Abstract
The hematopoietic stem cell (HSC) is arguably the most extensively characterized tissue stem cell. Since its identification by prospective isolation1, complex multi-parameter flow cytometric isolation of phenotypic subsets has facilitated studies on many aspects of HSC biology including, self-renewal2–4, differentiation, aging, niche5, and diversity6–8. Here we demonstrate by unbiased multi-step screening, identification of a single gene, Hoxb5 (homeobox B5 also known as Hox-2.1), whose expression in the bone marrow (BM) is limited to the long-term HSC (LT-HSC) in mice. Utilizing a single-color tri-mCherry reporter mouse driven by endogenous Hoxb5 regulation, only the Hoxb5-positive HSCs exhibit long-term reconstitution capacity after transplantation in primary transplant recipients, and critically, in secondary recipients. Only 7–35% of various previously defined immunophenotypic HSCs are LT-HSCs. Finally, by in situ imaging of mouse BM, we show that >94% of LT-HSC (Hoxb5+) are directly attached to VE-cadherin-positive cells, implicating a perivascular space as a near homogenous localization of the LT-HSC.
Prospective isolation of HSCs requires that the isolated cells are capable of long-term (LT) production of all blood cell types in primary irradiated hosts, as well as self renewal, such that the cells can transplant to secondary hosts to give rise to long-term multilineage repopulation. From the first enrichments and isolations of candidate HSCs1,9,10, this activity has been entirely contained in cell surface marker-defined cell populations, and more recently in fluorescent reporters11–13. However, the precise fraction of cells in those populations that are true LT-HSCs remains controversial.
To enable further purification of LT-HSC we sought to identify genes expressed exclusively in HSCs within cells resident in mouse BM, detectable by flow cytometry and in-situ fluorescence, and thus performed the following four-step screening (Fig. 1d).
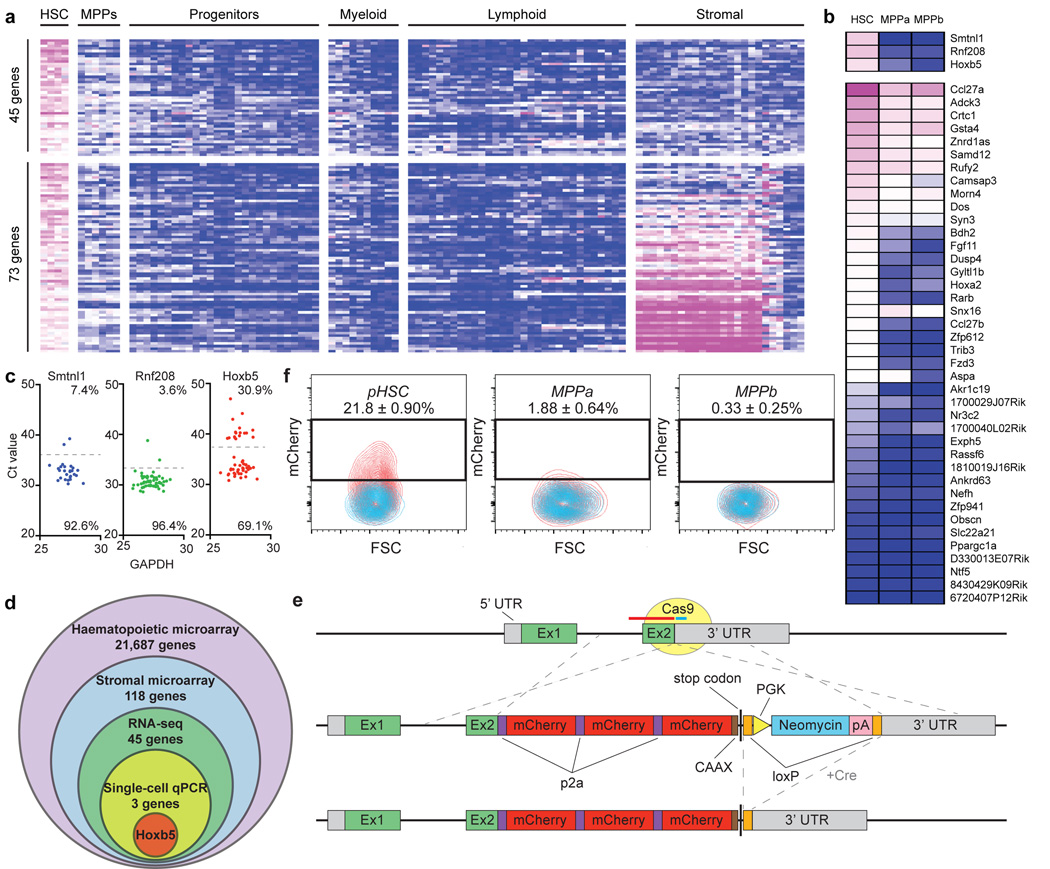
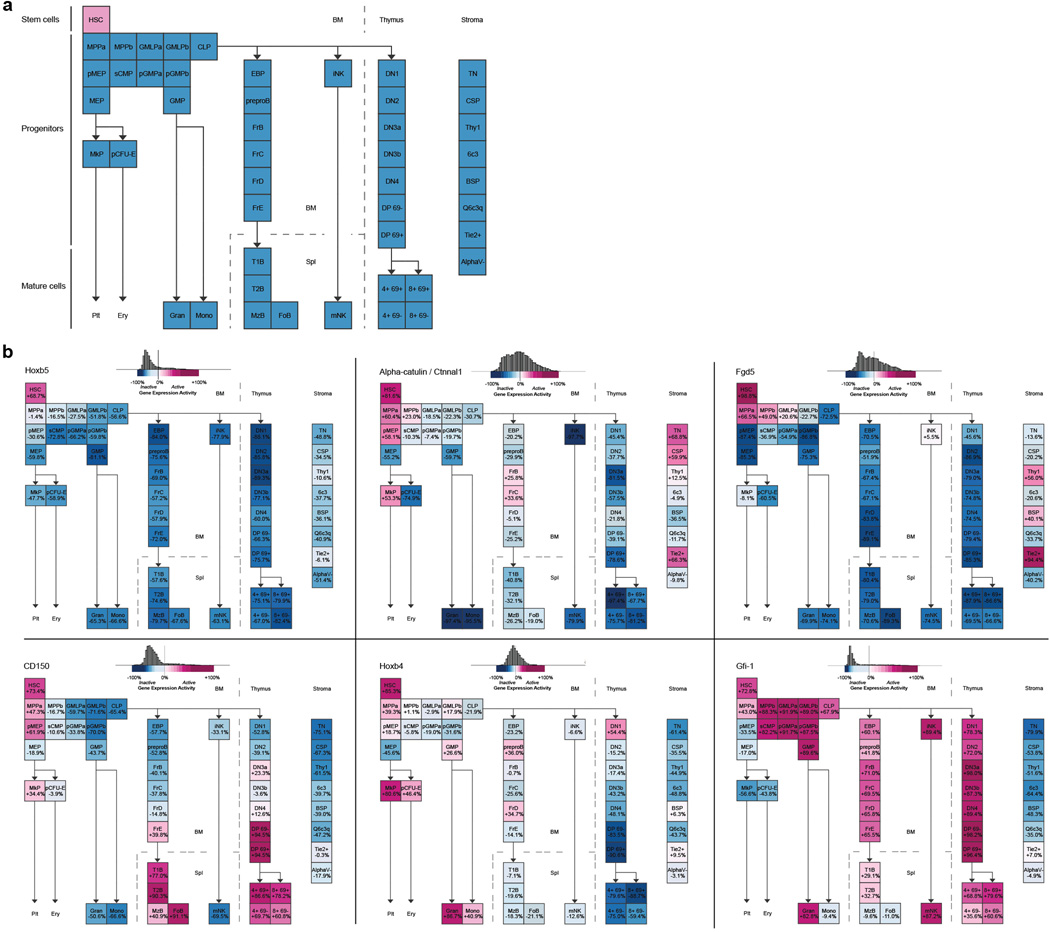
Figure 1. Multi-step unbiased screening identifies Hoxb5 as an LT-HSC marker.
a, Microarray heat map depicting relative expression (pink = high, blue = low) of HSC-specific genes in haematopoietic and stromal populations. Each row represents a gene; each subcolumn a replicate microarray; each labeled column a category of cell populations. The 45 genes in the top panel displayed limited activity in all non-HSC populations examined. b, Transcriptional profiling by RNA-seq of the 45 genes from (a). Three genes (top panel) exceeded the estimated threshold for detection (FPKM > 7.0) in HSCs while showing minimal expression (FPKM < 2.5) in MPPa and MPPb populations. c, Heterogeneity of expression for the three remaining candidate genes in HSCs as assessed by single-cell qPCR. d, Venn diagram reflecting the four-step screen that yielded Hoxb5 as an ideal candidate in the HSC transcriptome. e, Targeting strategy to generate a triple-mCherry Hoxb5 knock-in mouse reporter line (Hoxb5-tri-mCherry). f, Hoxb5 reporter expression (red) in immunophenotypic HSCs (pHSC) and MPPs compared to wild-type controls (blue). Values indicate the percentage of mCherry-positive cells ± S.D. in each fraction for n=3 mice.
First we compared microarray gene expression assays among 28 distinct populations of the hematopoietic system (Extended Data Fig. 1a and Supplementary Table 1). Using Gene Expression Commons14, we identified 118 candidate HSC-specific genes (Fig. 1a and Supplementary Table 2). Surprisingly, this list did not include all previously reported HSC-specific markers11–13 (Extended Data Fig. 1b, Supplementary Table 2). Second, to identify HSCs in situ, we needed to exclude candidates that also label BM non-hematopoietic cells, including stromal and endothelial cells15,16. Consequently, we excluded genes expressed in eight distinct non-hematopoietic BM populations, thereby narrowing the list to 45 candidates (Fig. 1a).
Next, we needed to ensure any candidates’ expression to be above threshold for detection in both flow cytometry and in-situ fluorescence. Therefore, we employed RNA-sequencing combined with a threshold gene standard to estimate the fragments per kilobase of transcript per million mapped reads (FPKM) value that could serve as a detection threshold. From 12-week-old mouse BM, we sorted and RNA-sequenced immunophenotypically defined (Lin−cKit+Sca1+CD150+CD34−/loFlk2−) HSCs (hereafter referred to as pHSC), multipotent progenitors subset A (MPPa) (Lin−cKit+Sca1+CD150+CD34+Flk2−), and multipotent progenitors subset B (MPPb) (Lin−cKit+Sca1+CD150−CD34+Flk2−) (Fig. 1b) to determine the FPKM value of candidate genes. Based on the Bmi-1-eGFP knock-in reporter17, we found that a single copy of eGFP is detectable at an estimated FPKM value of ~20. However, this high threshold would have excluded all candidates. Therefore, we designed a targeting construct (Fig. 1e) with three copies of mCherry, bringing the theoretical detection limit to ~7 FPKM. Lastly, to minimize aberrant detection we set threshold FPKM values for both MPPa and MPPb fractions to 2.5. Only three genes, Hoxb5, Rnf208, and Smtnl1 continued to qualify (Fig. 1b).
Given previous reports of heterogeneity within pHSC7,18–20, we analyzed single cells to determine whether our remaining candidates genes were heterogeneously expressed among pHSCs. We reasoned that an ideal pan-HSC candidate gene would label a significant fraction of pHSCs, with quantitative differences potentially reflecting HSC heterogeneity/diversity. We thus performed single-cell qPCR analysis of pHSCs, and evaluated expression of Hoxb5, Rnf208, and Smtnl1. Only Hoxb5 satisfied these criteria, exhibiting a bimodal expression in comparison to the unimodality of Rnf208 and Smtnl1 (Fig. 1c). Therefore, from the entire HSC transcriptome, only Hoxb5 satisfied this extensive unbiased screening (Fig. 1d).
We next sought to generate a Hoxb5 reporter with minimal disruption of endogenous Hoxb5 function. Thus we designed our targeting construct and CRISPR guide sequences to facilitate an in-frame knock-in to the endogenous Hoxb5 gene locus immediately 5′ of the sole endogenous stop codon. We utilized three tandem mCherry cassettes separated by P2A sequences, with the terminal mCherry carrying a CAAX membrane localization sequence (Hoxb5-tri-mCherry) (Fig. 1e).
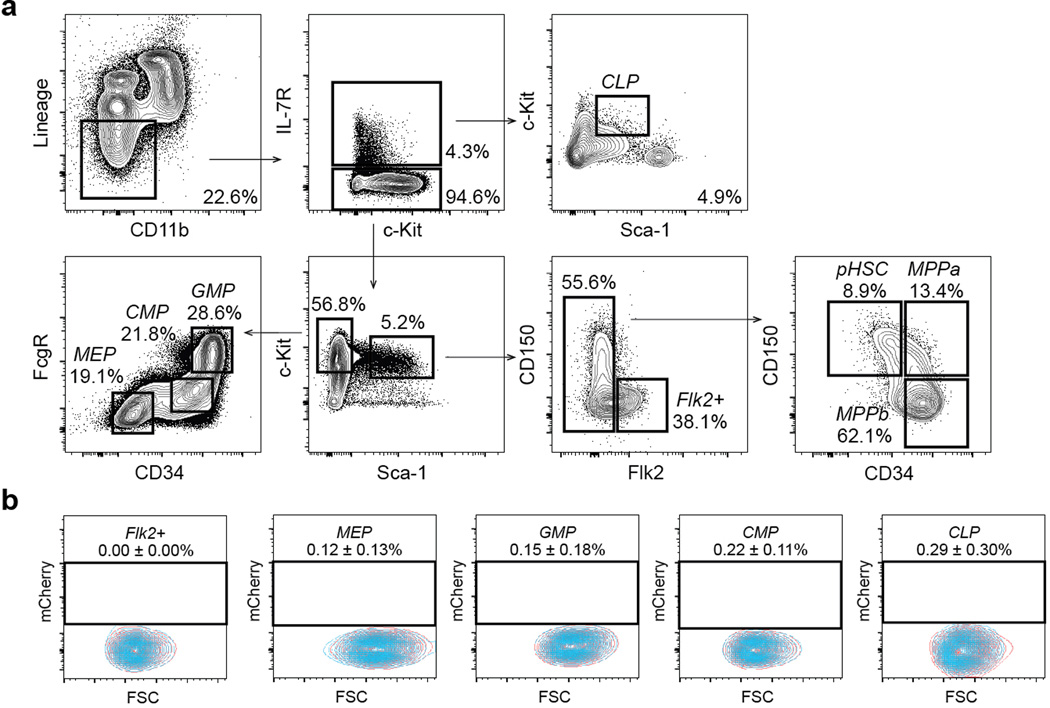
To evaluate the specificity of this reporter, we isolated whole BM cells from 12-week-old reporter mice and analyzed for mCherry-positive cells in the following immunophenotypic populations: pHSC, MPPa, MPPb, Flk2+ multipotent progenitor (Flk2+), megakaryocyte erythrocyte progenitor (MEP), granulocyte monocyte progenitor (GMP), common myeloid progenitor (CMP), common lymphoid progenitor (CLP) fractions, and differentiated cell populations (B cell, T cell, natural killer (NK) cell, neutrophil, eosinophil, monocyte, macrophage, dendritic cell, red blood cell, megakaryocyte), and in CD45 negative stromal fractions (Fig. 1f, Extended Data Fig. 2a–b, Extended Data Fig. 3, and data not shown). Consistent with our initial screen, (Fig. 1a–d), and using wild- type (WT) mice as a Fluorescence Minus One (FMO) threshold21, mCherry-labeled cells were highly enriched in the pHSC fraction (21.8% ± 0.90%), had a low frequency in the MPPa fraction (1.88% ± 0.64%), and background frequencies in the remaining fractions (Fig. 1f and data not shown). Interestingly, as only a minority of the events in the pHSC gate were mCherry positive, this suggested that either our reporter labeled only a subfraction of HSCs or that only a subfraction of pHSC gated events were indeed HSCs.
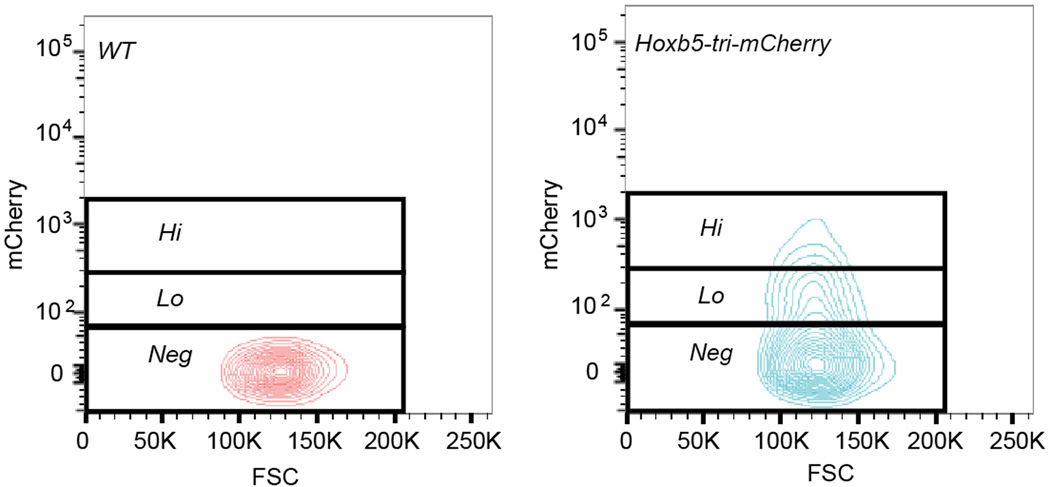
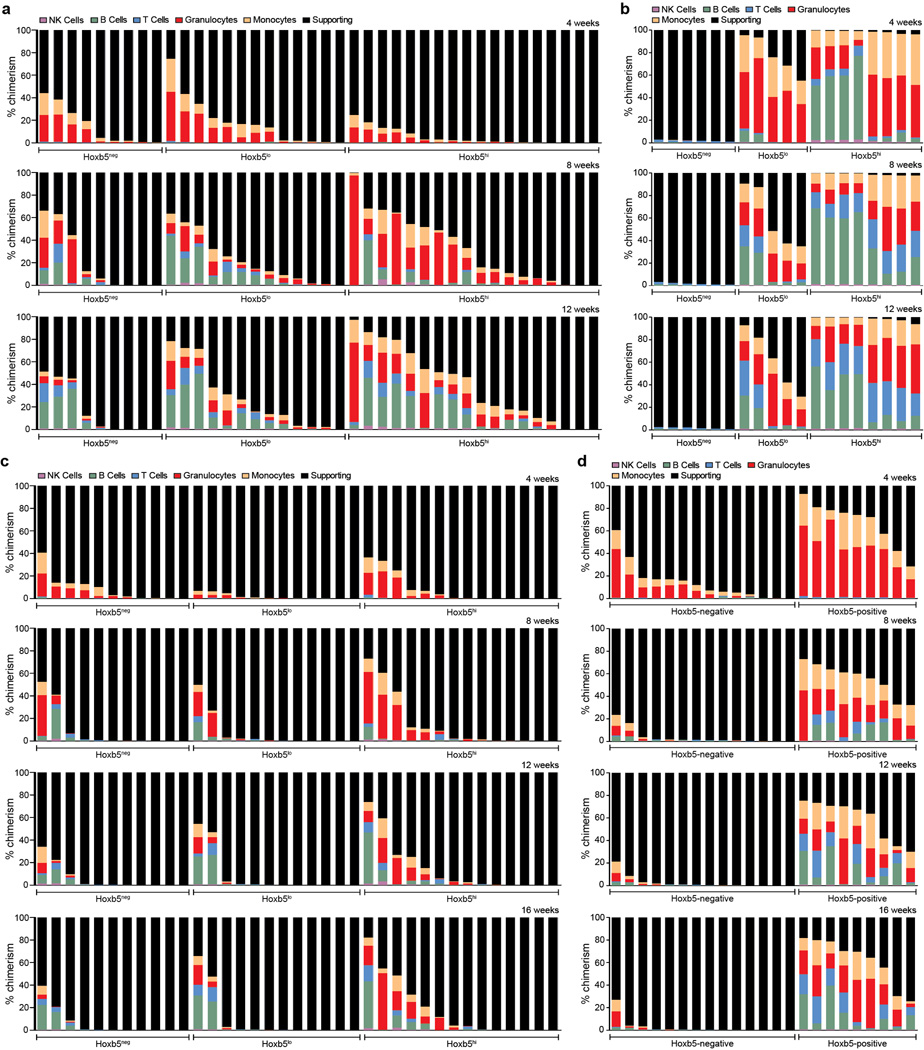
To distinguish between the two aforementioned possibilities and to validate whether Hoxb5 is a reporter of LT-HSCs, we sought to characterize Hoxb5-expressing cells by transplantation. In order to be inclusive of all events in the pHSC gate, we used WT FMO to define Hoxb5 negativity and divided the positive fraction into Hoxb5hi (top 5-percentile) and Hoxb5lo (14.1±7.7%) (Extended Data Fig. 4). Ten-cell and three-cell grafts of Hoxb5hi, Hoxb5lo, or Hoxb5neg pHSC were transplanted with supporting BM cells into irradiated mouse recipients. CD45.2+ was used to assess donor HSC contribution to haematopoietic lineages at 4-week intervals (Fig. 2a). Analysis of peripheral blood at 16 weeks post-transplantation of ten cells demonstrated the following: multilineage reconstitution was present in 78% of Hoxb5hi pHSC recipients, 70% of Hoxb5lo pHSC recipients, and 44% Hoxb5neg pHSC recipients (Fig. 2b and Extended Data Fig. 5a). Three-cell transplants exhibited similar kinetics (Extended Data Fig. 5c). Of note, the chimerism of the Hoxb5neg pHSCs decreased over time, most noticeably between 4 to 8 weeks and shifted toward a predominant lymphoid chimerism (Fig. 2b, d, and, Extended Data Fig. 5a), suggesting that either the Hoxb5-negative fraction was a lymphoid biased HSC or, more likely, a transiently self-renewing ST-HSC/MPP which had given rise to long-lived lymphocytes.
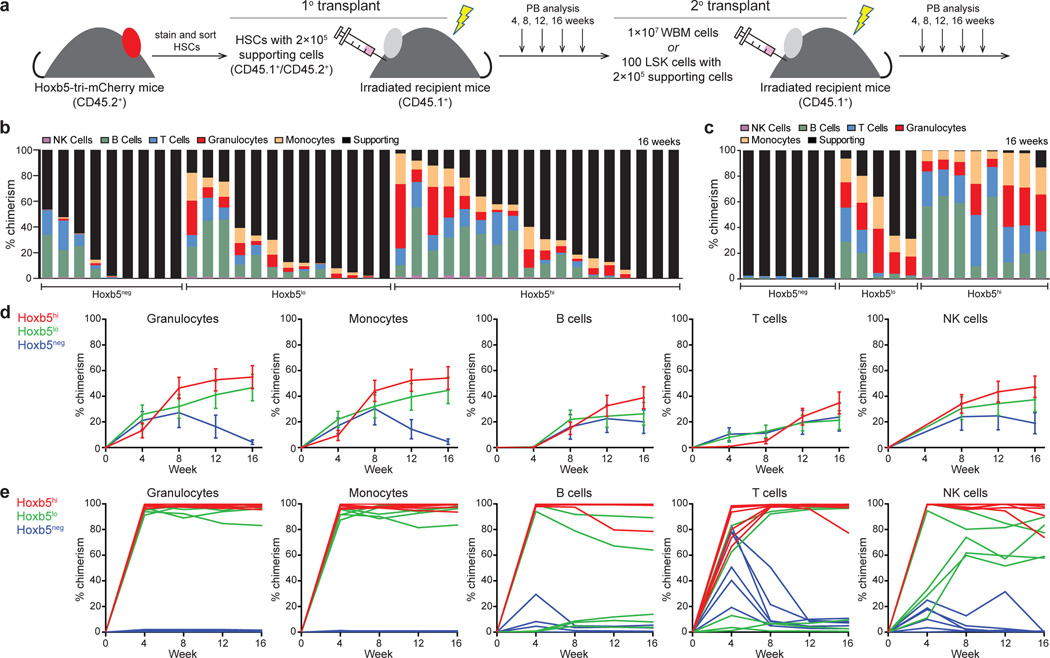
Figure 2. Hoxb5 distinguishes between LT-HSC and non-LT-HSC.
a, Experimental schematic for long-term haematopoietic reconstitution assays. CD45.1+ recipient mice were lethally irradiated and competitively transplanted with 10 or 3 Hoxb5-tri-mCherry HSCs and 2×105 CD45.1+/CD45.2+ supporting cells. For secondary transplants, 1×107 whole bone marrow (WBM) cells or 100 sorted LSK cells were transferred from primary recipient mice. b, 16 week chimerism in primary recipients receiving 10 Hoxb5neg (n=9 mice), Hoxb5lo (n=13 mice), or Hoxb5hi (n=18 mice) pHSCs. Each column represents an individual mouse. c, 16 week chimerism following WBM secondary transplant. d, Average donor lineage contribution in 10-cell primary transplants. Error bars denote S.D. e, Individual donor chimerism by lineage in WBM secondary recipients. Each line represents an individual mouse (n=6 mice for Hoxb5neg, n=5 mice for Hoxb5lo, and n=8 mice for Hoxb5hi).
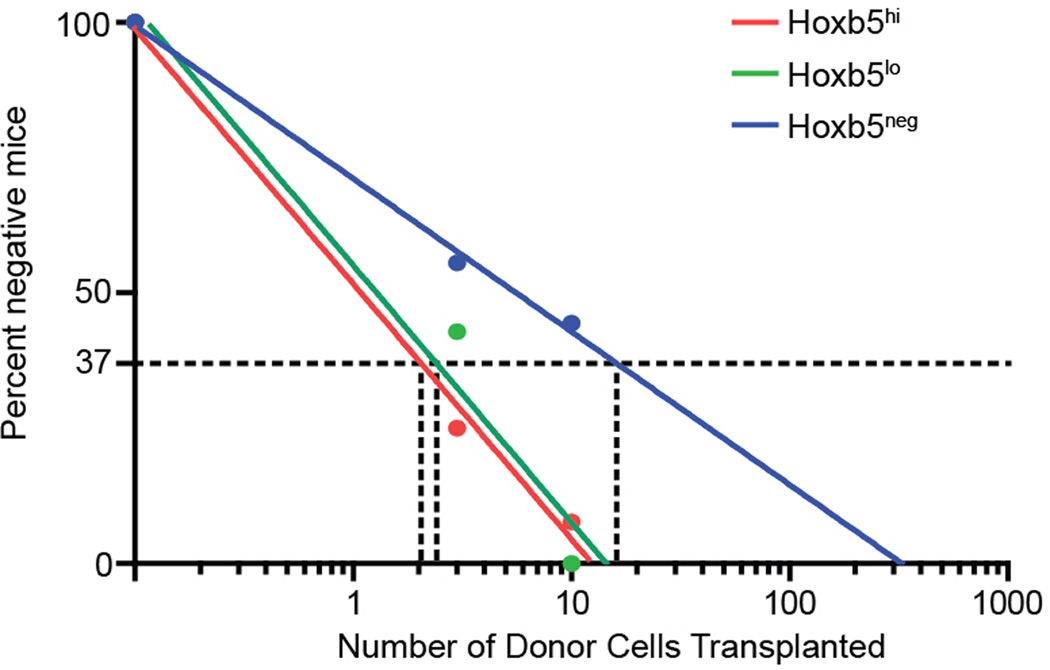
To evaluate between these two possibilities, we carried out a secondary transplantation from primary Hoxb5hi, Hoxb5lo, or Hoxb5neg recipients into lethally irradiated secondary recipients (Fig. 2a). At 16 weeks post-secondary transplantation, peripheral blood analysis revealed robust multilineage chimerism from all Hoxb5hi and Hoxb5lo recipients with minimal chimerism from the Hoxb5neg fraction (Fig. 2c, e, and Extended Data Fig. 5b). Furthermore, BM analysis of primary recipients for donor pHSC gate events revealed that the Hoxb5hi transplants contained Hoxb5hi, Hoxb5lo, and Hoxb5neg cells (100%, n=10 mice), while the Hoxb5neg transplants were negative (36%, n=4 mice) or devoid of donor cells (64%, n=7 mice). These results further suggested that the Hoxb5neg pHSCs were in fact transiently self-renewing ST-HSC/MPP. To determine if Hoxb5 positivity continued to distinguish the LT-HSC in a primary transplant, we normalized the number of donor cells used for secondary transplant by sorting 100 Lin−cKit+Sca1+ (LSK) donor cells from the primary Hoxb5hi recipients and transplanted them into irradiated secondary recipients (n=24 mice) (Fig. 2a). As with the primary BM transplantations, the chimerism was minimal in the Hoxb5-negative versus Hoxb5-positive recipients (Extended Data Fig. 5d). Limiting dilution analysis revealed that the frequency of LT/ST-HSC in primary hosts at 16 weeks is 1 in 2.1 for Hoxb5hi, 1 in 2.4 for Hoxb5lo, and 1 in 16.1 for Hoxb5neg (Extended Data Fig. 6). Taken together these results demonstrate that Hoxb5 labels the functional LT-HSC.
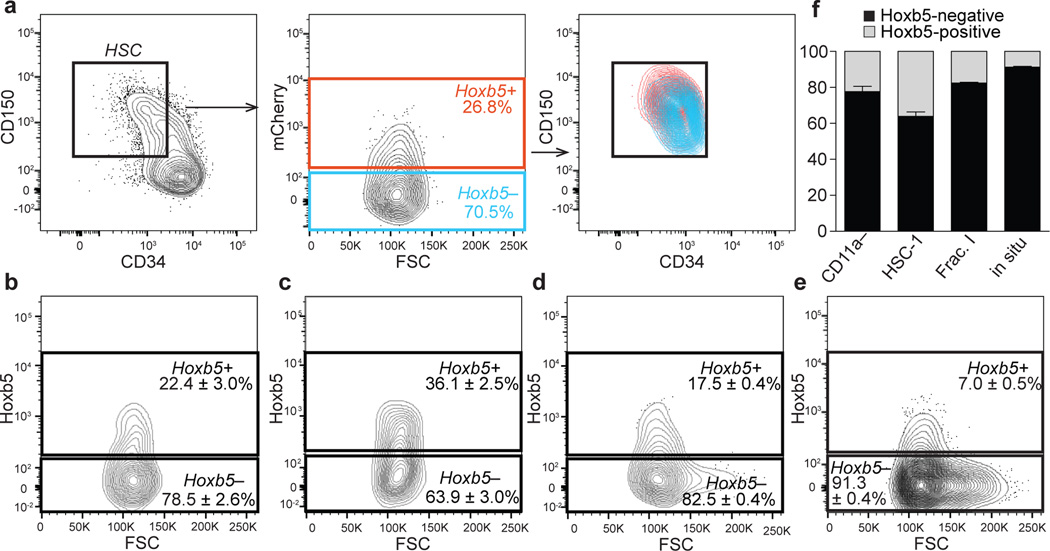
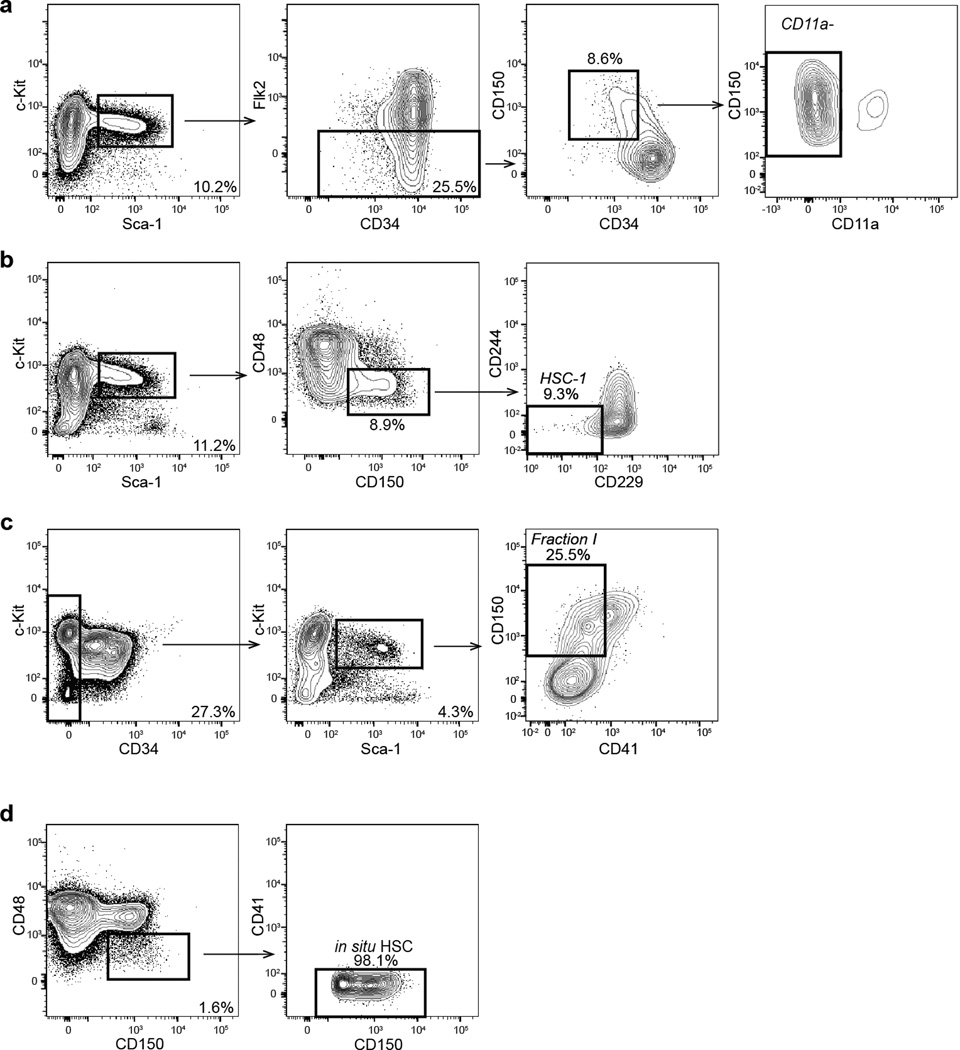
Given that Hoxb5 negative cells are non-LT-HSC, we sought to re-examine the specificity of past definitions (Fig. 3a,f), including recently reported refinements to the LT-HSC immunophenotype18–20 and the most widely used in situ definition over the past decade22. Of the CD11a negative HSC19, 78.5% ± 2.6% are Hoxb5 negative (Fig. 3b). Of the HSC-120 (Lin− cKit+Sca1+CD48−CD150+CD229−CD244−), 63.9% ± 3% are Hoxb5 negative (Fig. 3c). Of Fraction I18 (Lin−cKit+Sca1+ CD34−/loCD150+CD41−), 82.5% ± 0.4% are Hoxb5 negative (Fig. 3d). Surprisingly 91.3% ± 0.4% of Lin−CD48−CD41−CD150+22 cells are Hoxb5 negative (Fig. 3e). As this subset was initially utilized for localization of HSC in situ22, we decided to re-examine the in situ location of HSC with Hoxb5.
Figure 3. Hoxb5-negative cells contaminate previous HSC definitions.
Flow cytometry plots of 12-week-old C57BL/6 bone marrow depicting Hoxb5-tri-mCherry reporter activity in previously reported HSC immunophenotypes. a, pHSC (LSK CD150+CD34–/loFlk2−) Hoxb5+(red) and Hoxb5− (blue). b, CD11a− (LSK CD150+CD34–/loCD11a−)19. c, HSC-1 (LSK CD150+CD48–/loCD229–/loCD244−)20. d, Fraction I (LSK CD150+CD34–/loCD41−)18. e, CD150+CD48−CD41− cells, currently used to identify HSCs in situ22. Wild type FMO used to define Hoxb5 negativity for each panel. f, Summary percentage of Hoxb5-positive and negative cells in characterized HSC subfractions. Error bars denote S.D. (n=5 mice). Refer to Extended Data Fig. 7a–d for gating scheme.
Visualizing LT-HSCs in BM and identifying the cellular constituents and structures of the HSC niche remains a significant challenge. Despite this, multiple constituent cell types have been proposed, including mesenchymal stromal, endosteal osteoblasts, glia, endothelium, and pericytes5. In situ investigations are limited by several factors: the number of fluorescent colors; the difficulty of identifying HSCs surrounded by non-HSCs; and the ambiguity in translating fluorescently positive and negative cells from flow-cytometry to tissue sections.
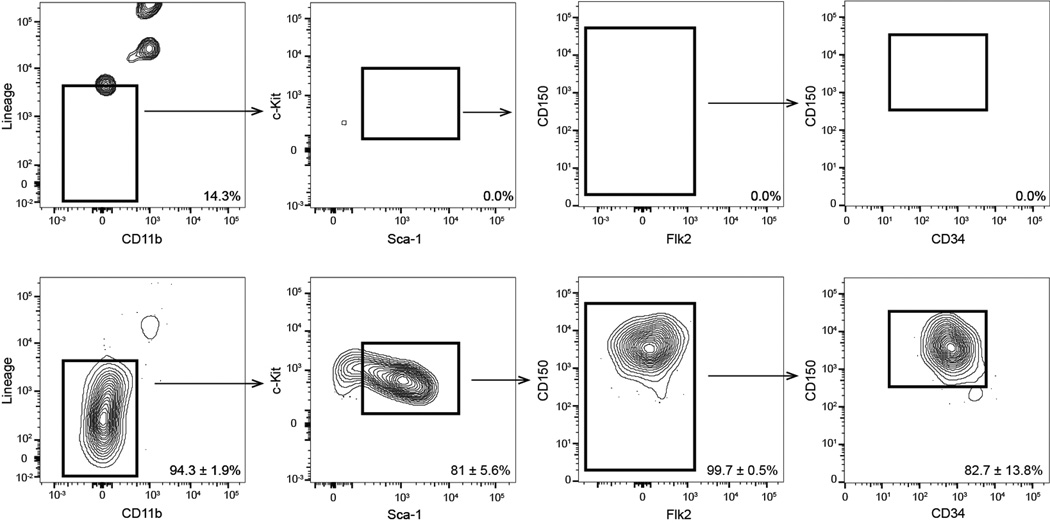
To address these issues, we employed the Hoxb5-tri-mCherry reporter. By flow cytometry we sought to determine the utility of Hoxb5 alone in identifying the LT-HSC. After logical exclusion of autofluorescence by comparison to WT control mice, we found that all Hoxb5 events reside within the cKit+ fraction and 62.0 ± 12.8% of all Hoxb5-positive events are located in the pHSC gate (Extended Data Fig. 8), representing an eight to nine fold enrichment compared to previous in situ labeling of HSCs (Lin−CD48−CD41−CD150+)22 (Fig. 3e).
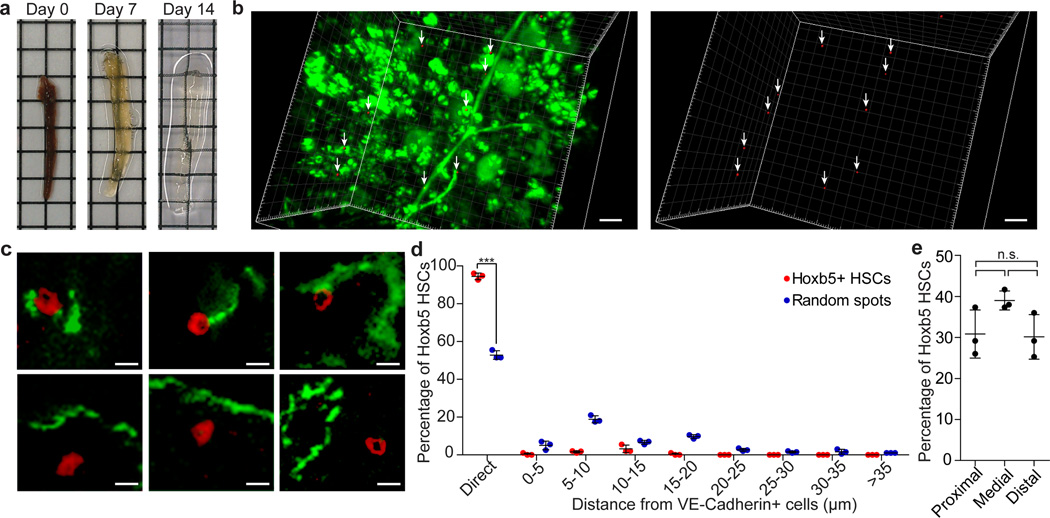
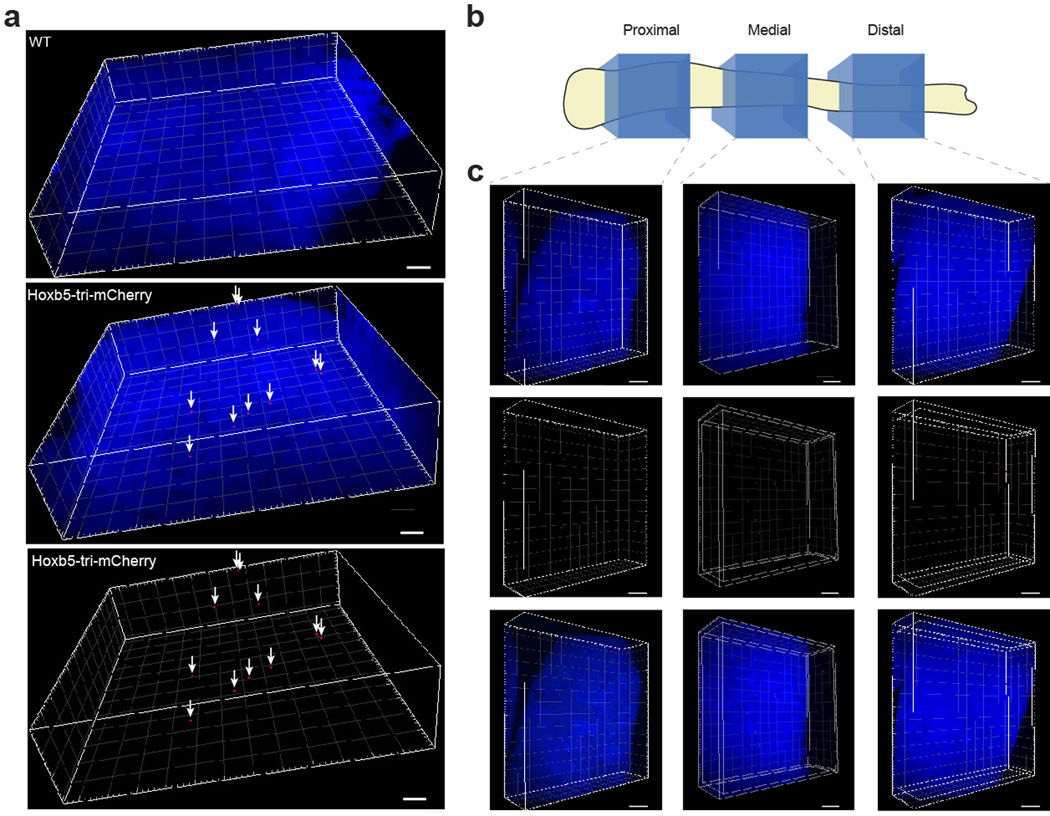
To reveal the three-dimensional HSC niche architecture, we applied the CUBIC technique23 to tibial BM, facilitating depletion of autofluorescent cellular components (Fig. 4a). Given that fluorescence intensity is preserved by CUBIC23, the specificity of LT-HSC in situ correlates with Hoxb5 intensity by flow cytometry. Therefore, based on the number of Hoxb5-positive LT-HSC in one tibia detected by flow cytometry (Extended Data Fig. 9a–b), we objectively gated the same number of Hoxb5-positive cells by intensity as in situ Hoxb5-positive (is-Hoxb5+). We observed a uniform distribution of is-Hoxb5+ cells along the longitudinal axis of the tibia (Fig. 4e and Extended Data 10a–c). The average percentage of is-Hoxb5+ was 30.8 ± 5.9 in the proximal epiphysis, 39.0 ± 2.4 in the diaphysis, and 30.1 ± 5.4 in the distal epiphysis per field (1.25 × 1.25 × 0.4mm3) (Fig. 4e).
Figure 4. LT-HSC exhibit near homogenous attachment to VE-cadherin positive cells.
a, Tissue preparation and representative images of tibial bone marrow plug after paraformaldehyde fixation (Day 0), treatment with Reagent-1 (Day 7, Day 14) 23. b, Localization of Hoxb5-positive cells (red and arrows) and VE-cadherin-positive cells (green) in 3D-reconstructed images. Scale bar 30µm c, Representative 2D images of direct (top panel) and non-direct (bottom panel) association of Hoxb5-positive cells (red) with VE-cadherin-positive cells (green). Scale bar 10µm. d, Frequency of Hoxb5-positive cells (n=287 cells, from n=3 mice) and random spots (n=600 spots, from n=3 mice) plotted against proximity to VE-cadherin-positive cells. unpaired student’s t-test (p<0.0001). e, Average number of Hoxb5-positive cells in proximal, medial, and distal regions of tibia (n=3 mice). unpaired student’s t-test.
To investigate the association of LT-HSC with vasculature, we injected anti-mouse VE-cadherin antibody into reporter mice and analyzed the BM with CUBIC23. We found 94.1 ± 1.9% of is-Hoxb5+ cells were directly attached to VE-cadherin-positive endothelial cells at their abluminal surface. In contrast, only 52.8 ± 2.3% of the random (Hoxb5−) spots were directly associated with VE-cadherin-positive cells. This implied a near homogeneous perivascular location for the LT-HSC niche (Fig. 4b–d).
Since the first success of enrichment of HSCs in 19881, many groups have attempted to identify surface markers to isolate the LT-HSC18–20,22,24,25. Identification of CD150, CD34, and CD48 enabled the field to isolate LT/ST-HSC from MPP. However, complete separation of LT-HSC from ST-HSC has never been fully accomplished. Our study demonstrates that Hoxb5 is the most specific single gene whose expression in adult mouse BM is limited to the LT-HSC. By the limiting dilution assay at least 1 in 2.1 of Hoxb5hi cells are LT-HSCs. However, this assay underestimates the functional potential of candidate HSCs. Transplantation, although the gold standard, may not take into account differences between sessile vs mobile HSCs26,27, cell cycle status28,29, expression of CD47, the ‘don’t eat me’ molecule, expressed highly on mobilized HSC but at low levels on sessile bone marrow HSC in un-manipulated mice 30, and irradiation, and their influence on engraftment efficiency.
The results presented here demonstrate that the LT-HSC compartment is near homogeneously perivascular. While other compartments have been implicated5, including less homogeneous association of candidate HSC with the vasculature 11, it is likely that our results differ due to the fraction of assayed populations that are LT-HSC (Extended Data Fig. 1b and Supplementary Table 3). In sum, these results suggest that the model of Hoxb5-tri-mCherry is the most rigorous yet published for LT-HSC, and will allow in situ lineage tracing to define their role in hematopoiesis in the absence of transplantation into irradiated hosts.
Methods
Mice
Eight- to twelve-week-old C57BL/6J male mice (Jackson Laboratory) served as wild-type controls. Eight- to twelve-week-old male B6.SJL-Ptprca Pepcb/BoyJ mice (Jackson Laboratory) served as recipients for transplantation assay. Supporting cells for competitive reconstitution assays were collected from B6.SJL-Ptprca Pepcb/BoyJ × C57BL/6J (F1 mice CD45.1+/CD45.2+). Mice were bred at our animal facility according to NIH guidelines. All animal protocols were approved by the Stanford University Administrative Panel on Laboratory Animal Care. Hoxb5-tri-mCherry (C57BL/6J background) mice were used as donor cells for transplantation as well as for analysis. Please see “Gene targeting” for mouse derivation.
Microarray data
All microarray data employed in this study are available at Gene Expression Commons (http://gexc.stanford.edu). Two to four microarray replicates were assessed for each distinct cell population14–16. The immunophenotype definition of each fraction is included in Supplementary Table 1.
RNA sequencing
Performed as previously described31. In brief, total RNA was isolated with trizol, treated with RQ1 RNase free DNase (Promega) to remove minute quantities of genomic DNA if present, and cleaned up using RNAeasy minelute columns (QIAGEN). cDNA libraries were prepared for pHSC, MPPa, and MPPb populations using Ovation RNA-Seq System V2 (NuGen) and sequenced separately using HiSeq 2500 (Illumina) to obtain 2 × 150 base pair paired-end reads. Raw transcriptome sequence data were mapped to M. musculus reference mRNAs using OLego32 to produce a reference-guided transcript assembly. Four replicates were sequenced for each population. Data accessible at GEO PRJNA305664
Single-cell qPCR
Single cells for qPCR were processed using the Single Cell-to-CT™ qRT-PCR Kit (Life Technologies) per manufacturer’s instructions. Cells were sorted directly into lysis solution in a 96-well plate, subjected to a reverse transcription reaction for cDNA synthesis, amplified for 14 cycles with pooled TaqMan Gene Expression Assays, and diluted with 1X TE buffer (pH 8.0). For real-time PCR, samples were amplified for 50 cycles of 3 seconds at 95°C followed by 30 seconds at 60°C using the following TaqMan probes: Mm00657672_m1 for Hoxb5, Mm03039759_s1 for Rnf208, Mm00470338_m1 for Smtnl1, and Mm99999915_g1 for Gapdh. All thermocycling was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Presence of a single cell was validated by a CT value of 30 or less for Gapdh.
Gene targeting
The targeting construct was cloned into pJYC (derived from pUC19, golden gate insert of TALE backbone vector33, and removal of LacZ sequence). Right and left homology arms of ~700bp were cloned by PCR from C57BL/6J genomic DNA, and the entire construct was sequence validated. BRUCE-4 ES cells (Millipore) derived from C57BL/6J mice were transfected with the targeting construct as well as bicistronic CRISPR vector px330 expressing Cas9 and a Hoxb5-specific single guide RNA (5’-GGCUCCUCCGGAUGGGCUCA-3’)34. Following homologous recombination, recombinant clones were positively selected for one week with neomycin (100µg/mL), transfected with an EF1α-Cre-puromycin vector, and selected for a second week with puromycin (1 µg/ml). Serially diluted ES colonies were then individually picked, expanded, and screened by PCR and qPCR for site-specific integration, exclusion of off-target effects, and correct copy number. After sequence validation of the targeted site, successfully targeted ES clones were used to establish chimeras. Chimeric mice were crossed with C57Bl/6 –Tyrc-2j/J female mice to establish germline transmission.
Flow cytometry and cell sorting
Flow cytometry and cell sorting were performed on a FACS Aria II cell sorter (BD Biosciences) and analyzed using FlowJo software (Tree Star). Bone marrow cells were harvested from bilateral tibias, femurs, humeri, and pelvises by crushing (unless otherwise specified) using a mortar and pestle in Ca2+ and Mg2+ free PBS supplemented with 2% heat inactivated bovine serum (Gibco) and 2mM EDTA. Cells were passed through 100um, 70um, and 40um strainers prior to analysis and sorting. To enrich HSCs and progenitor populations, cells were stained with APC-conjugated anti-c-Kit (2B8) and fractionated using anti-APC magnetic beads (Miltenyi Biotec) and LS columns (Miltenyi Biotec). c-Kit+ cells were then stained with combinations of antibodies for the following surface markers: Sca-1, Flk2, CD150, CD34, IL-7R, CD16/32, and the lineage markers Ter-119, B220, CD2, CD3, CD4, CD5, CD8a, Gr-1, CD11a, CD11b, CD41, CD48, CD229, and CD244. For lymphoid populations, bone marrow cells were stained with CD3, CD4, CD8a, CD11b, CD11c, Gr-1, NK-1.1, Ter-119, B220, c-Kit, and F4/80. For myeloid populations, cells were stained with CD3, CD3ε, CD11b, CD11c, CD19, Gr-1, NK-1.1, Ter-119, and F4/80. Antibody staining was performed at 4°C and incubated for 30 min. Stains with CD34 were incubated for 90 min. Prior to sorting or analysis, cells were stained with SYTOX Red Dead Cell Stain (Life Technologies) to assess viability per manufacturer’s recommendations. Transplanted cells were double-sorted for purity. Further details regarding flow cytometry reagents are provided in Supplementary Table 4.
Transplantations and peripheral blood analyses
B6.SJL-Ptprca Pepcb/BoyJ (Jackson Laboratory) recipient mice were lethally irradiated at a single-dose of 9.1 Gy. For reconstitution assays, donor cells were first combined with 2 × 105 whole bone marrow supporting cells (B6.SJL-Ptprca Pepcb/BoyJ × C57BL/6J F1 mice CD45.1+/CD45.2+) in 200µl of PBS with 2% FBS, then injected into the retro-orbital venous plexus. Peripheral blood analyses were performed at 4, 8, 12, and 16 weeks after primary and secondary transplantations. At each time point, 50 µl of blood was collected from the tail vein and added to 100µl of PBS with 2mM EDTA. Red blood cells were subsequently lysed using BD Pharm Lyse Buffer (BD Pharmingen) per manufacture’s protocol for 3 min on ice, followed by blocking with 5µg/ml Rat IgG. Leukocytes were stained with antibodies to (refer to Supplementary Table 4 for specifics regarding clone and colors), CD45.1 (FITC), CD45.2 (PE), CD11b (BUV395), Gr-1 (Alexa-Flour700), B220 (BV786), CD3 (BV421), TCRß (BV421), and NK-1.1 (PerCP-cy5.5). For each mouse, the percentage of donor chimerism in the peripheral blood was defined as the percentage of CD45.1− CD45.2+ cells among total CD45.1− CD45.2+ and CD45.1+ CD45.2+ cells. To control for variability in host response to lethal irradiation, mice with host chimerism of 50% or higher at 16 weeks post-transplantation were excluded from our analyses. The frequency of chimerism in peripheral blood was analyzed as follows. For evaluation of donor (CD45.1− CD45.2+) chimerism kinetics, after exclusion of recipient (CD45.1+ CD45.2−) fraction, the frequency of the donor fraction was calculated. Within the whole donor fraction, the frequency of each lineage (NK cell, B cell, T cell, Granulocyte, Monocyte) was determined. For evaluation of lineage contribution kinetics, after gating each lineage, the frequency of the donor fraction (CD45.1− CD45.2+) was calculated. Any recipients which were exhibited lower than 1% of chimerism were treated as negative to exclude ambiguous cases.
Limiting Dilution Analysis
The frequency of LT/ST-HSC was calculated using the transplantation data of 10-cell and 3-cell Hoxb5hi, Hoxb5lo, or Hoxb5neg pHSC transplants. Any mice showing long-term (>16week) multilineage reconstitution (>1% in each lineage) were counted as a positive recipient. A non-linear regression semi-log best fit line was used to calculate the frequency of LT/ST-HSCs at F0 = 0.368 (GraphPad Prism 6).
CUBIC bone marrow imaging
Bone clearing protocol was modified from the original Cubic protocol23. Specifically, tibias were collected and fixed in 4% PFA solution for two days, after which bone marrow plugs were extracted from the distal end by flushing method with a 25-gauge syringe. For nuclear staining, bone marrow plugs were immersed in DAPI/PBS solution at 37°C for three days with gentle shaking. For clearing, marrow plugs were immersed in ScaleCUBIC-1 (Reagent-1) at 37°C for two weeks with gentle shaking. The solution was changed every 48 hours. To visualize vasculature, 20 µg of Alexa488-conjugated anti-mouse VE-cadherin antibody (BV13) was administrated i.v. (retro-orbital) with tibias collected 30 minutes later. Processed plugs were embedded in 4-mm-diameter glass capillary with 2% agarose for imaging. Images were acquired using a ZEISS Z1 Lightsheet microscope (ZEISS) and reconstituted into three-dimensional (3D) image by using ZEN software (ZEISS). Acquired 3D images were analyzed with Imaris software (Bitplane). After exclusion of outliers including events of extraordinary size (>30µm) or intensity, all other mCherry+ (Hoxb5+) cells were analyzed in the tibia (n=287 cells in total from n=3 mice). The mCherry negative threshold was determined based on the intensity level of the wild-type control tibial plugs. These mCherry negative events from with intensities ranging from 0.002–75.998 (75.998 representing the upper bound of mCherry intensity) were transformed into an integer list of 75,000 values (75,000 representing the length of the list). 600 spots were then randomly selected from this integer list utilizing the randbetween(1,75,000) function in Excel 2015 (Microsoft). Using this list of random spots now identified by intensity, their location and distance to Ve-cadherin positive cells was measured using the Imaris software. All CUBIC imaging experiments were done in biological triplicates from three mice.
Genotyping
Genomic DNA from Hoxb5-tri-mCherry mice was isolated from tail biopsies using QuickExtract solution (EpiCentre). PCR amplification was performed using the same forward primer (F: 5’-GACGTATCGAGATCGCCCAC-3’) with two reverse primers to distinguish between the Hoxb5-tri-mCherry (R1: 5’-CCTTGGTCACCTTCAGCTTGG-3’) and wild-type (R2: 5’-AGATTGGAAGGGTCGAGCTG-3’) alleles.
Statistics
All the comparison analyses were done by unpaired Student’s t tests. Pearson’s chi-square test was done using online software (http://vassarstats.net/).
Extended Data
Extended Data Figure 1. GEXC expression of previously reported HSC markers in mouse bone marrow.
a, ideal expression pattern of HSC-specific genes (pink represents increased expression, blue represents decreased expression) b, Relative expression of Hoxb5 (top panel left), Alpha-catulin/Ctnnal1 (top panel middle), Fgd5 (top panel right), CD150/Slamf1 (bottom panel left), Hoxb4 (bottom panel middle), Gfi-1 (bottom panel right) in haematopoietic and stromal populations as determined by microarray.
Extended Data Figure 2. Gating Scheme for HSC and progenitors.
a, Representative flow cytometry gating to isolate pHSCs, MPPs, and oligopotent progenitors from mouse bone marrow. Panels gated as shown after exclusion of doublets and dead cells. b, Hoxb5 reporter expression (red) in Flk2+ MPPs, MEP, GMP, CMP, CLP compared to wild-type controls (blue). Values indicate the percentage of mCherry-positive cells ± S.D. in each fraction for n=3 mice.
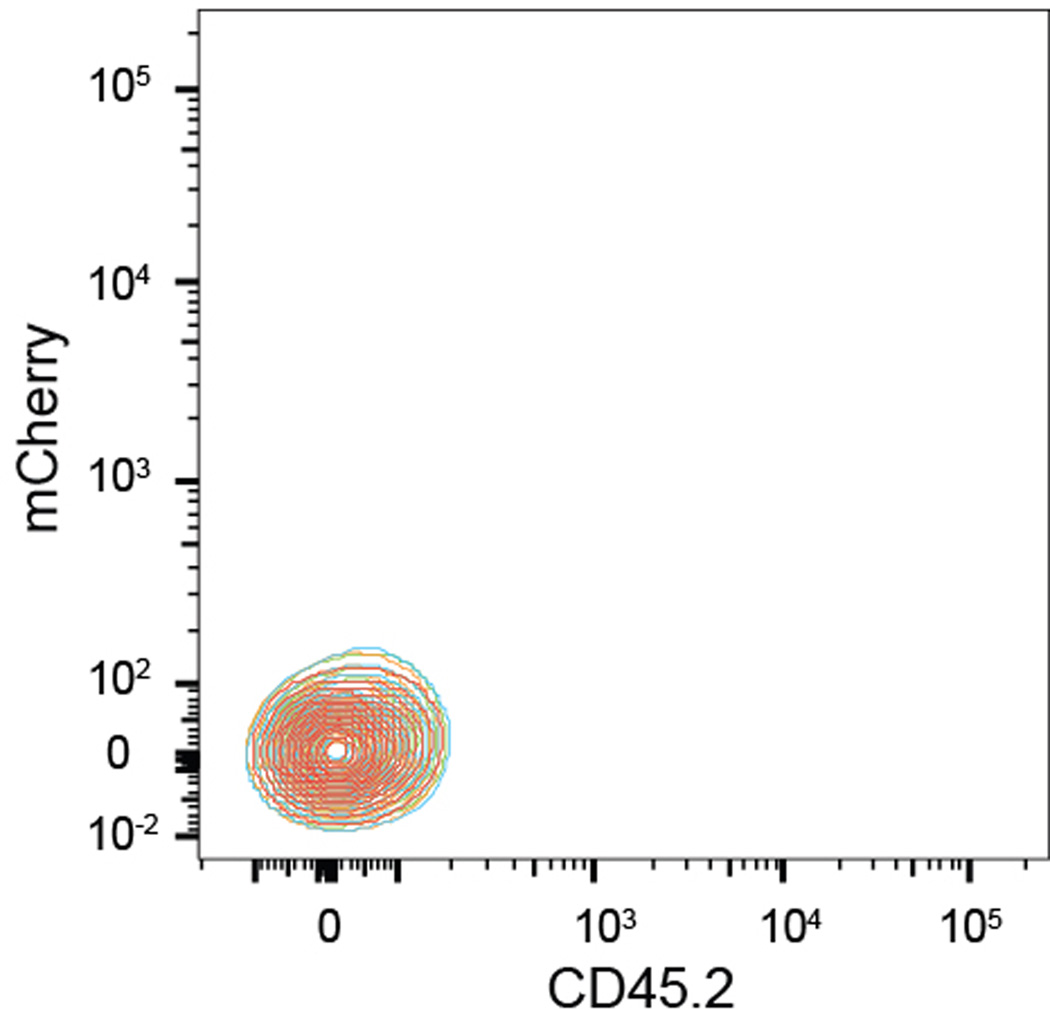
Extended Data Figure 3. Hoxb5 is not expressed in CD45- Bone Marrow.
Hoxb5 reporter expression in CD45-negative compartment within bone marrow. Wild type (Red) and three Hoxb5-tri-mCherry mice (blue, orange, and green, n=3 mice).
Extended Data Figure 4. FMO gating for Hoxb5 positive signal.
Representative flow cytometry gating to separate mCherry (Hoxb5) hi, lo, and negative populations in both wild-type and Hoxb5-tri-mCherry
Extended Data Figure 5. Hoxb5 distinguishes between LT-HSC and non-LT-HSC.
a, 4, 8, and 12-week reconstitution kinetics in primary recipients receiving 10 Hoxb5neg (n=9 mice), Hoxb5lo (n=13 mice), or Hoxb5hi (n=18 mice) pHSCs. Each column represents an individual mouse. b, 4, 8, and 12-week reconstitution kinetics following WBM secondary transplant. c, Reconstitution kinetics in primary recipients receiving three Hoxb5neg (n=11 mice), Hoxb5lo (n=12 mice), or Hoxb5hi (n=14 mice) pHSCs. Each column represents an individual mouse. d, Reconstitution kinetics following secondary transplant of 100 sorted Lin−cKit+Sca1+ (LSK) Hoxb5neg (n=14 mice) or Hoxb5pos (n=9 mice) cells and 2×105 supporting cells.
Extended Data Figure 6. Limiting dilution analysis of Hoxb5 positive and negative pHSCs.
Limiting dilution results of 10 cell and 3 cell transplants of Hoxb5hi (red, n=18 mice for 10 cell and n= 14 mice for 3 cell), Hoxb5lo (green, n=13 mice for 10 cell and n=12 mice for 3 cell), and Hoxb5neg (blue, n=9 mice for 10 cell and n=11 mice for 3 cell). Frequency of LT-HSC/ST-HSC by limit dilution for Hoxb5hi is 1 in 2.1, for Hoxb5lo is, 1 in 2.4 and Hoxb5neg is 1 in 16.1.
Extended Data Figure 7. Hoxb5-negative cells contaminate previous HSC definitions.
Representative HSC gating strategy for various HSC definitions after exclusion of doublets and dead cells. a, CD11a− (LSK CD150+CD34–/loCD11a−)21. b, HSC-1 (LSK CD150+CD48–/loCD229–/loCD244−)20. c, Fraction I (LSK CD150+CD34–/loCD41−)18. d, CD150+CD48−CD41− cells22 (n=5 mice).
Extended Data Figure 8. Specificity of Hoxb5 as a single marker for LT-HSC.
a, Flow cytometry plots of wild type (top row) and Hoxb5-tri-mCherry (bottom row) after excluding doublets, dead cells, autofluorescence, and gated on Hoxb5-positive events. Frequencies shown are percent gate ± S.D. in each fraction (n=3 mice).
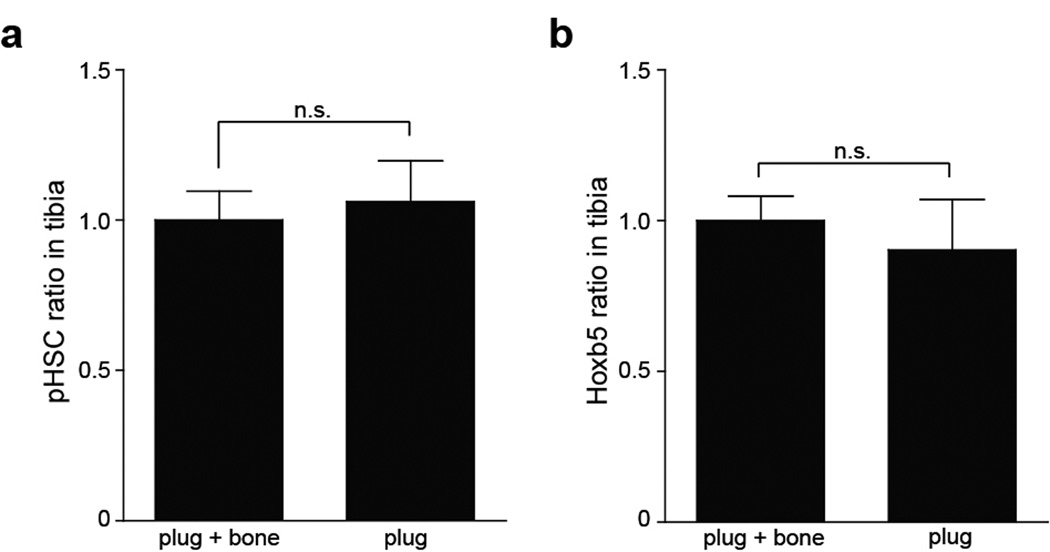
Extended Data Figure 9. Comparison of processing methods on pHSC and Hoxb5 LT-HSC yield.
Relative frequency of a, pHSCs and b, Hoxb5 LT-HSCs in tibial plugs (flushed) (n=6 mice) compared to tibial plugs plus bones (crushed) (n=6 mice).
Extended Data Figure 10. Hoxb5-positive HSC are evenly distributed in the tibia.
a, Distribution of Hoxb5-positive cells (red and arrows) in bone marrow in 3D-reconstructed images. Nuclei are counterstained with DAPI (blue) WT (left panel) Hoxb5-tri-mCherry (middle and right panel). Scale bar 100µm b, Cartoon representing the location of the proximal, medial, and distal sampling. c, Representative 3D-reconstructed images of Hoxb5-positive cells (red) in proximal (left column), medial (middle column), and distal (right column) regions of the tibia. Scale bar 150µm. Nuclei are counterstained with DAPI (blue) n=3 mice.
Supplementary Material
Acknowledgments
We would like to acknowledge Norma Neff and Gary Mantalas for advice regarding RNA sequencing; Brian Yu and Andrew Beel for for providing critical input on imaging data; Hidekazu Nishikii for advice regarding imaging data; Jun Seita for continued development of the GEXC; Seth Karten for help in editing the manuscript; Libuse Jerabek and Terry Storm for laboratory management; Aaron McCarty and Charlene Wang for animal care; Patty Lovelace and Jennifer Ho FACS Facility management; Hong Zeng and Yanfeng Li for collaboration in generating the mouse model; Yasuhiko Sato for technical assistance in Imaris software analysis. The authors would like to acknowledge ongoing support for this work: I.W. the NCI and the NHLBI of the NIH under award numbers R01 CA086065, U01 HL099999, and R01 HL058770; and by the Virginia and D. K. Ludwig Fund for Cancer Research; J.Y.C the Stanford University Medical Scientist Training Program (T32 GM007365) and the NHLBI Ruth L. Kirschstein National Research Service Award (F30-HL122096); M.M. the Human Frontier Science Program Long-Term Fellowships, the Uehara Memorial Foundation Research Fellowship, Toyobo Biotechnology Foundation Research Fellowship, and Kanzawa Medical Research Foundation Overseas study grants. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
AUTHOR CONTRIBUTIONS
M.M. and J.Y.C., conceived, performed, analyzed, and oversaw the experiments, with suggestions from I.L.W.. M.M. and J.Y.C. identified Hoxb5 as a LT-HSC marker, made and characterized the Hoxb5-tri-mCherry mouse. S.K.W. and K.S.K performed experiments and prepared figures with assistance of M.M. and J.Y.C. S.Y. generated Cubic data and evaluated the association with VE-Cadherin vasculature. R.S. helped design and perform RNAseq data analysis. M.M., J.Y.C., S.K.W., and K.S.K., wrote the manuscript. H.N. and R.S. commented on the manuscript. I.L.W. edited the manuscript and supervised the laboratory. The authors have no competing interests.
References
- 1.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 2.Guo W, et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008;453:529–533. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 4.Ito K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 5.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Beerman I, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida N, Weissman IL. Searching for hematopoietic stem cells: evidence that Thy-1.1lo Lin- Sca-1+ cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J. Exp. Med. 1992;175:175–184. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 11.Acar M, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazit R, et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 2014;211:1315–1331. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hills D, et al. Hoxb4-YFP reporter mouse model: A novel tool for tracking HSC development and studying the role of Hoxb4 in hematopoiesis. Blood. 2011;117:3521–3528. doi: 10.1182/blood-2009-12-253989. [DOI] [PubMed] [Google Scholar]
- 14.Seita J, et al. Gene expression commons: An open platform for absolute gene expression profiling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan CKF, et al. Identification and Specification of the Mouse Skeletal Stem Cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan CKF, et al. Clonal precursor of bone, cartilage, and hematopoietic niche stromal cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12643–12648. doi: 10.1073/pnas.1310212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosen N, et al. Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells. 2007;25:1635–1644. doi: 10.1634/stemcells.2006-0229. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto R, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Fathman JW, et al. Upregulation of CD11A on hematopoietic stem cells denotes the loss of long-term reconstitution potential. Stem cell reports. 2014;3:707–715. doi: 10.1016/j.stemcr.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat. Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 22.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 24.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 26.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 27.Forsberg EC, et al. Molecular signatures of quiescent, mobilized and leukemia-initiating hematopoietic stem cells. PLoS One. 2010;5:1–11. doi: 10.1371/journal.pone.0008785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleming WH, et al. Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. J Cell Biol. 1993;122:897–902. doi: 10.1083/jcb.122.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaiswal S, et al. CD47 Is Upregulated on Circulating Hematopoietic Stem Cells and Leukemia Cells to Avoid Phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moraga I, et al. Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands. Cell. 2015;160:1196–1208. doi: 10.1016/j.cell.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Anczuków O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.