Pyoverdine, a high-affinity ferric iron chelator synthesized by Pseudomonas fluorescens, impacts the growth/defense trade-off of Arabidopsis thaliana under iron deficient conditions.
Abstract
Pyoverdines are siderophores synthesized by fluorescent Pseudomonas spp. Under iron-limiting conditions, these high-affinity ferric iron chelators are excreted by bacteria in the soil to acquire iron. Pyoverdines produced by beneficial Pseudomonas spp. ameliorate plant growth. Here, we investigate the physiological incidence and mode of action of pyoverdine from Pseudomonas fluorescens C7R12 on Arabidopsis (Arabidopsis thaliana) plants grown under iron-sufficient or iron-deficient conditions. Pyoverdine was provided to the medium in its iron-free structure (apo-pyoverdine), thus mimicking a situation in which it is produced by bacteria. Remarkably, apo-pyoverdine abolished the iron-deficiency phenotype and restored the growth of plants maintained in the iron-deprived medium. In contrast to a P. fluorescens C7R12 strain impaired in apo-pyoverdine production, the wild-type C7R12 reduced the accumulation of anthocyanins in plants grown in iron-deficient conditions. Under this condition, apo-pyoverdine modulated the expression of around 2,000 genes. Notably, apo-pyoverdine positively regulated the expression of genes related to development and iron acquisition/redistribution while it repressed the expression of defense-related genes. Accordingly, the growth-promoting effect of apo-pyoverdine in plants grown under iron-deficient conditions was impaired in iron-regulated transporter1 and ferric chelate reductase2 knockout mutants and was prioritized over immunity, as highlighted by an increased susceptibility to Botrytis cinerea. This process was accompanied by an overexpression of the transcription factor HBI1, a key node for the cross talk between growth and immunity. This study reveals an unprecedented mode of action of pyoverdine in Arabidopsis and demonstrates that its incidence on physiological traits depends on the plant iron status.
Iron, the fourth most abundant element in the Earth’s crust, is an essential micronutrient for plant growth and development, notably through its involvement in major metabolic processes such as respiration and photosynthesis (Guerinot and Yi, 1994; Robin et al., 2008). Despite its abundance, iron is weakly bioavailable for organisms due to its poor solubility in soils under aerobic conditions. Therefore, plants and other organisms had to evolve mechanisms to efficiently assimilate iron from the soil. Bacteria, but also other microbes including fungi, produce iron chelators known as siderophores (Ahmed and Holmström, 2014). Siderophores are low-Mr molecules that exhibit a high affinity for ferric iron (Fe3+) and are synthesized in iron-limiting conditions. For instance, fluorescent Pseudomonas spp. produce yellow-green fluorescent siderophores termed pyoverdines (or pseudobactins) having a high affinity for Fe3+, with a stability constant of the ferric-pyoverdine complex around 1032 m−1 (Meyer and Abdallah, 1978; Albrecht-Gary et al., 1994). Once excreted, siderophores chelate ferric iron and are transported back as ferric siderophore complexes inside the bacterial cell, where ferric iron is released from the siderophore and further reduced to ferrous iron (Fe2+; Chu et al., 2010).
Higher plants evolved two main strategies to incorporate iron (Curie and Briat, 2003). Grass plants use a strategy similar to the bacterial one, based on the release of phytosiderophores (strategy II). Nongrass plants, like Arabidopsis (Arabidopsis thaliana), use the strategy I based on soil acidification through proton excretion by H+-ATPases, solubilization of Fe3+, and reduction of the Fe3+ chelate, mainly by the ferric chelate reductase FRO2. Fe2+ ions, which are more soluble than Fe3+, are subsequently taken up by root transporters, including IRON-REGULATED TRANSPORTER1 (IRT1). Once in the plants, iron has to be transported, utilized, or sequestrated because of its high reactivity and toxicity through the Fenton reaction. IRT1 and FRO2 expression is induced under iron-limiting conditions, and the transcription factor FIT1 is required for the proper regulation of the iron-uptake system in Arabidopsis (Colangelo and Guerinot, 2004). FIT1 does not act alone to orchestrate the iron deficiency response. Indeed, several studies highlighted the role of other basic helix-loop-helix (bHLH) transcription factors on the expression of genes involved in the adaptation to iron deficiency stress. These transcription factors were shown to act together with FIT1, such as bHLH38 and bHLH39 (Yuan et al., 2008), or alone, as reported for POPEYE, bHLH100, and bHLH101 (Long et al., 2010; Sivitz et al., 2012). Besides the FRO2/IRT1 system, Fe-efficient plants also respond to iron deficiency by enhancing the root secretion of coumarin compounds, improving plant iron nutrition (Fourcroy et al., 2014).
Because iron is central for many metabolic processes and not easily available, it is the focus of serious competition between organisms. In mammals, there are several examples in which the virulence of infectious bacteria has been correlated to their ability to assimilate host iron through their siderophores (Saha et al., 2013). In turn, the host fights back and develops strategies to avoid iron capture by bacteria, highlighting cross-regulatory interactions between iron homeostasis and immune responses (Ong et al., 2006; Nairz et al., 2014). Similar processes seem to operate in plants. For example, the production of siderophores by the pathogenic enterobacteria Dickeya dadantii, Erwinia amylovora, and Pseudomonas syringae pv tabaci was shown to facilitate infection in their hosts, pointing out their role as virulence factors (for review, see Franza and Expert, 2013). Consecutively, plants establish a strategy for keeping iron that consists in lowering iron availability by overexpressing iron-chelating proteins, notably ferritins, and in improving iron acquisition through IRT1 (Dellagi et al., 2005, 2009). As highlighted in animals, this iron-retention strategy used by plants to combat infections could be a key component of their immune responses. Supporting this concept, treatment of plant tissues or cell suspensions by bacterial siderophores was reported to trigger defense responses in tobacco (Nicotiana tabacum) and Arabidopsis (van Loon et al., 2008; Aznar et al., 2014).
On the other hand, siderophore secretion by bacteria is not always associated with pathogenicity and, in contrast, is even beneficial for plant health. In particular, different soil pseudomonad strains, commonly called plant growth-promoting rhizobacteria, were shown to suppress soil-borne diseases (Haas and Défago, 2005; Weller, 2007). Their biocontrol activities have been correlated to their capacity to release siderophores, which act as competitors reducing the iron availability for plant pathogens, including Fusarium oxysporum and Pythium ultimum (Lemanceau et al., 1992, 1993; Duijff et al., 1999; Bakker et al., 2007; Van Wees et al., 2008; Ahmed and Holmström, 2014). Importantly, siderophores produced by other bacterial species such as Bacillus subtilis also display biocontrol activities (Yu et al., 2011). Besides their ability to compete with soil pathogens for iron, siderophores produced by plant beneficial rhizobacteria also were shown to protect different plant species by eliciting induced systemic resistance (ISR). For example, purified pyoverdines from Pseudomonas putida WCS358 were shown to trigger ISR in Arabidopsis when using P. syringae pv tomato as the challenging pathogen. They were also efficient in inducing ISR against Botrytis cinerea in bean (Phaseolus vulgaris) and tomato (Solanum lycopersicum; Meziane et al., 2005). Accordingly, De Vleesschauwer et al. (2008) demonstrated that pyoverdine was the determinant responsible for Pseudomonas fluorescens WCS374-induced ISR in rice (Oryza sativa) against the leaf blast pathogen Magnaporthe oryzae. Investigations in tobacco cell suspensions indicated that the pyoverdine siderophores from P. putida WCS358 and P. fluorescens WCS417 strains are perceived by cells and mediate defense-related early signaling events (van Loon et al., 2008). However, these defense-related reactions were poorly correlated to the ability of the siderophores of interest to promote ISR. Therefore, how siderophores promote ISR remains poorly understood.
In addition to their protective function, siderophores produced by plant growth-promoting rhizobacteria were shown to enhance plant growth, probably by providing the plants with nutrients. Of particular interest here, Vansuyt et al. (2007) provided evidence that in its iron-chelated structure, pyoverdine from P. fluorescens strain C7R12 is assimilated by roots of Arabidopsis plantlets grown in vitro. This process was accompanied by an improvement of plant growth and chlorophyll content. This study also revealed that iron chelated to pyoverdine was incorporated in a more efficient way than iron chelated to EDTA. The mechanism by which siderophores enhance iron nutrition is little known. It has been proposed that microbial ferric siderophore could be reduced to release ferrous iron once transported in the apoplast of root cells, thus providing substrate for the iron transport system (Ahmed and Holmström, 2014). Also, ferric siderophore could provide iron to plant phytosiderophores (Crowley, 2006). In the case of pyoverdine, Vansuyt et al. (2007) provided another explanation by showing that, in Arabidopsis, pyoverdine is incorporated by plant roots and accumulates in shoots. Interestingly, this incorporation does not rely on the strategy I iron acquisition system. Taken together, these observations underline the importance of pyoverdine in plant-microbe interactions but call for further studies to better understand the mechanisms involved.
In this study, we further deciphered the mode of action of pyoverdine in plants. For this purpose, we investigated the incidence of pyoverdine from P. fluorescens C7R12 on the growth and defense capacities of Arabidopsis plantlets grown in iron-sufficient or iron-deficient medium. Pyoverdine was provided to the culture medium in its iron-free structure (apo-pyo), thus mimicking a situation in which it is produced by the bacterial strain. We observed that while apo-pyo displayed a remarkable growth-promoting effect on plants facing iron deficiency, it clearly impaired their resistance against B. cinerea. Contrary to a P. fluorescens C7R12 bacterial strain impaired in apo-pyo production, the wild-type C7R12 reduced the accumulation of anthocyanins in plants grown under iron-deficient conditions. Analysis of the underlying mechanisms showed that the apo-siderophore triggered a strong up-regulation of genes related to development and iron uptake/redistribution in planta. Accordingly, the growth-promoting effects of apo-pyo were shown to rely on IRT1 and FRO2 expression. Furthermore, we provide evidence that, in plants exposed to iron deficiency, the apo-siderophore modulates the expression of genes controlling the tradeoff between growth and immunity in favor of growth. Collectively, data from our work describe an unexpected mode of action of pyoverdine in Arabidopsis plants grown in iron deficiency.
RESULTS
Impact of Apo-Pyo on Arabidopsis Phenotype
Four-week-old plants were subjected to apo-pyo treatment in iron-deficient or iron-sufficient medium as described in Figure 1. For this purpose, after 4 weeks of hydroponic culture in a classical nutrient solution containing 25 µm Fe-EDTA, plants were subjected to 24 h of pretreatment in iron-sufficient (nutritive solution with 25 µm Fe-EDTA) or iron-deficient (nutritive solution without Fe-EDTA) medium. Then, each resulting batch was separated into two new batches containing fresh nutritive solution with or without 25 µm apo-pyo to obtain four different treatments: 25 µm apo-pyo in iron-sufficient (Fe 25 apo-pyo) or iron-deficient (Fe 0 apo-pyo) medium and the corresponding controls without apo-pyo (Fe 25 or Fe 0, according to the presence or absence or Fe-EDTA, respectively). Importantly, we measured the iron content of the culture media 6 h after apo-pyo addition (see Fig. 6A below). The corresponding data showed that apo-pyo addition did not result in iron enrichment in the medium, thus proving that the siderophore was purified in its iron-free structure and was not contaminated by ferri-pyo (pyoverdine chelated to iron).
Figure 1.
Experimental design. After 4 weeks of hydroponic culture in the nutritive solution containing 25 µm Fe-EDTA, plants were subjected to 24 h of pretreatment in iron-sufficient (nutritive solution with 25 µm Fe-EDTA) or iron-deficient (nutritive solution without Fe-EDTA) medium. Then, plants were treated for 7 d with 25 µm apo-pyo in iron-sufficient (Fe 25 apo-pyo) or iron-deficient (Fe 0 apo-pyo) medium. As controls, plants were cultivated in iron-sufficient (Fe 25) or iron-deficient (Fe 0) medium without apo-pyo.
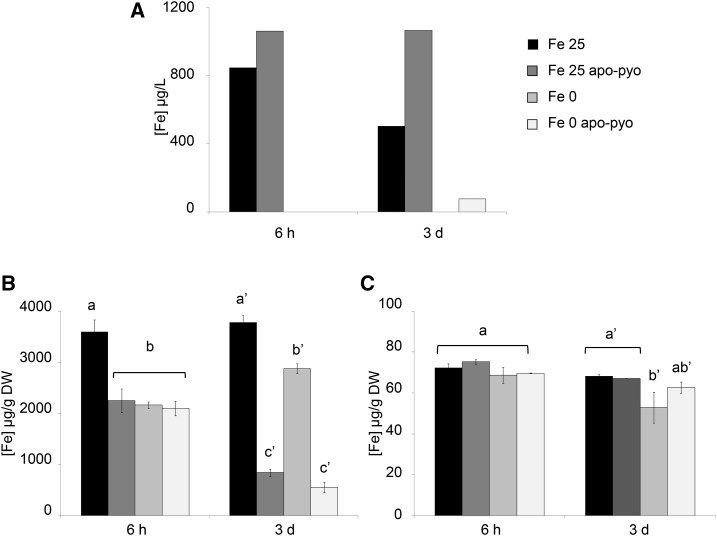
Figure 6.
Iron concentrations measured in the nutritive medium and tissues 6 h and 3 d after apo-pyo treatment in iron-sufficient or iron-deficient medium. A, Iron concentrations measured in the nutritive medium. B, Root iron contents. C, Shoot iron contents. Values are means of three measurements ± se from three independent experiments. Significant differences between the concentrations of iron for each time point were determined by one-way ANOVA/lsd method, with different letters signifying distinct statistical groups (P < 0.001 for B and P < 0.05 for C). DW, Dry weight.
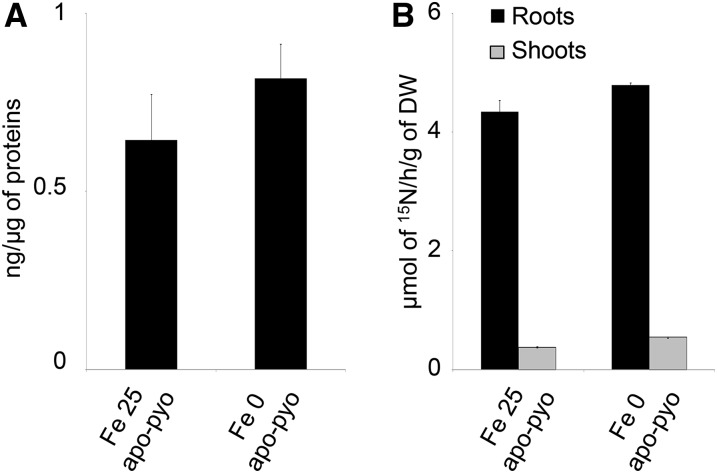
First, we checked that, in our experimental conditions, apo-pyo was efficiently assimilated by plants and potentially transported from roots to shoots. The presence of pyoverdine in root tissue was investigated by ELISA using rabbit polyclonal anti-pyoverdine antibodies. In root extracts of Fe 25 apo-pyo- and Fe 0 apo-pyo-treated plants, we detected 0.64 and 0.82 ng pyoverdine µg−1 protein, respectively (Fig. 2A). In shoots, we were not able to discriminate a positive signal from the background (data not shown). To strengthen the data, the presence of apo-pyo in plant tissues was further analyzed by isotope ratio mass spectrometry. Following the treatment of plants by [15N]apo-pyo, approximately 4.5 µmol 15N h−1 g−1 dry weight was measured in the roots of Fe 25 apo-pyo- and Fe 0 apo-pyo-treated plants, thus confirming the results from the ELISA. In addition, a lower signal (0.37 and 0.54 µmol 15N h−1 g−1 dry weight for Fe 25 apo-pyo and Fe 0 apo-pyo treatments, respectively) also was measured in the shoots (Fig. 2B). These data also showed that the concentration of apo-pyo assimilated by plants did not differ according to the concentration of iron in the culture medium.
Figure 2.
Detection of apo-pyo in Arabidopsis tissues. Plants were cultivated and treated by apo-pyo as indicated in Figure 1. A, Apo-pyo was detected in roots by ELISA using rabbit polyclonal anti-pyoverdine antibodies. Signal was expressed in ng of apo-pyo per µg of total root proteins. Each value represents the mean ± se of three measurements. Experiments were repeated three times with similar results. B, [15N]Apo-pyo was detected using isotope ratio mass spectrometry in roots and shoots. Each value represents the mean ± se of three measurements. Experiments were repeated three times with similar results. DW, Dry weight.
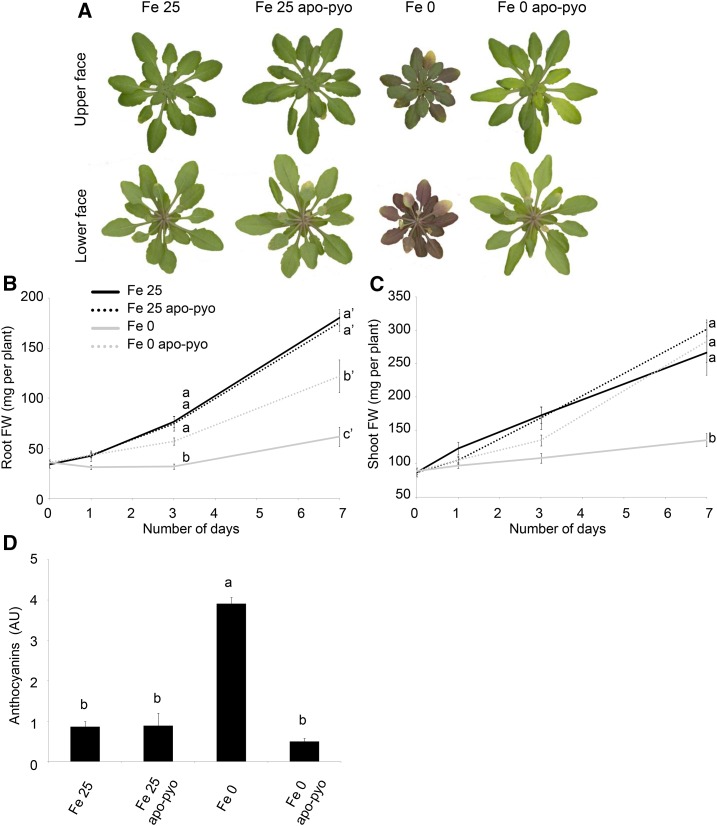
Next, the impact of apo-pyo treatment on the plant macroscopic phenotype was evaluated (Fig. 3). Fe 25- and Fe 25 apo-pyo-treated plants appeared bigger and greener compared with plants facing iron deficiency. As expected, plants facing Fe 0 treatment were severely affected after 7 d of culture: they showed reduced growth, and their leaf abaxial sides displayed a violet pigmentation probably due to anthocyanin production (Fig. 3A). In comparison with Fe 0-treated plants, Fe 0 apo-pyo-treated plants exhibited a distinct phenotype that resembled those of plants grown in iron-containing medium. Indeed, after 7 d of treatment with apo-pyo, their growth was considerably higher. Quantitatively, their root masses were between those of Fe 25/Fe 25 apo-pyo- and Fe 0-treated plants (Fig. 3B), and growth of their aerial parts was similar to that of Fe 25 plants (Fig. 3C). Furthermore, although we noticed a yellowing of several leaves, Fe 0 apo-pyo-treated plants did not show violet pigmentation compared with Fe 0-treated plants (Fig. 3A). Supporting this observation, the high production of anthocyanins measured in Fe 0 plants was not observed in Fe 0 apo-pyo-treated plants, which, in contrast, displayed an anthocyanin level similar to those of Fe 25- and Fe 25 apo-pyo-treated plants (Fig. 3D).
Figure 3.
Phenotypic analysis of Arabidopsis plantlets exposed to apo-pyo in iron-sufficient or iron-deficient conditions. Plants were cultivated and treated by apo-pyo as indicated in Figure 1. A, Rosette macroscopic phenotypes. Phenotypes were observed 7 d after the addition of apo-pyo. Results are representative of nine experiments. B, Root fresh weights. C, Shoot fresh weights. D, Anthocyanin contents. Each value represents the mean ± se of three measurements. Experiments were repeated at least three times with similar results. Significant differences between the treatments were determined by one-way ANOVA/lsd method, with different letters signifying distinct statistical groups (P < 0.01 for B and P < 0.0001 for C and D). AU, Arbitrary units; FW, fresh weight.
Collectively, these data indicate that apo-pyo is assimilated by Arabidopsis plants grown in hydroponic conditions and highlight a growth-promoting effect of the apo-siderophore in plants facing iron deficiency.
Transcriptomic Analysis of Apo-Pyo-Treated Plants
We initiated a transcriptomic analysis in order to provide, to our knowledge, a first and overall view of how apo-pyo impacts the growth of plants. For this purpose, a microarray analysis using CATMA arrays covering the entire transcriptome of Arabidopsis was performed from roots and shoots of plants 3 d after the addition of apo-pyo in the nutritive medium containing (Fe 25 apo-pyo condition) or not (Fe 0 apo-pyo condition) iron. This time point was chosen on the basis of Figure 3, B and C, demonstrating that the increase of growth of the Fe 0 apo-pyo plants was noticeable from 3 d. Changes in root and shoot gene expression were determined by comparing the transcriptomes of apo-pyo-treated plants with their respective controls not treated with the apo-siderophore (i.e. Fe 25 apo-pyo versus Fe 25 and Fe 0 apo-pyo versus Fe 0).
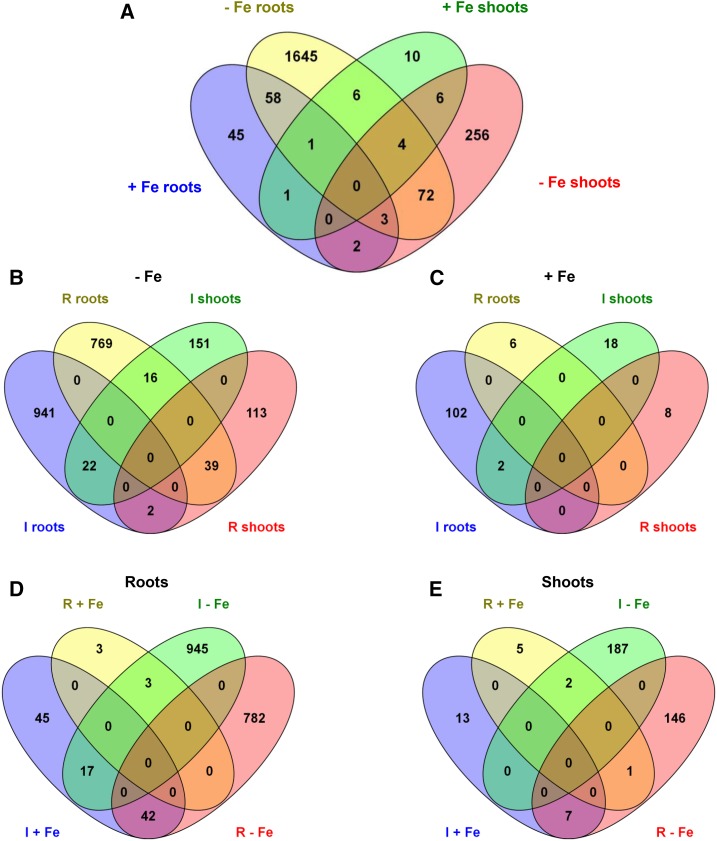
We first selected genes whose expression was modulated by a log2 ratio of +1.5 or greater or −1.5 or less (with adjusted P ≤ 0.05). This analysis revealed that 2,109 genes displayed significant differential expression in response to apo-pyo (Fig. 4A; listed in Supplemental Tables S1–S4). The incidence of apo-pyo on gene expression clearly differed according to the tissue and the presence or not of iron in the culture medium. Indeed, apo-pyo modulated the expression of 2,053 genes in iron-depleted medium (Fig. 4B) versus 136 genes in iron-containing medium (Fig. 4C). Furthermore, 1,837 genes showed a modified expression in the roots (Fig. 4D) versus 361 in the shoots (Fig. 4E). Regarding the percentages of induced/repressed genes, apo-pyo-induced genes represent 55% to 56% of the total apo-pyo-modulated genes when considering the roots (Fig. 4D), the shoots (Fig. 4E), or the Fe 0 condition (Fig. 4B). In contrast, in plants grown in the Fe 25 medium, 90% of the genes were induced in response to the apo-pyo treatment (Fig. 4C).
Figure 4.
Venn diagrams illustrating the transcriptome changes in response to apo-pyo in iron-deficient (−Fe) or iron-sufficient (+Fe) medium in roots or shoots (log2 ratio of +1.5 or greater or −1.5 or less; P ≤ 0.05). A, General view of the number of genes commonly regulated between the four comparisons. B, Focus on the number of genes induced (I) or repressed (R) in the iron-deficient medium. C, Focus on the number of genes induced or repressed in the iron-sufficient medium. D, Focus on the number of genes induced or repressed in the roots. E, Focus on the number of genes induced or repressed in the shoots. Diagrams were generated using http://bioinfogp.cnb.csic.es/tools/venny/index.html.
A closer analysis indicates that 79 of the apo-pyo-dependent genes (genes 1–79 in Supplemental Table S5) could be gathered in a first cluster corresponding to genes modulated under iron deficiency in both roots and shoots. Among these genes, 22 (28%) or 39 (49%) were induced or repressed in both tissues, respectively (Fig. 4B). A second cluster includes 63 genes (genes 2, 62, 75, and 80–138 in Supplemental Table S5) modulated in roots in iron-deficient and iron-sufficient media. Among this category, 45 (71%) were inversely regulated according to the presence or absence of iron in the culture medium. The third cluster groups 10 shoot genes (genes 49, 60, 61, 71, and 139–144 in Supplemental Table S5) that showed differential expression in iron-sufficient and iron-deficient medium, nine of them being inversely modulated in the two conditions. Therefore, a large part of the genes modulated by apo-pyo in roots and shoots of plants cultivated in the iron-deficient condition were similarly regulated. On the contrary, a majority of the apo-pyo-dependent genes modulated in both iron-sufficient and iron-deficient conditions were regulated in the opposite way. Using AmiGO (http://amigo1.geneontology.org/cgi-bin/amigo/term_enrichment), we identified these genes as mainly responsive to stress or stimulus.
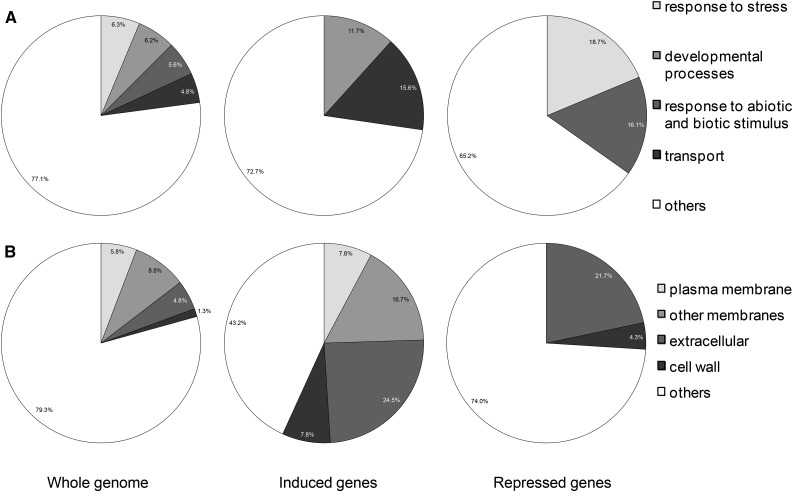
Next, we focused our analysis on the genes whose expression was highly induced (91 and 15 genes in roots and shoots, respectively) or highly repressed (94 and eight genes in roots and shoots, respectively) by apo-pyo in the iron-deficient condition (log2 ratio of +3 or greater or −3 or less; P ≤ 0.05). This choice was made based on the phenotypic analysis showing that apo-pyo deeply improved the growth of plants facing iron deficiency (Fig. 3). Genes were putatively assigned to functional categories based on the Gene Ontology annotations from The Arabidopsis Information Resource database (http://www.arabidopsis.org/tools/bulk/go/index.jsp; Fig. 5). Concerning the genes whose expression was induced by apo-pyo, an overrepresentation of genes related to developmental processes (11.7% compared with 6.2% in the whole genome) and, in particular, to cell differentiation was observed (Fig. 5A). These genes include α-EXPANSIN7 (AT1G12560) and α-EXPANSIN18 (AT1G62980), GLYCOSYL HYDROLASE9C1 (AT1G48930), CELLULOSE SYNTHASE-LIKE5 (AT4G15290), β-GALACTOSIDASE6 (AT5G63800), and CER4 (AT4G33790), which encodes an alcohol-forming fatty acyl-CoA reductase involved in cuticular wax biosynthesis. Moreover, this analysis revealed a predominance of genes involved in transport when comparing with the whole-genome categorization (15.6% compared with 4.8% in the whole genome). Accordingly, genes up-regulated by apo-pyo encode proteins preponderantly located in the plasma membrane, in endomembranes, in the cell wall, and also extracellular proteins (Fig. 5B). These four cellular component categories represent 56.8% of the genes induced by apo-pyo, while they represent only 20.6% of the whole genome. Importantly, this category includes genes related to iron transport, including IRT1 (AT4G19690) and IRT2 (AT4G19680; Supplemental Table S6). Strengthening this evidence, other genes encoding proteins involved in iron homeostasis also were identified as up-regulated by apo-pyo in roots facing iron deficiency (Supplemental Table S6). Among these, FRO2 (AT1G01580) and the transcription factor bHLH39 (also named ORG3; AT3G56980) belonged to the most induced genes. Another transcription factor, bHLH100 (AT2G41240), was highly induced in roots and shoots. Although to a lower extent, NAS4, encoding NICOTIANAMINE SYNTHASE4, also was found to be up-regulated. We recently showed that the corresponding protein, which catalyzes the synthesis of the iron chelator/in planta transporter nicotianamine, plays a key role in the plant response to iron deficiency (Koen et al., 2014). To check the microarray data, we tested the level of accumulation of four transcripts among the most modulated by apo-pyo in iron-deficient medium by reverse transcription-quantitative PCR (RT-qPCR). The results, presented in Supplemental Figure S1, were in agreement with the CATMA transcriptomic data. In particular, we noticed a major induction of bHLH39 and AT2G38240 (encoding a 2-oxoglutarate- and Fe2+-dependent oxygenase superfamily protein) in the shoots and of FRO2, bHLH39, and IRT1 in the roots in response to apo-pyo.
Figure 5.
Overrepresented functional categories of induced and repressed genes relative to the whole genome by annotation for Gene Ontology (GO). A, GO Biological process. B, GO cellular component. Induced or repressed genes refer to genes whose expression was highly modulated (log2 ratio of +3 or greater or −3 or less; P ≤ 0.05) by apo-pyo in iron-deficient medium in roots and shoots.
Regarding the genes whose expression was deeply repressed by apo-pyo in roots and shoots of plants grown in the iron-deficient condition, an overrepresentation of genes involved in the stress response (18.7% compared with 6.3% in the whole genome) and in abiotic or biotic stimulus (16.1% compared with 5.6% in the whole genome; Fig. 5A) was observed. For instance, we identified the lipid transfer protein LTP3 (AT5G59320), which is predicted to encode a pathogenesis-related protein (Supplemental Table S7). As described for up-regulated genes, down-regulated genes encoding extracellular proteins (21.7% versus 4.8% in the whole genome) and proteins located in the cell wall (4.3% versus 1.3% in the whole genome) were well represented compared with the data for the whole genome (Fig. 5B). It also should be specified that AtFER1, encoding FER1 involved in iron sequestration, was found to be strongly down-regulated in roots of iron-deprived plants (Supplemental Tables S6 and S7). These data further support the assumption that apo-pyo impacts the iron homeostasis of plants grown in the iron-deficient condition.
The functional categorization of the root genes whose expression was strongly modulated by apo-pyo treatment (log2 ratio of +3 or greater or −3 or less; P ≤ 0.05) in plants grown in the iron-deficient condition was further assessed using AmiGO and the MapMan program. This analysis clearly confirmed an incidence of apo-pyo in the expression of genes involved in development and biotic stresses (Supplemental Fig. S2A). The genes gathered in the biotic stress category encode proteins that play roles in hormone signaling, in cell wall dynamics, and in signaling processes (Supplemental Fig. S2B). To complete this investigation, we performed a more systemic analysis by including root genes regulated by apo-pyo to a lower extent (log2 ratio of +1.5 or greater or −1.5 or less; P ≤ 0.05) in plants facing iron deficiency. We noticed a repression of genes encoding resistance genes (R genes), transcription factors ERF, WRKY, and MYB, salicylic acid (SA)-related genes (such as AT5G24210, belonging to the lipase class 3 protein family), and abscisic acid (ABA)-related genes (such as the lipid transfer protein LTP3; Supplemental Fig. S2C; Supplemental Table S7). The observation that genes related to the SA and ABA pathways were repressed prompted us to compare our transcriptomic data with those recorded for hormone perturbations in Genevestigator. The online analysis revealed that many of the root genes modulated by apo-pyo in the iron-deficient condition were inversely expressed in Arabidopsis exposed to SA or ABA (Supplemental Fig. S3). These data reinforce the possibility that apo-pyo might repress the SA/ABA pathways. We also compared our transcriptomic signature with microarray analyses searching for genes modulated in response to elicitors and biotic stresses. This comparison highlighted that most of the apo-pyo-modulated root genes of plants facing iron deficiency were regulated in an opposite way than in response to the pathogens Phytophthora parasitica, Sclerotinia sclerotiorum, and P. syringae pv maculicola (Supplemental Fig. S4). These results support the hypothesis that apo-pyo could display biological properties related to plant defense responses.
Collectively, the transcriptomic analysis revealed that apo-pyo treatment of plants facing iron deficiency impacted the expression of numerous genes. Notably, apo-pyo positively regulated the expression of genes related to development and iron acquisition (uptake/transport). On the opposite side, several genes encoding proteins involved in iron sequestration (FER) but also in the plant response to biotic stresses were repressed.
Perturbations of Iron Homeostasis Induced by Apo-Pyo
The impact of apo-pyo on the expression levels of genes related to iron homeostasis led us to investigate its impact on the plant iron content in our different conditions (for the experimental design, see Fig. 1) 6 h and 3 d after the addition of apo-pyo.
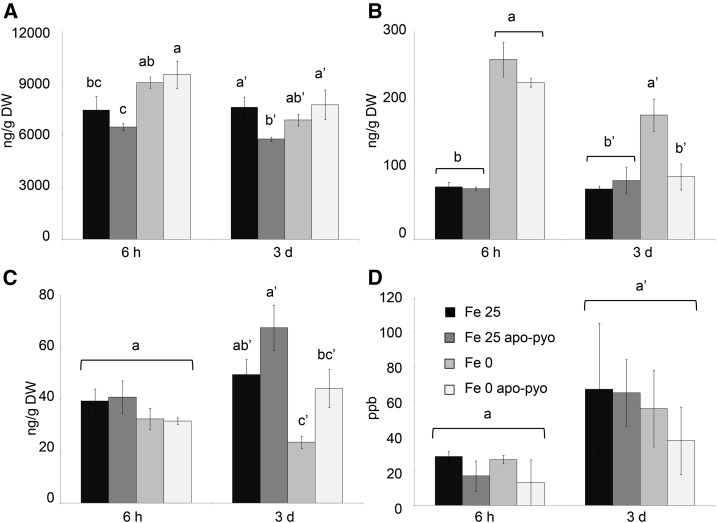
First, we measured the iron content in the nutritive medium. In the Fe 25 medium, a decrease of the iron content occurred between 6 h and 3 d after the treatment, indicating that plants imported part of the iron present in the medium. When apo-pyo was added in the same medium, the concentration of iron did not decrease with time (Fig. 6A). The measurement of iron contents in roots further supported these data (Fig. 6B). Indeed, compared with the Fe 25 treatment, the addition of apo-pyo in the medium containing 25 µm iron (Fe 25 apo-pyo condition) strongly reduced the root iron concentration at 6 h and 3 d (Fig. 6B). In contrast, the iron content in roots of plants grown in the Fe 25 µm condition without apo-pyo remained almost constant over time. As expected, this first set of data indicates that apo-pyo negatively impacts the dynamic of iron import from the medium by plant roots, probably by direct chelation of iron in the medium. Accordingly, the expression of IRT1 and FRO2 was repressed 3 d after the addition of apo-pyo in the iron-containing medium (Supplemental Table S6). The comparison of the Fe 0 and Fe 0 apo-pyo conditions also was relevant. In the absence of apo-pyo, the level of iron at 6 h in the roots of Fe 0-treated plants was reduced compared with Fe 25-treated plants (Fig. 6B). Therefore, it is likely that plants already perceived the lack of iron in the medium. In comparison, the addition of apo-pyo did not further alter the root iron content of plants grown in the iron-deficient condition. In contrast, at day 3, whereas the iron content of the Fe 0 plants was almost similar to that measured at 6 h, Fe 0 apo-pyo-treated plants showed a strong reduction in the iron content. Taken as a whole, these results indicate that the addition of apo-pyo in the culture medium containing or missing iron triggered a reduction of the root iron content.
Completing this analysis, the iron content in shoots also was measured (Fig. 6C). Six hours after the treatments, the iron content was similar in all conditions. After 3 d, while the apo-pyo treatments induced a strong reduction of the root iron content in culture medium containing or not containing iron (Fig. 6B), the same treatments did not impact the iron concentration in shoots (Fig. 6C). In contrast, the iron content in shoots of plants grown in medium lacking iron without apo-pyo (Fe 0 treatment) decreased slightly but significantly.
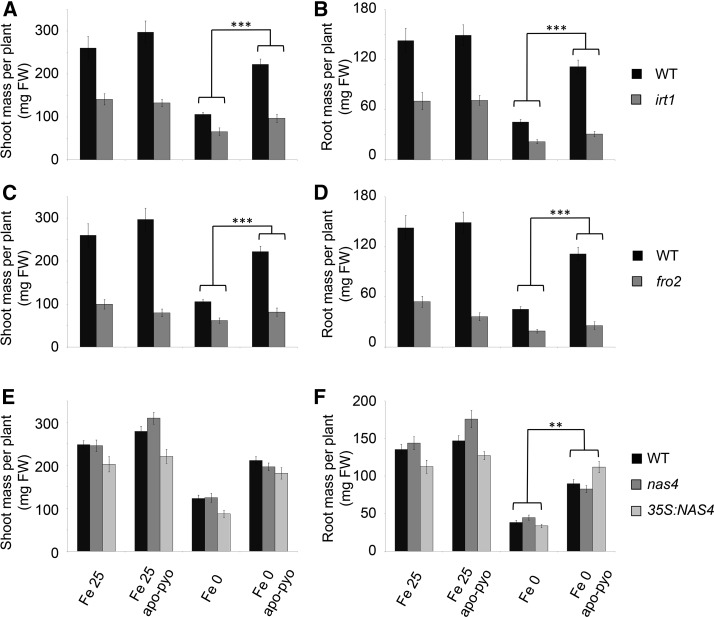
To further investigate the incidence of apo-pyo treatment on iron homeostasis, we performed a functional analysis of six key iron-related genes found to be modulated by apo-pyo: IRT1, FRO2, FER1, NAS4, bHLH100, and bHLH39/ORG3. As discussed above, with the exception of FER1, which was repressed, the expression of these genes was up-regulated in the roots in the Fe 0 apo-pyo condition. Furthermore, FRO2 and IRT1 expression was down-regulated in the Fe 25 apo-pyo condition (see above; Supplemental Table S6). Therefore, we studied whether corresponding mutants could be affected in their response to apo-pyo in iron-deficient or iron-sufficient medium by measuring their shoot and root masses after 7 d of treatment, as described in Figure 3, B and C. The response to apo-pyo was not modified in the bhlh100 and bhlh39/org3 mutants or in the FER1 overexpressor (ov. fer.) regardless of the iron content in the culture medium (Supplemental Fig. S5, A–D). In contrast, compared with the wild type, the irt1 and fro2 knockout mutants showed an impaired response to apo-pyo in iron-deficient medium in shoots (Fig. 7, A and C) and in roots (Fig. 7, B and D). Indeed, the growth-promoting effect triggered by apo-pyo in the wild type was strongly reduced in both mutants in this culture condition, highlighting the involvement of IRT1 and FRO2 in this process. Regarding NAS4, whereas the knockout nas4 mutant did not show a modified response to apo-pyo compared with the wild type (Fig. 7, E and F), the growth-promoting effect of the siderophore was significantly more pronounced in roots of plants overexpressing NAS4 (35S:NAS4) grown in iron deficiency (Fig. 7F). Similar to wild-type plants, apo-pyo treatment did not affect the shoot and root masses of all the mutants and overexpressors tested when grown in iron-sufficient conditions. Collectively, these data highlight an involvement of IRT1, FRO2, and, to a lower extent, NAS4 in the growth effect of apo-pyo in plants exposed to iron deficiency.
Figure 7.
Growth phenotypes of mutants impaired in genes involved in iron homeostasis in response to apo-pyo in iron-sufficient or iron-deficient medium after 7 d of treatment. Plants were treated as described in Figure 1. A, C, and E, Fresh shoot masses measured in wild-type (WT) and irt1 (A), fro2 (C), and nas4 and 35S:NAS4 (E) plants. B, D, and F, Fresh root masses measured in wild-type and irt1 (B), fro2 (D), and nas4 and 35S:NAS4 (F) plants. Each value represents the mean ± se of almost 12 measurements from almost three independent biological experiments. Significant differences between the shoot and root masses of the wild type and the mutants in response to apo-pyo and/or iron deficiency were determined by two-way ANOVA/Tukey’s honestly significant difference method (**, P < 0.01 and ***, P < 0.0001). FW, Fresh weight.
Incidence of Apo-Pyo in SA, ABA, Indole-3-Acetic Acid, and Ethylene Synthesis/Accumulation
The finding that apo-pyo modulated the expression of genes related to SA and ABA signaling and increased the growth of plants in medium deprived of iron prompted us to analyze the concentration of several hormones in our distinct conditions, namely SA, ABA, indole-3-acetic acid (IAA; the major endogenous auxin), and ethylene. Hormone measurements were performed 6 h and 3 d after apo-pyo treatment (for the experimental design, see Fig. 1).
In plants grown in iron-containing medium, at 3 d, we only measured a slight decrease of SA content by apo-pyo (Fe 25 apo-pyo versus Fe 25; Fig. 8A). Concerning ABA, iron deficiency induced a clear increase of its concentration at 6 h and 3 d (Fe 0 condition versus Fe 25 condition; Fig. 8B). At 6 h, the addition of apo-pyo did not influence the ABA content in iron-containing or iron-missing culture medium. However, after 3 d, the higher level of ABA measured in the shoots of plants grown in iron-deficient conditions (Fe 0) was not observed in plants grown in the same medium but supplied with apo-pyo (Fe 0 apo-pyo). In this latter condition, the shoot concentration of ABA was similar to those found in the shoots of Fe 25- and Fe 25 apo-pyo-treated plants. Therefore, apo-pyo appeared to negatively impact ABA synthesis/accumulation in the iron-deficient condition at 3 d, the time point where apo-pyo begins to rescue the growth of plants facing this deficiency (Fig. 3). In the case of IAA, there was no difference after 6 h regardless of the treatment. However, after 3 d, iron deficiency induced a reduction of the content of IAA (Fe 25 versus Fe 0 treatment; Fig. 8C). The presence of apo-pyo in Fe 25 or Fe 0 medium had no significant additive effect on the IAA level. In addition, none of the treatments significantly affected the ethylene content of the plantlets (Fig. 8D).
Figure 8.
Analysis of hormone contents in the shoots (A–C) or in the air (D) 6 h and 3 d after apo-pyo treatment in iron-sufficient or iron-deficient medium. A, SA contents. B, ABA contents. C, IAA contents. D, Ethylene contents. Values are means of at least three measurements ± se from at least three independent experiments. Significant differences between the concentrations of hormones for each time point were determined by one-way ANOVA/lsd method, with different letters signifying distinct statistical groups (P < 0.05 for A and C and P < 0.01 for B). DW, Dry weight; ppb, parts per billion (nL L−1).
Involvement of Apo-Pyo in the Growth/Defense Balance
We and others recently reported that iron deficiency induces an increased resistance of Arabidopsis plants against the fungal pathogen B. cinerea (Kieu et al., 2012; Koen et al., 2014). The findings that apo-pyo negatively regulated the levels of ABA and the expression of defense-related genes of plants in the iron-deficiency condition led us to check whether the siderophore could impact the plant resistance to this pathogen. For this purpose, leaves of plants exposed to our different conditions (for the experimental design, see Fig. 1) were inoculated with B. cinerea at day 3, and the disease symptoms were measured 4 d after the inoculation. As expected and as reported previously, the average of the necrotic lesion areas produced by B. cinerea infection was reduced in plants that suffered from iron deficiency compared with plants grown in the Fe 25 medium (22 versus 50 mm2; Fig. 9). Interestingly, the induced resistance conferred by iron deficiency was partly impaired in response to apo-pyo. Indeed, compared with the condition without apo-pyo, plants grown in iron-deficient medium in the presence of the siderophore were more sensitive to the fungal infection (necrotic lesion areas about 36 mm2). In contrast, no effect of apo-pyo was observed in plants grown in the iron-sufficient condition.
Figure 9.
Evaluation of the sensitivity to B. cinerea of plants treated by apo-pyo in iron-sufficient or iron-deficient medium. Each value represents the mean of the area of the necrotic lesion ± se of 30 measurements. Experiments were repeated four times with similar results. Significant differences between the mean lesion areas were determined by one-way ANOVA followed by one-way ANOVA/lsd method, with different letters signifying distinct statistical groups (P < 0.05).
To further understand how apo-pyo impacts the growth and defense of plants grown under iron deficiency, we analyzed the involvement of four genes highly modulated by the siderophore and shown previously to display important functions in development and/or defense. These genes are LTP3, CER4, AT5G59320 (named PLTP in this article), encoding a protein involved in lipid transport and binding and whose transcripts were shown to be induced in leaves of Arabidopsis plants subjected to a combination of drought and heat stresses (Rizhsky et al., 2004), and AT3G21460 (named GRX in this article), which encodes a glutaredoxin family protein involved in cell redox homeostasis (Finkemeier et al., 2005). The growth-promoting effects of apo-pyo observed in plants facing iron deficiency were not altered in the mutants impaired in the expression of the genes of interest (Supplemental Fig. S5, A, B, E, and F).
We completed this analysis by investigating whether apo-pyo could impact the balance between growth and defense, as the siderophore had a positive effect on growth but a negative effect on the resistance to B. cinerea in plants cultivated in iron deficiency. Recently, it was demonstrated that this tradeoff can be based on cross talk operating between growth- and immune-related signaling pathways (Bai et al., 2012; Fan et al., 2014; Malinovsky et al., 2014). Notably, it has been shown that such cross talk could involve two transcription factors, BZR1 and HBI1, the second being a partner of the tripartite module PRE1-IBH1-HBI1 that regulates cellular elongation upstream from such external or endogenous signals as brassinosteroids and temperature (Fan et al., 2014). IBH1 interacts with and inhibits HBI1, which is a positive regulator of cellular elongation. In particular, HBI1 activates ATEXPA8 and ATEXPA1, two genes encoding expansins (cell wall-loosening enzymes), by direct binding to their promoters. The effects of IBH1 on HBI1 are inhibited by PRE1, which sequestrates IBH1. We first checked the modulation of these different genes in a microarray experiment (Supplemental Table S8). It appeared that HBI1 and PRE1 were highly up-regulated by apo-pyo in the shoots of Fe 0 apo-pyo- versus Fe 0-treated plants, while the inhibitor IBH1 was slightly down-regulated. We also noticed an induction of the expression of ATEXPA1 and ATEXPA8. Moreover, other genes encoding expansins also were activated in response to apo-pyo in roots and/or shoots and could be involved in the siderophore-induced promotion of growth in iron deficiency.
As no HBI1-deficient mutant is available (Malinovsky et al., 2014), we tested the phenotype of HBI1-ox plants, which overexpress HBI1, and HBI1(L214E)-ox plants, which display a dominant-negative effect on HBI1 in our different conditions (for the experimental design, see Fig. 1). In the absence of apo-pyo, HBI1-ox plants had higher growth that wild-type plants in Fe 25 medium (Fig. 10). However, in response to iron deprivation, the shoot (Fig. 10A) and root (Fig. 10B) masses of HBI1-ox plants were similar to those of wild-type plants. Finally, the growth-promoting effects of apo-pyo observed in wild-type plants facing iron deficiency were not statistically different in HBI1-ox and HBI1(L214E)-ox plants.
Figure 10.
Growth phenotypes of HBI1-ox and HBI1(L214E)-ox mutants in response to apo-pyo in iron-sufficient or iron-deficient medium after 7 d of treatment. Plants were treated as described in Figure 1. A, Fresh shoot masses. B, Fresh root masses. Each value represents the mean ± se of almost 12 measurements from almost three independent biological experiments. Significant differences between the shoot and root masses of wild-type (WT) and mutants plants in response to apo-pyo and/or iron deficiency were determined by two-way ANOVA/Tukey’s honestly significant difference method (*, P < 0.05). FW, Fresh weight.
Comparison of Apo-Pyo Effects with Those Induced by P. fluorescens C7R12 Bacterial Inoculation
Four-week-old plants were subjected to apo-pyo treatment as described in Figure 1 or inoculated with the wild-type P. fluorescens strain C7R12 or with the mutant strain PL1 in iron-deficient or iron-sufficient medium. PL1 is a pyoverdine minus C7R12 mutant obtained by random insertion of the transposon Tn5 in the pyoverdine synthetase gene pvsB (Mirleau et al., 2000). PL1 is unable to produce pyoverdine but produces ornicorrugatine, another siderophore.
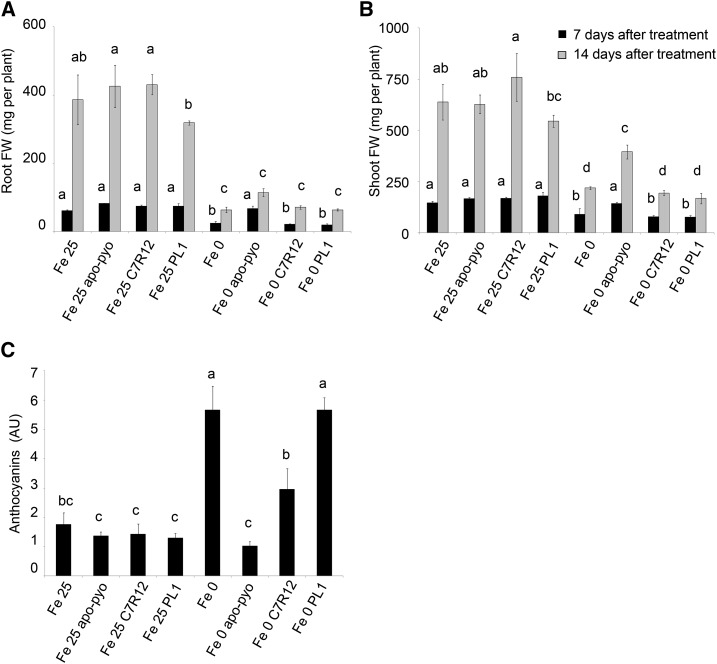
First, we compared the macroscopic phenotypic effects of the purified siderophore with those of bacterial inoculation with the C7R12 or PL1 strain. As described for apo-pyo (Fig. 3), no effect of C7R12 or PL1 was visible in iron-sufficient medium after 7 d (Supplemental Fig. S7A) or 14 d (Supplemental Fig. S7B) of treatment. Indeed, root and shoot weights of the inoculated plants were equivalent to those of noninoculated plants (Fig. 11, A and B). Importantly, in contrast to the treatment with purified apo-pyo, in iron-deficient conditions, the C7R12 strain producing apo-pyo did not partially restore the growth of plants (Fig. 11, A and B; Supplemental Fig. S7). However, compared with plants grown in iron-deficient medium without apo-pyo treatment (Fe 0-treated plants), the C7R12 strain triggered a reduction of the violet pigmentation after 14 d of treatment in the iron-deprived medium (Supplemental Fig. S7B). Accordingly, the production of anthocyanins measured in C7R12-treated plants facing iron deficiency was intermediate between those measured in Fe 0- and Fe 0 apo-pyo-treated plants (Fig. 11C). Highlighting the putative involvement of apo-pyo in this process, the reduction of the anthocyanin level observed in C7R12-treated plants was not found when plants suffering from iron deficiency were inoculated with the PL1 strain. Indeed, PL1-treated plants displayed an anthocyanin concentration similar to that of Fe 0-treated plants (Fig. 11C).
Figure 11.
Phenotypic analysis of Arabidopsis plantlets exposed to apo-pyo or inoculated with C7R12 or PL1 and grown in iron-sufficient or iron-deficient conditions. Plants were cultivated and treated by apo-pyo as indicated in Figure 1. Inoculation with C7R12 or PL1 was performed as described in “Materials and Methods.” A, Root fresh weights. B, Shoot fresh weights. C, Anthocyanin contents. Each value represents the mean ± se of three measurements. Experiments were repeated three times with similar results. Significant differences between the treatments were determined by one-way ANOVA/lsd method, with different letters signifying distinct statistical groups (P < 0.05). AU, Arbitrary units; FW, fresh weight.
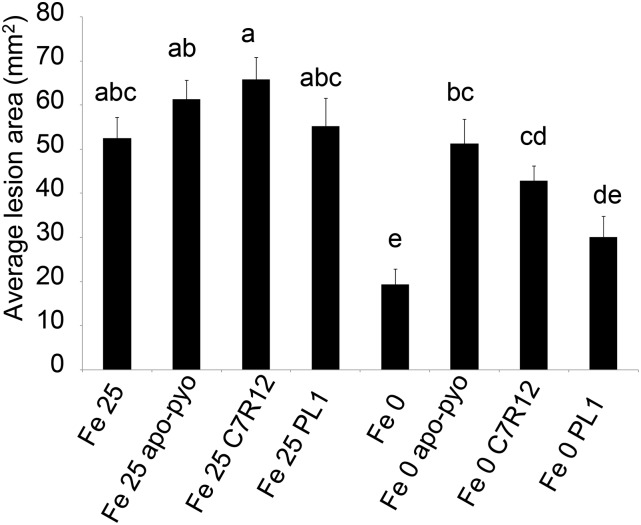
We also compared the sensitivity to B. cinerea of the plants treated with apo-pyo and those inoculated with the C7R12 or PL1 strain. No effect of both strains was observed in the Fe 25 medium, as described previously for apo-pyo (Fig. 12). In the medium lacking iron, C7R12-inoculated plants (Fe 0 C7R12) were more sensitive to the pathogen than Fe 0-treated plants, their higher sensitivity being intermediate between those of Fe 0 apo-pyo-treated plants and Fe 0 PL1-treated plants. These data strongly suggest that the production of apo-pyo by the wild-type strain was responsible for the higher sensitivity of the plants to B. cinerea in iron-deficient medium.
Figure 12.
Sensitivity to B. cinerea of plants treated by apo-pyo or inoculated with C7R12 or PL1 in iron-sufficient or iron-deficient medium. Each value represents the mean of the area of the necrotic lesion ± se of 30 measurements. Experiments were repeated three times with similar results. Significant differences between the mean lesion areas were determined by one-way ANOVA followed by one-way ANOVA/lsd method, with different letters signifying distinct statistical groups (P < 0.05).
DISCUSSION
Most studies dealing with the effects of bacterial siderophores on plants were performed in nonlimiting iron nutrition medium and/or using ferri-siderophores (Vansuyt et al., 2007; Aznar et al., 2014). Here, we analyzed the biological effects of a nonchelated siderophore, apo-pyo, on plants that were maintained in a medium depleted or not of iron. This enabled us to mimic a situation in which the two partners, bacterial strain and host plant, are subjected to iron deficiency and attempt to acquire iron from the rhizosphere. As a matter of fact, siderophores are known to be produced by bacteria only in iron stress conditions (Meyer and Abdallah, 1978), as in the rhizosphere environment in which pyoverdine synthesis was evidenced using a reporter gene (Loper and Henkels, 1997; Duijff et al., 1999).
We first showed that apo-pyo was effectively taken up by roots when plants were cultivated in hydroponic solution. This phenomenon was shown previously to occur in plants cultivated in vitro and treated by ferri-pyo in a medium deprived of iron (Vansuyt et al., 2007). Moreover, using [15N]apo-pyo, we were also able to detect 15N in the shoots. The fact that apo-pyo was not detected by ELISA in the shoots could be explained by the lesser sensitivity of this technique compared with 15N detection by mass spectrometry or, eventually, by a degradation or a chemical modification that would impair its recognition by antibodies, as suggested previously by Vansuyt et al. (2007). In both cases, this result indicates that apo-pyo or a derivative is a systemic compound. Moreover, the quantity of apo-pyo assimilated by plants did not differ whether or not the plants were precultivated 24 h in the presence of iron. This observation indicates that plants may not adapt the rate of apo-pyo assimilation according to the availability of iron in the rhizosphere.
Apo-pyo had a striking macroscopic effect on plants facing iron deficiency. Compared with nontreated plants grown without iron, it induced a remarkable promotion of growth together with a reduction of anthocyanin accumulation. We checked that the iron-free medium was still deprived of iron once apo-pyo was added, thus ruling out the possibility that the growth phenotype observed in the presence of the siderophore was due to iron contamination. A promotion of growth was observed previously by Vansuyt et al. (2007) in iron-deficient medium following the addition of ferri-pyo to the roots of Arabidopsis. This promotion was significantly higher with ferri-pyo than with Fe-EDTA and was associated with a higher increase in the iron content of the plants.
To strengthen these data, we performed similar experiments using the wild-type C7R12 strain and the corresponding PL1 strain impaired in apo-pyo production. Similar to purified apo-pyo treatment, in the iron deficiency condition, the C7R12 strain triggered a reduction of anthocyanin accumulation when compared with Fe 0-treated plants. This phenomenon was due to apo-pyo production, since it was not visible in response to PL1 inoculation. However, the promotion of growth induced by the purified apo-pyo in iron-deficient conditions was not observed in Fe 0 C7R12-inoculated plants. This discrepancy may be explained by different factors. First, it is conceivable that the concentration of the apo-pyo released by the bacteria may be different from that of the purified apo-pyo (25 µm). Second, besides apo-pyo, other compounds produced by the bacteria and recognized by the plant, such as lipopolysaccharides or exopolysaccharides, as well as molecules secreted into plant host cells, also may account for the effects of C7R12, thus making the situation much more complex. Indeed, it has been shown that numerous plant-beneficial pseudomonads such as P. fluorescens C7R12 harbor type 3 secretion system (T3SS)-like genes related to the HYPERSENSITIVE RESPONSE AND PATHOGENICITY1 T3SS family (Mazurier et al., 2004). This T3SS contributes to the plant-bacteria interactions and to the bacterial effects in plants. The involvement and role of this system in the Arabidopsis-C7R12 bacteria interaction remain to be determined.
To investigate the mechanisms underlying apo-pyo effects, we choose to perform a large-scale transcriptomic analysis. It appeared that apo-pyo modulated the expression of around 2,100 genes (log2 ratio of +1.5 or greater or −1.5 or less; P ≤ 0.05). The number of genes modulated by apo-pyo and their induction or repression status clearly depended on the tissue and/or the presence or not of iron in the hydroponic solution. First, root genes rather than shoot genes were preferentially modulated. This result was expected, as apo-pyo was applied in the nutrient solution, directly in contact with the roots, and once assimilated by plants accumulated preferentially in roots rather than in shoots. Second, a great part of the apo-pyo-dependent genes was modulated in iron-deficient rather than in iron-sufficient conditions. Therefore, the effect of apo-pyo on gene expression appeared to be dependent on the plant iron status. Furthermore, the higher number of apo-pyo-modulated genes seen in plants grown in the iron-deficient medium corroborates the strong phenotypic effect triggered by the siderophore in this condition.
A focus on the apo-pyo-regulated genes in plants grown in iron-free medium showed that the most induced genes encoded proteins involved in development and iron acquisition processes, while the most repressed genes were related to iron sequestration and defense responses. Interestingly, Aznar et al. (2014) recently reported data from a transcriptomic analysis performed on Arabidopsis plantlets cultivated in iron-sufficient conditions in which the apo-form of the siderophore deferrioxamine (DFO; 1 mm) was infiltrated in leaves. The major categories of genes modulated by DFO in leaves were related to biotic and abiotic stresses, including the immune response, whereas in roots, the DFO target genes were associated with heavy metal homeostasis. Importantly, no effect on gene modulation was observed when Fe-DFO complexes were used instead of iron-free DFO, and the physiological responses induced by DFO were similarly observed in response to a synthetic iron chelator. Therefore, the effects of DFO were due, or partly due, to its iron chelation ability. Inevitably, these data raise the question of the incidence of apo-pyo on plant iron homeostasis and whether its effects also could be related to its chelating properties. The transcriptomic analysis as well as the quantification of the iron content in apo-pyo-treated plants provided first answers. In plants grown in iron-containing medium, the addition of apo-pyo did not affect the shoot iron content but triggered a strong reduction of the root iron concentration accompanied by a repression of IRT1 and FRO2 expression. Furthermore, many of the genes regulated by apo-pyo in roots also were found to be modulated by iron deficiency in roots of Arabidopsis in the study of Schuler et al. (2011; listed in Supplemental Table S9), and, using Genevestigator, we identified a profile close to those of two iron deficiency treatments (Supplemental Fig. S6). Therefore, apo-pyo triggered an iron deficiency-like response in plants grown in the iron-sufficient condition. As expected, we provided evidence that this process was due mainly to the chelation by apo-pyo of the iron present in the culture medium. Intriguingly, the resulting reduction of iron availability did not impact the growth of the plants. This observation suggests that, although the apo-pyo treatment impacted the root iron content, the iron concentration of plants grown in iron-containing medium was probably sufficient to ensure normal growth. This assumption does not exclude the possibility that part of the siderophore imported by the plants could be internalized in its iron-chelated structure and contribute to iron assimilation. If this is the case, this mechanism poorly accounts for the plant iron nutrition.
In plants facing iron deficiency, apo-pyo also triggered a strong reduction of the root iron content compared with nontreated plants grown in iron-free medium. However, the reduction of shoot iron concentration observed in the latter was less pronounced in the apo-pyo-treated plants. It is striking that, in plants grown in the iron-deficient condition, apo-pyo promoted an important induction of genes related to iron assimilation, such as IRT1, FRO2, and ORG3, as well as a down-regulation of AtFER1. These data deeply contrast with those obtained in apo-pyo-treated plants grown in iron-sufficient conditions and highlight that, in plants facing iron deficiency, apo-pyo strongly promotes the iron-uptake machinery. Furthermore, in addition to the iron-uptake process, apo-pyo also induced a strong expression of genes encoding proteins related to in planta iron redistribution, such as NAS4 and OPT3 (Supplemental Table S6; Koen et al., 2014; Zhai et al., 2014). As plants were grown in iron-free conditions, it is reasonable to assume that apo-pyo provided in the culture could be imported by plants in its iron-free structure and, once internalized in roots, could chelate cellular iron. Accordingly, due to their high affinity for iron, bacterial siderophores including pyoverdine are able to efficiently compete with host iron-binding proteins for iron acquisition (Saha et al., 2013). This process could account for the ability of apo-pyo to induce the overexpression of the plant iron-import system. Whatever the scenario, we provide evidence that the induction of the iron-import machinery contributes to the growth-promoting effects of apo-pyo in plants facing iron deficiency. Indeed, the positive effect of apo-pyo on growth was severely impaired in mutants deficient in IRT1 and FRO2 expression. Furthermore, the positive effect of apo-pyo on growth was further induced in 35S:NAS4 plants overproducing NA, a nonproteinogenic amino acid that contributes to iron long-distance circulation in phloem and transfer within the cells and that is required for the proper establishment of the plant response to iron deficiency (Koen et al., 2014).
The analysis of the content of ABA in plants exposed to apo-pyo also supports the findings that apo-pyo had a major impact on iron homeostasis. First, the hormone accumulated in the shoots of plants suffering from iron deficiency in the presence or not of apo-pyo within 6 h. Second, after 3 d of apo-pyo treatment, plants grown in iron-free medium showed a drastic reduction of shoot ABA level compared with nontreated plants cultivated in the same medium. Therefore, the production of ABA appeared to be more transient in plants exposed to apo-pyo. Accordingly, the transcriptomic analysis revealed that, in the iron-free condition, apo-pyo repressed the expression of root genes related to ABA signaling. Our data should be discussed in light of the investigation of Lei et al. (2014). Those authors reported that iron deficiency induced a rapid ABA accumulation (within 6 h) in the roots of Arabidopsis. In that study, ABA was shown to help alleviate iron deficiency by promoting root iron reutilization and transport from the roots to the shoots. This process involved the ABA-dependent regulation of genes encoding proteins related to the reutilization of the iron stored in the vacuoles, such as AtNRAMP3, and to long-distance transport of iron, including AtFRD3, AtYSL2, and AtNAS1. In our study, the observations that, in iron deficiency, the apo-pyo-treated plants displayed an induced expression of root genes related to iron in planta transport/redistribution suggest that the mechanism described by Lei et al. (2014) could operate efficiently and, therefore, more transiently in response to the siderophore. However, none of the ABA-dependent genes described by those authors were found to be modulated by apo-pyo after 3 d of treatment, thus minimizing this possibility.
Recent investigations have reported that the plant iron status conditions its resistance against microorganisms. Notably, it has been found that iron deficiency activates defense-related processes and confers resistance against microbial pathogens, including D. dadantii and B. cinerea (Kieu et al., 2012; Koen et al., 2014). Several studies also demonstrated that siderophores produced by microorganisms trigger an immune response that could be linked to their capacity to chelate iron from the host (Aznar and Dellagi, 2015; Aznar et al., 2015). In the same vein, we recently demonstrated that β-aminobutyric acid, a priming agent that confers enhanced resistance to numerous pathogens, is a powerful iron chelator triggering a transient iron deficiency in plants. This process could bring the plant to a defense-ready state, contributing to an efficient resistance (Koen et al., 2014). Further strengthening the concept that plant immunity is tightly linked to iron homeostasis, several genes, including those encoding the transcription factor MYB72 (AT1G56160) and the β-glucosidase BGLU42 (AT5G36890), were shown to function in the cross talk of root signaling pathways regulating rhizobacteria-mediated ISR and the plant response to iron deficiency (Zamioudis et al., 2014, 2015). In our investigation, in accordance with the studies listed above, we confirmed that plants cultivated in iron deficiency displayed a higher resistance to B. cinerea. This higher resistance was reduced by the apo-pyo treatment but also by a C7R12 bacterial inoculation. In response to apo-pyo, this negative effect was correlated to a general down-regulation of genes encoding proteins involved in immunity with few exceptions. In particular, the expression of BGLU42 required for rhizobia-induced ISR was slightly induced in apo-pyo-treated plants (Supplemental Table S6). Several works demonstrated that pyoverdines act as determinants in ISR against numerous pathogens. The induction of ISR is not systematic and depends on the specific pathosystem and the type of pyoverdine. For instance, some pyoverdines, such as those of C7R12, induce resistance against the rice blast, but some do not. De Vleesschauwer and Höfte (2009) suggested that these differences can be explained by the induction of iron deficiency by certain pyoverdines. In our case, despite the induction of an iron deficiency-like response in the Fe 25 medium, apo-pyo did not induce ISR in Arabidopsis against B. cinerea. Other factors are probably involved in the induction of ISR.
We observed that pyoverdine did not favor the resistance against B. cinerea but rather partially impaired it in plants grown in the iron-deficient condition. The interpretation of this process should take into account the drastic growth-promoting activity of apo-pyo. Indeed, the antagonistic effect of apo-pyo on growth and resistance observed in plants facing iron deficiency strongly suggests that the siderophore impacts the tradeoff between growth and immunity. As recently highlighted in several studies, cross talk between growth and immune signaling operates in plants. According to the physiological contexts, notably pathogen challenge or light conditions, such cross talk prioritizes one process over the other (for review, see Lozano-Durán and Zipfel, 2015). Clearly, our data indicate that, in plants grown in iron-deficient conditions and in response to apo-pyo, growth is prioritized over immunity. Further supporting this assumption, the microarray analysis revealed that apo-pyo modulates the expression of the PRE1-IBH1-HBI1 module in shoots of plants facing iron deficiency. As discussed in “Results,” HBI1 is a key node for the cross talk between growth and immunity and, once activated, negatively regulates defense responses while prioritizing growth (Fan et al., 2014; Malinovsky et al., 2014). HBI1 effects are antagonized by IBH1. In turn, through its binding to HBI1, PRE1 prevents its inhibitory effect (Bai et al., 2012). Supporting a role of HBI1 in the mediation of the tradeoff between growth and immunity, its overexpression was shown to trigger the repression of the expression of genes involved in immunity and to enhance the susceptibility of the plants to the bacterium P. syringae (Fan et al., 2014; Malinovsky et al., 2014). We found that both HBI1 and PRE1 expression in shoots were strongly up-regulated in response to apo-pyo, whereas the expression of IBH1 was slightly repressed. Therefore, these expression profiles reflect a physiological situation in which the balance between growth and immunity is weighted in favor of growth. To strengthen this possibility, we checked whether the growth-promoting effect of apo-pyo in plants grown in the iron-deficient condition was enhanced in the HBI1-overexpressing line HBI1-ox. We did not observe such an effect. This finding could be explained by the fact that the level of HBI1 expression induced by apo-pyo in wild-type plants was particularly high.
CONCLUSION
To conclude, this investigation provides evidence that the effects of pyoverdine depend on iron availability and, consequently, on the plant iron status. Furthermore, it highlights that, in plants facing iron deficiency, the apo-siderophore modulates the tradeoff between growth and immunity in favor of growth, this mechanism being dependent on the expression of the iron uptake-related genes IRT1 and FRO2. More generally, this work indicates that microorganism-derived siderophores impact the cross-regulatory interactions operating on host plants between iron homeostasis, immune function, and growth.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 ecotype was used as the wild-type control for phenotype comparisons for all mutants and transgenic lines described in this study. The transfer DNA mutants bhlh100 (SALK_074568C), bhlh39 (SALK_025676C), cer4 (SALK_000575C), grx (SALK_001139C), ltp3 (SALK_095248C), and pltp (SALK_063856) were obtained from the Nottingham Arabidopsis Stock Centre. Homozygosity for the mutant alleles was checked using specific primers (Supplemental Table S10). The following lines were described previously: irt1 (Vert et al., 2002), fro2 (also named frd1-1; Robinson et al., 1999), ov. fer. (Vansuyt et al., 2007), HBI1-ox and HBI1(L214E)-ox (Malinovsky et al., 2014), and 35S:NAS4 and nas4 (Koen et al., 2013).
Seeds were surface sterilized by immersion in a solution containing 1.2% (v/v) bleach and 50% (v/v) ethanol for 3 min, rinsed three times with 100% ethanol, air dried, and placed for 3 d at 4°C in the dark in 0.15% (w/v) agar. Then, they were sown in seed holders filled with 0.75% (w/v) agar and put in a cover placed above the nutrient solution container (Araponics). The nutritive medium named Fe 25 nutrient solution [0.25 mm Ca(NO3)2, 0.5 mm KNO3, 1 mm KH2PO4, 1 mm MgSO4, 50 µm H3BO3, 19 µm MnCl2, 10 µm ZnCl2, 1 µm CuSO4, 0.02 µm Na2MoO4, and 25 µm Fe-Na-EDTA] was changed twice per week. Plants were allowed to grow in a climate-controlled growth chamber for 4 or 5 weeks before the treatments in the following conditions: 10-h day (200 µE m−2 s−1 light intensity, 20°C)/14-h night (18°C) with 70% relative humidity.
Chemicals
All chemicals were purchased from Sigma-Aldrich unless stated otherwise.
Production and Purification of Apo-Pyo and [15N]Apo-Pyo
Pseudomonas fluorescens C7R12 was isolated from a soil naturally suppressive to soil-borne disease and is an efficient biocontrol agent against pathogenic Fusarium oxysporum (Lemanceau and Alabouvette, 1991). P. fluorescens was grown on a King’s B (KB; King et al., 1954) agar plate for 2 d at 25°C. Several colonies were sown in succinate medium and incubated with shaking (180 rpm) for 1 d at 25°C (Meyer and Abdallah, 1978). A few milliliters of this culture was used to inoculate the main culture (650 mL in a 3-L Erlenmeyer flask) that was incubated for 4 d in the same conditions as described above. After incubation, bacterial cells were removed by two centrifugations at 4,800g for 30 min (4°C). The pool of resulting supernatants was adjusted at pH 6 with 6 n HCl and then mixed with an Amberlite XAD-4 resin (Carson et al., 2000), 300 mL of resin being mixed under agitation with 3.9 mL of the supernatant for 4 h. After loading, the resin was washed with ultrapure water, and apo-pyo was eluted from the column overnight with 100% methanol at 4°C. The eluate was concentrated with a rotary evaporator in order to obtain 2 to 3 mL of solution. The obtained solution was adsorbed on a Sep-Pak C18 Vac 6-cc cartridge containing 500 mg of octadecylsilane (Waters; Nagata et al., 2013). After loading, the column was washed with 0.1 m EDTA followed by pH 4 acidified water (1% formic acid), and apo-pyo was eluted with aqueous 80% methanol, concentrated with a rotary evaporator, and lyophilized prior to storage of the powder at −20°C. The same protocol was used to produce [15N]pyoverdine with a supply of 98% (15NH4)2SO4 to the succinate medium.
Apo-Pyo Treatments
After 4 weeks of culture (or 5 weeks for infection experiments), plant roots were rinsed using Fe 0 nutrient solution (hydroponic solution described above without Fe-Na-EDTA), and plants were separated in four batches corresponding to four treatments (Fig. 1): Fe 25, pretreatment for 1 d in Fe 25 nutrient solution and treatment for 7 d in Fe 25 nutrient solution; Fe 25 apo-pyo, pretreatment for 1 d in Fe 25 nutrient solution and treatment for 7 d in Fe 25 nutrient solution supplemented with 25 µm purified apo-pyo; Fe 0, pretreatment for 1 d in Fe 0 nutrient solution and treatment for 7 d in Fe 0 nutrient solution; and Fe 0 apo-pyo, pretreatment for 1 d in Fe 0 nutrient solution and treatment for 7 d in Fe 0 nutrient solution supplemented with 25 µm purified apo-pyo.
Bacterial Inoculations
Wild-type C7R12 and Tn5 mutant PL1 (Mirleau et al., 2000) were grown on agar slant tubes containing KB medium for 72 h at 25°C. Bacterial inoculants were produced on KB agar plates and incubated at 25°C for 48 h. Bacteria were scraped from the medium and suspended in sterile distilled water. The bacterial density of the suspensions was determined using a calibration curve assessed by turbidity (λ = 600 nm). A 1.4-mL inoculum of C7R12 or PL1 was introduced in the plant nutrient medium to obtain a bacterial density of 106 colony-forming units per mL of solution.
ELISA Procedure for the Detection of Pyoverdine in Roots
Primary antibody production was described by Vansuyt et al. (2007).
Roots of three plants per condition of treatment were washed 10 min with mQ water; two times 10 min with the following solution: 5 mm EDTA, 1 mm KCl, 5 mm Na2S2O4, and 0.5 mm CaSO4, pH 6; and finally, 10 min with mQ water. Roots were frozen and ground with a pestle and mortar in liquid nitrogen. Proteins were extracted with 15 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 100 mm NaCl, 10% glycerol, 0.5 mm EDTA, 0.5 mm EGTA, 1 mm dithiothreitol, 10 mm phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail (UltraCruz Protease Inhibitor Cocktail Tablet; Santa Cruz Biotechnology) following the manufacturer’s instructions. After centrifugation of the samples (14,000g, 15 min, and 4°C), the protein concentration of the supernatants was estimated according to the procedure of Bradford (1976). Root protein extracts were diluted to obtain 10 µg of proteins in 100 μL of Tris-buffered saline (TBS) and then loaded in each well of a microtitration plate (NUNC Maxisorp). These extracts were coated directly overnight at 4°C. After three washes of 5 min with 200 µL of TBS-Tween 20 (0.1%, v/v), each well was saturated with 100 µL of TBS-Tween 20 with 5% (v/v) milk for 1 h. Anti-pyoverdine antibodies were diluted 100-fold in TBS-Tween 20 with 5% (v/v) milk, and 100 μL per well was added in each well for 2 h. After another three washes of 5 min with 200 µL of TBS-Tween 20 (0.1%, v/v), anti-rabbit secondary antibodies coupled to alkaline phosphatase were diluted 30,000-fold in 50 mm Na2CO3, pH 9.6, and 100 μL per well was added for 1 h. Three washes of 5 min with 200 µL of TBS were performed, and 100 µL of 1 mg mL−1 p-nitrophenyl phosphate dissolved in 10% diethanolamine HCl, pH 9.8, and 0.5 mm MgCl2 was added in each well. Absorbance was measured at 405 nm for 18 h in a microplate reader (Thermomax; Molecular Devices).
Detection of [15N]Apo-Pyo
After the treatments, roots or shoots of four plants were pooled for each condition. Roots were washed as described for the ELISA procedure. Root or shoot tissues were dried at 60°C for 2 d and ground with a pestle and mortar. 15N was detected and quantified in the tissues by the stable isotope analytical platform at the Biochimie et Physiologie Moléculaire des Plantes.
RNA Preparation for Transcriptomic Analysis
For three independent experiments, root and shoot tissues were collected separately after the treatments, immediately frozen in liquid nitrogen, and ground with a TissueRuptor (Qiagen) after the addition of 175 µL of RNA lysis buffer (Promega) per 30 mg of tissues. Total RNA extraction was carried out using the SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. RNAs were solubilized in 100 µL of RNase-free water. Their concentration and purity were determined using a NanoDrop 2000 (Thermo Scientific). For each experiment, RNA samples from four plants per condition were pooled in order to obtain 20 µL of RNAs at 220 ng µL−1. RNA integrity numbers were determined with the 2100 Bioanalyzer (Agilent Technologies) and considered sufficient for microarray if they were between 8.5 and 9.8 for the shoot RNAs and between 7 and 7.8 for the root RNAs.
Microarray Experiment
Microarray analysis was carried out at the Institute of Plant Sciences Paris-Saclay using the CATMA version 6.2 array based on Roche-NimbleGen technology. A single high-density CATMA version 6.2 microarray slide contains 12 chambers, each containing 219,684 primers representing all the Arabidopsis genes: 30,834 probes corresponding to CDS The Arabidopsis Information Resource version 8 annotation (including 476 probes of mitochondrial and chloroplast genes) + 1,289 probes corresponding to EUGENE software predictions. Moreover, it included 5,352 probes corresponding to repeat elements, 658 probes for microRNA/MIR, 342 probes for other RNAs (ribosomal RNA, tRNA, snRNA, and soRNA), and finally, 36 controls. Each long primer is in triplicate in each chamber for robust analysis and in both strands. For each comparison, one technical replicate with fluorochrome reversal was performed for each biological replicate (i.e. four hybridizations per comparison). The labeling of complementary RNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products) was performed as described by Lurin et al. (2004). The hybridization and washing were performed according to NimbleGen Arrays User’s Guide version 5.1 instructions. Two-micrometer scanning was performed with an InnoScan900 scanner (InnopsysR), and raw data were extracted using MapixR software (InnopsysR).
Statistical Analysis of Microarray Data
For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). For each array, a global intensity-dependent normalization using the LOESS procedure (Yang et al., 2002) was performed to correct the dye bias. The differential analysis is based on the log ratios averaging over the duplicate probes and over the technical replicates. Hence, the numbers of available data for each gene equal the numbers of biological replicates and are used to calculate the moderated Student’s t test (Smyth, 2004). Under the null hypothesis, no evidence that the specific variances vary between probes is highlighted by Limma; consequently, the moderated t statistic is assumed to follow a standard normal distribution.
To control the false discovery rate, adjusted P values found using the optimized false discovery rate approach of Storey and Tibshirani (2003) were calculated. We considered as being differentially expressed the probes with an adjusted P ≤ 0.05. Analysis was done with R software. The function SqueezeVar of the library limma was used to smooth the specific variances by computing empirical Bayes posterior means. The library kerfdr was used to calculate the adjusted P values.
Microarray Data Deposition
Microarray data from this article were deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE71163) and at CATdb (http://urgv.evry.inra.fr/CATdb/; project no. INRA13-03_pyo) according to Minimum Information About a Microarray Experiment standards.
RT-qPCR Analyses
RNA samples were reverse transcribed using the ImpromII Reverse Transcriptase kit (Promega) with a mix of anchored oligo(dT)17 and random hexamer primers according to the manufacturer’s specifications. The resulting complementary DNAs (cDNAs) were subjected to a 20-fold dilution with water, and 5 µL of each cDNA sample was assayed by RT-qPCR in a LightCycler (ViiA 7 Real-Time PCR System; Thermo Fischer) using GoTaq qPCR Master Mix (Promega) according to the manufacturer’s instructions. Expression levels were calculated relative to the housekeeping genes AT5G08290 (encoding the mitosis protein YLS8) and AT4G26410 (encoding an uncharacterized conserved protein) using the relative standard curve method. These two genes were shown to be among the five most reliable reference genes for normalization purposes in Arabidopsis Columbia-0 (Wang et al., 2014). For each sample, the target quantity of the gene of interest was determined by interpolating the value from the standard curve made from a cDNA pool, which enables one to take into consideration the efficiency of amplification. The value was then divided by the target quantity of the housekeeping gene. For a list of primers used for RT-qPCR, see Supplemental Table S11.
Determination of Iron Concentrations
After the treatments, roots or shoots of four plants were pooled for each condition. Roots were washed 10 min with mQ water; two times 10 min with the following solution: 5 mm EDTA, 1 mm KCl, 5 mm Na2S2O4, and 0.5 mm CaSO4, pH 6; and finally, 10 min with mQ water. Root or shoot tissues were dried at 60°C for 2 d. Mineralization of the samples was performed in Pyrex tubes previously washed 3 h with 0.1 n HCl and 1 h with 5 mm EDTA and rinsed three times with mQ water. Once dried, the samples were ground and solubilized in 300 µL of ultra-pure nitric acid (67% Normaton for trace analysis; VWR BDH Prolabo). After heating of the samples (1 h at 80°C and 1 h at 100°C), 100 µL of ultra-pure nitric acid and 200 µL of hydrogen peroxide were added in the tubes to obtain a 10-mL solution in mQ water. The concentrations of iron were determined by inductively coupled plasma-atomic emission spectroscopy (Vista Pro; Varian) by Welience (Pôle de Chimie Moléculaire, Université De Bourgogne).
Measurements of Anthocyanins
Shoots from three plants per treatment were harvested and immediately frozen in liquid nitrogen. After grinding, anthocyanin content was determined using the protocol described by Teng et al. (2005).
ABA, SA, JA, and IAA Content Analysis
Shoots from five plants per treatment were harvested, dried, and ground. For each sample, 10 mg of freeze-dried powder was extracted with 0.8 mL of acetone:water:acetic acid (80:19:1, v/v/v). ABA, SA, JA, and IAA stable labeled isotopes used as internal standards were prepared as described by Le Roux et al. (2014). Two nanograms of each standard was added to the sample. The extract was shaken vigorously for 1 min, sonicated for 1 min at 25 Hz, shaken for 10 min at 4°C in a Thermomixer (Eppendorf), and then centrifuged (8,000g, 4°C, and 10 min). The supernatants were collected, and the pellets were reextracted twice with 0.4 mL of the same extraction solution, then vigorously shaken (1 min) and sonicated (1 min at 25 Hz). After the centrifugations, the three supernatants were pooled and dried (final volume of 1.6 mL).
Each dry extract was dissolved in 140 µL of acetonitrile:water (50:50, v/v), filtered, and analyzed using a Waters Acquity ultra-performance liquid chromatograph coupled to a Waters Xevo triple quadrupole mass spectrometer TQS. The compounds were separated on a reverse-phase column (Uptisphere C18 UP3HDO; 100 × 2.1-mm × 3-µm particle size; Interchim) using a flow rate of 0.4 mL min−1 and a binary gradient: 0.1% (v/v) acetic acid in water (A) and acetonitrile with 0.1% acetic acid (B). The following binary gradient was used (time, % A): 0 min, 98%; 3 min, 70%; 7.5 min, 50%; 8.5 min, 5%; 9.6 min, 0%; 13.2 min, 98%; and 15.7 min, 98%. Mass spectrometry was conducted in electrospray and multiple reaction monitoring scanning modes, in positive mode for IAA, and in negative ion mode the other hormones. Relevant instrumental parameters were set as follows: capillary, 1.5 kV (negative mode); source block and desolvation gas temperatures, 130°C and 500°C, respectively. Nitrogen was used to assist the cone and desolvation (150 and 800 L h−1, respectively), and argon was used as the collision gas at a flow of 0.18 mL min−1. The parameters used for multiple reaction monitoring quantification of the different hormones are described by Leroux et al. (2014) for the other hormones.
Samples were reconstituted in 140 µL of 50:50 (v/v) acetonitrile:water per 1 mL of injected volume. The limit of detection and the limit of quantification were extrapolated for each hormone from calibration curves and samples using the Quantify module of MassLynx software, version 4.1.
Ethylene Content Analysis
Plants were placed in 30-mL sealed vials containing 10 mL of growth medium with the different treatments as described in Figure 1. Each plantlet weight during the incubation time was, on average, 125 ± 6 mg (over three biological replicates), and all plants were surrounded by the same headspace volume. Incubation time was set for 6 h or 3 d. After incubation, 1 mL of headspace gas was sampled and analyzed by gas chromatography (2-m × 3-mm 80/100 alumina column; injector at 110°C, N2 vector gas in an isocratic oven at 70°C, and flame ionization detector at 250°C).
Plant Inoculation with Botrytis cinerea
B. cinerea BMM strain was grown on 39 g L−1 potato dextrose agar plates (BD Biosciences) at 23°C for 10 d in the dark until sporulation. Spores were harvested in sterile mQ water and filtered through glass wool to remove hyphae. Then, they were diluted in 6 g L−1 potato dextrose broth medium (BD Biosciences) for inoculation. Five-week-old plants were treated as described previously. Then, 3 d after the addition of 25 µm pyoverdine or water, Fe 25-, Fe 25 apo-pyo-, Fe 0-, and Fe 0 apo-pyo-treated plants were inoculated with B. cinerea. Inoculation was achieved by the application of 5-µL droplets of spore suspensions at a concentration of 5 × 104 spores mL−1 to the upper surface of five leaves per plant with five plants per condition and per experiment minimum. The inoculated plants were incubated at 100% relative humidity under 16-h days (23°C) and 8-h nights (20°C). Symptom development was scored 7 d after inoculation by measuring the lesion area using Optimas version 6.0 (Adept Turnkey).
Statistics
One-way ANOVA was performed using XLSTAT software (Addinsoft) followed by a multiple comparison procedure with the Fisher’s lsd method (at least P < 0.05). Two-way ANOVA was performed using VassarStats online software (http://vassarstats.net/) followed by a multiple comparison procedure with Tukey’s honestly significant difference method (at least P < 0.05).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: IRT1, AT4G19690; FRO2, AT1G015800; FER1, AT5G01600; NAS4, AT1G56430; bHLH100, AT2G41240; bHLH39, AT3G56980; CER4, AT4G33790; GRX, AT3G21460; LTP3, AT5G59320; PLTP, AT5G59330; and HBI1, AT2G18300.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR validation of the results obtained in the microarray analysis.
Supplemental Figure S2. Mapman analysis of genes modulated by apo-pyo in the roots of plants facing iron deficiency.
Supplemental Figure S3. Comparison between the profile of expression of the most induced and repressed genes by apo-pyo in the roots in iron-deficiency medium (log 2 ratio ≥ + 3 or ≤ − 3) to the profiles of other transcriptomic data (most different perturbations).
Supplemental Figure S4. Comparison between the profile of expression of the most induced and repressed genes by apo-pyo in the roots in iron-deficiency medium (log 2 ratio ≥ + 3 or ≤ − 3) to the profiles of other transcriptomic data (most different perturbations).
Supplemental Figure S5. Growth phenotypes of mutants overexpressing or impaired in genes involved in iron homeostasis (bhlh100, bhlh39, ov. fer.) or defense responses or/and growth (cer4, grx, ltp3, and pltp) in response to apo-pyo in iron sufficient or in iron deficient medium.
Supplemental Figure S6. Rosette macroscopic phenotype of A. thaliana plantlets exposed to apo-pyo or inoculated with the C7R12 or PL1 strains in iron-sufficient or iron-deficient conditions.
Supplemental Figure S7. Comparison between the profile of expression of the genes modulated by apo-pyo in the roots in iron-containing medium (log 2 ratio ≥ + 1.5 or ≤= − 1.5) to the profiles of other transcriptomic data (most similar perturbations).
Supplemental Table S1. Genes modulated by apo-pyo (log 2 ratio ≥ + 1.5 or ≤ −1.5) in iron containing medium in the shoots (+ Fe shoots).
Supplemental Table S2. Genes modulated by apo-pyo (log 2 ratio ≥ 1.5 or ≤ −1.5) in iron deficient medium in the shoots (− Fe shoots).
Supplemental Table S3. Genes modulated by apo-pyo (log 2 ratio ≥ 1.5 or ≤ −1.5) in iron containing medium in the roots (+ Fe roots).
Supplemental Table S4. Genes modulated by apo-pyo (log 2 ratio ≥ 1.5 or ≤ −1.5) in iron deficient medium in the roots (− Fe roots).
Supplemental Table S5. Clustering of the genes modulated by apo-pyo in 2 or 3 conditions.
Supplemental Table S6. Genes related to iron homeostasis modulated by apo-pyo (Log 2 ratio ≥ 1.5 or ≤ −1.5) in iron deficient conditions in roots or in shoots.
Supplemental Table S7. Genes related to defense modulated by apo-pyo (Log 2 ratio ≥ 1.5 or ≤ −1.5) in iron deficient conditions in roots or in shoots.
Supplemental Table S8. Genes related to the trade-off growth/defense mediated by HBI1 modulated by apo-pyo (Log 2 ratio ≥ 1.5 or ≤ −1.5) in iron deficient conditions in roots or in shoots.
Supplemental Table S9. Comparison between the root genes modulated by apo-pyo in iron-containing medium (Log 2 ratio ≤ −1.5 or ≥ + 1.5) and the genes modulated by iron deficiency in roots in the study of Schuler et al. (2011).
Supplemental Table S10. List of the primers used for the characterization of the mutants lines.
Supplemental Table S11. List of the primers used in the RT-qPCR analyses.
Supplementary Material
Acknowledgments
We thank Carine Fournier for the preparation of B. cinerea spores; Pascal Tillard and Alain Gojon from the Analytical Platform of the Biochimie et Physiologie Moléculaire des Plantes; Cyril Zipfel for the gift of HBI1-ox and HBI1(L214E)-ox lines; and Olivier Lamotte, Valérie Nicolas-Francès, Claire Rosnoblet, and Hoai-Nam Truong for careful reading of the article.
Glossary
- ISR
induced systemic resistance
- RT-qPCR
reverse transcription-quantitative PCR
- SA
salicylic acid
- ABA
abscisic acid
- IAA
indole-3-acetic acid
- T3SS
type 3 secretion system
- DFO
deferrioxamine
- KB
King’s B
- TBS
Tris-buffered saline
- cDNA
complementary DNA
Footnotes
This work was supported by the University of Burgundy (BQR project), the Burgundy State (PARI AGRALE 8 project), and the Ministère de l’Education Nationale de la Recherche et de la Technologie.
References
- Ahmed E, Holmström SJ (2014) Siderophores in environmental research: roles and applications. Microb Biotechnol 7: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht-Gary AM, Blanc S, Rochel N, Ocaktan AZ, Abdallah MA (1994) Bacterial iron transport: coordination properties of pyoverdin Paa, a peptidic siderophore of Pseudomonas aeruginosa. Inorg Chem 33: 6391–6402 [Google Scholar]
- Aznar A, Chen NW, Rigault M, Riache N, Joseph D, Desmaële D, Mouille G, Boutet S, Soubigou-Taconnat L, Renou JP, et al. (2014) Scavenging iron: a novel mechanism of plant immunity activation by microbial siderophores. Plant Physiol 164: 2167–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar A, Chen NW, Thomine S, Dellagi A (2015) Immunity to plant pathogens and iron homeostasis. Plant Sci 240: 90–97 [DOI] [PubMed] [Google Scholar]
- Aznar A, Dellagi A (2015) New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J Exp Bot 66: 3001–3010 [DOI] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker PAHM, Pieterse CMJ, van Loon LC (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97: 239–243 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carson KC, Meyer JM, Dilworth MJ (2000) Hydroxamate siderophores of root nodule bacteria. Soil Biol Biochem 32: 11–21 [Google Scholar]
- Chu BC, Garcia-Herrero A, Johanson TH, Krewulak KD, Lau CK, Peacock RS, Slavinskaya Z, Vogel HJ (2010) Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23: 601–611 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley DE. (2006) Microbial siderophores in the plant rhizosphere. In Barton LL, Abadίa J, eds, Iron Nutrition in Plants and Rhizospheric Microorganisms. Springer, Dordrecht, The Netherlands, pp 169–189 [Google Scholar]
- Curie C, Briat JF (2003) Iron transport and signaling in plants. Annu Rev Plant Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Dellagi A, Rigault M, Segond D, Roux C, Kraepiel Y, Cellier F, Briat JF, Gaymard F, Expert D (2005) Siderophore-mediated upregulation of Arabidopsis ferritin expression in response to Erwinia chrysanthemi infection. Plant J 43: 262–272 [DOI] [PubMed] [Google Scholar]
- Dellagi A, Segond D, Rigault M, Fagard M, Simon C, Saindrenan P, Expert D (2009) Microbial siderophores exert a subtle role in Arabidopsis during infection by manipulating the immune response and the iron status. Plant Physiol 150: 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Djavaheri M, Bakker PAHM, Höfte M (2008) Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol 148: 1996–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Höfte M (2009) Rhizobacteria-induced systemic resistance. Adv Bot Res 51: 223–281 [Google Scholar]
- Duijff BJ, Recorbet G, Bakker PAHM, Loper JE, Lemanceau P (1999) Microbial antagonism at the root level is involved in the suppression of fusarium wilt by the combination of nonpathogenic Fusarium oxysporum Fo47 and Pseudomonas putida WCS358. Phytopathology 89: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, et al. (2014) The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26: 828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Goodman M, Lamkemeyer P, Kandlbinder A, Sweetlove LJ, Dietz KJ (2005) The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J Biol Chem 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Alvarez-Fernández A, Briat JF (2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201: 155–167 [DOI] [PubMed] [Google Scholar]
- Franza T, Expert D (2013) Role of iron homeostasis in the virulence of phytopathogenic bacteria: an ‘à la carte’ menu. Mol Plant Pathol 14: 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319 [DOI] [PubMed] [Google Scholar]
- Kieu NP, Aznar A, Segond D, Rigault M, Simond-Côte E, Kunz C, Soulie MC, Expert D, Dellagi A (2012) Iron deficiency affects plant defence responses and confers resistance to Dickeya dadantii and Botrytis cinerea. Mol Plant Pathol 13: 816–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Koen E, Besson-Bard A, Duc C, Astier J, Gravot A, Richaud P, Lamotte O, Boucherez J, Gaymard F, Wendehenne D (2013) Arabidopsis thaliana nicotianamine synthase 4 is required for proper response to iron deficiency and to cadmium exposure. Plant Sci 209: 1–11 [DOI] [PubMed] [Google Scholar]
- Koen E, Trapet P, Brulé D, Kulik A, Klinguer A, Atauri-Miranda L, Meunier-Prest R, Boni G, Glauser G, Mauch-Mani B, et al. (2014) β-Aminobutyric acid (BABA)-induced resistance in Arabidopsis thaliana: link with iron homeostasis. Mol Plant Microbe Interact 27: 1226–1240 [DOI] [PubMed] [Google Scholar]
- Lei GJ, Zhu XF, Wang ZW, Dong F, Dong NY, Zheng SJ (2014) Abscisic acid alleviates iron deficiency by promoting root iron reutilization and transport from root to shoot in Arabidopsis. Plant Cell Environ 37: 852–863 [DOI] [PubMed] [Google Scholar]
- Lemanceau P, Alabouvette C (1991) Biological control of fusarium diseases by fluorescent Pseudomonas and non-pathogenic Fusarium. Crop Prot 10: 279–286 [Google Scholar]
- Lemanceau P, Bakker PAHM, De Kogel WJ, Alabouvette C, Schippers B (1992) Effect of pseudobactin 358 production by Pseudomonas putida WCS358 on suppression of fusarium wilt of carnations by nonpathogenic Fusarium oxysporum Fo47. Appl Environ Microbiol 58: 2978–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanceau P, Bakker PAHM, De Kogel WJ, Alabouvette C, Schippers B (1993) Antagonistic effect of nonpathogenic Fusarium oxysporum Fo47 and pseudobactin 358 upon pathogenic Fusarium oxysporum f. sp. dianthi. Appl Environ Microbiol 59: 74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C, Del Prete S, Boutet-Mercey S, Perreau F, Balagué C, Roby D, Fagard M, Gaudin V (2014) The hnRNP-Q protein LIF2 participates in the plant immune response. PLoS ONE 9: e99343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Henkels MD (1997) Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl Environ Microbiol 63: 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Zipfel C (2015) Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci 20: 12–19 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim SK, Zipfel C (2014) Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol 164: 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier S, Lemunier M, Siblot S, Mougel C, Lemanceau P (2004) Distribution and diversity of type III secretion system-like genes in saprophytic and phytopathogenic fluorescent pseudomonads. FEMS Microbiol Ecol 49: 455–467 [DOI] [PubMed] [Google Scholar]
- Meyer JM, Abdallah MA (1978) The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol 107: 319–328 [Google Scholar]
- Meziane H, van der Sluis I, van Loon LC, Höfte M, Bakker PA (2005) Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol Plant Pathol 6: 177–185 [DOI] [PubMed] [Google Scholar]
- Mirleau P, Delorme S, Philippot L, Meyer J, Mazurier S, Lemanceau P (2000) Fitness in soil and rhizosphere of Pseudomonas fluorescens C7R12 compared with a C7R12 mutant affected in pyoverdine synthesis and uptake. FEMS Microbiol Ecol 34: 35–44 [DOI] [PubMed] [Google Scholar]
- Nagata T, Oobo T, Aozasa O (2013) Efficacy of a bacterial siderophore, pyoverdine, to supply iron to Solanum lycopersicum plants. J Biosci Bioeng 115: 686–690 [DOI] [PubMed] [Google Scholar]
- Nairz M, Haschka D, Demetz E, Weiss G (2014) Iron at the interface of immunity and infection. Front Pharmacol 5: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong ST, Ho JZ, Ho B, Ding JL (2006) Iron-withholding strategy in innate immunity. Immunobiology 211: 295–314 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin A, Vansuyt G, Hinsinger P, Meyer JM, Briat JF, Lemanceau P (2008) Iron dynamics in the rhizosphere: consequences for plant health and nutrition. Adv Agron 99: 183–225 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Saha R, Saha N, Donofrio RS, Bestervelt LL (2013) Microbial siderophores: a mini review. J Basic Microbiol 53: 303–317 [DOI] [PubMed] [Google Scholar]
- Schuler M, Keller A, Backes C, Philippar K, Lenhof HP, Bauer P (2011) Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biol 11: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Bakker PA, van der Heijdt WH, Wendehenne D, Pugin A (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol Plant Microbe Interact 21: 1609–1621 [DOI] [PubMed] [Google Scholar]
- Vansuyt G, Robin A, Briat JF, Curie C, Lemanceau P (2007) Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol Plant Microbe Interact 20: 441–447 [DOI] [PubMed] [Google Scholar]
- Van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11: 443–448 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang J, Jiang J, Chen S, Guan Z, Liao Y, Chen F (2014) Reference genes for normalizing transcription in diploid and tetraploid Arabidopsis. Sci Rep 4: 6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller DM. (2007) Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97: 250–256 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Ai C, Xin L, Zhou G (2011) The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur J Soil Biol 47: 138–145 [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Hanson J, Pieterse CM (2014) β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol 204: 368–379 [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Korteland J, Van Pelt JA, van Hamersveld M, Dombrowski N, Bai Y, Hanson J, Van Verk MC, Ling HQ, Schulze-Lefert P, et al. (2015) Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. Plant J 84: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung HI, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.