PvdN (PA2394) from P. aeruginosa is a nonribosomal peptide synthetase tailoring enzyme that is involved in pyoverdine biosynthesis. PvdN has been identified as a periplasmic protein that is pyridoxal 5′-phosphate-dependent and shows homology to class I aminotransferases. Here, the crystal structure of PvdN is reported at 1.22 Å resolution.
Keywords: PvdN, pyoverdine, nonribosomal peptide synthetase, Pseudomonas aeruginosa
Abstract
The Gram-negative pathogen Pseudomonas aeruginosa uses a nonribosomal peptide synthetase (NRPS) biosynthetic cluster for the production of a peptide siderophore. In addition to four multimodular NRPS proteins, the biosynthetic pathway also requires several additional enzymes involved in the production of nonproteinogenic amino acids and maturation of the peptide product. Among the proteins that are required for the final steps in pyoverdine synthesis is PvdN, a pyridoxal phosphate-dependent enzyme that catalyzes an uncharacterized step in pyoverdine production. This study reports the high-resolution structure of PvdN bound to a PLP cofactor solved by multi-wavelength anomalous dispersion (MAD). The PvdN model shows high structural homology to type I aspartate aminotransferases and also contains positive density that suggests an uncharacterized external aldimine.
1. Introduction
Natural products derived from nonribosmal peptide sythetases (NRPSs) are composed of a wide array of bioactive metabolites that are essential for host survival in specific environments and therefore of interest to both the medicinal and biotechnology fields (Fischbach & Walsh, 2009 ▸). While the peptide backbones of these natural products are the purview of the large covalently linked domains of the NRPSs themselves, much of the diversity is provided by standalone tailoring enzymes that modify this backbone through a multitude of reactions including methylation, epimerization, hydroxylation and heterocyclization (Walsh et al., 2001 ▸).
PLP aminotransferases utilize a vitamin B6-derivative cofactor as an electron sink, allowing them to carry out multiple reaction scenarios. They are classified into five main classes based on structural features (Hester et al., 1999 ▸; Schneider et al., 2000 ▸), with the most common being fold type I or the aspartate aminotransferase (AAT) superfamily. This superfamily has recently been shown to be active in PKS/NRPS assemblies (Milano et al., 2013 ▸) and to play an integral part in introducing structural modifications into secondary metabolites.
In Pseudomonas aeruginosa, multiple NRPS and tailoring enzymes synthesize the siderophore pyoverdine. The final step for the siderophore precursor is transport to the periplasm, where maturation and diversification occurs and the molecule is exported out of the cell (Schalk & Guillon, 2013 ▸). We have previously determined the structure of PvdQ, a periplasmic acylase that removes a transiently installed fatty acid from the pyoverdine precursor (Drake & Gulick, 2011 ▸). Another of the periplasmic tailoring enzymes (PA2394, PvdN) is an essential gene in pyoverdine biosynthesis (Voulhoux et al., 2006 ▸) that shows homology to other pyridoxal 5′-phosphate (PLP)-dependent aminotransferases.
To continue our studies of pyoverdine maturation and to further characterize PvdN, we present its structure at 1.2 Å resolution.
2. Materials and methods
2.1. Macromolecule production
PvdN (PA2394) was PCR-amplified from P. aeruginosa genomic DNA using primers to incorporate restriction sites at the 5′ and 3′ ends of the gene (Table 1 ▸). The gene was then subcloned into a modified pET-15b vector. To allow the PvdN protein to be properly directed to the periplasm, our cloning strategy used the pET-15b NcoI site, resulting in the removal of the plasmid-encoded N-terminal 5×His tag. For proper NcoI digestion, the second residue of PvdN was mutated from an asparagine to an aspartic acid (AAC to GAC). All sequences were confirmed by DNA sequencing. The final plasmid was transformed into Escherichia coli BL21(DE3) cells for protein production.
Table 1. Macromolecule-production information.
| Source organism | P. aeruginosa (PAO1) |
| DNA source | P. aeruginosa (PAO1) |
| Forward primer (5′ to 3′)† ‡ | CATGCCATG GACGACCGTCGTACCTTTCTCAAGCAGC |
| Reverse primer (5′ to 3′)† | GGAATTCCATATGTCAGGCGAGTTGCGGGGCGAG |
| Cloning vector | pET15b, TEV-modified |
| Expression vector | pET15b, TEV-modified |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced‡ § | M D DRRTFLKQAGILAAGLPLLSAAQSLRAEGVPSDAGNKWKALRQQFDLDPQYLHFANFLLTSHPRPVREAIERLRVRFDRNPGEAVDWHREEIWKYEDEARAWAGRYFAVQPGQVALTGSTTDGLAAIYGGLLVQPGKEILTSSHEHYSTYTTLEYRHKRMGTQVREFPLFKDPHRVSADEILSSIAAQIRPQTRVLGMTWVQSGSGVKLPIREIGKLVRELNQKRDEQDRIIYVVDGVHGFGVEDVSFADFDCDYFIAGTHKWLFGPRGTGVIIARSEQLQEHLVPSIPTFSRADNFGTLMTPGGYHAFEHRLALGTAFELHLQLGKAEVQARIHQLNAYLKQRLGEHPKVRLVTPTSPELSSGFTFFRVEGRDCEAVAKHLMAHRVISDAVDRDVGPVVRLAPSLLNDEAEIDRVLEILAPQLA |
Restriction sites (NcoI for the forward primer and NdeI for the reverse primer) are underlined.
The designed point mutation is shown in bold.
The predicted periplasmic signaling sequence is underlined.
For native protein production, the cells were grown in lysogeny broth (LB) at 37°C and aerated at 250 rev min−1 until an optical density (OD) at 595 nm of 0.6 was reached, induced with 750 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) and then grown overnight at 16°C while shaking at 250 rev min−1. Selenomethionine-derivatized (SeMet) protein was grown in selenomethionine-labeling autoinduction medium (Studier, 2005 ▸) at 25°C with shaking at 225 rev min−1 for 60 h. The periplasmic proteins were then harvested.
In brief, the method consisted of centrifugation at 5000g for 10 min at 10°C and resuspension at room temperature in 10 mM Tris–HCl pH 8.0 at 40 ml g−1, centrifugation at 3500g for 10 min at 10°C and resuspension at room temperature in 500 mM sucrose, 30 mM Tris–HCl pH 8.0, 2 mM Na2EDTA at 40 ml g−1, centrifugation at 4000g for 10 min at 10°C and resuspension at 4°C in 10 mM Tris–HCl pH 8.0 at 25 ml g−1, and centrifugation at 5000g for 10 min at 4°C. The supernatant was then combined with a stock solution of 1.5 M Tris–HCl pH 8.0 to a final concentration of 50 mM and further clarified by centrifugation. All of the remaining purification steps were performed at 4°C.
The periplasmic lysate was then subjected to 60% ammonium sulfate precipitation with solid ammonium sulfate. The lysate was equilibrated for 1 h and the precipitate was harvested by centrifugation and resuspended in 50 mM Tris–HCl pH 8.0. The protein was passed over a 5 ml HiTrap desalting column (GE Healthcare) in 50 mM Tris–HCl pH 8.0. The fractions were pooled based on their OD280 and then run over a 5 ml HiTrap Q FF column (GE Healthcare) using the same buffer. The protein was found in the initial flowthrough by SDS–PAGE analysis and these fractions were pooled, concentrated and dialyzed overnight against gel-filtration buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 0.2 mM TCEP). Gel filtration was performed on a Superdex 200 HiLoad 16/60 column and the protein was concentrated to ∼8 mg ml−1 using an extinction coefficient at 280 nm of 48 268 M −1 cm−1. The isolated protein showed a distinct absorption peak at 420 nm attributable to the presence of PLP cofactor (data not shown). A typical yield for the purified enzyme was 0.86 mg per gram of cell pellet.
2.2. Crystallization
PvdN crystals were optimized from leads first identified from an in-house sparse-matrix screen. Crystals of the native protein were grown at 20°C by hanging-drop vapor diffusion using a 1:1 mixture of protein and precipitant consisting of 10–14% PEG 8000, 100–200 mM glycine, 50 mM MES pH 6.0. Native crystals were mounted in nylon loops and cryoprotected by transferring them through three precipitant solutions containing increasing amounts of ethylene glycol (8, 16 and 24%) for approximately 60 s each and were then stored in liquid nitrogen. The final cryoprotectant solution consisted of 24% ethylene glycol, 16% PEG 8000, 200 mM glycine, 50 mM MES pH 6.0. The crystallization conditions for SeMet crystals were optimized to 6–10% PEG 8000, 200–300 mM glycine, 50 mM MES pH 6.0 and followed the same cryoprotection protocol. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Hanging-drop vapor diffusion |
| Plate type | 24-well Linbro |
| Temperature (K) | 287 |
| Protein concentration (mg ml−1) | 8 |
| Buffer composition of protein solution | 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 0.2 mM TCEP |
| Composition of reservoir solution | 10–14% PEG 8000, 100–200 mM glycine, 50 mM MES pH 6.0 |
| Volume and ratio of drop | 4 µl, 1:1 |
| Volume of reservoir (µl) | 600 |
2.3. Data collection and processing
X-ray diffraction data were collected at 100 K on beamline 9-2 at the Stanford Synchotron Radiation Lightsource (SSRL), Menlo Park, California, USA.
A single heavy-atom-derivatized (SeMet) crystal provided phase information through three unique data sets (peak, inflection and remote).
A native data set was collected by merging a low-resolution scan and high-resolution scan from the same crystal. The low-resolution data were collected first at a distance of 300 mm with 1.5° oscillations over 180° and 1 s exposures. This data set was scaled and merged with a high-resolution scan of the same crystal collected at a distance of 150 mm with 0.5° oscillations over 180° and 2 s exposures.
Data were processed in space group P21 using HKL-2000 (Otwinowski & Minor, 1997 ▸).
Data-collection and processing statistics for the SeMet and the high-resolution native crystal are summarized in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the highest resolution shell.
| SeMet (peak) | SeMet (inflection) | SeMet (remote) | Native | |
|---|---|---|---|---|
| Diffraction source | Beamline 9-2, SSRL | Beamline 9-2, SSRL | Beamline 9-2, SSRL | Beamline 9-2, SSRL |
| Wavelength (Å) | 0.9791 | 0.9794 | 0.9565 | 0.9791 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Detector | MARmosaic 325 | MARmosaic 325 | MARmosaic 325 | MARmosaic 325 |
| Crystal-to-detector distance (mm) | 300 | 300 | 300 | 150 |
| Rotation range per image (°) | 2 | 2 | 2 | 0.5 |
| Total rotation range (°) | 180 | 180 | 180 | 180 |
| Exposure time per image (s) | 2 | 2 | 2 | 2 |
| Space group | P21 | P21 | P21 | P21 |
| a, b, c (Å) | 62.4, 98.9, 63.2 | 62.4, 98.9, 63.2 | 62.4, 98.9, 63.2 | 62.52, 99.00, 63.05 |
| α, β, γ (°) | 90, 102.32, 90 | 90, 102.32, 90 | 90, 102.32, 90 | 90, 102.32, 90 |
| Resolution range (Å) | 49.90–2.30 | 49.90–2.30 | 49.90–2.30 | 29.41–1.22 (1.26–1.22) |
| Total No. of reflections | 127844 | 126968 | 127216 | 1042761 |
| No. of unique reflections | 33502 | 33480 | 33539 | 221938 (21937) |
| Completeness (%) | 100.0 | 99.90 | 99.90 | 99.88 (98.99) |
| 〈I/σ(I)〉 | 12.3 | 12.2 | 11.5 | 22.92 (3.30) |
| R merge (%) | 8.6 | 8.4 | 9.4 | 3.7 |
| Overall B factor from Wilson plot (Å2) | 14.21 |
2.4. Structure solution and refinement
Initial assessment of the heavy-atom data set using the Matthews coefficient predicted the presence of two copies in the asymmetric unit with a solvent content of 38%, providing ten selenium sites.
The heavy-atom substructure was determined with SOLVE (Terwilliger & Berendzen, 1999 ▸), which found eight of the ten heavy-atom sites. This was then subjected to solvent flattening and automated chain tracing using RESOLVE (Terwilliger, 2000 ▸), fitting 517 of the 854 residues in the asymmetric unit. The resulting model was fed into Buccaneer (Cowtan, 2006 ▸), which fitted 650 of the 854 residues in the asymmetric unit.
The phases from Buccaneer were merged with the higher resolution native data set and submitted to ARP/wARP (Langer et al., 2008 ▸). The resulting model was used for continued cycles of manual model building with Coot (Emsley & Cowtan, 2004 ▸), REFMAC5 (Murshudov et al., 2011 ▸), PHENIX (Adams et al., 2010 ▸) and the PDB_REDO server (Joosten et al., 2014 ▸). Residues 1–38 were not present in either subunit. The periplasmic signaling sequence for PvdN is predicted to undergo proteolytic cleavage between residues 29 and 30 (Petersen et al., 2011 ▸). The N-terminal residues 30–38 were not modelled and were presumably in a disordered state. Refinement statistics are summarized in Table 4 ▸.
Table 4. Structure refinement.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 29.41–1.22 (1.26–1.22) |
| Final R cryst | 0.1396 (0.1963) |
| Final R free | 0.1583 (0.2067) |
| No. of non-H atoms | |
| Protein | 6426 |
| Ligand | 52 |
| Water | 780 |
| Total | 7143 |
| R.m.s. deviations | |
| Bonds (Å) | 0.006 |
| Angles (°) | 0.880 |
| Average B factors (Å2) | |
| Overall | 18.89 |
| Protein | 18.01 |
| Ligand | 18.50 |
| Water | 27.46 |
| Ramachandran plot | |
| Favored regions (%) | 98.0 |
| Additionally allowed (%) | 2.0 |
3. Results and discussion
3.1. Structure overview
The protein is a tightly integrated homodimer consisting of a dimer interface with ∼2300 Å2 of solvent-accessible surface buried per monomer or roughly 14% of the total surface area (Figs. 1 ▸ and 2 ▸ a). Each subunit is organized into two distinct α/β domains, colloquially referred to as the small (α) and large (β) domain (Figs. 1 ▸ and 2 ▸ b). The shared surface area is symmetrical and distributed across both the small and large domains. While the active-site residues and PLP cofactors are mostly monomer-specific, the dimer interface helps create a substrate pocket or cleft composed of residues from both monomers and across subdomains.
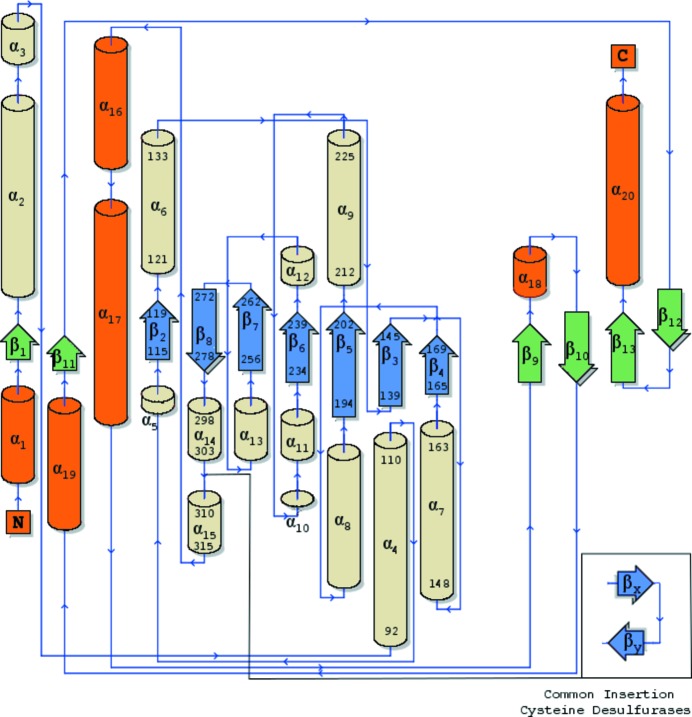
Figure 1.
Topology diagram of secondary-structure elements in PvdN. Small domain components are shown with orange α-helices and green β-sheets, while large domain elements have wheat α-helices and blue β-sheets. Residue numbers are labeled for the secondary-structure elements detailed in the text. The inset displays a common insertion in the cysteine desulfurase family. The diagram was created using the PDBSum server at https://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html (Laskowski, 2009 ▸).
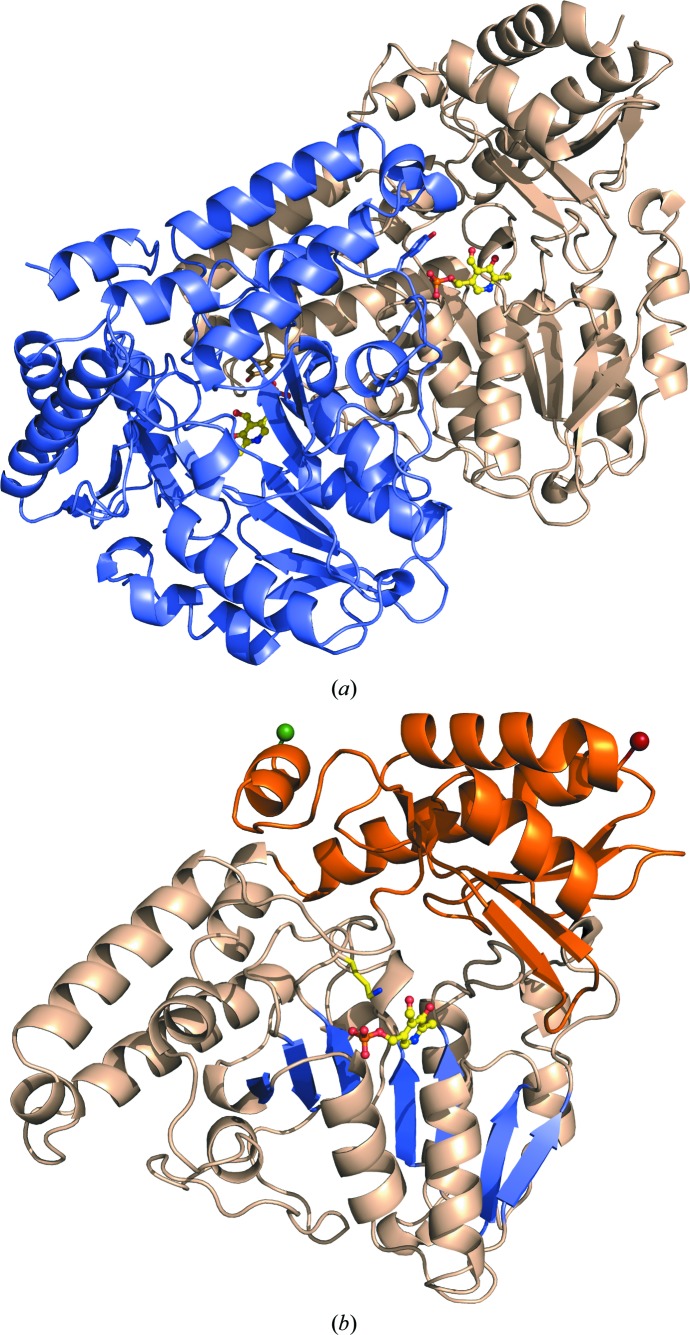
Figure 2.
Three-dimensional structure of PvdN. (a) The secondary structure is depicted for both subunits in the homodimer, with one subunit in wheat and the other in light blue. The PLP cofactor is shown in ball-and-stick representation with yellow C atoms, blue N atoms, red O atoms and orange P atoms. The tyrosine caps (Tyr308) contributing to the ligand pocket from each adjacent monomer are highlighted as sticks. (b) A single subunit of the homodimer with the N-terminal residue as a green sphere and the C-terminal residue as a red sphere. The small domain is shown in orange and the large domain in wheat. The seven-stranded β-sheet (β2–β8) in the large domain is highlighted in blue, with the PLP cofactor shown in ball-and-stick representation (yellow C atoms, blue N atoms, red O atoms and orange P atoms). The active-site lysine (Lys264) off the loop connecting β7 and β8 is shown extended towards the PLP.
The small domain contains a four-stranded antiparallel β-sheet trapped against the large domain with three α-helices. The domain is composed of residues from both the N-terminus (38–58) and C-terminus (328–427). The large domain displays the canonical seven-stranded β-sheet (β2–β8) and is flanked by α-helices, most notably α4 and α9 on the solvent-exposed surface and helices α6 and α7 at the dimer interface. This central sheet, along with the dimer-flanking helices, also house the active site and PLP cofactor. The conserved active-site lysine (Lys264) that is PLP-associated is found on a small loop connecting β7 and β8 (Fig. 2 ▸).
The substrate-binding cavity and bound PLP cofactor are located at the base of the substrate pocket and provide solvent access from two unobstructed channels. The larger of the channels leads to the 5′-phosphate of the PLP and has walls composed from both subunits including a tyrosine (Tyr308) cap from the dimer large domain extending off the loop connecting α14 and α15 of the neighboring subunit. A smaller channel formed only from the residues of the subunit concerned provides exposure to the solvent for the C2 substituent of the PLP aromatic ring.
3.2. Active site
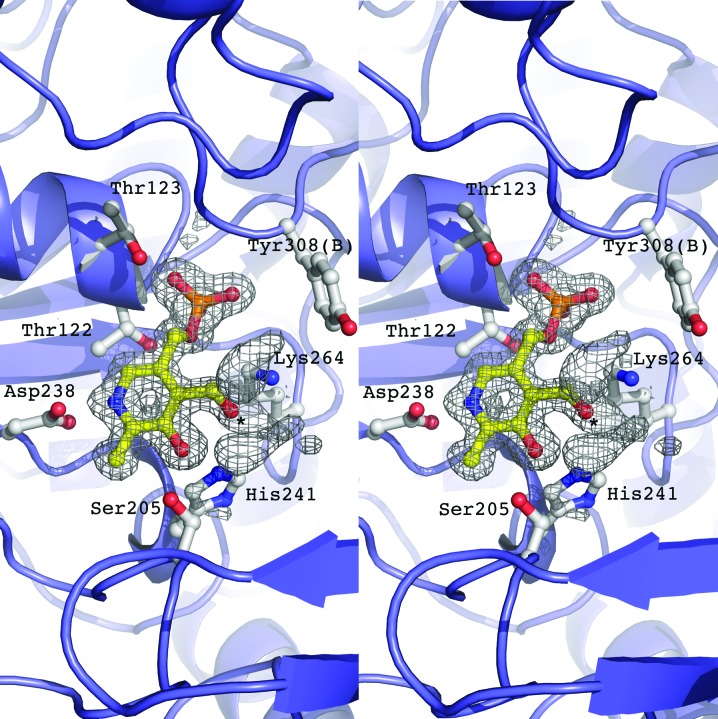
A PLP external aldimine is modeled into the active site of both monomers (Fig. 3 ▸) in the homodimer and the interacting residues are symmetrical between monomers. The aromatic ring is anchored between a π-stacking interaction with His148 and hydrophobic contacts of Val240 with the aromatic substituent and of Thr201 with the C2 substituent. This forces the pyridine N atom of the PLP to face inwards towards the large domain and places the pyridine N atom 2.57 Å from the conserved aspartic acid (Asp238). Additionally, this turns the C4 atom of the PLP towards the dimer interface and places it in proximity to the lysine (Lys264) that normally participates in the covalent linkage for an enzyme aldimine. The phosphate tail of the PLP is then kinked back towards the central sheet, sharing hydrogen bonds with residues from its associate monomer and the amino group of the tyrosine cap (Tyr308) contributed from the dimer.
Figure 3.
Stereoimage of the catalytic active site of PvdN. Residues that create a hydrogen-bonding network with the PLP cofactor are depicted. The electron density is calculated with coefficients of the form F o − F c generated prior to the inclusion of the cofactor in the active site and is contoured at 2.5σ. Positive density is present off the PLP O4 atom (denoted by an asterisk), suggesting an external aldimine.
Whereas many crystal structures of PLP-dependent enzymes are observed with an internal aldimine covalently bound via a Schiff-base linkage to the conserved lysine (Lys264 in PvdN), the current structure contains electron density suggestive of an external aldimine with partial occupancy. The PLP ligand was incorporated during bacterial growth and remained in the active site even after the purification and crystallization protocols, showing that a covalent adduct had been formed with the amine N atom on the unknown substrate and replacing the N atom of Lys264. The external aldimine conformation is confirmed by the electron-density data (Fig. 3 ▸), which clearly show positive density off the O4 atom of the PLP. This adduct is stabilized by interaction with both the PLP hydroxyl and probable salt links with two arginines (Arg396 and Arg403) that protrude into the substrate-binding pocket from the large domain. This adduct was not built into the structure directly as the exact nature of the amino acid has not been elucidated.
3.3. Structure comparison
Sequence-similarity searches place PvdN as a class V aminotransferase with a twin-arginine translocation sequence for periplasmic localization. Included in this class are cysteine desulfurases and cystine lyases that have been shown to play crucial roles in iron–sulfur (Fe–S) cluster biosynthetic systems (Johnson et al., 2005 ▸). A structure-homology search with DALI (Holm & Rosenström, 2010 ▸) indeed shows a cystine lyase (C-DES; PDB entry 1n31; Kaiser et al., 2003 ▸) to be the most similar structure, with a Z-score of 43.9, with the majority of other high-similarity structures also being cysteine desulfurases.
PvdN shares a homodimer quaternary structure with C-DES and has a tertiary structure that is almost identical, with a root-mean-square displacement (r.m.s.d.) across all Cα atoms of 1.89 Å. This is not surprising as all fold type I aminotransferases contain the small/large domain architecture, with the large domain housing the seven-stranded β-sheet and associated active site. However, the active sites of the structurally related cysteine desulfurases/cystine lyases, including C-DES, show large differences from that of PvdN. Cysteine desulfurases generally have a catalytic cysteine residue or utilize a cystine substrate in conjunction with a PLP cofactor for sulfur mobilization. PvdN does not have an active-site catalytic cysteine nor the mobile loop on which this residue is normally situated. Although the true substrate of PvdN is not yet known, the mechanism for sulfur mobilization using a cystine substrate has been examined (Kaiser et al., 2003 ▸). This mechanism in C-DES relies on two conserved arginines (Arg360 and Arg369) and a tryptophan (Trp251), contributed from the dimer, for proper positioning of the cystine substrate. PvdN only shares Arg369 with the cystine lyases, with no other significant shared active-site residues. Interestingly, the dimer-contributed tryptophan in C-DES sits adjacent to an insertion between α14 and α15 that is common to the cysteine desulfurases/cystine lyases and that is lacked by PvdN (Fig. 1 ▸, inset).
The exact role of PvdN in pyoverdine biosynthesis has yet to be resolved. Although it is interesting to note that the closest structural homologs are cysteine desulfurases/cystine lyases involved in Fe–S synthetic clusters, no literature yet points to these systems or proteins being involved in pyoverdine biogenesis. Analysis of the active site in PvdN and associated structural elements also appears to contradict this homology and points to a more traditional aminotransferase mechanism of action.
Supplementary Material
PDB reference: PvdN, 5i90
Acknowledgments
This research was supported in part by grants from the National Institutes of Health (GM116957, AMG). Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, NCRR, Biomedical Technology Program and the National Institute of General Medical Sciences.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Drake, E. J. & Gulick, A. M. (2011). ACS Chem. Biol. 6, 1277–1286. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fischbach, M. A. & Walsh, C. T. (2009). Science, 325, 1089–1093. [DOI] [PMC free article] [PubMed]
- Hester, G., Stark, W., Moser, M., Kallen, J., Marković-Housley, Z. & Jansonius, J. N. (1999). J. Mol. Biol. 286, 829–850. [DOI] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Johnson, D. C., Dean, D. R., Smith, A. D. & Johnson, M. K. (2005). Annu. Rev. Biochem. 74, 247–281. [DOI] [PubMed]
- Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. (2014). IUCrJ, 1, 213–220. [DOI] [PMC free article] [PubMed]
- Kaiser, J. T., Bruno, S., Clausen, T., Huber, R., Schiaretti, F., Mozzarelli, A. & Kessler, D. (2003). J. Biol. Chem. 278, 357–365. [DOI] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A. (2009). Nucleic Acids Res. 37, D355–D359. [DOI] [PMC free article] [PubMed]
- Milano, T., Paiardini, A., Grgurina, I. & Pascarella, S. (2013). BMC Struct. Biol. 13, 26. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. (2011). Nature Methods, 8, 785–786. [DOI] [PubMed]
- Schalk, I. J. & Guillon, L. (2013). Environ. Microbiol. 15, 1661–1673. [DOI] [PubMed]
- Schneider, G., Käck, H. & Lindqvist, Y. (2000). Structure, 8, R1–R6. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Voulhoux, R., Filloux, A. & Schalk, I. J. (2006). J. Bacteriol. 188, 3317–3323. [DOI] [PMC free article] [PubMed]
- Walsh, C. T., Chen, H., Keating, T. A., Hubbard, B. K., Losey, H. C., Luo, L., Marshall, C. G., Miller, D. A. & Patel, H. M. (2001). Curr. Opin. Chem. Biol. 5, 525–534. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PvdN, 5i90