The expression, purification and crystallization of the DNA-binding Myb domain of LUX ARRHYTHMO from A. thaliana in complex with DNA are reported. The binding affinity of the Myb domain for DNA was measured and was found to be in the low-nanomolar range, suggesting that a single Myb domain is able to bind to its cognate DNA with very high affinity.
Keywords: Myb domain, DNA binding, protein–DNA complex, LUX ARRHYTHMO, Arabidopsis thaliana
Abstract
LUX ARRHYTHMO (LUX) is a Myb-domain transcription factor that plays an important role in regulating the circadian clock. Lux mutations cause severe clock defects and arrhythmia in constant light and dark. In order to examine the molecular mechanisms underlying the function of LUX, the DNA-binding Myb domain was cloned, expressed and purified. The DNA-binding activity of the Myb domain was confirmed using electrophoretic mobility shift assays (EMSAs), demonstrating that the LUX Myb domain is able to bind to DNA with nanomolar affinity. In order to investigate the specificity determinants of protein–DNA interactions, the protein was co-crystallized with a 10-mer cognate DNA. Initial crystallization results for the selenomethionine-derivatized protein and data-set collection statistics are reported. Data collection was performed using the MeshAndCollect workflow available at the ESRF.
1. Introduction
The Myb family, named after avian myeloblastosis virus protein, where it was first described (Klempnauer et al., 1982 ▸), is characterized by one to four or more imperfect Myb-domain repeats and is subdivided into different groups based on sequence homology and the number of Myb domains. The canonical Myb domain is a ∼52-amino-acid primarily α-helical domain with a helix–turn–helix motif which folds into a three-helix bundle. The third helix lies in the major groove of DNA, providing most of the interaction surface. The majority of Myb domains from all kingdoms of life have three regularly spaced bulky hydrophobic residues, most often tryptophans, with an 18- or 19-amino-acid spacing (Kanei-Ishii et al., 1990 ▸). In LUX, however, the second and third tryptophan residues are replaced by a proline (Pro171) and leucine (Leu192) based on sequence alignments of representative Myb domains (Du et al., 2015 ▸). The GARP subfamily of Myb-domain proteins (named after the founding members GOLDEN2, ARR-B and Psr1; Riechmann et al., 2000 ▸) possesses a plant-specific signature sequence SH(A/L)QK(F/Y) in the DNA-binding helix 3. Previous NMR structural studies of the GARP transcription factor ARR10 from Arabidopsis (which shares 60% sequence identity with the LUX Myb domain) revealed that the GARP subfamily contains the highly conserved three-helix bundle fold characteristic of Myb domains (Hosoda et al., 2002 ▸). ARR10 had a relatively low DNA-binding affinity in the high-nanomolar to low-micromolar range, and the NMR structure was not solved in complex with DNA, so the interacting residues could not be fully mapped (Hosoda et al., 2002 ▸). Thus, the widely divergent DNA sequences bound by the GARP-like Myb domains leave open the question of which residues are responsible for DNA-sequence specificity. In order to understand DNA-binding specificity and affinity, we expressed, purified and performed electrophoretic mobility shift assays (EMSAs) of the Myb domain of LUX. These results indicate that LUX is able to bind its cognate DNA with low-nanomolar affinity. Co-crystallization of selenomethionine-substituted LUX Myb domain with a 10-mer DNA was performed and yielded small crystals that diffracted to 2.8 Å resolution. A full data set in space group P1 was collected from a mesh scan comprising a collection of small needle-like crystals. Currently, crystal optimization is being performed in order to increase the resolution of the data and to allow the collection of full data sets from a single crystal for MAD phasing.
2. Materials and methods
2.1. Macromolecule production and DNA-binding assays
The LUX Myb domain (residues 139–200; AT3G46640.1, ecotype Columbia) was cloned into the expression vector pESPRIT2 (Hart & Tarendeau, 2006 ▸; Guilligay et al., 2008 ▸) using the AatII and NotI restriction sites. The plasmid contains an N-terminal 6×His tag followed by a TEV protease cleavage site. Protein expression for selenomethionine incorporation followed standard protocols as described below. Selenomethionine-derivatized (SeMet) LUX Myb domain was produced according to Doublié (2007 ▸). Briefly, SeMet LUX Myb domain was produced in M9 minimal medium using a non-auxotrophic Escherichia coli strain [Rosetta2 (DE3) pLysS cells]. Overnight-grown Luria–Bertani precultures were spun down, washed with M9 medium and used to inoculate 1 l M9 cultures supplemented with 37 mg ml−1 chloramphenicol, 50 mg ml−1 kanamycin, 2 mM MgSO4, 0.4%(w/v) glucose and 0.1 mM MgCl2. The cells were grown at 310 K until an OD600 of 1.2 was reached. Amino acids (100 mg l−1 lysine, threonine and phenylalanine, 50 mg l−1 leucine, valine, isoleucine and l-selenomethionine) were added and the temperature was reduced to 293 K. After 16 h, the cells were harvested by centrifugation at 8000g and 277 K for 30 min. The cells were resuspended in 200 mM CAPS pH 10.5, 500 mM NaCl, 1 mM TCEP (buffer A) supplemented with Benzonase and protease inhibitors (Roche). The cells were lysed by sonication and the cell debris was pelleted at 45 000g and 277 K for 30 min. The supernatant containing the LUX Myb domain was applied onto a 1 ml Ni–NTA column (Qiagen) pre-equilibrated with buffer A. The column was then washed with 15 CV of wash buffer (buffer A + 10 mM imidazole) and subsequently eluted with buffer B (buffer A + 200 mM imidazole). Fractions of interest were pooled and dialysed overnight at 277 K against dialysis buffer (50 mM CAPS pH 9.7, 500 mM NaCl, 1 mM TCEP) in the presence of 2%(w/w) TEV protease in order to cleave the N-terminal 6×His tag. Protein samples were then concentrated and buffer-exchanged against buffer C (50 mM CAPS pH 9.7, 100 mM NaCl, 1 mM TCEP) before being applied onto a 1 ml heparin column (GE Healthcare) and eluted against a 25 CV salt gradient (buffer D: buffer C + 1 M NaCl). Fractions of interest were pooled after the heparin column, buffer-exchanged with buffer C and concentrated to approximately 10 mg ml−1; the concentration was estimated from the UV absorbance at 280 nM using an extinction coefficient calculated by ProtParam (Gasteiger et al., 2005 ▸). All column chromatography was performed at 277 K. The purity of the final fractions was estimated based on SDS–PAGE analysis, with a final purity of greater than 95%. The yield was approximately 1–1.5 mg per litre of culture. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | A. thaliana |
| DNA source | A. thaliana |
| Forward primer | AATTGACGTCAGGGGAAAACACTTAAACGAC |
| Reverse primer | AATTGCGGCCGCTTATTTGAGGTAAAGCCTATAC |
| Expression vector | pESPRIT2 |
| Expression host | E. coli |
| Complete amino-acid sequence of the construct produced† | MGHHHHHHDYDIPTTENLYFQGRQGKTLKRPRLVWTPQLHKRFVDVVAHLGIKNAVPKTIMQLMMNVEGLTRENVASHLQKYRLYLK |
Selenomethione is represented by an underlined M.
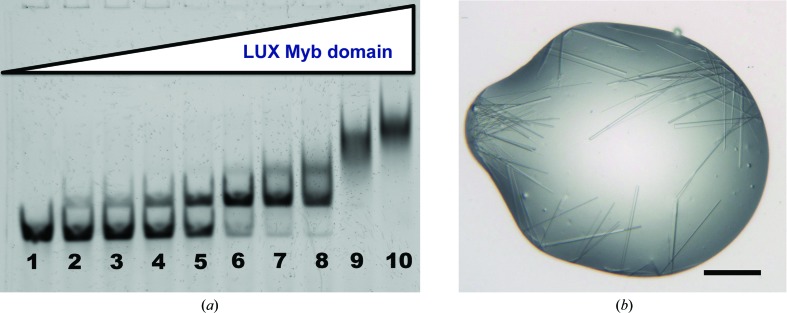
The DNA-binding affinity of the LUX Myb domain was tested via electrophoretic mobility shift assays (EMSAs). A 36 bp DNA oligomer (5′-ATGATGTCTTCTCAAGATTCGATAAAAATGGTGTTG-3′) from the PRR9 promoter containing a LUX DNA-binding site (underlined) was used in the assay. The dsDNA oligomer was Cy5-labelled (Eurofins Genomics) for visualization of the protein–DNA complex. The protein concentration was varied from 0 to 1000 nM (0, 2.5, 5.0, 15, 30, 60, 120, 250, 500 and 1000 nM) using a constant DNA concentration of 10 nM in the reaction. Protein and DNA were incubated at room temperature for 40 min in binding buffer [10 mM Tris pH 7.0, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP, 3% glycerol, 28 ng µl−1 herring sperm DNA (Roche), 20 µg ml−1 BSA, 2.5% CHAPS, 1.27 mM spermidine] and then run on a 8% polyacrylamide gel using 0.5× TBE buffer under nondenaturing conditions at 277 K. The gel was scanned using a ChemiDoc scanner (Bio-Rad). The apparent K d was calculated by quantifying the intensity of the shifted band and the free DNA using the ImageJ software (Schneider et al., 2012 ▸) and plotting the amount of bound DNA versus the protein concentration. The experiment was performed in triplicate. The K d was calculated as 36.7 ± 2.9 nM (Fig. 1 ▸).
Figure 1.
DNA binding and crystallization of the LUX Myb domain. (a) EMSA of the LUX Myb domain bound to a 36 bp fragment of its cognate PRR9 promoter DNA. The protein concentration ranged from 0 to 1000 nM (0, 2.5, 5.0, 15, 30, 60, 120, 250, 500 and 1000 nM in lanes 1–10, respectively) with a constant concentration of 10 nM for the DNA. Based on the band shifts, the binding affinity was calculated to be 36.7 ± 2.9 nM (three replicates). (b) Selenomethionine-derivatized LUX Myb-domain protein in complex with a 10-mer DNA. Crystals grew as small needles with widths ranging from 1 to 5 µm. The black scale bar at the bottom right is approximately 100 µm in length.
2.2. Crystallization
A protein–DNA complex was prepared with SeMet LUX Myb domain using a 1:1.2 protein:DNA molar ratio. The 10-mer dsDNA sequence (forward oligo 5′-TAGATACGCA-3′, reverse oligo 5′-ATGCGTATCT-3′) was ordered as single-stranded oligomers (Eurofins). Equimolar concentrations of the two oligomers were mixed, heated to 368 K for 5 min and annealed on the benchtop, and were used without further purification. Crystallization experiments were carried out by the vapour-diffusion method at 293 K using sitting drops with a 1:1 ratio of protein–DNA complex:precipitant with a final protein concentration of ∼6 mg ml−1. Crystallization was performed by the EMBL High Throughput Crystallization Facility, Grenoble, France. All crystallization experiments were performed in 200 nl sitting drops using a Cartesian PixSys 4200 crystallization robot (Genomic Solutions, UK) using Greiner CrystalQuick plates (flat bottom, untreated) and imaged with a Rock Imager (Formulatrix, USA) at 277 K (Dimasi et al., 2007 ▸). Suitable well diffracting crystals were grown after 2–4 d in 0.1 M bis-tris propane pH 6.5, 20% PEG 3350, 0.2 M sodium malonate. Crystals grew as clusters of needles. Multiple needles were harvested in the same micromesh (MiTeGen) and were flash-cooled in liquid nitrogen without additional cryoprotection. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Vapour diffusion, sitting drop |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 6 |
| Buffer composition of protein solution | 50 mM CAPS pH 9.7, 100 mM NaCl, 1 mM TCEP |
| Composition of reservoir solution | 0.1 M bis-tris propane pH 6.5, 20% PEG 3350, 0.2 M sodium malonate |
| Volume and ratio of drop | 0.2 µl, 1:1 |
| Volume of reservoir (µl) | 50 |
2.3. Data collection and processing
Diffraction data were collected at 100 K on beamline ID23-EH2 (Flot et al., 2010 ▸) at the European Synchrotron Radiation Facility (ESRF), Grenoble, France using the MeshAndCollect protocol (Zander et al., 2015 ▸). In this data-collection protocol, a mesh scan is performed in order to produce a diffractive map of the sample (Bowler et al., 2010 ▸). The best diffracting regions of the sample were thereby identified and oscillation data sets of 10° were collected at the best diffracting positions. These data sets were then processed with XDS (Kabsch, 2010 ▸). Merging the data from all of these mini-data sets yielded a data set of very poor quality. The identification of a subset of mini-data sets was therefore performed using a genetic algorithm (M. Nanao, script available upon request). Merging the resultant 36 partial data sets using XSCALE (Kabsch, 2010 ▸) yielded a complete data set to 2.8 Å resolution. The mosaicity for the partial data sets ranged from 0.1 to 0.8°, with an overall average of 0.29°. The anomalous signal was, however, not sufficiently strong for de novo determination of the phases. Because the data were not ultimately used for SAD phasing, the final processing was performed assuming that the Freidel pairs were equivalent. Data-collection and processing statistics are given in Table 3 ▸. Phasing by molecular replacement with MOLREP (Vagin & Teplyakov, 2010 ▸) and Phaser (McCoy et al., 2007 ▸) using Myb-domain structures from the plant proteins RAD (PDB entry 2cjj; 17% sequence identity; Stevenson et al., 2006 ▸) and ARR10 (PDB entry 1irz; 60% sequence identity; Hosoda et al., 2002 ▸) did not yield any convincing solutions. This is likely to be owing to these structures lacking DNA. As the DNA accounts for approximately half of the total molecular weight of the complex, it is not surprising that molecular replacement was unsuccessful.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Synchrotron |
| Wavelength (Å) | 0.873 |
| Temperature (K) | 100 |
| Detector | Pilatus3 2M |
| Crystal-to-detector distance (mm) | 338.3 |
| Rotation range per image (°) | 0.1 |
| Total rotation range (°) | 360 [36 noncontinuous 10° oscillations] |
| Exposure time per image (s) | 0.1 |
| Space group | P1 |
| a, b, c (Å) | 32.98, 70.95, 67.03 |
| α, β, γ (°) | 103.9, 92.4, 91.0 |
| Mosaicity (°) | 0.29 |
| Resolution range (Å) | 68–2.8 (2.87–2.80) |
| Total No. of reflections | 55177 (3381) |
| No. of unique reflections | 13659 (861) |
| Completeness (%) | 94.1 (82.9) |
| Multiplicity | 4.0 (3.9) |
| 〈I/σ(I)〉 | 8.0 (4.95) |
| R r.i.m. † | 0.49 (1.2) |
| Overall B factor from Wilson plot (Å2) | 19.4 |
The redundancy-independent merging R factor R r.i.m. was estimated by multiplying the conventional R merge value by the factor [N/(N − 1)]1/2, where N is the data multiplicity.
3. Results and discussion
Most Myb transcription factors possess 2–4 Myb-domain repeats, with the R2R3 Myb-domain proteins forming the largest family in plants (Lipsick, 1996 ▸). Transcription factors with multiple Myb domains are able to exploit avidity and site spacing to appropriately bind their cognate DNA sequences and regulate their target genes. LUX, in contrast, is part of a smaller group of single Myb-domain proteins which are largely monomeric. Our results demonstrate, however, that the LUX Myb domain is able to bind to its cognate DNA with low-nanomolar affinity (Fig. 1 ▸ a). The closely related protein ARR10 has the same highly conserved plant-specific motif in DNA-recognition helix 3 and, like LUX, is monomeric; however, it recognizes a different sequence to the LUX Myb domain and exhibits a much lower affinity for DNA (500–1000 nM; Hosoda et al., 2002 ▸). Our results indicate that multiple Myb domains or additional DNA-binding domains are not necessary for specific and low-nanomolar DNA binding by at least some members of the GARP subfamily, which was an open question based on previous structural and biochemical studies. The crystal structure of LUX, when phased, will provide important information as to the specificity determinants of protein–DNA binding as well as revealing how the affinities of two highly related Myb domains from plant transcription factors can vary by orders of magnitude in their ability to bind DNA.
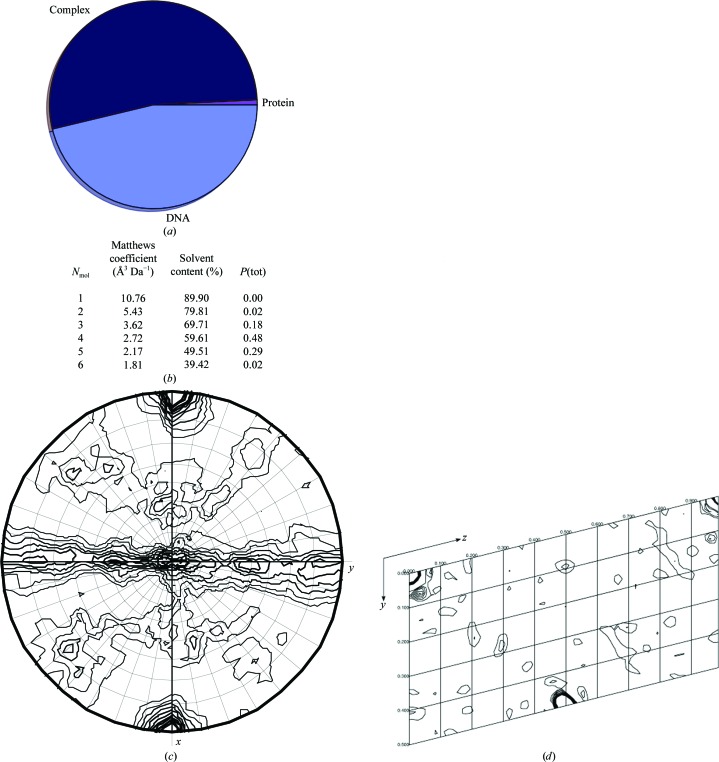
Optimization of protein purification with high-pH buffers was critical for producing sufficient LUX Myb domain for crystallization trials. High-throughput robotic screening yielded multiple conditions with needle-like clusters of crystals. While these crystals diffracted to an acceptable resolution, manual setup of crystallization optimization screens will be required to produce sufficiently large crystals for data collection and phasing (Figs. 1 ▸ b and 2 ▸). We demonstrate that the selenomethionine-derivatization method produces sufficient quantities of LUX Myb-domain protein for crystallization and that the protein–DNA complex will crystallize. In order to confirm the likelihood that a protein–DNA complex, as opposed to protein alone, was present in the crystal, DIBER was used to calculate the likelihood of a complex being present (Fig. 3 ▸ a; Chojnowski et al., 2012 ▸). These results showed a highest probability of a complex being present in the crystals, with a lower probability of DNA alone. Examination of the Matthews coefficient (Fig. 3 ▸ b) indicates the highest probability of the presence of four Myb protein–DNA complexes per asymmetric unit (assuming a molecular weight of 14 kDa for the complex; Matthews, 1968 ▸; Kantardjieff & Rupp, 2003 ▸; Winn et al., 2011 ▸). Examination of the self-rotation function showed strong peaks corresponding to two twofold noncrystallographic rotations (Fig. 3 ▸ c). The Patterson map generated using FFT (Read & Schierbeek, 1988 ▸) from the CCP4 suite (Winn et al., 2011 ▸) gives one strong non-origin Patterson peak, suggesting a noncrystallographic translation (Fig. 3 ▸ d). These tests (the self-rotation function and Patterson analysis) are indicative of clear noncrystallographic symmetry and translations between the multiple molecules in the asymmetric unit. It is most likely that the DNA forms chains in the crystal parallel to the a axis, with regularly spaced Myb domains bound on opposite faces of the DNA, resulting in the observed translational and rotational symmetry. Each DNA oligomer corresponds to the length of the a axis (3.3 Å × 10 bp = 33 Å). In addition, analysis of the local intensity averages using DIBER suggests regular helices oriented parallel to the a axis (Chojnowski et al., 2012 ▸). Each chain would in turn be related by a noncrystallographic twofold rotation.
Figure 2.
Representative diffraction from crystals (space group P1) of the LUX Myb–DNA complex collected on the ESRF ID23-EH2 microfocus beamline. The inset at the top left shows a heat map of the diffraction intensity, with crosses marking the regions automatically selected for data collection and with lighter colours indicating more intense diffraction. Resolution rings are displayed on the diffraction image.
Figure 3.
Characterization of the crystal contents. (a) Probability of crystal contents as calculated using DIBER (Chojnowski et al., 2012 ▸). The probability of protein alone is shown in purple, that of complex is in dark blue and that of DNA alone is in light blue. (b) Matthews coefficients for 1–6 molecules in the asymmetric unit. Four molecules is the most probable content (Matthews, 1968 ▸; Kantardjieff & Rupp, 2003 ▸; Winn et al., 2011 ▸). (c) Self-rotation function calculated using MOLREP (Vagin & Teplyakov, 2010 ▸) for 180°. The radius of integration was 16.5 Å and the full resolution range was used. The convention for orthonormal coordinate systems defines the orthonormal axes, with the crystallographic a axis parallel to x and the reciprocal axis, c*, parallel to z. (d) Patterson section generated using MapSlicer (Winn et al., 2011 ▸) with the v and w axes displayed at u = 0 with default display parameters and the full resolution data used. A peak is observed at 0, 0.5, 0.5. The origin peak height was 79 and the non-origin translational peak height was 40.
The initial studies presented here provide a demonstration that serial crystallography methods and automatic MeshAndCollect data-collection protocols are acceptable ways to collect complete data sets from multiple crystals even for low-symmetry space groups. By exploiting the highly automated data-collection protocols that are now becoming routinely available at many synchrotrons and combining these with methods to select the most isomorphous partial data sets, even multiple needle clusters can provide sufficient diffraction-quality crystals for further characterization and targeted optimization.
Acknowledgments
We would like to acknowledge Darren Hart and Philippe Mas for the use of the pESPRIT2 vector, Philip Wigge for providing us with LUX ARRHYTHMO DNA and the staff of ESRF beamline ID23-2 and the HTX laboratory of the EMBL Grenoble Outstation for their support during experiments. Funding was from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement No. 283570) and ATIP-Avenir (CZ).
References
- Bowler, M. W., Guijarro, M., Petitdemange, S., Baker, I., Svensson, O., Burghammer, M., Mueller-Dieckmann, C., Gordon, E. J., Flot, D., McSweeney, S. M. & Leonard, G. A. (2010). Acta Cryst. D66, 855–864. [DOI] [PubMed]
- Chojnowski, G., Bujnicki, J. M. & Bochtler, M. (2012). Bioinformatics, 28, 880–881. [DOI] [PMC free article] [PubMed]
- Dimasi, N., Flot, D., Dupeux, F. & Márquez, J. A. (2007). Acta Cryst. F63, 204–208. [DOI] [PMC free article] [PubMed]
- Doublié, S. (2007). Methods Mol. Biol. 363, 91–108. [DOI] [PubMed]
- Du, H., Liang, Z., Zhao, S., Nan, M.-G., Tran, L.-S. P., Lu, K., Huang, Y.-B. & Li, J.-N. (2015). Sci. Rep 5, 11037. [DOI] [PMC free article] [PubMed]
- Flot, D., Mairs, T., Giraud, T., Guijarro, M., Lesourd, M., Rey, V., van Brussel, D., Morawe, C., Borel, C., Hignette, O., Chavanne, J., Nurizzo, D., McSweeney, S. & Mitchell, E. (2010). J. Synchrotron Rad. 17, 107–118. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Guilligay, D., Tarendeau, F., Resa-Infante, P., Coloma, R., Crepin, T., Sehr, P., Lewis, J., Ruigrok, R. W., Ortin, J., Hart, D. J. & Cusack, S. (2008). Nature Struct. Mol. Biol. 15, 500–506. [DOI] [PubMed]
- Hart, D. J. & Tarendeau, F. (2006). Acta Cryst. D62, 19–26. [DOI] [PubMed]
- Hosoda, K., Imamura, A., Katoh, E., Hatta, T., Tachiki, M., Yamada, H., Mizuno, T. & Yamazaki, T. (2002). Plant Cell, 14, 2015–2029. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kanei-Ishii, C., Sarai, A., Sawazaki, T., Nakagoshi, H., He, D.-N., Ogata, K., Nishimura, Y. & Ishii, S. (1990). J. Biol. Chem. 265, 19990–19995. [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Klempnauer, K. H., Gonda, T. J. & Bishop, J. M. (1982). Cell, 31, 453–463. [DOI] [PubMed]
- Lipsick, J. S. (1996). Oncogene, 13, 223–235. [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Read, R. J. & Schierbeek, A. J. (1988). J. Appl. Cryst. 21, 490–495.
- Riechmann, J. L. et al. (2000). Science, 290, 2105–2110. [DOI] [PubMed]
- Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. (2012). Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed]
- Stevenson, C. E. M., Burton, N., Costa, M. M. R., Nath, U., Dixon, R. A., Coen, E. S. & Lawson, D. M. (2006). Proteins, 65, 1041–1045. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. (2011). Acta Cryst. D67, 235–242.
- Zander, U., Bourenkov, G., Popov, A. N., de Sanctis, D., Svensson, O., McCarthy, A. A., Round, E., Gordeliy, V., Mueller-Dieckmann, C. & Leonard, G. A. (2015). Acta Cryst. D71, 2328–2343. [DOI] [PMC free article] [PubMed]