a concentration of 500 µM in the same buffer and was titrated into the sample cell with (22–31) × 1.2–1.5 µl injections at 180 s intervals. The temperature difference or heat release was measured and integrated using NITPIC v.1.0.3 (Keller et al., 2012 ▸). Isotherm fitting was then performed by SEDPHAT v.10.58d (Houtman et al., 2007 ▸). The association constant K a is directly accessible from the slope of the binding curve. Based on this value, the dissociation constant K d as well as the Gibbs free energy ΔG° can be determined using the equations ΔG° = −RTln(K a) and K d = 1/K a, where R is the gas constant (8.3144 J mol−1 K−1) and T is the temperature of the experiment in kelvin (298.15 K). In the displacement/competitive titration, the association constant of the weak-binding fragment (K a-weak) to EP was determined according to Zhang & Zhang (1998 ▸) using the equation K a-weak = [(K a-tight/K a-app) − 1] × (1/[M]weak), where [M]weak is the concentration of the weak binder, K a-app is the apparent binding constant of SAP114 in the presence of the fragment and K a-tight is the association constant of the tight binder (SAP114). Based on the equations shown above, the dissociation constant of the weak binder (K d-weak) as well as the binding free energy of the fragment were calculated. The error in the latter quantity was derived via error propagation using the standard deviation of three K a-tight measurements. For the reference ligand three thermograms were recorded, while for each fragment only one was obtained. Furthermore, the ligand efficiency (LE) of each fragment was determined via the equation LE = −ΔG°/non-H atom count. Unfortunately, the low solubility of fragments 51 and 91 did not allow their investigation by ITC experiments. Since the difference between the binding isotherms obtained for the reference ligand and the titration displacing fragment 53 was insignificant, we refrained from specifying any exact thermodynamic parameters for this ligand, but rather state that this molecule is a very weak EP binder (K d > 10 mM). Finally, fragment 50 cannot be displaced by SAP114 since it binds remotely from the substrate-binding cleft. Consistently, no significant difference was observed between the reference and displacement titration for this fragment.
3. Results and discussion
3.1. Library assembly and composition
The initial version of the assembled fragment library is presented in Supplementary Table S1. The library contains compounds that can be roughly divided into five chemical classes. These classes are (i) buffer ingredients (e.g. HEPES), (ii) carbohydrates and their derivatives (e.g. N-acetyl-d-glucosamine), (iii) nucleotides (e.g. ADP), (iv) amino acids, amino-acid-like molecules and peptide-like fragments (e.g. 7-aminoheptanoic acid) and (v) various organic compounds of drug-like character (e.g. theobromine). While classes (i)–(iv) each comprise about 10% of the total library content, class (v) makes up the remaining 60% and is therefore the largest and most diverse of the five classes. 57 of the 96 fragments belong to this class. They represent a large variety of chemical compounds, such as cyclic molecules, aromatic compounds, drugs and other bioactive substances. The average molecular weight of the 96 library compounds is 213 Da (Table 3 ▸), which falls well within the definition range of fragments, with a definite bias towards the lower end. However, there are also some compounds that are significantly larger than the average. The reason for including a small number of very large compounds (e.g. NAD+) is that one of the aims of the library is to also enable functional analysis. In this context, ligands such as NAD+ are important probe compounds to analyze biological systems. Another important feature of the library is its affordability. On average, each of the 96 library compounds costs about 110 Euro per 100 mg. Consequently, the material cost for a single crystallographic fragment-screening campaign with this library is much less than 1000 euro (assuming fragment consumption in the low-milligram range). This should be well within the reach of most research groups in academia. Relevant average physicochemical parameters of the library are detailed in Table 3 ▸. With an average of three hydrogen-bond donors, three hydrogen-bond acceptors and three rotatable bonds, the library roughly follows the ‘rule of three’ (Congreve et al., 2003 ▸), although this was not intended by design. Naturally, any individual compound may differ significantly from these average values and thus should be considered separately. It also needs to be mentioned that neither the octanol–water partition coefficient logP nor the topological polar surface area (tPSA) were criteria for the library design. Yet, the most desired property of the library is the proven capability of its compounds to bind to proteins, which is why all compounds were selected based on their presence in the PDBeChem database.
Table 3. Properties of the 96-compound fragment library described in this work.
| Parameters | Minimum | Maximum | Average |
|---|---|---|---|
| MW (Da)† | 101 | 743 | 213 |
| No. of C atoms | 3 | 21 | 8 |
| No. of N atoms | 0 | 7 | 2 |
| No. of O atoms | 1 | 16 | 4 |
| No. of hydrogen-bond donors | 0 | 9 | 3 |
| No. of hydrogen-bond acceptors | 0 | 14 | 3 |
| No. of rings | 0 | 5 | 1 |
| No. of rotatable bonds | 0 | 13 | 3 |
| LogP (calculated)† | −6.7 | 3.0 | −1.0 |
| tPSA (Å2)† | 35 | 365 | 96 |
Calculated by MOE, version 2014.09 (Chemical Computing Group Inc., 2010 ▸).
3.2. Validation of the library
The assembled library was validated by screening all compounds against the aspartic protease endothiapepsin (EP) from Cryphonectria parasitica. This choice of target was made because EP has previously been used as a target in a fragment-screening experiment (Köster et al., 2011 ▸) and it is also the subject of a larger study currently being carried out in the group of G. Klebe at Philipps University Marburg, Germany. Applying the assembled library in a fragment-screening campaign against EP yielded an appreciable hit rate of 9.4%. The hit rate here is defined as the number of complex structures observed divided by the number of fragments soaked. This demonstrates that the library is capable of delivering hit rates that are comparable to or higher than those observed in other studies (Supplementary Table S2). Moreover, the library has already been validated a priori owing to the selection of compounds from PDBeChem entries. In addition to the validation experiment described here, the library has already been shared with other research groups and further fragment-screening campaigns are under way. Preliminary results (data not shown) indicated that the hit rates in these campaigns are similar to those described here.
3.3. Analysis of the nine fragment-complex crystal structures
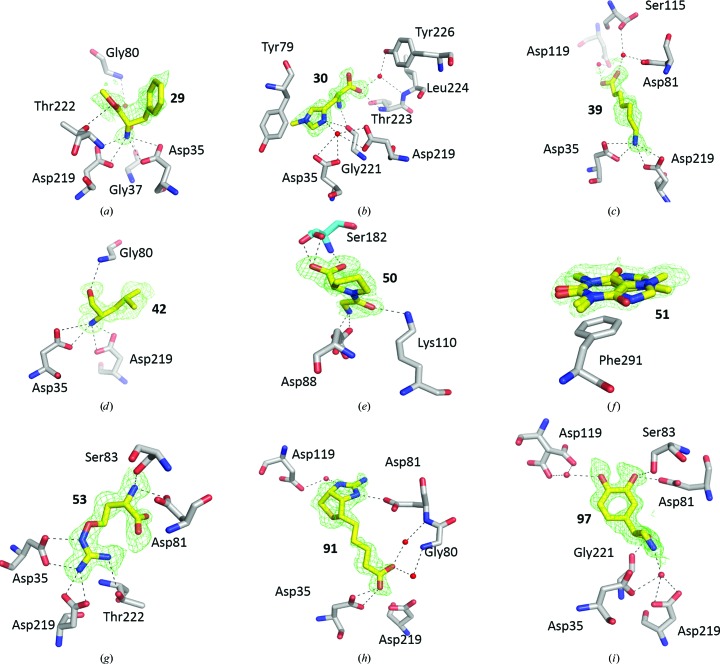
Individual soaking experiments of the 96 compounds into monoclinic EP crystals and subsequent structural analysis revealed that nine fragments (fragments 29, 30, 39, 42, 50, 51, 53, 91 and 97; see Supplementary Table S1) were bound to EP (Fig. 1 ▸). All nine complex structures are of high quality, as is shown by indicators such as the ISa value (Diederichs, 2010 ▸) of the data (average 42.0), the data resolution (1.03–1.44 Å) and the R free value (12.6–16.4%). Detailed statistics of data collection, data processing and refinement are listed in Tables 1 ▸ and 2 ▸. The refined occupancy of fragments was between 50 and 95% and the corresponding real-space correlation coefficients (RSCCs) range from 0.84 to 0.93 (Table 4 ▸). All fragments were reasonably well defined in the electron density. Despite the fact that the fragments are expected to be weak-binding ligands, according to the validation categorization of Deller & Rupp (2016 ▸) three of the nine fragments were in the top category (RSCC 1.0–0.9), meaning that they fit the density well. The remaining six fragments fall into the second category (0.9–0.8) where the ligand fits the density at least partially (Deller & Rupp, 2016 ▸). None of the observed fragments exhibited an RSCC of below 0.8. Nevertheless, these rather high RSCC values demonstrate convincingly that all nine EP–fragment complex structures are reliable and that the presence of the fragment is well established. In the following, the binding of each of the fragments to EP will be described in brief.
Figure 1.
Binding of fragments to EP. Shown is the unbiased (mF o − DF c, αc) difference electron density (contoured at 2σ) of the fragments (identified by their fragment number; see Supplementary Table S1) soaked into monoclinic EP crystals superimposed onto the refined fragment coordinates (shown as yellow sticks) and the neighbouring EP amino acids (shown as grey sticks). Ser182 in (e) is from a symmetry-related molecule and is therefore coloured light blue. Hydrogen-bond interactions up to a donor–acceptor distance of 3.2 Å are shown as black dashed lines. Water molecules are shown as small red spheres. (a)–(i) show the binding of fragment 29, fragment 30, fragment 39, fragment 42, fragment 50, fragment 51, fragment 53, fragment 91 and fragment 97, respectively.
Table 4. Occupancies and RSCCs of the nine fragment hits.
| Fragment | Occupancy | RSCC |
|---|---|---|
| 29 | 0.71 | 0.87 |
| 30 | 0.50 | 0.87 |
| 39 | 0.77 | 0.84 |
| 42 | 0.95 | 0.88 |
| 50 | 0.71 | 0.91 |
| 51† | 0.77 | 0.84 |
| 53 | 0.81 | 0.90 |
| 91 | 0.77 | 0.93 |
| 97 | 0.59 | 0.89 |
Fragment 51 has been modelled in two conformations: A and B. The refined occupancy values were 0.38 for conformation A and 0.39 for conformation B. The RSCC is identical for both conformations.
3.4. EP in complex with l-phenylalanine methyl ester (fragment 29; Fig. 1 ▸ a)
The primary amino N atom of fragment 29 displaces the catalytic water molecule of EP and engages in hydrogen bonds to the carboxylate O atoms of the two catalytic aspartate residues. The shortest distances observed are to Asp35 OD1 (2.80 Å) and Asp219 OD2 (2.92 Å), although the other two carboxylate O atoms are also within hydrogen-bonding distance (3.06 and 3.17 Å, respectively). A further observed interaction is a hydrogen bond mediated by water 508 between the amino N atom of the fragment and Gly37 O. The ester O atom of the fragment interacts with Thr222 OG1, and the carbonyl O atom of the fragment engages in a water-mediated hydrogen bond (water 531) to Gly80 N. The phenyl group of the fragment is located in the S1 pocket of the enzyme.
3.5. EP in complex with 1-methyl-l-histidine (fragment 30; Fig. 1 ▸ b)
The unmethylated N atom of the imidazole ring of fragment 30 forms two hydrogen bonds to the catalytic water 501 and to Gly221 O (2.73 and 3.09 Å, respectively). The amino group of the fragment also forms a hydrogen bond to Gly221 O (2.74 Å), and the carboxyl O atom of the fragment is hydrogen-bonded to Thr223 N (2.96 Å). The carboxylate O atom of the fragment is bound to the protein via two water-mediated hydrogen bonds (water 629) to Tyr226 OH and Leu224 N. The imidazole ring is located in the S1 pocket almost parallel to the phenyl ring plane of Tyr79.
3.6. EP in complex with 7-aminoheptanoic acid (fragment 39; Fig. 1 ▸ c)
The N atom of fragment 39 displaces the catalytic water 501 and is bound to the carboxylate O atoms of the catalytic aspartates. The carboxylate group of the fragment engages in two water-mediated hydrogen bonds to the protein: via water 630 to Asp81 OD2 (2.95 Å) and Ser115 OG (2.85 Å) and via water 696 to Asp119 OD2. The nonpolar carbon chain (C2-C3-C4) is stretched across the S1 pocket.
3.7. EP in complex with (S)-2-amino-4-methyl-1-pentanol/(S)-(+)-leucinol (fragment 42; Fig. 1 ▸ d)
The N atom of fragment 42 displaces the catalytic water and binds to the carboxylate O atoms of the catalytic aspartates. The hydroxyl O atom of the fragment forms a hydrogen bond to Gly80 N (3.07 Å), which is one of the flap residues of EP. Both the N atom and the hydroxyl O atom of the fragment interact with Asp35 OD2 via a water-mediated hydrogen bond. Interestingly, fragment 42 is the only one of the fragments bound to the active site of EP which is oriented in the direction of the S1′ pocket.
3.8. EP in complex with 1-(2-aminoacetyl)-pyrrolidine-2-carboxylic acid/glycylproline/Gly-Pro (fragment 50; Fig. 1 ▸ e)
In contrast to the other bound fragments, fragment 50 was observed to bind outside the active site, between symmetry-related EP molecules in the crystal (Supplementary Fig. S1). The glycyl moiety or N-terminal part of the fragment interacts mainly with one EP molecule. Hydrogen bonds are formed between the amino N atom of the fragment and Asp88 OD2 (2.65 Å) and between the carbonyl O atom of the fragment and Lys110 NZ (2.83 Å). The prolyl moiety or C-terminal part of the fragment interacts mainly with the neighbouring EP molecule, with hydrogen bonds to Ser182 OG (2.65 Å) and to Ser182 N (2.79 Å). In addition, one of the carboxylate atoms of the fragment interacts with Gly72 N (2.95 Å), thus contributing to the stabilization of the crystal lattice (Supplementary Fig. S1).
3.9. EP in complex with 1,3,7-trimethylxanthine/caffeine (fragment 51; Fig. 1 ▸ f)
Fragment 51 was also observed to bind remotely from the catalytic centre of EP. However, at a distance of about 13 Å from the catalytic aspartate residues, it is still within the large peptide-recognition cleft of EP. The planar fragment 51 binds in two orientations (related by a 180° rotation about an axis in the plane of the molecule) exclusively through a π-stacking interaction with the phenyl ring of Phe291. No polar interactions between the fragment and EP were observed.
3.10. EP in complex with l-α-amino-γ-(guanidinooxy)-n-butyric acid/l-canavanine (fragment 53; Fig. 1 ▸ g)
The guanidinium group of the fragment was found to bind to the two catalytic aspartates as well as to the hydroxyl group of Thr222. One of the guanidinium N atoms (N2) displaces the catalytic water of EP. The amino N atom of the fragment interacts with Ser83 OG (2.84 Å) and Asp81 OD2 (2.64 Å), whereas the carboxylate group of the fragment shows no obvious interaction with the protein except for a rather surprising stacking with the carboxylate group of Asp81 in a mutually reversed orientation. The central part of the fragment (O-C1-C2) is stretched across the S1 pocket of the enzyme.
3.11. EP in complex with 2-iminobiotin (fragment 91; Fig. 1 ▸ h)
Notably, the carboxylate group of fragment 91 interacts with the carboxylate group of solely the catalytic aspartate Asp35 (2.52 and 3.13 Å between the O1 atom of the fragment and Asp35 OD1 and OD2, respectively), thereby leaving no room for the catalytic water molecule to bind. This interaction between the two carboxylate groups requires that one of them is protonated, which is not inconceivable at a crystallization pH of 4.6. Since the carboxylate group of the extended biotin side chain should exhibit a higher pK a value, it is this carboxylate that is more likely to pick up the proton. The carboxylate group of the fragment also forms two water-mediated hydrogen bonds (waters 520 and 537) to Gly80 N and Asp81 N of EP. The guanidinium moiety of the fragment is hydrogen-bonded to Asp81 OD2 (2.62 Å) and via a water molecule (water 732) to Asp119 OD2. The aliphatic C atoms C6–C8 of the fragment occupy the S1 pocket.
3.12. EP in complex with (R)-4-(2-amino-1-hydroxyethyl)-1,2-benzenediol/(−)-norepinephrine (fragment 97; Fig. 1 ▸ i)
The amino group of fragment 97 interacts with both catalytic aspartates. Remarkably, this time the interaction is via the catalytic water molecule 501. The hydroxyl group of the fragment is hydrogen-bonded to Gly221 O (2.69 Å). Both hydroxyl groups of the catechol ring interact either directly with the protein (O2 to Ser83 OG, 3.09 Å; O2 to Asp81 OD2, 2.28 Å) or indirectly via water molecule 696 (O1 of the fragment to Asp119 OD2). The middle part of fragment 97 (C, C1 and C2) occupies the S1 pocket.
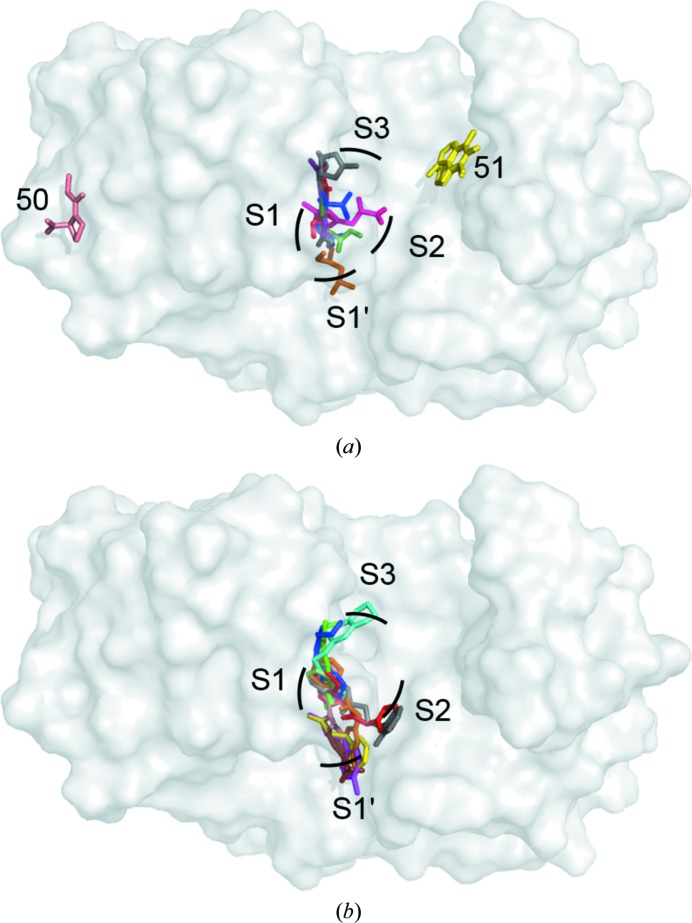
3.13. Binding modes
A closer look at the observed binding modes for each of the nine fragments shows that they can be roughly grouped into four classes (Fig. 2 ▸ a). The first type of binding was realised by fragments 29, 39, 42, 53 and 91. Here, the fragments bind to the catalytic centre of EP. One most likely protonated and positively charged N atom of the fragment displaces the catalytic water molecule and engages in direct hydrogen-bonding interactions with the catalytic aspartate residues of EP. The remaining parts of the fragments typically extend into the S1 or S1′ pocket. A second mode of binding is realised by fragments 30 and 97, which also bind close to the catalytic centre of EP. In contrast to the first mode, however, the catalytic water is retained and the fragments interact with the catalytic aspartates indirectly via the catalytic water molecule. Similar to the first mode, the additional parts of the fragments address mainly the S1 or S1′ pocket. The binding modes of the remaining two fragments are completely different from the first two. Fragment 51 binds without any obvious polar interactions 13 Å away from the catalytic centre, while fragment 50 binds between two symmetry-related molecules of the EP crystal lattice (Supplementary Fig. S1).
Figure 2.
Superposition of EP–fragment complexes. (a) All fragments reported in this study; (b) all fragments reported in Köster et al. (2011 ▸). The binding pockets S1, S2, S3 and S1′ of the active-site cleft of EP are indicated, as are compounds 50 and 51 (in a), which bind remotely from the catalytic centre of EP. EP itself is depicted as a surface representation. Fragments are shown as sticks in different colours.
3.14. Comparison with other EP–fragment structures in the PDB
In a previous study on fragment binding to EP (Köster et al., 2011 ▸), 11 fragments (out of 55 soaked compounds identified by an enzymatic pre-screening assay carried out for the complete library of 361 fragments) were observed to bind to EP. A superposition of these 11 fragments (Fig. 2 ▸ b) shows that they all bind very close to the catalytic centre of EP. In this sense, the observed binding poses closely match binding modes 1 and 2 described here. No binding of fragments was observed outside the immediate vicinity of the catalytic aspartate residues. Most of the fragments of the previous study address the S1 and S1′ pockets of EP, although some of the larger molecules also extend partly into the S2 and S3 pockets of the protein. Among the fragments which bind utilizing either modes 1 or 2, there is one (fragment 30) which appears to be substantially different from those presented in Köster et al. (2011 ▸). Fragment 30 (purple in Fig. 2 ▸ a) extends partially into the S2 pocket, but addresses this pocket from a different direction.
The observed binding of fragment 50 between symmetry-related molecules results from the formation of an additional binding pocket upon ordering of the EP molecules in the monoclinic crystal lattice. It is possible that the binding of a fragment to this part of the surface of EP may point to another hitherto undiscovered functional role of this surface patch.
3.15. Determination of binding affinities by isothermal titration calorimetry (ITC)
The crystallographic fragment hits obtained by screening against EP were further analyzed using displacement ITC (Wang, 2015 ▸; Zhang & Zhang, 1998 ▸). The results are summarized in Table 5 ▸. For five of the nine hits K d values of between 3 and 7 mM were obtained, while one compound exhibited a K d of above 10 mM and three further fragments could not be analyzed owing to their low water solubility or to their remote binding position not interfering with the binding pose of the displacement ligand, respectively. These results not only confirm the expected weak, millimolar, binding affinities of the crystallographic fragment hits, but more importantly that the binding observed in the crystalline state can be translated to the solution state. Consequently, some of the observed hits can be used as a starting point for the development of stronger binders and possibly inhibitors. This is underlined by the finding that fragments 39 and 42 are characterized by ligand efficiencies (LEs) of 1.41 and 1.68 kJ mol−1 atom−1, which are clearly above 1.25 kJ mol−1 atom−1 (or 0.3 kcal mol−1 atom−1) which is considered to be a threshold for promising starting points in fragment-based drug development (Congreve et al., 2008 ▸).
Table 5. ITC analysis of fragment binding to EP.
Fragments 50, 51 and 91 could not be investigated using displacement ITC. The errors in ΔG°, K d and LE were estimated via error propagation using the standard deviation of three measurements of the displacement ligand. For details, see §2.7.
| Fragment | ΔG° (kJ mol−1) | K d (mM) | LE† (kJ mol−1 atom−1) |
|---|---|---|---|
| 29 | −13.7 ± 0.1 | 4.0 ± 0.2 | 1.05 ± 0.01 |
| 30 | −12.4 ± 0.2 | 6.8 ± 0.6 | 1.03 ± 0.02 |
| 39 | −14.1 ± 0.1 | 3.3 ± 0.2 | 1.41 ± 0.01 |
| 42 | −13.4 ± 0.2 | 4.5 ± 0.3 | 1.68 ± 0.02 |
| 53 | >−11.4 | >10.0 | <0.95 |
| 97 | 14.1 ± 0.1 | 3.4 ± 0.2 | 1.18 ± 0.01 |
The ligand efficiency (LE) was calculated dividing −ΔG° by the number of non-H atoms in the fragment.
3.16. Comparison to the results from other fragment-screening campaigns reported in the literature
When comparing the reported results obtained from screening different fragment libraries, it is very important to take into account whether the crystallographic experiment was preceded by a pre-screening step using some other biophysical technique. Although pre-screening is not infallible, it helps to increase hit rates in a crystallographic experiment (Schiebel et al., 2015 ▸). In this work, the assembled fragment library was screened against EP without any prior pre-screening. For fair and meaningful comparison, Supplementary Table S2 shows only the results of several purely crystallographic fragment-screening campaigns reported in the literature. The number of fragments that were used for screening in these other fragment-screening campaigns varied widely from 80 to 800. The used libraries were either unbiased or general-purpose libraries, such as that described here, or target-specific libraries. In all studies reported so far except one, cocktails of 4–10 fragments were used for crystallographic screening. The hit rate varied between 0.5 and 10%. Potentially, the use of cocktails led to hit rates smaller than that reported here. The reason is that in cocktails the concentration of individual compounds is usually much lower than for individually tested compounds, so that the weakest binders escape discovery. Compared with the observed hit rates from other campaigns (Supplementary Table S2), the hit rate of 9.4% observed in this study is certainly on the promising side. This may be owing to the high concentration of each individual fragment during the soaking procedure, but it may also highlight that the library presented here is indeed useful and can clearly compete with any of the other libraries described so far. In summary, we have introduced a new library and successfully validated it using crystallographic fragment screening. The results show the applicability of the library to the protein target EP. Moreover, the identified fragment hits may be used as starting points for fragment-based drug discovery (FBDD).
3.17. Experimental considerations
During the screening of the target described here (as well as several other targets; data not shown), a number of challenges related to manual handling during soaking experiments and observations of the behaviour of different targets were encountered. Based on this experience, some important difficulties that can arise are discussed as well as possible solutions. A very important boundary condition is the solubility of the fragments (Schiebel et al., 2015 ▸). For preparing a fragment stock solution, the fragment should be soluble in DMSO, because DMSO is mostly tolerated well by protein crystals during the soaking experiment. Sometimes it may be required, however, to add further solvents such as water or methanol to improve the solubility. In the case where there is no way to improve the fragment solubility, the final choice may be to add the fragment as a suspension to the protein crystals. The next issue to consider is the actual soaking experiment. Fragment stock solution may be added to the crystallization drop or a crystal may be placed into a manufactured solution of the fragment stock in the crystallization solution. In both cases, however, the fragments may crystallize or precipitate, thus making it difficult to identify the protein crystals. Further, the fragment may cause the crystal to crack or even to dissolve during the soaking experiments, which typically last 24 h or longer. In such cases shorter soaking times should be tried. In some cases the space group of the crystals might also change after exposure to certain fragments, although no such case was encountered in the fragment-screening campaign described here. Yet another point to consider is cryoprotection of the crystals before cryocooling to 100 K. Depending on the choice of cryoprotectant, the cryoprotectant itself may influence the outcome of the experiments. Some cryoprotectants, e.g. glycerol, are fragments themselves and can compete with other fragments for binding. In order to avoid washing out of the fragment by the cryoprotectant, it is important to have the fragments present in the cryosolution. In conclusion, the exact experimental design of a fragment-screening campaign depends predominantly on the target itself and on the chemical and physicochemical boundary conditions set by the required solutions. Another important question one might ask is: what is the diffraction resolution that is required in order to be able to reliably identify the presence of a ligand in a crystal structure and its pose? In this aspect, EP is certainly a favourable case as a model system because it diffracts to very high resolution (1.03–1.44 Å). However, using the compound library introduced here, a number of fragment-screening campaigns have already been performed against various targets. In our experience, the presence of ligands can be reliably established at resolutions of 3.0 Å or even slightly lower (data not shown).
3.18. Time requirements for one fragment-screening campaign
The total time required for a complete fragment-screening campaign is about one week, provided that the supply of well diffracting crystals is not a limiting factor. For each fragment, it is advisable to use two crystals. This means that for about 100 fragments, 200 crystals need to be soaked. The manual transfer of the 200 crystals into the respective soaking solution requires about 8–10 h of work. Obviously, this time estimate strongly depends on the crystal-fishing skills of the experimenter, as well as on the adherence of the crystals to the bottom of the well (sitting drop) and/or whether some kind of skin forms around the drop. After a soaking time of 24 h, it takes a further 8–10 h for manual crystal fishing and cryocooling of the crystals. If the cryoprotectant is not yet present in the soaking solution, the cryocooling will take additional time, sometimes even requiring a gentle multi-bath transfer from the soaking to the cryobuffer conditions. Finally, the data collection of 200 crystals can be carried out in 24–48 h if at least a semi-automated beamline equipped with an automated sample changer is available, such as for instance beamline BL14.1 of the Helmholtz-Zentrum Berlin (Mueller et al., 2012 ▸, 2015 ▸). By employing automated data processing (Krug et al., 2012 ▸; Sparta et al., 2016 ▸) and a (semi)-automated refinement pipeline, initial results may be available quickly after the termination of the data-collection experiment.
4. Summary and outlook
In summary, we have presented a novel, diverse and affordable 96-compound library for fragment screening. This library has been tested and validated against the aspartic protease endothiapepsin and a remarkably high hit rate of 9.4% was achieved. Various binding poses have been observed, including two which had not been observed before. Further screening campaigns using the library against different protein targets are currently under way. The library itself is under constant development and improvement in order to facilitate handling and to optimize its affordability, without compromising its diversity. Moreover, as preliminary experiments indicate that natural product scaffolds (such as those from AnalytiCon Discovery, Potsdam, Germany) may lead to even higher hit rates, we plan to complement our library with more such fragments. The library is available to any academic group wishing to carry out a crystallographic fragment-screening campaign.
5. Related literature
The following references are cited in the Supporting Information for this article: Amano et al. (2015 ▸), Davies et al. (2009 ▸), Howard et al. (2006 ▸), Murray et al. (2007 ▸), Drinkwater et al. (2010 ▸), Perryman et al. (2010 ▸), Bauman et al. (2012 ▸), Bauman, Patel, Baker et al. (2013 ▸), Bauman, Patel, Dharia et al. (2013 ▸), Grøftehauge et al. (2013 ▸), Tiefenbrunn et al. (2014 ▸), Hartshorn et al. (2005 ▸) and Verlinde et al. (2009 ▸).
Supplementary Material
PDB reference: endothiapepsin, complex with fragment 29, 4y38
PDB reference: complex with fragment 30, 4y3j
PDB reference: complex with fragment 39, 4ze6
PDB reference: complex with fragment 42, 4y3y
PDB reference: complex with fragment 50, 4y48
PDB reference: complex with fragment 51, 4y4d
PDB reference: complex with fragment 53, 4y4g
PDB reference: complex with fragment 91, 4zea
PDB reference: complex with fragment 97, 4y4j
Supporting Information.. DOI: 10.1107/S2053230X16004623/hv5327sup1.pdf
Acknowledgments
This study was supported by the BMBF Project Frag2Xtal (No. 05K13M1) and by the European Research Council (ERC) of the European Union (grant 268145-DrugProfilBind). The authors are grateful to the Chemical Computing Group Inc. and to Chemaxon LLC for granting no-cost academic licenses for their software.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Amano, Y., Tanabe, E. & Yamaguchi, T. (2015). Bioorg. Med. Chem. 23, 2310–2317. [DOI] [PubMed]
- Baker, M. (2013). Nature Rev. Drug Discov. 12, 5–7. [DOI] [PubMed]
- Bauman, J. D., Patel, D. & Arnold, E. (2012). Top. Curr. Chem. 317, 181–200. [DOI] [PMC free article] [PubMed]
- Bauman, J. D., Patel, D., Baker, S. F., Vijayan, R. S. K., Xiang, A., Parhi, A. K., Martinez-Sobrido, L., LaVoie, E. J., Das, K. & Arnold, E. (2013). ACS Chem. Biol. 8, 2501–2508. [DOI] [PMC free article] [PubMed]
- Bauman, J. D., Patel, D., Dharia, C., Fromer, M. W., Ahmed, S., Frenkel, Y., Vijayan, R. S. K., Eck, J. T., Ho, W. C., Das, K., Shatkin, A. J. & Arnold, E. (2013). J. Med. Chem. 56, 2738–2746. [DOI] [PMC free article] [PubMed]
- Chemical Computing Group Inc. (2010). Molecular Operating Environment (MOE), Montreal, Canada.
- Congreve, M., Carr, R., Murray, C. & Jhoti, H. (2003). Drug Discov. Today, 8, 876–877. [DOI] [PubMed]
- Congreve, M., Chessari, G., Tisi, D. & Woodhead, A. J. (2008). J. Med. Chem. 51, 3661–3680. [DOI] [PubMed]
- Davies, D. R., Mamat, B., Magnusson, O. T., Christensen, J., Haraldsson, M. H., Mishra, R., Pease, B., Hansen, E., Singh, J., Zembower, D., Kim, H., Kiselyov, A. S., Burgin, A. B., Gurney, M. E. & Stewart, L. J. (2009). J. Med. Chem. 52, 4694–4715. [DOI] [PMC free article] [PubMed]
- Deller, M. C. & Rupp, B. (2016). J. Comput. Aided Mol. Des. 29, 817–836. [DOI] [PMC free article] [PubMed]
- Diederichs, K. (2010). Acta Cryst. D66, 733–740. [DOI] [PubMed]
- Dimitropoulos, D., Ionides, J. & Henrick, K. (2006). Curr. Protoc. Bioinformatics, Unit 14.3. 10.1002/0471250953.bi1403s15. [DOI] [PubMed]
- Drinkwater, N., Vu, H., Lovell, K. M., Criscione, K. R., Collins, B. M., Prisinzano, T. E., Poulsen, S. A., McLeish, M. J., Grunewald, G. L. & Martin, J. L. (2010). Biochem. J. 431, 51–61. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fink, T. & Reymond, J.-L. (2007). J. Chem. Inf. Model. 47, 342–353. [DOI] [PubMed]
- Grøftehauge, M. K., Therkelsen, M. Ø., Taaning, R., Skrydstrup, T., Morth, J. P. & Nissen, P. (2013). Acta Cryst. F69, 1060–1065. [DOI] [PMC free article] [PubMed]
- Hajduk, P. J. & Greer, J. (2007). Nature Rev. Drug Discov. 6, 211–219. [DOI] [PubMed]
- Hartshorn, M. J., Murray, C. W., Cleasby, A., Frederickson, M., Tickle, I. J. & Jhoti, H. (2005). J. Med. Chem. 48, 403–413. [DOI] [PubMed]
- Hopkins, A. L., Groom, C. R. & Alex, A. (2004). Drug Discov. Today, 9, 430–431. [DOI] [PubMed]
- Houtman, J. C. D., Brown, P. H., Bowden, B., Yamaguchi, H., Appella, E., Samelson, L. E. & Schuck, P. (2007). Protein Sci. 16, 30–42. [DOI] [PMC free article] [PubMed]
- Howard, N., Abell, C., Blakemore, W., Chessari, G., Congreve, M., Howard, S., Jhoti, H., Murray, C. W., Seavers, L. C. & van Montfort, R. L. (2006). J. Med. Chem. 49, 1346–1355. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Keller, S., Vargas, C., Zhao, H., Piszczek, G., Brautigam, C. A. & Schuck, P. (2012). Anal. Chem. 84, 5066–5073. [DOI] [PMC free article] [PubMed]
- Köster, H. (2012). PhD thesis. Fachbereich Pharmazie, Philipps-Universität Marburg.
- Köster, H., Craan, T., Brass, S., Herhaus, C., Zentgraf, M., Neumann, L., Heine, A. & Klebe, G. (2011). J. Med. Chem. 54, 7784–7796. [DOI] [PubMed]
- Krug, M., Weiss, M. S., Heinemann, U. & Mueller, U. (2012). J. Appl. Cryst. 45, 568–572.
- Kuhnert, M., Köster, H., Bartholomäus, R., Park, A. Y., Shahim, A., Heine, A., Steuber, H., Klebe, G. & Diederich, W. E. (2015). Angew. Chem. Int. Ed. 54, 2849–2853. [DOI] [PubMed]
- Kumar, K. D., Gurusaran, M., Satheesh, S. N., Radha, P., Pavithra, S., Thulaa Tharshan, K. P. S., Helliwell, J. R. & Sekar, K. (2015). J. Appl. Cryst. 48, 939–942.
- Mueller, U., Darowski, N., Fuchs, M. R., Förster, R., Hellmig, M., Paithankar, K. S., Pühringer, S., Steffien, M., Zocher, G. & Weiss, M. S. (2012). J. Synchrotron Rad. 19, 442–449. [DOI] [PMC free article] [PubMed]
- Mueller, U., Förster, R., Hellmig, M., Huschmann, F. U., Kastner, A., Malecki, P., Pühringer, S., Röwer, M., Sparta, K., Steffien, M., Ühlein, M., Wilk, P. & Weiss, M. S. (2015). Eur. Phys. J. Plus, 130, 141–152.
- Murray, C. W., Callaghan, O., Chessari, G., Cleasby, A., Congreve, M., Frederickson, M., Hartshorn, M. J., McMenamin, R., Patel, S. & Wallis, N. (2007). J. Med. Chem. 50, 1116–1123. [DOI] [PubMed]
- Perryman, A. L., Zhang, Q., Soutter, H. H., Rosenfeld, R., McRee, D. E., Olson, A. J., Elder, J. E. & Stout, C. D. (2010). Chem. Biol. Drug Des. 75, 257–268. [DOI] [PMC free article] [PubMed]
- Rees, D. C., Congreve, M., Murray, C. W. & Carr, R. (2004). Nature Rev. Drug Discov. 3, 660–672. [DOI] [PubMed]
- Schiebel, J. et al. (2015). ChemMedChem, 10, 1511–1521.
- Schulz, M. N. & Hubbard, R. E. (2009). Curr. Opin. Pharmacol. 9, 615–621. [DOI] [PubMed]
- Sparta, K., Krug, M., Heinemann, U., Mueller, U. & Weiss, M. S. (2016). J. Appl. Cryst. 49, 10.1107/S1600576716004416.
- Tiefenbrunn, T., Forli, S., Happer, M., Gonzalez, A., Tsai, Y., Soltis, M., Elder, J. H., Olson, A. J. & Stout, C. D. (2014). Chem. Biol. Drug Des. 83, 141–148. [DOI] [PMC free article] [PubMed]
- Verlinde, C. L. M. J. et al. (2009). Curr. Top. Med. Chem. 9, 1678–1687. [DOI] [PMC free article] [PubMed]
- Wang, X. (2015). Masterarbeit. Institute of Pharmaceutical Chemistry, Philipps-Universität Marburg.
- Zhang, Y.-L. & Zhang, Z.-Y. (1998). Anal. Biochem. 261, 139–148. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: endothiapepsin, complex with fragment 29, 4y38
PDB reference: complex with fragment 30, 4y3j
PDB reference: complex with fragment 39, 4ze6
PDB reference: complex with fragment 42, 4y3y
PDB reference: complex with fragment 50, 4y48
PDB reference: complex with fragment 51, 4y4d
PDB reference: complex with fragment 53, 4y4g
PDB reference: complex with fragment 91, 4zea
PDB reference: complex with fragment 97, 4y4j
Supporting Information.. DOI: 10.1107/S2053230X16004623/hv5327sup1.pdf