A Vigna umbellata formate dehydrogenase, VuFDH, confers tolerance to Al and low pH by regulating formate metabolism in plants.
Abstract
Formate dehydrogenase (FDH) is involved in various higher plant abiotic stress responses. Here, we investigated the role of rice bean (Vigna umbellata) VuFDH in Al and low pH (H+) tolerance. Screening of various potential substrates for the VuFDH protein demonstrated that it functions as a formate dehydrogenase. Quantitative reverse transcription-PCR and histochemical analysis showed that the expression of VuFDH is induced in rice bean root tips by Al or H+ stresses. Fluorescence microscopic observation of VuFDH-GFP in transgenic Arabidopsis plants indicated that VuFDH is localized in the mitochondria. Accumulation of formate is induced by Al and H+ stress in rice bean root tips, and exogenous application of formate increases internal formate content that results in the inhibition of root elongation and induction of VuFDH expression, suggesting that formate accumulation is involved in both H+- and Al-induced root growth inhibition. Over-expression of VuFDH in tobacco (Nicotiana tabacum) results in decreased sensitivity to Al and H+ stress due to less production of formate in the transgenic tobacco lines under Al and H+ stresses. Moreover, NtMATE and NtALS3 expression showed no changes versus wild type in these over-expression lines, suggesting that herein known Al-resistant mechanisms are not involved. Thus, the increased Al tolerance of VuFDH over-expression lines is likely attributable to their decreased Al-induced formate production. Taken together, our findings advance understanding of higher plant Al toxicity mechanisms, and suggest a possible new route toward the improvement of plant performance in acidic soils, where Al toxicity and H+ stress coexist.
Aluminum (Al) is the most abundant metal in the earth’s crust, and occurs primarily in the form of aluminosilicates or oxides that are nontoxic to plants. However, when the soil pH drops below 5.5, soluble forms of ionic Al, mainly Al3+, are released into soil solution, inhibiting root growth and function, which in turn causes crop yield loss by impaired root absorption of soil water and mineral nutrients. Thus, Al toxicity is one of the major constraints limiting plant growth and productivity in acid soils, which comprise about 50% of potential arable lands worldwide (Kochian, 1995; Kochian et al., 2004).
Although it has long been recognized that the primary visible symptom of Al toxicity syndrome is inhibition of root elongation, the underlying mechanisms of Al root toxicity have remained ambiguous (Kochian, 1995). Because Al is such a reactive element, it may simultaneously target multiple sites in root cells, including cell wall, plasma membrane, and cytoplasm (Zheng and Yang, 2005). On the other hand, many plant species have evolved sophisticated mechanisms to deal with the toxic effects of specific aspects of Al toxicity. Two main types of Al tolerance mechanism have been proposed (Ma et al., 2001). One is an exclusion mechanism that prevents Al from entering the root apex (both symplasm and apoplasm) and the other is internal tolerance relying on the detoxification and sequestration of Al within cells. Thus far, the most well-documented Al tolerance mechanism is an exclusion strategy based on exudation of Al-chelating organic acids (mainly citrate, malate, and oxalate) from root apices into the rhizosphere (Ryan et al., 2001; Kochian et al., 2004).
Because the importance of mitochondrial respiration in regulation of organic acid metabolism and maintenance of redox homeostasis, metabolic engineering for transgenic breeding of Al-tolerant plants has previously been exploited. For example, genes coding for citrate synthase have been introduced into tobacco (Nicotiana tabacum; de la Fuente et al., 1997), Arabidopsis (Koyama et al., 2000), canola (Brassica napus; Anoop et al., 2003), and alfalfa (Medicago sativa; Barone et al., 2008), and genes coding for malate dehydrogenase have been introduced into tobacco (Wang et al., 2010) and alfalfa (Tesfaye et al., 2001). Similarly, genes related to protection from oxidative stress including manganese superoxide dismutase, dehydroascorbate reductase, peroxidase, and glutathione s-transferase have also been introduced into plants (Ezaki et al., 2000; Basu et al., 2001; Yin et al., 2010). Recently, Panda et al. (2013) reported that a transgenic tobacco cell line over-expressing the alternative oxidase gene NtAOX1 displayed decreased respiration inhibition and reduced reactive oxygen species production, and consequently better growth capacity, suggesting that mitochondrial respiratory electron transport is responsible for Al stress. However, the gains in Al tolerance through metabolic engineering in these transgenic plants appear to be limited (Ryan et al., 2011).
Recently, new (to our knowledge) insight into the importance of metabolism in plant Al-tolerance mechanism is emerging. For example, transcriptional profiling studies revealed that genes related to metabolism were differentially expressed in response to Al stress in Arabidopsis, rice (Oryza sativa), barrel clover (Medicago truncatula), maize (Zea mays), and rice bean [Vigna umbellata; (Zhang et al., 2007; Chandran et al., 2008; Kumari et al., 2008; Fan et al., 2014; Mattiello et al., 2014; Wang et al., 2014)]. In addition, proteomic analysis has indicated that proteins of metabolic function are differentially regulated in response to Al stress in roots of tobacco, soybean (Glycine max), and rice (Zhou et al., 2009; Zhen et al., 2007; Wang et al., 2014). These changed properties of metabolic genes and proteins suggest that the maintenance of normal metabolism plays an important role in alleviating the toxic effects of Al stress. Furthermore, comparative transcriptomic analysis of the effects of various rhizotoxic ions on Arabidopsis roots indicated that changes in the expression of genes related to metabolic functions are an Al-specific response (Zhao et al., 2009). Therefore, metabolic change appears to be a specific autonomous adaptive mechanistic response to Al stress, rather than to the consequences of Al toxicity.
Compared with the progress made in the role of mitochondrial respiratory metabolism in plant Al stress response (Nunes-Nesi et al., 2014), much less is known concerning other metabolisms. Ślaski et al. (1996) demonstrated that up-regulation of 6-phosphogluconate dehydrogenase enzyme activity occurs in an Al-resistant wheat (Triticum aestivum) cultivar, but not in an Al-sensitive control, suggesting that an active switch from glycolysis to the oxidative pentose P-pathway under Al stress is an Al-tolerance mechanism. Similarly, transcriptional analyses in rice bean root apices showed that genes involved in the fatty acid oxidation and oxidative pentose P-pathways were up-regulated, while genes involved in glycolysis were down-regulated in response to Al stress (Fan et al., 2014). Screening of Arabidopsis Al-hypersensitive mutant als7-1 has led to the identification of SLOW WALKER2, a putative nucleolar localized ribosomal biogenesis factor, which is involved in the indirect regulation of s-adenosyl-Met recycling and endogenous spermine biosynthesis (Nezames et al., 2013). Al-dependent accumulation of putrescine has also been reported to be involved in Al tolerance in both wheat and red kidney bean (Phaseolus vulgaris) with tolerance mechanisms being different (Wang et al., 2013; Yu et al., 2015). Thus, our understanding on how metabolism contributes to Al tolerance is still in its infancy.
Previously, a rice bean gene (VuFDH) encoding formate dehydrogenase (VuFDH) was shown to be up-regulated in rice bean root apices in response to both low (5 μM) or high (25 μM) concentrations of Al, suggesting that VuFDH may play a pivotal role in response to Al stress (Fan et al., 2014). FDHs catalyze the oxidation of formate into CO2, reducing NAD+ to NADH in the process. In higher plants, FDHs are mainly localized in the mitochondrial matrix (Halliwell, 1974; Oliver, 1981; Colas des Francs-Small et al., 1993). These observations raised questions as to whether VuFDH is involved in formate catabolism in rice bean roots, and whether the induction of VuFDH by Al is an Al stress response whose function is to reduce Al toxicity.
In this study, we isolated a full-length VuFDH cDNA, and found that the expression of VuFDH in rice bean root tips is enhanced greatly not only by Al (as previously shown; Fan et al., 2014) but also by low pH stress. We also showed that Al induces the rapid accumulation of formate in rice bean root apices, an accumulation that may contribute to Al-induced root growth inhibition. In accord with this latter possibility, over-expression of VuFDH in tobacco resulted in increased tolerance of Al and low pH. Our results therefore suggest that reduction in stress-induced formate levels may provide a (to our knowledge) novel and previously unappreciated route toward improved plant tolerance of soil acidity and Al toxicity.
RESULTS
Cloning and Sequence Analysis of VuFDH from V. umbellata
On the basis of a previously identified EST sequence (Fan et al., 2014), a full-length VuFDH cDNA was isolated from rice bean via PCR-based methods (including 5′-and 3′-RACE; GenBank: KR494281; Supplemental Fig. S1). The VuFDH coding region is 1146 bp in length, and encodes a protein of 381 amino acids. As predicted by PROSITE (http://prosite.expasy.org/scanprosite), VuFDH contains three d-isomer-specific 2-hydroxyacid dehydrogenase signatures (Supplemental Fig. S2). Sequence analysis by SignalP server 3.0 (http://www.cbs.dtu.dk/services/SignalP/) indicates that VuFDH contains a cleavable signal peptide of 24 amino acids (Supplemental Fig. S2). Although they are not extremely conserved between proteins, mitochondrial signal peptides share common features, being abundant in hydroxylated (Ser), basic (Arg), and hydrophobic (Ala, Leu) residues (Supplemental Fig. S2; Ambard-Bretteville et al., 2003). There is a mitochondrial targeting sequence (SRNLHA) that is highly conserved within the same family, but seems less conservable between different families (Supplemental Fig. S2). Phylogenetic relationship analysis indicated that VuFDH was most closely clustered with PvFDH from red kidney bean and GmFDH from soybean. However, VuFDH was loosely related to Arabidopsis AtFDH, and they were clustered into different clades (Supplemental Fig. S3).
VuFDH Functions as a Formate Dehydrogenase
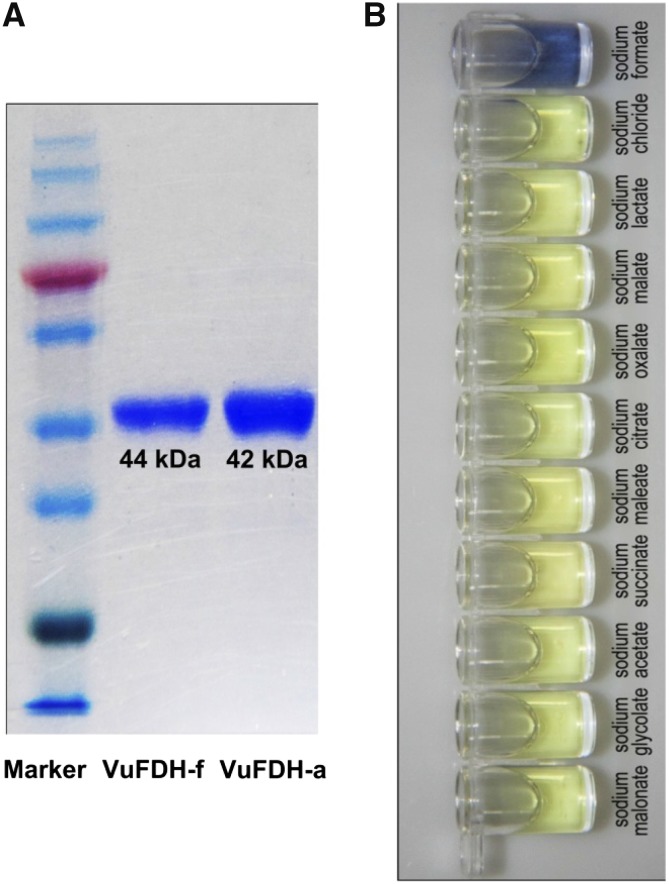
To examine if formate is indeed the substrate for VuFDH, we assayed the activity of a recombinant VuFDH protein with a range of potential substrates. Upon blue-native polyacrylamide gel electrophoresis (PAGE) and sodium dodecyl-sulfate PAGE, respectively, the appearance of a single band for both His-tagged full-length VuFDH (VuFDHf) and His-tagged VuFDH without signal peptide (VuFDHa) indicated that the recombinant protein was sufficiently pure to permit further analyses (Fig. 1A). A preparation of the recombinant protein was therefore tested against 10 different sodium carboxylic salts and the negative control sodium chloride (at pH 7.0 and using 50 mm substrate). Substrate oxidation reaction catalyzed by FDH results in the concomitant production of NADH that couples the reduction reaction of NBT to form methyl hydrazine with blue color. As shown in Figure 1B, His-VuFDH displayed high catalytic activity (as reported by the density of the blue reporter color) only with formate. Activities against other related compounds (such as oxalate, malonate, succinate, malate, glycolate, acetate, lactate, maleate, citrate, and chloride), were much lower (Fig. 1B). Thus, VuFDH is a highly specific formate dehydrogenase.
Figure 1.
Biochemical analysis of the recombinant VuFDH protein. A, SDS-PAGE gel showing the HisTrap FF affinity-purified His-tagged VuFDH protein stained with Coomassie Blue. Marker, molecular weight markers; VuFDH-f, His-tagged full-length VuFDH; VuFDH-a, His-tagged VuFDH lacking the signal peptide. B, Substrate specificity of VuFDH. Formate dehydrogenase activities were determined at pH 7.0, with various substrates at concentrations of 50 mm.
VuFDH Is a Mitochondrial Protein
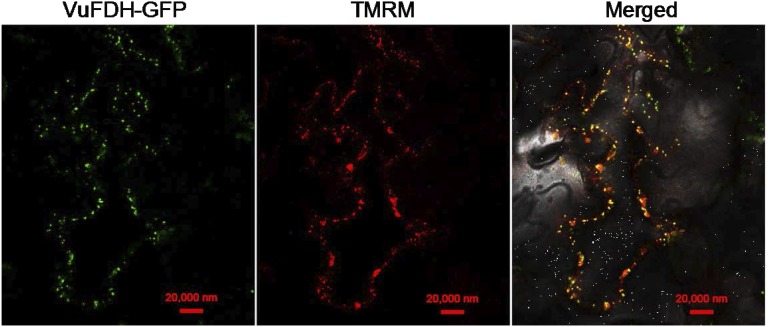
Most FDHs in higher plants are localized to mitochondria (Halliwell, 1974; Oliver, 1981; Colas des Francs-Small et al., 1993). In order to determine the subcellular localization of VuFDH, we used 35S::VuFDH::GFP fusion constructs to be transiently expressed in N. benthamiana leaves and to examine whether the GFP signal is colocalized with the mitochondria-staining fluorescence dye TMRM (tetramethyl rhodamine methyl ester). The result showed that VuFDH::GFP colocalized with TMRM in cells of N. benthamiana leaves (Fig. 2). We also constructed transgenic Arabidopsis plants over-expressing a VuFDH-GFP fusion protein under the control of the CaMV 35S promoter. The results showed that GFP fluorescence was detected as numerous scattered small spots in cells of both leaves and roots, a pattern characteristic of mitochondrial localization (Nelson et al., 2007), and were colocalized with the staining of TMRM mitochondria dye (Supplemental Fig. S4). Taken together, these results suggest that VuFDH is a mitochondrial protein.
Figure 2.
Subcellular localization of VuFDH in N. benthamiana leaves. Constructs expressing VuFDH-GFP fusion protein were transiently expressed in N. benthamiana leaves. A mitochondria-specific fluorescence dye, TMRM, was used to stain mitochondria. Bar: 20 μm.
Root VuFDH Expression Levels Are Increased by Al and Low pH Stress
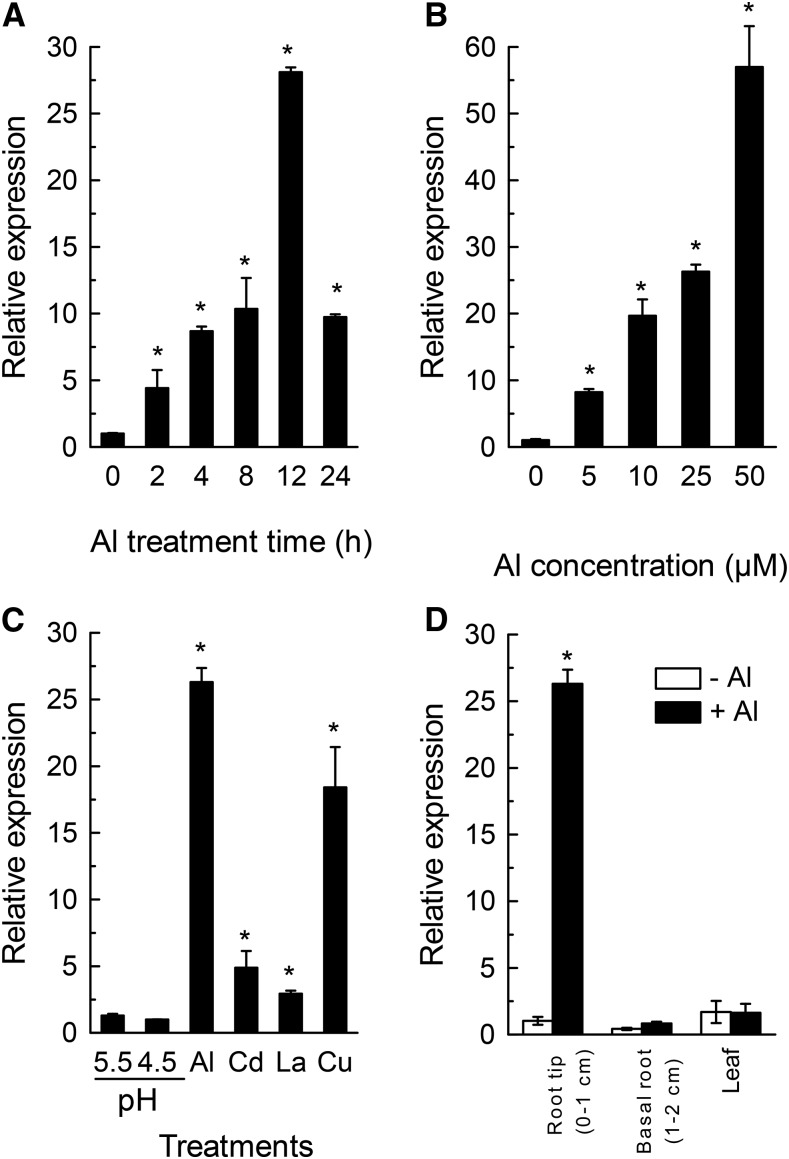
We have previously found that the expression of VuFDH was up-regulated by Al stress (Fan et al., 2014). In order to characterize comprehensively the expression of VuFHD in response to Al stress, we here used quantitative real-time PCR (qRT-PCR) to investigate the expression pattern of VuFDH. In a time-course experiment, as shown in Figure 3A, the expression of VuFDH was found to have increased within 2 h of exposure to 25 μM Al, and to be dramatically increased as the treatment was prolonged, although this increase had fallen after 12 h of exposure. In addition, in a dose-response experiment, the expression of VuFDH increased with increasing Al concentrations after 12 h of exposure (Fig. 3B).
Figure 3.
Rice bean VuFDH expression patterns. A, Time-dependent VuFDH expression in rice bean root tips (0–1 cm). The roots were exposed to 25 μm Al for various times. B, Dose-dependent VuFDH expression in rice bean root tips (0–1 cm). The roots were exposed to various concentrations of Al for 12 h. C, pH- and metal-dependent VuFDH expression in rice bean root tips (0–1 cm). The seedlings were grown at pH 5.5 and subjected to low pH (4.5) or low pH with various metals for 12 h. D, Tissue-specific expression of VuFDH. Seedlings were exposed to 0 or 25 μm Al for 12 h. All data were normalized relative to VuFDH expression in the absence of Al at pH 4.5. The expression was determined by RT-PCR and 18S rRNA was used as an internal control. Values are means ± sd (n = 3). The asterisk indicates significant differences between treatment and control (pH 4.5 without Al stress).
To examine whether this up-regulation of VuFDH expression is Al-stress-specific, we next compared the effects of Al with that of other metals (e.g. Cd, La, and Cu) and low pH. The expression of VuFDH was similar between pH 5.5 and pH 4.5. The expression level due to Al stress was much higher than that due to the stressful effects of the other metals, but Cu stress also notably increased VuFDH expression (Fig. 3C).
We also investigated the spatial patterning of VuFDH expression in either the presence or absence of Al. In the absence of Al, VuFDH is expressed in both leaves and roots and the expression level is slightly higher in leaves and root tip than that in basal root (Fig. 3D). In the presence of Al, root tip expression of VuFDH was up-regulated by more than 25-fold, while that of both basal root and leaves was not significantly affected by Al stress (Fig. 3D).
To further investigate the tissue-specific localization of VuFDH, a 0.95-kb DNA sequence upstream of the start codon was isolated. This promoter fragment was fused to a β-glucuronidase (GUS) reporter gene and transformed into Arabidopsis wild-type plants. Under normal growth conditions with pH 5.5, GUS activity was mainly expressed in leaf and the stele of mature roots, but not in the root apex (Supplemental Fig. S5). However, when the pH of the culture solution was dropped to 5.0, GUS activity was induced in the entire root elongation zone. In contrast, GUS activity levels were not affected by this decrease in solution pH in other parts of the plant (Supplemental Fig. S5). In the combined presence of Al and pH 5.0, the root GUS staining intensity was greater than in pH 5.0 alone, and this staining was also found to be further extended into the root apex (Supplemental Fig. S5).
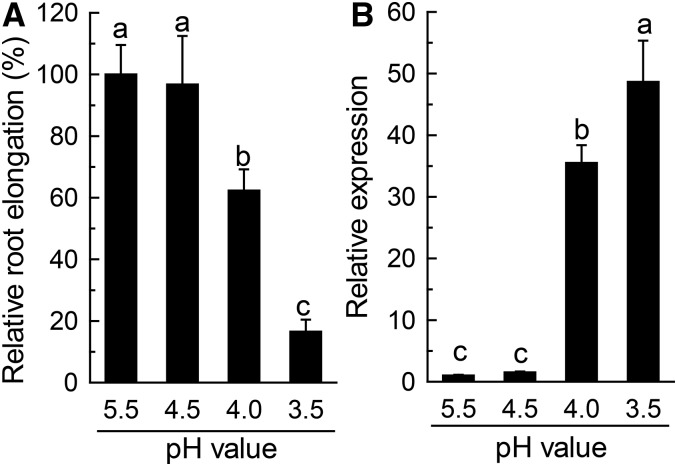
The differential behavior of VuFDH expression with respect to low pH stress between rice bean and Arabidopsis led us to hypothesize that a pH value of 4.5 is not sufficiently low to affect root growth of rice bean, but pH value of 5.0 is able to affect Arabidopsis root growth (Yang et al., 2014; Fan et al., 2015). In order to examine whether the expression of VuFDH is actually sensitive to low pH stress, we compared the effects of low pH stress on root elongation and VuFDH expression. As expected, root elongation was not affected at pH 4.5 in comparison with pH 5.5 (Fig. 4A). When pH was further dropped to 4.0, at which root elongation was inhibited by 40% (Fig. 4A), the expression of VuFDH increased significantly (Fig. 4B). Decreasing pH to 3.5 resulted in a further inhibition of root elongation, in concert with the further increase of VuFDH expression (Fig. 4).
Figure 4.
The effect of low pH stress on root elongation and VuFDH expression in rice bean. A, Relative root elongation. Seedlings were grown in nutrient solution with different pH values for 12 h. Root elongation was measured with a ruler before and after treatment (n = 12). B, VuFDH expression. After treatments, root tips (0–1 cm) were excised for RNA extraction and qRT-PCR analysis of VuFDH expression (n = 3). Different letters indicate significant differences between treatments.
Formate Accumulation Contributes to Al- and Low pH-Induced Root Growth Inhibition
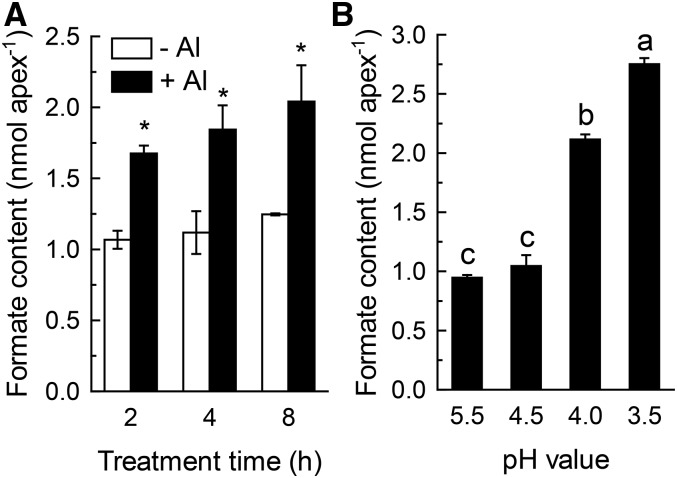
VuFDH has formate dehydrogenase activity, and can catalyze the oxidation of formate into CO2, with the accompanying reduction of NAD+ to NADH. Both low pH- and Al-dependent expression of VuFDH suggests a possible role for VuFDH in the oxidation of stress-induced formate production. To test this hypothesis, we measured via ion chromatography the internal formate content change in response to Al and low pH stress (Supplemental Fig. S6). Our results showed that the internal formate content of root apices is maintained at a relatively constant level in the absence of Al. However, Al stress causes a significant increase in the formate content of rice bean root tips within 8 h of the onset of exposure, and that this increase actually begins relatively rapidly (within 2 h; Fig. 5A). Formate content also increases significantly when pH is lower than 4.5 (Fig. 5B), which is in concert with low pH-dependent root growth inhibition (Fig. 4A). In a parallel experiment, our results showed that formate secretion rate from excised root apex decreased dramatically over time, irrespective of being treated with or without Al (Supplemental Fig. S7). Although the secretion rate during the first 2 h of treatment in Al-stressed root apex was greater than that in Al-free root apex, there was no statistical difference (Supplemental Fig. S7). Thus, the secretion of formate from rice bean root apex could be the consequence of leakage from cut damage rather than an active Al-dependent process. These findings suggest that rice bean root-tip formate accumulation is significantly influenced by Al and low pH stress, and that Al-induced formate accumulation is a relatively early Al stress response in the rice bean root tip.
Figure 5.
The effect of Al stress and H+ stress on rice bean root tip formate content. A, Al-induced formate accumulation. Seedlings were exposed to nutrient solution containing 0 or 25 μm AlCl3 for different times. B, H+ stress-induced formate accumulation. Seedlings were exposed to nutrient solution with different pH values for 12 h. After treatment, the root tips were homogenized thoroughly in deionized water for formate content analysis. Data are expressed as mean ± sd (n = 3). Asterisks in (A) and different letters in (B) indicate significant differences between treatments at P < 0.05.
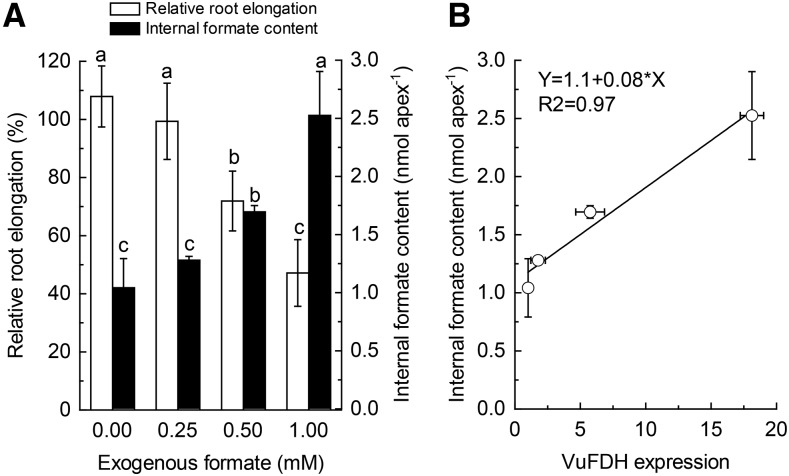
In order to further test the relationship between formate levels and root growth, we next tested the effect of exogenous formate application on rice been root elongation, internal formate content, and VuFDH expression. In normal growth conditions (pH 4.5), exogenous formate application resulted in a dose-dependent inhibition of root elongation, although formate concentration of 0.25 mm caused no significant root growth inhibition (Fig. 6A). On the contrary, internal formate content increased with increase of exogenous formate, which was in concert with root growth inhibition (Fig. 6A). We further found that internal formate content is positively correlated with VuFDH expression (Fig. 6B), suggesting that formate accumulation causes induction of VuFDH expression.
Figure 6.
The effect of exogenous formate on rice bean root growth, internal formate accumulation, and VuFDH expression. A, The effect of formate on rice bean root elongation. Seedlings were exposed to nutrient solution (pH 4.5) containing different concentrations of exogenous formate for 12 h. Root elongation was measured with a ruler before and after treatment (n = 12). After treatment, root tips (0–1 cm) were excised for internal formate quantification (n = 3). In a parallel experiment, root tips (0–1 cm) were excised for RNA extraction and qRT-PCR analysis of VuFDH expression (n = 3). B, Correlation between internal formate content and VuFDH expression. Different letters indicate significant differences between treatments at P < 0.05.
Over-Expression of VuFDH in Transgenic Tobacco Confers Al and Low pH Tolerance
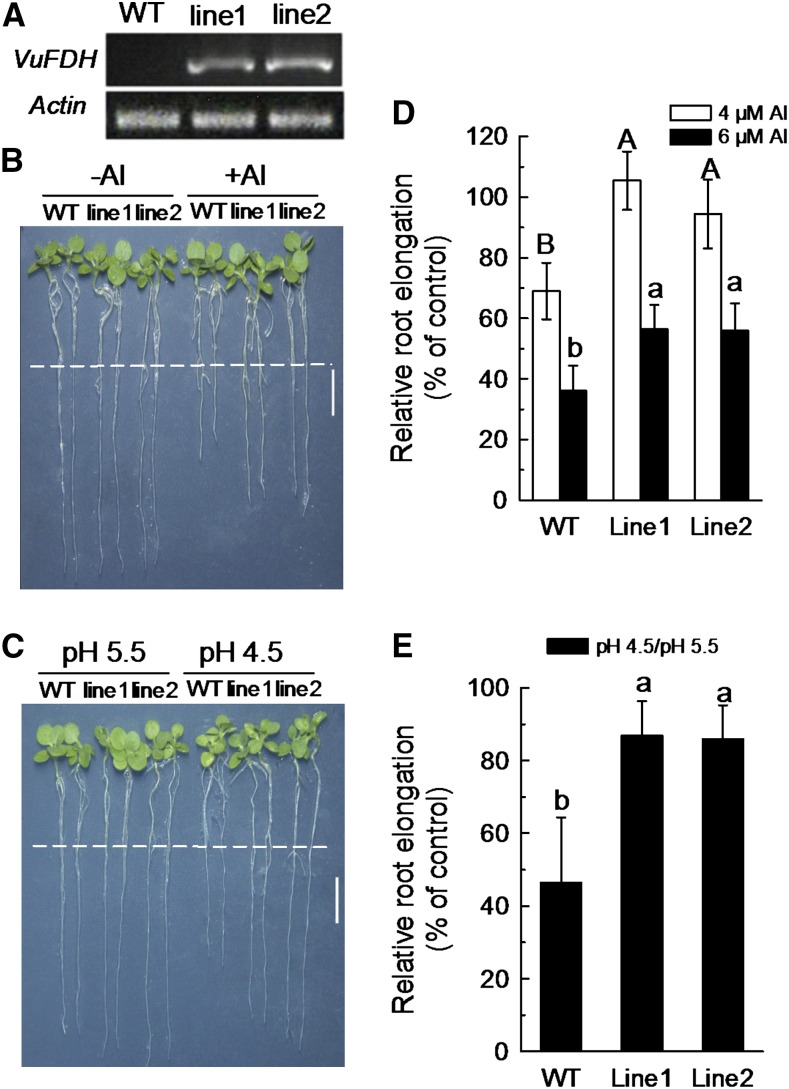
To further characterize the role of VuFDH in Al and low pH response, a 35S::VuFDH construct was introduced into tobacco plants. We selected two independent homozygous T2 transgenic lines (lines 1 and 2) for phenotypic and physiological analysis. Semi-qRT-PCR analyses showed that VuFDH was highly expressed in both transgenic lines (Fig. 7A).
Figure 7.
Over-expression of VuFDH enhances Al and H+ tolerance. A, Detection of expression of VuFDH in the wild-type and VuFDH over-expression lines. RT-PCR analysis was performed to detect the mRNA expression of VuFDH (32 cycles) and the internal control NtACTIN (29 cycles). B, Representative seedlings showing difference in Al sensitivity between the wild-type and the over-expression lines. Seedlings were grown in the 1:30 strength Hoagland nutrient solution containing 0, 4 or 6 μm AlCl3 at pH 5.0 for 6 d. C, Representative seedlings showing difference in H+ sensitivity between the wild-type and the over-expression lines. Seedlings were grown in the 1:30 strength Hoagland nutrient solution at pH 5.5 or 4.5 for 6 d. D, Relative root elongation of wild-type and the transgenic lines grown as described in (B). Data are means ± sd (n = 15). E, Relative root elongation between wild-type and the transgenic lines grown as described in (C). Data are means ± sd (n = 15). Dashed white lines in (B) and (C) indicate the root tip position at the beginning of treatment. Different letters indicate significant differences between treatments at P < 0.05. Bar: 1 cm in (B) and (C).
In a test of Al tolerance, both wild-type and transgenic plants were grown hydroponically either in the presence or absence of Al (at different concentrations). While 6 d of exposure to 4 μm Al inhibited the root growth of transgenic line2 by about 5% (and while transgenic line1 was not detectably affected), the root growth of wild-type control plants was inhibited by approximately 30% (Fig. 7D). Increase of the Al concentration to 6 μm resulted in inhibition of the root growth of wild-type control plants by approximately 65%, while root growth of both line1 and line2 was inhibited by approximately 45% (Fig. 7, B and D). These observations suggest that over-expression of VuFDH in tobacco confers increased Al tolerance.
As VuFDH expression is greatly induced by low pH (Fig. 4B), we next investigated the low pH tolerance of transgenic tobacco plants over-expressing VuFDH. We found no significant difference among the root growths of any of wild-type, transgenic line1, or line2 plants grown at pH 5.5 for 6 d (Fig. 7C). However, when the culture solution pH was decreased to 4.5 for 6 d, the root growth of transgenic plants was inhibited by approximately 15% (in both transgenic lines 1 and 2), while that of wild-type plants was inhibited by approximately 55% (Fig. 7, C and E). These results suggest that VuFDH is involved in H+ stress tolerance as well as in Al stress tolerance.
VuFDH Increases Al and Low pH Tolerance by Decreasing Formate Accumulation
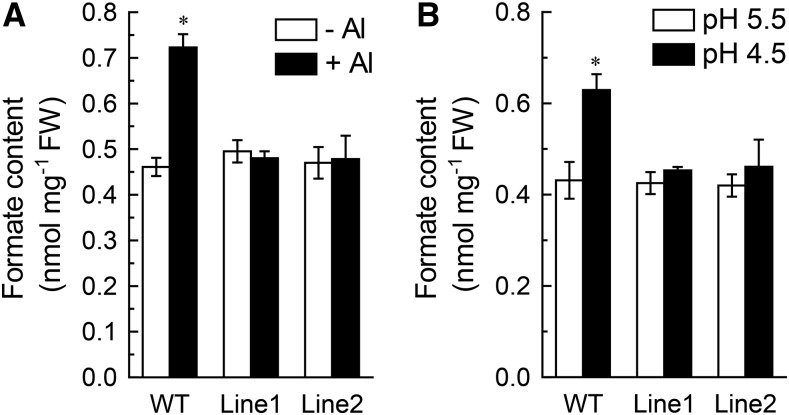
To determine if the increased Al and low pH tolerance of transgenic tobacco plants over-expressing VuFDH is associated with a decrease in formate accumulation, we compared the Al- and low pH-induced formate production of transgenic plants to that of wild-type plants. In the absence of Al stress (Fig. 8A) or in the conditions of pH 5.5 (Fig. 8B), there are no detectable differences between the formate contents of wild type and both of the transgenic lines. However, while both Al stress and low pH stress significantly increased the accumulation of formate in wild-type roots, it had no detectable effect on formate accumulation in the roots of both transgenic lines.
Figure 8.
The effect of Al and H+ stress on formate content in wild-type and over-expression tobacco lines. A, Al-induced accumulation of formate. The plants of wild-type and two independent transgenic lines were exposed to 1:30 strength Hoagland nutrient solution with 0 or 4 μm Al for 24 h. B, H+ stress-induced accumulation of formate. The plants of wild-type and two independent transgenic lines were exposed to 1:30 strength Hoagland nutrient solution with pH adjusted to either 5.5 or 4.5 for 24 h. After treatment, root tips were homogenized thoroughly in deionized water for formate content analysis. Data are mean ± sd (n = 3). Asterisk indicates significant differences between treatments at P < 0.05.
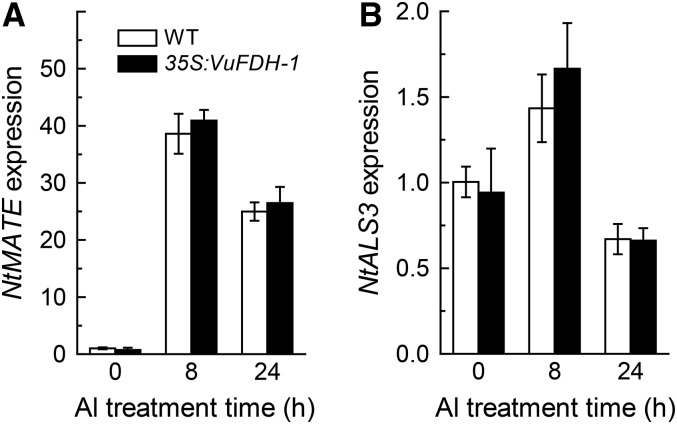
In tobacco, the decreased Al tolerance of Sensitive to Proton Rhizotoxicity1 RNAi transgenic plants is related to the conferred down-regulation of Multidrug and Toxic Compound Extrusion (NtMATE) and Aluminum Sensitive3 (NtALS3; Ohyama et al., 2013). To determine if the increased Al tolerance of VuFDH over-expressing lines is associated with changes in NtMATE or NtALS3 expression, we next compared the expression of these genes in VuFDH-over-expressing line1 with that in wild-type plants. However, there was no detectable difference between the expression of either of these genes in wild-type versus transgenic line1, either in the presence or absence of Al, although the expression of both was induced by Al (Fig. 9, A and B). Taken together, these observations indicate that VuFDH affects Al resistance mainly via decreases in formate content, and not via effects on the expression of NtMATE or NtALS3.
Figure 9.
The effect of Al stress on the expression of Al-tolerance gene expression in tobacco. A, NtMATE expression. B, NtALS3 expression. The plants of wild-type and independent transgenic line1 were exposed to 1:30 strength Hoagland nutrient solution containing 4 μm Al for different times. The expression was determined by RT-PCR and NtACTIN was used as an internal control. Data are means ± sd (n = 3).
DISCUSSION
In plants, FDHs have been reported to be involved in stress responses, because the expression of FDHs was responsive to a wide range of abiotic and biotic stresses (Hourton-Cabassa et al., 1998; David et al., 2010). Recently, Choi et al. (2014) reported that FDHs play important role in hypersensitive response-like cell death and defense responses to bacterial pathogens in both peppers (Capsicum annuum) and Arabidopsis. However, the role of FDHs in stress tolerance remains largely unknown. In this study, we demonstrated that rice bean VuFDH is involved in both Al and low pH stress tolerance as evidenced by the improvement of Al and H+ stress tolerance in transgenic tobacco over-expressing VuFDH (Fig. 7). While it remains to be investigated whether our transgenic tobacco plants are also resistant to the attack of bacterial pathogens, the expression of Arabidopsis for Aluminum-activated Malate Transporter1 has been documented to be involved in both Al tolerance and pathogen resistance (Rudrappa et al., 2008; Lakshmanan et al., 2012; Kobayashi et al., 2013). In our previous study, a pathogenesis-related gene was found to be up-regulated by Al stress in rice bean (Fan et al., 2014). Thus, Al may act as an elicitor of a pathogenesis-related transduction pathway. In addition to Aluminum-activated Malate Transporter1, VuFDH could be another good target for studies aimed at elucidating the complex nature between abiotic (i.e. Al tolerance) and biotic (i.e. defense response) stress tolerance in plants.
We found that the expression of VuFDH is induced by Al and H+ stress in the root apex of rice bean (Figs. 3 and 4). In barley roots, the expression of FDH is induced by iron deficiency, an induction that appears to be a secondary effect of the oxygen deficiency resulting from iron deficiency (Suzuki et al., 1998). It has also been proposed that glycolysis may play an important role in the FDH expression in response to abiotic stresses (Hourton-Cabassa et al., 1998). Similarly, we have previously shown that genes involved in glycolysis are repressed, but that genes related to anaerobic respiration are promoted by Al stress in rice bean root apices (Fan et al., 2014), implying that hypoxia and glycolysis may be responsible for rice bean VuFDH expression in response to Al stress. However, the exact mechanisms regulating FDH expression during stress conditions are not known. Here we suggested that formate is the mediator that induces VuFDH expression in response to Al and H+ stress. This conclusion is supported by the following lines of evidence. First, both Al and H+ stress resulted in the accumulation of formate (Fig. 5), and the rapid accumulation of formate in root apices under Al stress raises the possibility that formate itself could be a direct activator of VuFDH expression induction. Second, the specificity of the VuFDH enzyme with respect to formate as a substrate (versus its relative inactivity with succinate, malate, glycolate, acetate, lactate, etc.) further strengthens this hypothesis (Fig. 1). Finally, exogenous formate application experiment provided evidence that VuFDH expression is positively correlated with internal formate accumulation (Fig. 6). In line with our hypothesis, it has also been reported that the spraying of formate onto leaves can effectively induce the expression of FDH (Hourton-Cabassa et al., 1998).
Al toxicity and H+ stress are two coexisting factors limiting plant growth in acid soils. It has previously been suggested that H+ and Al rhizotoxicities induce root damage by different mechanisms (Koyama et al., 1995; Kinraide, 2003), although H+ toxicity also causes a severe inhibition of root growth resembling that due to Al toxicity. However, dysfunction of an Arabidopsis gene encoding C2H2-type zinc finger transcription factor, AtSTOP1, resulted in both Al and H+ hypersensitivity (Iuchi et al., 2007). Recently, we isolated and characterized VuSTOP1, a homolog of AtSTOP1, with respect to Al and H+ stress tolerance (Fan et al., 2015). Interestingly, similar to the expression pattern of VuFDH, the expression of VuSTOP1 is also regulated by both Al and H+ stress. These results suggest that Al toxicity and H+ stress are interrelated at transcriptional levels. Here we found that formate accumulation is common to both Al and H+ stress (Fig. 5). In addition, exogenous formate inhibits root elongation, and actually the inhibition of root elongation was correlated with the increase of internal formate accumulation and VuFDH expression induction (Fig. 6). Furthermore, the enhanced Al and H+ stress tolerance of transgenic tobacco plants is associated with the decrease of formate accumulation (Fig. 8). In accord with our results, Li et al. (2002) has previously found that formate is toxic to Arabidopsis root growth, and that over-expression of an FDH enzyme in Arabidopsis conferred enhanced tolerance to the effects of exogenous formate on root growth. Therefore, formate accumulation could be a link between Al toxicity and H+ stress at the metabolic level. While a question remains open as to the regulatory mechanisms of Al and H+ stress-induced formate production, it is also unclear whether VuSTOP1 is involved in regulation of VuFDH under Al and H+ stress, and this possibility needs further investigation.
FDHs catalyze the oxidation of the formate into carbon dioxide, coupled with NAD+ reduction to NADH. Therefore, in addition to the formate catabolism, H+ may also be consumed due either to formation of NADH or carbonate. It is well known that H+ stress is associated with cytosolic acidosis, but whether Al stress is also related to cytosolic acidosis is a matter of debate (Shavrukov and Hirai, 2016). Sawaki et al. (2009) reported that genes and metabolites involved in γ-aminobutyric acid shunt and biochemical pH stat were down-regulated by the dysfunction of AtSTOP1 in Arabidopsis, implying that cellular acidification may be a common process in response to Al and H+ stress. However, Bose et al. (2010) reported that H+ stress caused intercellular acidification, while Al stress in combination with H+ stress had opposite effects in Arabidopsis root apex. Using NMR technique, we found that H+ stress caused cellular acidification of rice bean root apex, while Al stress could not (data not shown). Thus, it is formate catabolism, but not H+ consumption, that contributes to the Al- and low-pH-stress tolerance mediated by VuFDH expression.
We found that VuFDH has a mitochondrial targeting sequence (Supplemental Figs. S1 and S2), and is localized to the mitochondria (Fig. 2; Supplemental Fig. S4). This result is in accord with previous reports concerning the localization of FDH proteins in various higher plant tissues (Halliwell, 1974; Oliver, 1981; Colas des Francs-Small et al., 1993). Question then arises concerning the nature of the pathways to formate formation in the mitochondrial matrix under Al and H+ stress. In plants, various formate production pathways including photorespiration, glycolysis, cell wall synthesis, or degradation have been proposed (Igamberdiev et al., 1999; Hanson et al., 2000), but direct experimental evidence is limited. Recently, a previously unknown oxalyl-CoA synthetase, ACYL-ACTIVATING ENZYME3 (AAE3), was identified to be involved in oxalate degradation to form formate, which is further degraded by FDH in Arabidopsis (Foster et al., 2012). Furthermore, FDH is coexpressed with AAE3 and regulated by AAE3. Interestingly, a gene encoding PCAS (peroxisomal-coenzyme A synthetase) was found to be up-regulated significantly in root apices of rice bean, and PCAS has high amino-acid sequence homology with Arabidopsis AAE3 (Fan et al., 2014). Thus, it is likely that formate accumulation originates from degradation of oxalate mediated by PCAS. The role of PCAS in oxalate degradation and formate accumulation requires further investigation.
Transgenic approaches have been identified as potentially powerful methods to increase the Al tolerance of plants in acidic soils (Ryan et al., 2011; Kochian et al., 2015). Thus far, a number of genes involved in different biological processes (including organic acid metabolism, stress response, and organic acid transport) have been successfully introduced into plants, with the resultant transgenic plants showing enhanced Al tolerance (Ryan et al., 2011). In this study we have demonstrated that over-expression of VuFDH, a gene involved in formate metabolism, enhanced not only the Al tolerance but also H+ stress tolerance of transgenic tobacco plants (Fig. 7). While the prevailing ideas on transgenic modification of plant Al tolerance have focused on increasing the efflux of organic acids from root cells (thus increasing chelation of external Al), little attention has been paid to the possible carbon-use efficiency penalties of such a strategy (Liu et al., 2012). In fact, root exudation is known to represent a significant carbon loss to the plant (Whipps, 1990). In this study, the expression of NtMATE did not detectably differ between wild-type and transgenic plants (Fig. 9A), suggesting that the enhanced Al tolerance of these transgenic plants is not associated with enhanced citrate efflux (an important Al resistance mechanism in tobacco; Delhaize et al., 2001). In addition, NtALS3 is a tobacco homolog of OsSTAR2 of rice. Although the detailed function of NtALS3 remains to be characterized, OsSTAR2 interacts with OsSTAR1 to form a complex that transports UDP-Glc to the apoplast, thus protecting the cell wall from Al damage (Huang et al., 2009). Thus the apparently unchanged expression of NtALS3 in wild-type versus transgenic plants (Fig. 9B) makes it unlikely that the increased Al tolerance of our transgenic plants is due to the exudation of chelating substances out of cells, a process that in any case is potentially detrimental to plant carbon-use efficiency. Our findings therefore not only suggest that detoxification by VuFDH of the formate that accumulates during Al stress conditions plays an important role in the overall Al tolerance mechanism, but that such detoxification also provides an alternative route toward the increase of plant Al tolerance through genetic engineering, a route that does not incur increased carbon loss.
In summary, we have characterized rice bean VuFDH, and shown that it is a formate dehydrogenase that mediates oxidation of the formate that is produced in conditions of Al- and low pH-stresses. To the best of our knowledge, this is the first report of the production of formate in response to Al- and low pH-stress and of the significance of formate detoxification in enhancing plant Al- and low pH-tolerance. We have shown that tobacco plants over-expressing rice bean VuFDH display an increased ability to tolerate both Al stress and H+ stress. Our findings contribute to the further understanding of stress-induced formate toxicity and provide a potential new solution to the improvement of plant performance in acidic soils, where Al toxicity and H+ stresses coexist.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice bean (Vigna umbellata) was used in this study. Seeds were soaked in deionized water overnight, then wrapped within four layers of moist gauze and germinated at 26°C in the dark. The germinated seeds were cultured in 0.5 mm CaCl2 (pH 4.5) solution for 3 d. The solution was renewed daily. Seedlings of similar size were transplanted into nutrient solution of the following composition: CaSO4 (200 μm), CaCl2 (200 μm), MgSO4 (100 μm), KNO3 (400 μm), NH4NO3 (300 μm), NaH2PO4 (5 μm), H3BO3 (3 μm), MnCl2 (0.5 μm), ZnSO4 (0.4 μm), CuSO4 (0.2 μm), Fe-EDTA (10 μm), and (NH4)6Mo7O24 (1 μm).The solution was adjusted to pH 4.5 with HCl, and renewed daily. After 2 d of culture, the plants were subjected to the following treatments. The nutrient solution was used as the control treatment solution. For the time-course experiment, seedlings were exposed to 25 μm AlCl3 for 0, 2, 4, 8, 12, or 24 h. For the Al concentration dependence experiment, seedlings were exposed to 0, 5, 10, 25, or 50 μm AlCl3 for 12 h. For other treatments, the seedlings were exposed to nutrient solution (pH 4.5) containing 25 μm AlCl3, 20 μm CdCl2, 10 μm LaCl3, or 0.5 μm CuCl2 or in different pH conditions for 12 h. For the exogenous formate experiment, the seedlings were exposed to nutrient solution (pH 4.5) containing 0, 0.25, 0.5, or 1.0 mm sodium formate for 12 h. All experiments were performed in an environmentally controlled growth room with a 12 h/30°C d and a 12 h/22°C night regime, a light intensity of 300–350 μmol photons m−2 s−1, and a relative humidity of 60%.
Cloning of VuFDH and Over-Expression of VuFDH in Tobacco
Total RNA isolated from 25 μm Aluminum (Al)-treated rice bean root apices (0–1 cm) was used for the synthesis of 5′-RACE-ready and 3′-RACE-ready cDNA. Gene-specific primers used for the 5′-and 3′-RACE amplification (Supplemental Table S1) were obtained from a differentially expressed cDNA library from rice bean root tips grown under Al stress (Fan et al., 2014). All steps were performed according to the manufacturer’s protocol (Clontech Laboratories, Madison, WI).
The RACE products were purified, and sequenced. The VuFDH coding region was amplified by PCR using primer pairs (Supplemental Table S1) and ligated into a modified pCAMBIA1300 vector under the control of the CaMV 35S promoter, then transformed into Agrobacterium tumefaciens (strain GV1301). Tobacco (Nicotiana tabacum) plants were transformed as described by Horsch et al. (1985). Transgenic lines carrying VuFDH were selected by PCR using the primers described above.
Gene Expression Analysis
Total RNA was isolated using the Column Plant RNAOUT kit (Tiandz, Dalian, China). One microgram of total RNA was synthesized into first-strand cDNA using Primescript reverse transcriptase (Clontech Laboratories/Takara Bio USA, Madison, WI). Gene expression levels were determined by quantitative reverse-transcription (RT)-PCR using the SYBR Premix Ex Taq kit (Clontech Laboratories/Takara Bio USA) on a LightCycler 480 machine (Roche Diagnostics, Indianapolis, IN). Expression levels were normalized relative to the expression level of the 18S rRNA (as internal control in rice bean) or NtACTIN (as internal control in tobacco). The primers used were listed in Supplemental Table S1. The reaction conditions were 45 cycles at 95°C for 15 s, 56°C for 10 s, and 72°C for 15 s. For all experiments, quantitative RT-PCRs were performed as triplicates on three different RNA samples isolated independently from each tested condition.
Construction of the VuFDH-GFP Fusion and Subcellular Localization of VuFDH
To construct the VuFDH-GFP fusion-protein-expressing constructs, a VuFDH cDNA fragment containing KpnI and BamHI restriction sites, but not the stop codon, was amplified by PCR using the primers (Supplemental Table S1). The amplified cDNA fragment was then cloned in-frame in front of the GFP coding region in the modified pCAMBIA1300 vector, thus placing VuFDH-GFP under the control of the 35S promoter. Subcellular localization was investigated by over-expressing 35S:VuFDH-GFP transiently in tobacco (N. benthamiana) leaves or stably in Arabidopsis (transformation according to the floral dip method; Clough and Bent, 1998). GFP fluorescence was observed using confocal laser scanning microscopy (LSM710; Carl Zeiss, Jena, Germany).
Evaluation of the Sensitivity of Transgenic Plants to Al and Low pH Stress
Seeds from transgenic and wild-type tobacco lines were first surface-sterilized with 15% (v/v) sodium hypochlorite for 5 min, and then washed four times with deionized water. Then, seeds were sown onto MS plates containing 3% (w/v) Suc and 0.8% (w/v) agar (pH 5.7). After incubation in a refrigerator at 4°C for 3 d, the seeds were then placed in a growth chamber in 12-h light/12-h dark conditions at 23°C. When the length of the primary root had reached approximately 1 cm, the seedlings were transferred to the 1:30 strength Hoagland nutrient solution without NH4H2PO4 and with 1 mm CaCl2. For Al sensitivity evaluations, the seedlings were grown in nutrient solution containing 0, 4, or 6 μm AlCl3 at pH 5.0 for 6 d. For low pH tolerance evaluations, the seedlings were grown in nutrient solution at pH 4.5 or pH 5.5 for 6 d. The solution was renewed every 2 d. Al sensitivity was evaluated by relative root elongation expressed as (root elongation with Al treatment/root elongation without Al) × 100. Low pH tolerance was evaluated by relative root elongation expressed as (root elongation at pH 4.5 /root elongation at pH 5.5) × 100.
β-Glucuronidase Analysis
The VuFDH promoter was obtained by genome walking using the Genome Walker Universal Kit (Clontech Laboratories). In brief, four genome walker libraries were constructed by digesting separate aliquots of DNA with four different restriction enzymes (DraI, EcoRV, PvuII, and StuI), followed by ligation to a genome walker adaptor. The outer/inner adaptor primer provided by the kit and two VuFDH gene-specific primers (Supplemental Table S1) were used to perform the nested PCR. The amplified fragments were subsequently cloned into the pMD19-T vector (Clontech Laboratories/Takara Bio USA). Sequences extending upstream of the cDNA sequence were isolated as the 5′-upstream regions of the gene.
An approximately 0.95-kb VuFDH promoter fragment was amplified from this genomic DNA using primers (Supplemental Table S1). For GUS analysis, the obtained VuFDH upstream sequence was subcloned into a pCAMBIA1301 vector as a fusion to the β-glucuronidase (GUS) gene and finally transformed into Arabidopsis wild-type (Col-0) plants by Agrobacterium-mediated transformation.
For histochemical staining of GUS activity, homozygous T3 plants were used. GUS staining was performed according to Jefferson et al. (1987), with or without exposure to 10 μm AlCl3 at pH 5.0 or pH 5.5 for 24 h. Seedlings were observed and photographed with a model no. AZ100 microscope (Nikon, Melville, NY).
Determination of Formate
Formate extracted from plant materials or collected from root exudates were filtered (0.22 μm) before analysis. Formate was detected by an ion chromatography apparatus (ICS 3000; Dionex, Sunnyvale, CA) equipped with a model no. AS11 IonPac anion-exchange analytical column (4 × 250 mm; Dionex) and a guard column (4 × 50 mm). The mobile phase was deionized water at a flow rate of 1.0 ml min−1.
Purification of His-Tagged FDH Proteins
Escherichia coli strain BL21 (DE3)-competent cells (Tiangen Biotech, Beijing, China) were transformed with the N-terminal His-tagged pET-28a (+) vector (Novagen/Merck Millipore, Darmstadt, Germany) with a bacterial expression vector containing VuFDH and spread on Luria-Bertani medium plates with 100 mg/l kanamycin. Positive clones were PCR-tested. The primers for detection of full-length VuFDH and VuFDH without the signal peptide were listed in Supplemental Table S1. Positive clones were then incubated in Luria-Bertani medium supplemented with 100 mg/l kanamycin at 37°C until an OD600 of 0.6 was reached. To induce expression, 1 mm IPTG was added, and the culture was grown for an additional 6 h at 28°C with shaking at 200 rpm. The cells were harvested by centrifugation and resuspended in binding buffer, and the suspension was subsequently homogenized by 1 h of 200 W sonication (Vibra Cell VC 505 Sonicator; Sonics & Materials, Newtown, CT). Cell debris was subsequently removed with 10-min centrifugation at 12,000 rpm, in order to protect the columns. Protein purification was performed using HisTrap FF affinity columns (GE Healthcare, Washington, NY) following the manufacturer’s instructions. To obtain maximum purity, we used 40 mm and 500 mm imidazole for binding buffer and elution buffer, respectively; and no reducing agents, denaturing agents, detergents. or other additives were used during purification. The protein solutions were desalted by ultrafiltration with Amicon Ultra 15 ml 30 KD Tubes (Millipore, Billerica, MA). The sizes of the native protein and of the monomer were assessed by blue-native polyacrylamide gel electrophoresis (Fiala et al., 2011) and sodium dodecyl-sulfate-polyacrylamide gel electrophoresis, respectively.
Determination of FDH Enzyme Activity
FDH activity was visualized on non-denaturing polyacrylamide gels following the method of Uotila and Koivusalo (1979). The size and purity of the protein were assessed by running a sample on an sodium dodecyl-sulfate-polyacrylamide gel, and the purified protein was then incubated in darkness for 30 min at room temperature in the following solution: 100 mm sodium P-buffer, pH 7.0, 50 mm substrate, 0.8 mm NAD+, 0.03 mg ml−1 phenazine methosulfate, and 0.4 mg ml−1 NBT. Substrates tested for FDH activity were oxalate, malonate, succinate, malate, glycolate, acetate, lactate, maleate, citrate, chloride, and formate.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: V. umbellata VuFDH (KR494281).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nucleotide and deduced amino-acid sequences of rice bean VuFDH cDNA.
Supplemental Figure S2. Alignment of rice bean VuFDH.
Supplemental Figure S3. Phylogram of FDH proteins.
Supplemental Figure S4. Subcellular localization of VuFDH in Arabidopsis leaves and roots.
Supplemental Figure S5. GUS activity analysis of transgenic VuFDHp::GUS Arabidopsis plants.
Supplemental Figure S6. Ion chromatography quantification of formate in extracts of plant materials.
Supplemental Figure S7. The effect of Al stress on formate exudation from root tips of rice bean.
Supplemental Table S1. Primer sequences used in the study.
Footnotes
Articles can be viewed without a subscription.
References
- Ambard-Bretteville F, Small I, Grandjean O, Colas des Francs-Small C (2003) Discrete mutations in the presequence of potato formate dehydrogenase inhibit the in vivo targeting of GFP fusions into mitochondria. Biochem Biophys Res Commun 311: 966–971 [DOI] [PubMed] [Google Scholar]
- Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ (2003) Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol 132: 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone P, Rosellini D, Lafayette P, Bouton J, Veronesi F, Parrott W (2008) Bacterial citrate synthase expression and soil aluminum tolerance in transgenic alfalfa. Plant Cell Rep 27: 893–901 [DOI] [PubMed] [Google Scholar]
- Basu U, Good AG, Taylor GJ (2001) Transgenic Brassica napus plants overexpressing aluminium-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant Cell Environ 24: 1269–1278 [Google Scholar]
- Bose J, Babourina O, Shabala S, Rengel Z (2010) Aluminum-dependent dynamics of ion transport in Arabidopsis: specificity of low pH and aluminum responses. Physiol Plant 139: 401–412 [DOI] [PubMed] [Google Scholar]
- Chandran D, Sharopova N, Ivashuta S, Gantt JS, Vandenbosch KA, Samac DA (2008) Transcriptome profiling identified novel genes associated with aluminum toxicity, resistance and tolerance in Medicago truncatula. Planta 228: 151–166 [DOI] [PubMed] [Google Scholar]
- Choi DS, Kim NH, Hwang BK (2014) Pepper mitochondiral FORMATE DEHYDROGENASE1 regulates cell death and defense responses against bacterial pathogens. Plant Physiol 166: 1298–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Rémy R (1993) Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol 102: 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P, des Francs-Small CC, Sévignac M, Thareau V, Macadré C, Langin T, Geffroy V (2010) Three highly similar formate dehydrogenase genes located in the vicinity of the B4 resistance gene cluster are differentially expressed under biotic and abiotic stresses in Phaseolus vulgaris. Theor Appl Genet 121: 87–103 [DOI] [PubMed] [Google Scholar]
- de la Fuente JM, Ramírez-Rodríguez V, Cabrera-Ponce JL, Herrera-Estrella L (1997) Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science 276: 1566–1568 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Hebb DM, Ryan PR (2001) Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiol 125: 2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki B, Gardner RC, Ezaki Y, Matsumoto H (2000) Expression of aluminum-induced genes in transgenic arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol 122: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Lou HQ, Gong YL, Liu MY, Cao MJ, Liu Y, Yang JL, Zheng SJ (2015) Characterization of an inducible C2 H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol 208: 456–468 [DOI] [PubMed] [Google Scholar]
- Fan W, Lou HQ, Gong YL, Liu MY, Wang ZQ, Yang JL, Zheng SJ (2014) Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant Cell Environ 37: 1586–1597 [DOI] [PubMed] [Google Scholar]
- Fiala GJ, Schamel WWA, Blumenthal B (2011) Blue native polyacrylamide gel electrophoresis (BN-PAGE) for analysis of multiprotein complexes from cellular lysates. J Vis Exp 48: 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Kim HU, Nakata PA, Browse J (2012) A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. (1974) Oxidation of formate by peroxisomes and mitochondria from spinach leaves. Biochem J 138: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Gage DA, Shachar-Hill Y (2000) Plant one-carbon metabolism and its engineering. Trends Plant Sci 5: 206–213 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hourton-Cabassa C, Ambard-Bretteville F, Moreau F, Rémy R, Colas de Francs-Small C, Davy de Virville J (1998) Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol 116: 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Bykova NV, Kleczkowski LA (1999) Origins and metabolism of formate in higher plants. Plant Physiol Biochem 37: 503–513 [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104: 9900–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinraide TB. (2003) Toxicity factors in acidic forest soils: attempts to evaluate separately the toxic effects of excessive Al3+ and H+ and insufficient Ca2+ and Mg2+ upon root elongation. Eur J Soil Sci 54: 323–333 [Google Scholar]
- Kobayashi Y, Kobayashi Y, Sugimoto M, Lakshmanan V, Iuchi S, Kobayashi M, Bais HP, Koyama H (2013) Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol 162: 732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV. (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 237–260 [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Kochian LV, Piñeros MA, Liu J, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Plant Biol 66: 23.1–23.28 [DOI] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D (2000) Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol 41: 1030–1037 [DOI] [PubMed] [Google Scholar]
- Koyama H, Toda T, Yotoka S, Dawair Z, Hara T (1995) Effects of aluminum and pH on root growth and cell viability in Arabidopsis thaliana strain Landsberg in hydroponic culture. Plant Cell Physiol 36: 201–205 [Google Scholar]
- Kumari M, Taylor GJ, Deyholos MK (2008) Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol Genet Genomics 279: 339–357 [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160: 1642–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Moore M, Bonham-Smith PC, King J (2002) Overexpression of formate dehydrogenase in Arabidopsis thaliana resulted in plants tolerant to high concentrations of formate. J Plant Physiol 159: 1069–1076 [Google Scholar]
- Liu J, Luo X, Shaff J, Liang C, Jia X, Li Z, Magalhaes J, Kochian LV (2012) A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance. Plant J 71: 327–337 [DOI] [PubMed] [Google Scholar]
- Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6: 273–278 [DOI] [PubMed] [Google Scholar]
- Matsumoto H. (2000) Cell biology of aluminum toxicity and tolerance in higher plants. Int Rev Cytol 200: 1–46 [DOI] [PubMed] [Google Scholar]
- Mattiello L, Begcy K, da Silva FR, Jorge RA, Menossi M (2014) Transcriptome analysis highlights changes in the leaves of maize plants cultivated in acidic soil containing toxic levels of Al3+. Mol Biol Rep 41: 8107–8116 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nezames CD, Ochoa V, Larsen PB (2013) Mutational loss of Arabidopsis SLOW WALKER2 results in reduced endogenous spermine concomitant with increased aluminum sensitivity. Funct Plant Biol 40: 67–78 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Santos Brito D, Inostroza-Blancheteau C, Fernie AR, Araújo WL (2014) The complex role of mitochondrial metabolism in plant aluminum resistance. Trends Plant Sci 19: 399–407 [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Ito H, Kobayashi Y, Ikka T, Morita A, Kobayashi M, Imaizumi R, Aoki T, Komatsu K, Sakata Y, Iuchi S, Koyama H (2013) Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol 162: 1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DJ. (1981) Formate oxidation and oxygen reduction by leaf mitochondria. Plant Physiol 68: 703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda SK, Sahoo L, Katsuhara M, Matsumoto H (2013) Overexpression of alternative oxidase gene confers aluminum tolerance by altering the respiratory capacity and the response to oxidative stress in tobacco cells. Mol Biotechnol 54: 551–563 [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P, Delhaize E, Jones D (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52: 527–560 [DOI] [PubMed] [Google Scholar]
- Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 62: 9–20 [DOI] [PubMed] [Google Scholar]
- Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, Koyama H (2009) STOP1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol 150: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavrukov Y, Hirai Y (2016) Good and bad protons: genetic aspects of acidity stress responses in plants. J Exp Bot 67: 15–30 [DOI] [PubMed] [Google Scholar]
- Ślaski JJ, Zhang G, Basu U, Stephens JL, Taylor GJ (1996) Aluminum resistance in wheat (Triticum aestivum) is associated with rapid, Al-induced changes in activities of glucose-6-phosphate dehydrogenase and 6-phoshogluconate dehydrogenase in root apices. Physiol Plant 98: 477–484 [Google Scholar]
- Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa NK, Yoshimura E, Mori S (1998) Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol 116: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127: 1836–1844 [PMC free article] [PubMed] [Google Scholar]
- Uotila L, Koivusalo M (1979) Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys 196: 33–45 [DOI] [PubMed] [Google Scholar]
- Wang H, Liang W, Huang J (2013) Putrescine mediates aluminum tolerance in red kidney bean by modulating aluminum-induced oxidative stress. Crop Sci 53: 2120–2128 [Google Scholar]
- Wang Q-F, Zhao Y, Yi Q, Li K-Z, Yu Y-X, Chen L-M (2010) Overexpression of malate dehydrogenase in transgenic tobacco leaves: enhanced malate synthesis and augmented Al-resistance. Acta Physiol Plant 32: 1209–1220 [Google Scholar]
- Wang ZQ, Xu XY, Gong QQ, Xie C, Fan W, Yang JL, Lin QS, Zheng SJ (2014) Root proteome of rice studied by iTRAQ provides integrated insight into aluminum stress tolerance mechanisms in plants. J Proteomics 98: 189–205 [DOI] [PubMed] [Google Scholar]
- Whipps JM. (1990) Carbon economy. In Lynch JM, ed, The Rhizosphere. John Wiley & Sons, Essex, UK, pp 59–97 [Google Scholar]
- Yang Z-B, Geng X, He C, Zhang F, Wang R, Horst WJ, Ding Z (2014) TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 26: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang S, Eltayeb AE, Uddin MI, Yamamoto Y, Tsuji W, Takeuchi Y, Tanaka K (2010) Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 231: 609–621 [DOI] [PubMed] [Google Scholar]
- Yu Y, Jin C, Sun C, Wang J, Ye Y, Lu L, Lin X (2015) Elevation of arginine decarboxylase-dependent putrescine production enhances aluminum tolerance by decreasing aluminum retention in root cell walls of wheat. J Hazard Mater 299: 280–288 [DOI] [PubMed] [Google Scholar]
- Zhang J, He Z, Tian H, Zhu G, Peng X (2007) Identification of aluminium-responsive genes in rice cultivars with different aluminium sensitivities. J Exp Bot 58: 2269–2278 [DOI] [PubMed] [Google Scholar]
- Zhao CR, Ikka T, Sawaki Y, Kobayashi Y, Suzuki Y, Hibino T, Sato S, Sakurai N, Shibata D, Koyama H (2009) Comparative transcriptomic characterization of aluminum, sodium chloride, cadmium and copper rhizotoxicities in Arabidopsis thaliana. BMC Plant Biol 9: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Qi JL, Wang SS, Su J, Xu GH, Zhang MS, Miao L, Peng XX, Tian D, Yang YH (2007) Comparative proteome analysis of differentially expressed proteins induced by Al toxicity in soybean. Physiol Plant 131: 542–554 [DOI] [PubMed] [Google Scholar]
- Zheng SJ, Yang JL (2005) Target sites of aluminum phytotoxicity. Biol Plant 49: 321–331 [Google Scholar]
- Zhou S, Sauvé R, Thannhauser TW (2009) Proteome changes induced by aluminium stress in tomato roots. J Exp Bot 60: 1849–1857 [DOI] [PubMed] [Google Scholar]