OsABF1, which is a drought inducible transcription factor, may function reduandantly with OsbZIP40 to regulate rice heading date in response to the alteration of ambient water availability.
Abstract
The molecular mechanisms underlying photoperiod or temperature control of flowering time have been recently elucidated, but how plants regulate flowering time in response to other external factors, such as water availability, remains poorly understood. Using a large-scale Hybrid Transcription Factor approach, we identified a bZIP transcriptional factor, O. sativa ABA responsive element binding factor 1 (OsABF1), which acts as a suppressor of floral transition in a photoperiod-independent manner. Simultaneous knockdown of both OsABF1 and its closest homologous gene, OsbZIP40, in rice (Oryza sativa) by RNA interference results in a significantly earlier flowering phenotype. Molecular and genetic analyses demonstrate that a drought regime enhances expression of the OsABF1 gene, which indirectly suppresses expression of the Early heading date 1 (Ehd1) gene that encodes a key activator of rice flowering. Furthermore, we identified a drought-inducible gene named OsWRKY104 that is under the direct regulation of OsABF1. Overexpression of OsWRKY104 can suppress Ehd1 expression and confers a later flowering phenotype in rice. Together, these findings reveal a novel pathway by which rice modulates heading date in response to the change of ambient water availability.
Flowering time (or heading date) and drought resistance are two major yield traits in crops, especially rice (Oryza sativa). As global climatic change looms, drought has become the biggest abiotic stress to limit crop yields. Breeders have capitalized on naturally occurring genetic variations to improve or maintain crop yield in times or areas of drought by different strategies (Eisenstein, 2013). Manipulation of floral transition has been a promising way to maximize crop yield during dry periods. This strategy has been successful due to extensive identification of genetic loci and elucidation of molecular mechanisms that control flowering time under diverse or unpredictable environments.
Heading date in rice is influenced by many environmental cues such as day length (photoperiod), temperature, nutrition, and water availability. Molecular mechanisms that underlie photoperiod regulation of flowering time have already been characterized, probably because day length is more predictable than other environmental factors during seasonal changes. Rice is a facultative short-day plant that flowers earlier in short days (SDs) than in long days (LDs). Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) are two paralogous genes in rice encoding “florigen” molecules expressed in the phloem of leaves and transported to the shoot apical meristem to promote flowering (Tamaki et al., 2007; Komiya et al., 2008, 2009; Tsuji et al., 2013). Heading date 1 (Hd1, counterpart of Arabidopsis [Arabidopsis thaliana] CONSTANS, CO), Days to heading 2 (DTH2, encoding another CO-like protein), and Early heading date 1 (Ehd1) regulate expression of these two “florigen” genes. Hd1 acts as a transcription activator for Hd3a in SDs but as a repressor in the presence of functional O. sativa Phytochrome B in LDs (Hayama et al., 2003; Ishikawa et al., 2011). DTH2 acts in LDs to suppress flowering (Wu et al., 2013). Ehd1 encodes a rice-specific B-type response factor that acts in both LDs and SDs to promote Hd3a and RFT1 mRNA expression, thus inducing floral initiation (Doi et al., 2004; Komiya et al., 2009; Sun et al., 2014). Many repressors or activators, including Ehd2, Ehd3, Ehd4, Ghd7, OsMADS51, OsLFL1, and OsPPR37/DTH7, regulate the expression and activity of Ehd1 itself in day length-dependent or -independent manners (Doi et al., 2004; Kim et al., 2007; Peng et al., 2007; Xue et al., 2008; Wei et al., 2010; Matsubara et al., 2011; Koo et al., 2013; Tsuji et al., 2013; Yan et al., 2013; Gao et al., 2014; Kwon et al., 2015). It appears that Ehd1 is under the convergent regulation of SD activators, LD repressors, and photoperiod-independent regulators to control the heading date of rice in response to the internal development “clock” and environmental cues.
Water availability is another critical environmental factor affecting flowering time. Plants from xeric or mesic areas may utilize self-appropriate flowering strategies to cope with water deficits (Chaves et al., 2003). Many plants (Arabidopsis, Phyllostachys heterocycla, Mimulus guttatus, etc.) accelerate flowering to complete life cycle under drought conditions (Peng et al., 2013; Wolfe and Tonsor, 2014; Kooyers et al., 2015), a phenomenon referred to as drought escape (DE). Because drought triggers phytohormone abscisic acid (ABA) biosynthesis and signal transduction, it has been proposed that ABA mediates DE response. Consistent with this hypothesis, an ABA biosynthesis knockout mutant, aba1-6, was shown to flower later under LDs (Riboni et al., 2013). External application of ABA also delays flowering (Cheng et al., 2002; Achard et al., 2006; Domagalska et al., 2010; Wang et al., 2013), suggesting a dual effect of ABA on floral transition. Interestingly, drought accelerates flowering of Arabidopsis only under LDs but not SDs. Genetic studies have revealed that GIGANTEA (GI) knockout abolishes the drought-induced early flowering behavior of Arabidopsis under LDs (Han et al., 2013; Riboni et al., 2013), but the mechanism of GI-mediated photoperiod-dependent DE response has not been elucidated. The biological networks connecting drought perception, ABA signaling, and flowering gene regulation also remain unclear. In contrast to Arabidopsis species, rice cultivars tend to delay flowering in response to drought treatment (Wopereis et al., 1996; Ndjiondjop et al., 2010), but the underlying mechanism is less characterized.
Genome-wide investigations of transcription profiles have revealed that drought influences plant development and physiological responses by inducing hierarchical regulation of gene expression (Ding et al., 2012; Su et al., 2013; Huang et al., 2014; Maruyama et al., 2014). Several classes of transcription factors (TFs) play critical roles in orchestrating ABA-dependent or -independent drought responses (Jin et al., 2014; Todaka et al., 2015). One such class of TFs consists of a subgroup of bZIP proteins, which contain a basic region for DNA binding and a Leu zipper motif for dimerization. The DNA binding region preferentially interacts with ABA-responsive elements (ABREs), which are predominantly located in the promoter regions of ABA-inducible genes (Jakoby et al., 2002; Nijhawan et al., 2008). A great deal of genetic evidence shows that these ABRE-associated bZIP TFs are involved in drought tolerance response (Kang et al., 2002; Oh et al., 2005; Xiang et al., 2008; Lu et al., 2009; Hossain et al., 2010a, 2010b; Tang et al., 2012). Until now, none had been shown to regulate rice floral transition in response to drought stress. Here, we revealed that the bZIP TF, O. sativa ABA responsive element binding factor 1 (OsABF1), functions redundantly with OsbZIP40 to delay floral transition upon water deficit, which established a direct link between external water availability and plant developmental program.
RESULTS
OsABF1 Is a Negative Regulator of Floral Initiation
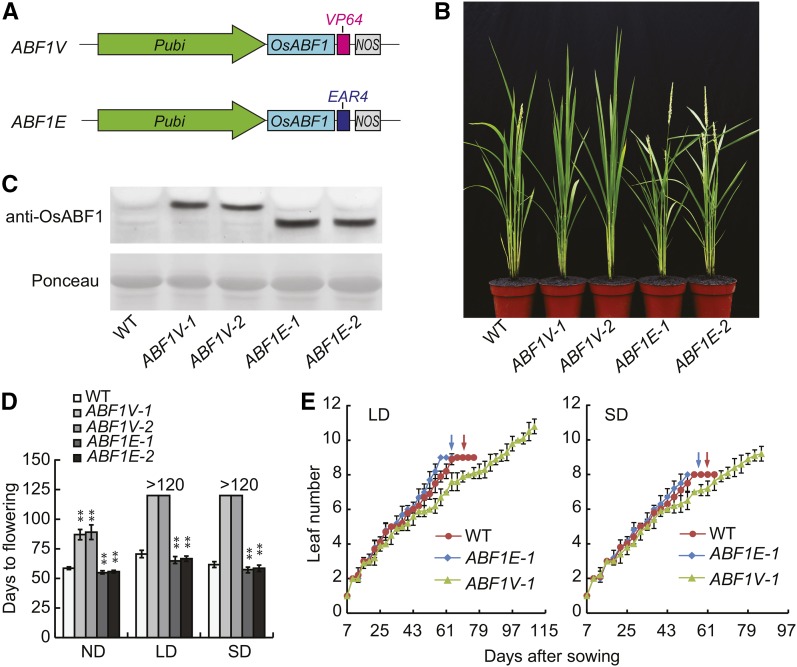
Previously, we used a large-scale Hybrid Transcription Factor (HTF) strategy to examine roles of different TFs in plant growth and development (Zhao et al., 2015). In the HTF approach, TF coding sequences are fused with a universal transcriptional activation module VP64 (tetrameric repeats of VP16) or a repression module EAR4 (tetrameric repeats of EAR). The HTF strategy aims to probe for quantifiably distinct transgenic phenotypes by activating transcription in the TF-VP64 group or by repressing transcription in the TF-EAR4 group. Through surveying the phenotypes of about 50,000 transgenic lines, we identified a pair of HTFs, named OsABF1-VP64 (ABF1V) and OsABF1-EAR4 (ABF1E; Fig. 1A). Respective overexpression of ABF1V and ABF1E in rice under the control of the maize ubiquitin promoter (Pubi) results in flowering time phenotypes that are effectively opposite to one another (Fig. 1B). Hereafter, the Pubi:OsABF1-VP64 and Pubi:OsABF1-EAR4 transgenic lines are designated as ABF1V and ABF1E, respectively. In total, 52 ABF1Vs and 9 ABF1Es were obtained, with 36 ABF1Vs and 8 ABF1Es recapitulated for at least two generations demonstrating late or early flowering phenotypes, respectively. The overexpression of ABF1V or ABF1E in these transgenic lines was verified by immunoblotting probed with anti-OsABF1 antibody (Fig. 1C; two representative lines of each construct were shown). The statistical results showed that ABF1Vs were flowering later while ABF1Es were flowering earlier than wild type under the nature day (ND, summer in Beijing), LD, or SD conditions (Fig. 1D). To be noted, ABF1Vs were still not flowering for 120 d in the controlled growth chamber under LD/SD treatment. The different severity of flowering time phenotype under LDs/SDs or NDs is probably because the light intensity or temperature in the controlled chamber is lower than that in nature field. Moreover, the number of main shoot leaves of ABF1V-1 was significantly more abundant than that of ABF1E-1 or wild type at maturation stage under both LDs and SDs (Fig. 1E).
Figure 1.
OsABF1 HTFs lead to opposite flowering time phenotypes. A, Diagrams of the OsABF1 HTF constructions. ABF1V, Pubi:OsABF1-VP64; ABF1E, Pubi:OsABF1-EAR4. B, Representative flowering image of indicated genotypes under NDs in summer at Beijing. C, Protein expression analysis of ABF1V or ABF1E in indicated lines. Kita-ake wild-type plants (WT) were used as control. The immunoblot was probed with anti-OsABF1 antibody. The ponceau staining bands of Rubisco large subunit was used as loading control. D, Flowering time of each genotype under different day length conditions. Mean values ± sd were shown. The value of each genotype was compared to that of wild type (Student’s t tests, **P < 0.01, n = 20). LDs (14 h light/10 h dark); SDs (10 h light/14 h dark). >120 indicates ABF1V transgenic plants cultured in growth chambers were not flowering after sowing for 120 d. E, Leaf number of each genotype grown under LDs or SDs in growth chambers. Red and blue arrows indicate the days of flowering of wild type and ABF1E-1, respectively.
To further test the role of OsABF1 in flowering regulation, we made a Pubi:OsABF1-3Flag construct (Supplemental Fig. S1A) and obtained 32 independent OsABF1 overexpression lines (ABF1Fs) that displayed a late flowering phenotype under ND conditions (Supplemental Fig. S1B). The overexpression of ABF1F in transgenic plants was validated by immunoblotting probed with anti-Flag antibody (Supplemental Fig. S1C, two representative lines were shown). Statistical analysis revealed that both ABF1F-1 and ABF1F-2 flower significantly later than wild type under ND, LD, or SD conditions (Supplemental Fig. S1D), demonstrating that OsABF1 acts as a negative regulator of floral initiation.
OsABF1 Acts as a Transcription Activator
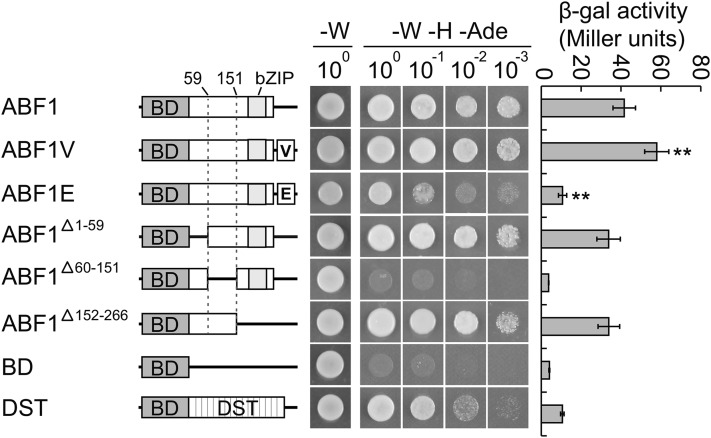
To gain insight into how ABF1V and ABF1E result in opposite flowering time phenotypes, we performed a Yeast One-Hybrid assay to test the transcriptional activity of OsABF1, ABF1V, or ABF1E. The result showed that OsABF1 is a transcriptional activator with an activation domain located between amino acids 60 and 151 of the protein (Fig. 2). Significant alteration in activity was observed when OsABF1 was fused with different effector modules (Fig. 2, left and middle). Transcriptional activation activity was increased by approximately 50% when OsABF1 was fused with VP64, while fusion with EAR4 resulted in repression of transcriptional activation activity by about 70% (Fig. 2, right). This result suggested that the opposite phenotypes of ABF1Vs and ABF1Es were due to the bilateral alteration of OsABF1 transcriptional activity when fused with VP64 or EAR4, respectively.
Figure 2.
Yeast one-hybrid assay of transcriptional activation activity. Left, Diagrams of yeast one-hybrid bait constructs comprising OsABF1 (ABF1) fragments fused with the GAL4 DNA binding domain (BD), VP64 (V), or EAR4 (E) as indicated. BD and BD fused with DST were used as negative and positive controls, respectively. Middle, Plate auxotroph assays showing transcriptional activation activity of each protein. W, Trp; H, His; Ade, adenine. Right, Quantitative yeast one-hybrid assays define transcriptional activation activity of each protein. Values are means of β-galactosidase activity ± sd, and the value of ABF1V or ABF1E was compared to that of ABF1 (Student’s t tests, **P < 0.01, n = 3).
OsABF1 Delays Floral Initiation through Inhibition of Ehd1
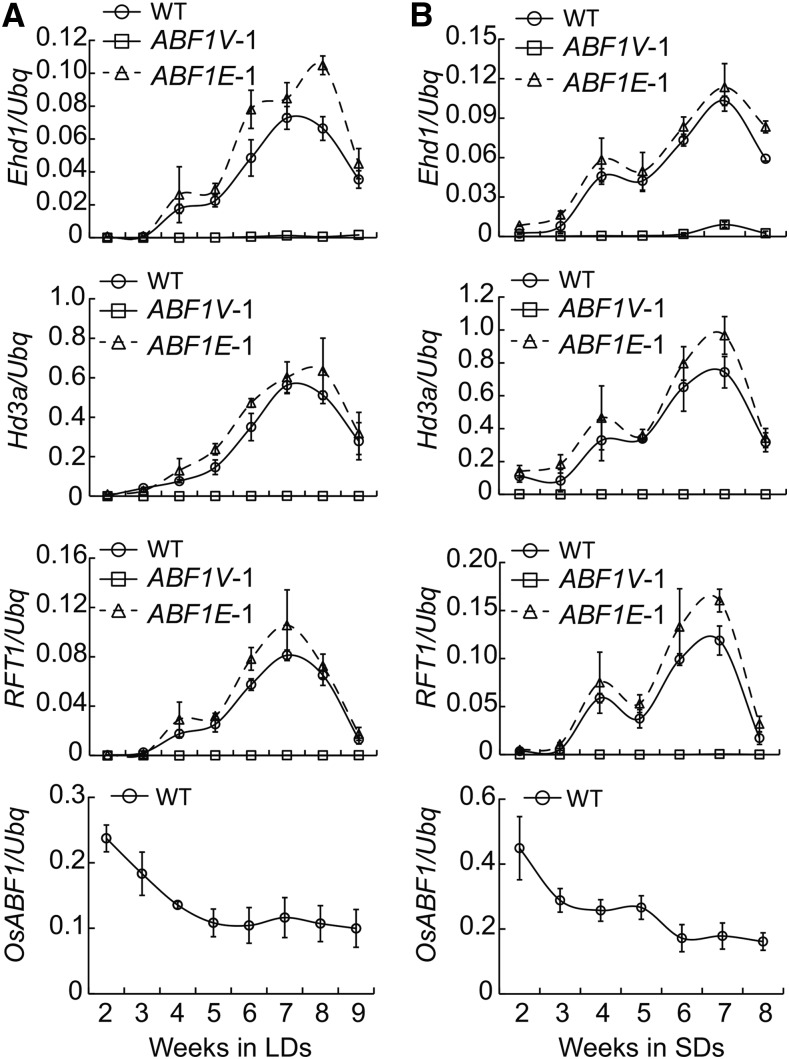
Next, we sought to identify genes that confer opposite flowering phenotypes in ABF1V versus ABF1E genotypes by investigating the mRNA expression profiles of 16 well-documented flowering-associated genes in rice in a time-course manner under both SDs and LDs (Supplemental Figs. S2 and S3). The result indicated that transcription levels of Ehd1, Hd3a, and RFT1 clearly vary between the genotypes and that variation is not apparent in the other flowering-associated genes. Expression levels of these three genes increased in ABF1E compared with wild type, but sharply decreased to an undetectable level in ABF1V. We then used incrementally aged plant groups to show that Ehd1, Hd3a, and RFT1 expression continually increased over time from week 3, but then decreased around weeks 7 and 8 in wild type and ABF1E. Again, we saw greater expression levels in ABF1E compared to wild type under both LDs and SDs, but wild-type expression levels also increased over time (Fig. 3). Expression levels in ABF1V were undetectable throughout the entire growth period (Fig. 3). Such expression patterns are consistent with the opposite flowering phenotype of ABF1Vs and ABF1Es. Interestingly, expression of the OsABF1 gene incrementally decreased with age under both LDs and SDs. This negative correlation between OsABF1 and Ehd1 mRNA levels suggests that OsABF1 may delay flowering by suppressing Ehd1 transcription.
Figure 3.
Transcriptional levels of indicated flowering-associated genes in each genotype at different developmental stages. A, Plants were grown in LDs. Samples were the latest fully expended leaves that were collected just at the end of dark period. Three biological replicates of qRT-PCR experiment were performed using Ubq gene as the internal control and the representative results were shown. Values were shown as mean ± sd (n = 3). B, Plants were grown in SDs and qRT-PCR experiments were performed as in A.
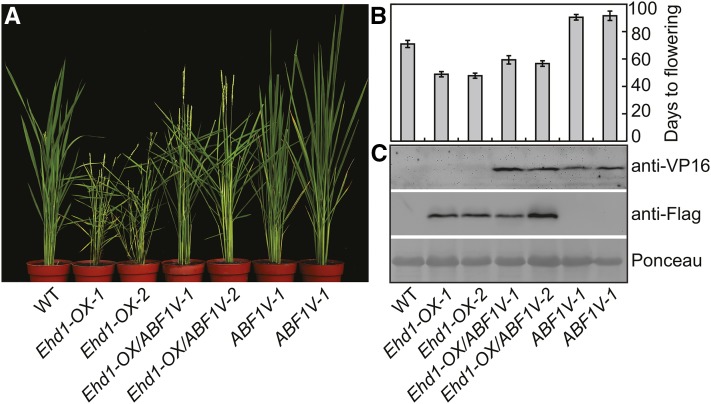
To test if ABF1V suppresses flowering through down-regulation of Ehd1 expression, we generated 12 and 18 Pubi:Ehd1-Flag transgenic lines in wild-type background (Ehd1-OX) and ABF1V-1 background (Ehd1-OX/ABF1V), respectively. Among them, 8 Ehd1-OX and 11 Ehd1-OX/ABF1V lines at T1 generation were flowering earlier than wild type under NDs (two representative lines of each genotype were shown in Fig. 4, A and B), demonstrating that overexpression of Ehd1 can efficiently suppress the later flowering phenotype of ABF1V. To measure and compare the flowering times between Ehd1-OX and Ehd1-OX/ABF1V, two independent lines of each genotype expressing equal levels of Ehd1-Flag protein were selected for the investigation (Fig. 4C). The results showed that Ehd1-OX/ABF1V lines were flowering about 4 d later than Ehd1-OX lines grown in NDs. This slight flowering difference might be due to the dosage effect of Ehd1 mRNA, because the transcription of endogenous Ehd1 was suppressed in Ehd1-OX/ABF1V but not in Ehd1-OX. Alternatively, ABF1V may not repress flowering in a manner entirely dependent on the Ehd1 pathway. Furthermore, we tested if ABF1E accelerates flowering through enhancing Ehd1 expression. In total, 18 and 15 Ehd1-RNAi transgenic lines in wild-type background (Ehd1R) and ABF1E-1 background (Ehd1-RNAi/ABF1E) were generated, respectively. Among them, 15 Ehd1R and 12 Ehd1R/ABF1E lines at T1 generation were similarly flowering later than wild type under NDs (two representative lines of each genotype were shown in Supplemental Fig. S4), demonstrating that knockdown of Ehd1 can efficiently suppress the early flowering phenotype of ABF1E. Taken together, our molecular and genetic results support the hypothesis that OsABF1 regulates flowering time through the Ehd1-mediated flowering pathway.
Figure 4.
Overexpression of Ehd1 suppresses the late flowering phenotype of ABF1V. A, The flowering phenotypes of indicated genotypes grown in NDs. All plants are in Kita-ake background. Ehd1-OX indicates the Pubi:Ehd1-Flag transgenic line in T1 generation. Ehd1-OX/ABF1V-1 and Ehd1-OX/ABF1V-2 were two independent T1 lines generated by stacking transformation of Pubi:Ehd1-Flag construct into ABF1V-1 homozygotes line. B, Statistical analysis of flowering time of indicated genotypes as in A. C, Protein expression analysis of OsABF1V or Ehd1-Flag in each genotype. OsABF1V or Ehd1-Flag was detected using anti-VP16 antibody or anti-Flag antibody, respectively. The ponceau staining band of Rubisco large subunit was used as loading control.
OsABF1 Acts Redundantly with OsbZIP40 to Delay Rice Flowering upon Drought Stress
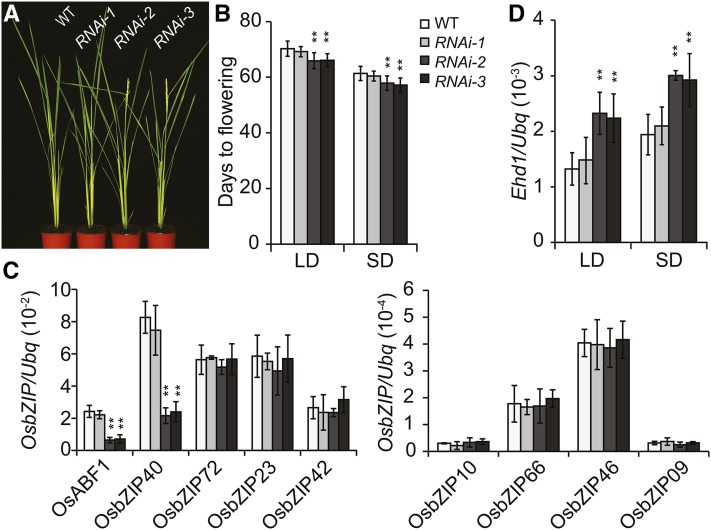
To test if OsABF1 loss of function affects rice flowering time, we obtained a new T-DNA insertion mutant Osabf1-3 (Supplemental Fig. S5A). Gene expression analysis showed that the transcription of OsABF1 is undetectable in the Osabf1-3 mutant (Supplemental Fig. S5B). Nevertheless, Osabf1-3 mutant plants displayed no obvious flowering time phenotype under all experimental conditions (NDs, LDs, and SDs), which is probably due to functional redundancy. To test this possibility, an RNA interference (RNAi) approach was applied to knockdown OsABF1 and its closest homologous gene OsbZIP40 simultaneously (Fig. 5; Supplemental Fig. S6; Nijhawan et al., 2008). A total of 15 RNAi lines were obtained, and 11 of them showed early flowering phenotype at T1 generation under both SDs and LDs (Fig. 5, A and B). Gene expression analysis revealed that the mRNA levels of both OsABF1 and OsbZIP40 significantly decreased in the RNAi effective lines (RNAi-2 or RNAi-3) showing early flowering phenotype, but not in wild type or in the RNAi ineffective line (RNAi-1). Expression levels of other OsABF1 homologous genes were not altered, demonstrating that just OsABF1 and OsbZIP40 were specifically silenced in the RNAi effective lines (Fig. 5C). Consistent with this result, the level of Ehd1 mRNA was significantly up-regulated in the RNAi effective lines (Fig. 5D).
Figure 5.
Simultaneous knockdown of OsABF1 and OsbZIP40 by RNAi approach results in early flowering phenotype. A, Representative flowering image of indicated genotypes under SDs. B, Flowering days of each genotype grown under LDs or SDs. Mean values ± sd are shown. The value of each genotype was compared to that of wild type (Student’s t tests, **P < 0.01, n = 20). C, qRT-PCR analysis of OsABF1 or the homologous genes in indicated lines. Values were shown as mean ± sd (Student’s t tests, **P < 0.01, n = 3). D, Expression analysis of Ehd1 in indicated genotypes grown under LDs or SDs for 4 weeks. Samples were the latest fully expended leaves that were collected just at the end of dark period. Values were shown as mean ± sd (Student’s t tests, **P < 0.01, n = 3).
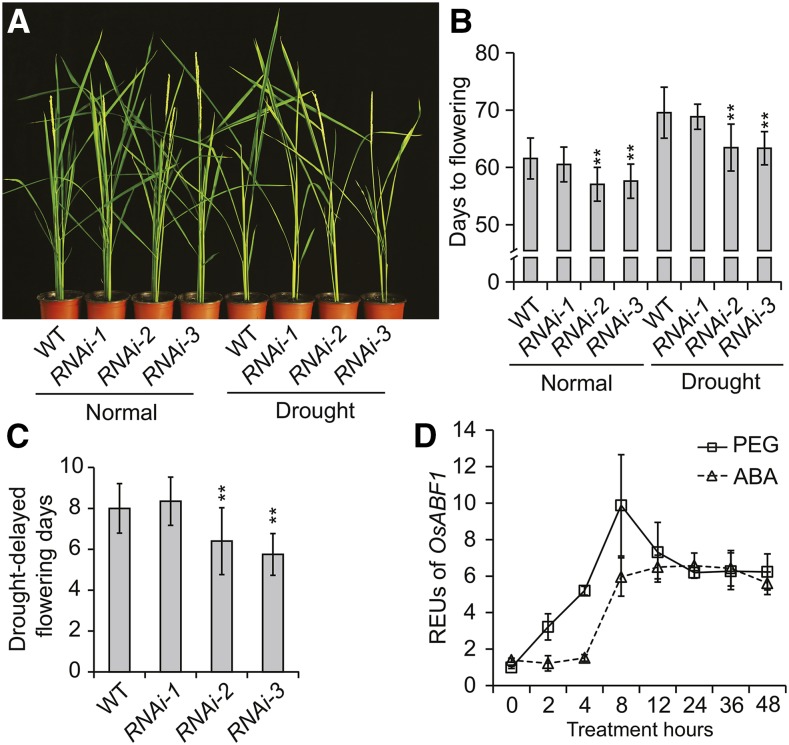
Given that polyethylene glycol (PEG) (or ABA) treatment can trigger and enhance the expression of OsABF1 (Fig. 6A; Amir Hossain et al., 2010), we tested the expression of Ehd1 in the similar conditions. The results demonstrated that the mRNA level of Ehd1 rapidly decreased upon dehydration treatment (Supplemental Fig. S5C), which is in sharp contrast to the enhanced expression pattern of OsABF1 mRNA (Fig. 6A). Moreover, the expression level of Ehd1 was significantly higher in Osabf1-3 than in wild type (Factorial ANOVA, P < 0.001, n = 3; Supplemental Fig. S5C), suggesting that OsABF1 may act as a drought-inducible suppressor of floral transition. To test this hypothesis, we compared the flowering time phenotype of indicated genotypes under normal and drought conditions (Fig. 6B). The results demonstrated that RNAi effective lines (RNAi-2, RNAi-3) were flowering earlier than wild type and RNAi ineffective line (RNAi-1) under both conditions (Fig. 6C). Moreover, drought treatment delayed flowering in wild type and RNAi ineffective line more efficiently than the RNAi effective lines (Fig. 6D), demonstrating that OsABF1-RNAi attenuates drought delay of flowering. Together, these results suggested that OsABF1 and OsbZIP40 function redundantly in mediating drought inhibition of flowering through the Ehd1 pathway.
Figure 6.
Flowering phenotype of OsABF1-RNAi lines in response to drought treatment. A, Dynamic transcription of OsABF1 in wild type under PEG or ABA treatment for the indicated periods. The seedlings were grown in continuous light for 3 weeks and then subjected to PEG or ABA treatment. The Relative Expression Units (REUs) were calculated by the formula: [(OsABF1/ubq) of each time point]/ [(OsABF1/ubq) of time point 0]. B, Representative flowering image of indicated genotypes under normal or drought conditions. RNAi-1 is an RNAi infective transgenic line used for negative control. C, Flowering days of each genotype as in A. Mean values ± sd were shown. The value of each genotype was compared to that of wild type under similar growth condition (Student’s t tests, **P < 0.01, n = 20). D, The value of drought-delayed flowering days of each genotype was calculated with the flowering days under drought regime minus that under normal conditions. The significant difference between each genotype and wild type was calculated (Student’s t tests, **P < 0.01, n = 20).
OsABF1 May Delay Flowering by Directly Activating OsWRKY104 Gene
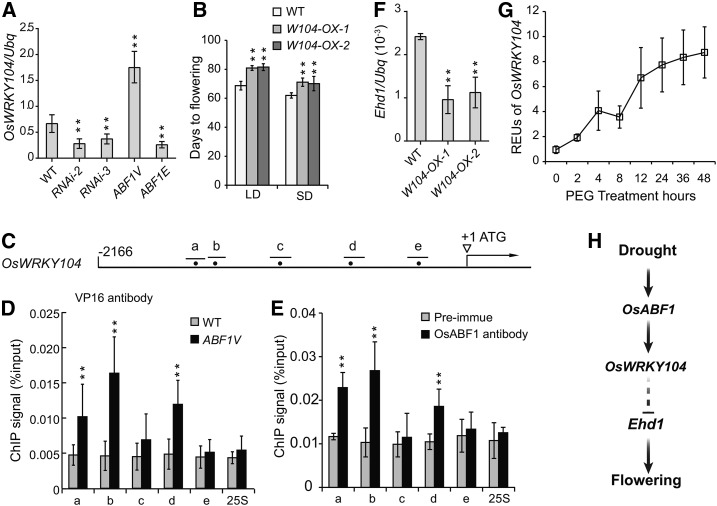
Given that OsABF1 is a transcriptional activator and that Ehd1 transcription is up-regulated in the Osabf1-3 mutant and OsABF1-RNAi lines, Ehd1 is unlikely under the direct regulation of OsABF1. We speculate that OsABF1 may activate the expression of an unknown TF that acts as a repressor of Ehd1 in response to drought treatment. Therefore, we interrogated the transcriptional profiles of rice TF genes in response to drought treatment (NCBI database, GEO accession no. GSE65022) and found 271 drought inducible TF genes (DTFs). By overlapping analysis with the heading dates of the large HTF transgenic population (Zhao et al., 2015), we found 29 DTFs that could confer abnormal flowering time phenotype. We further tested these DTF expression levels in different genotypes and found that a gene named OsWRKY104 was up-regulated in ABF1V, but down-regulated in ABF1E and OsABF1-RNAi lines (Fig. 7A; Supplemental Fig. S7, D and E). Moreover, in contrast to the early flowering phenotype of EAR4-OsWRKY104 (EWRKY104) HTF transgenic lines (Supplemental Fig. S7), overexpression of OsWRKY104 (W104-OX) resulted in a later flowering phenotype (Fig. 7B; Supplemental Fig. S8). Next, we tested if OsWRKY104 is a direct target gene of OsABF1. Chromatin immunoprecipitation (ChIP) using ABF1V transgenic plants and anti-VP16 antibody showed that ABF1V robustly binds to a, b, and d sites that contain the ACGT core sequence of the ABRE motif (Ono et al., 1996; Hobo et al., 1999; Nijhawan et al., 2008; Fig. 7D). Furthermore, we performed ChIP experiments using wild-type plants and the anti-OsABF1 antibody. The results also showed similar robust binding signals at a, b, and d sites (Fig. 7E), validating that OsWRKY104 is a direct target gene under the regulation of OsABF1. All the above results together with the fact that Ehd1 expression is down-regulated in W104-OX lines (Fig. 7F) but up-regulated in EWRKY104 lines (Supplemental Fig. S7F) suggest that OsABF1 suppresses Ehd1 expression at least partially through activating OsWRKY104 transcription in response to drought stress (Fig. 7, G and H).
Figure 7.
OsWRKW104 is a direct target of OsABF1that delays flowering. A, qRT-PCR analysis of OsWRKW104 expression in indicated lines. The means ± sd (Student’s t tests, **P < 0.01, n = 3) are shown. B, Flowering days of OsWRKW104 overexpression lines. The means ± sd (n = 15) are shown. C, A diagram representing the promoter region of OsWRKW104 gene. The bars represent the distribution of DNA fragments containing ACGT core as indicated by dots. D, Verification of ABF1V direct binding sites in the OsWRKW104 promoter by ChIP-qPCR analysis. ChIP samples were prepared using ABF1V and wild-type plants, precipitated with anti-VP16 antibody, and subjected to qPCR analysis. Results of ChIP-qPCR were quantified by normalization of the immunoprecipitation signal with the corresponding input signal. The binding to 25S rDNA was used as negative control. The means ± sd (n = 3) are shown. E, Verification of OsABF1 binding sites in the OsWRKW104 promoter by ChIP-qPCR analysis. ChIP samples were prepared using wild-type rice and precipitated with anti-OsABF1 antibody or with the preimmune serum as negative control. The means ± sd (n = 3) are shown. F, qRT-PCR analysis of Ehd1 expression in OsWRKW104 overexpression lines. Values were shown as mean ± sd (n = 3). G, Dynamic transcription of OsWRKW104 in wild type under PEG treatment for the indicated periods. The seedlings were grown in continuous light for 3 weeks and then subjected to PEG treatment. The REUs were calculated by the formula: [(OsWRKW104/ubq) of each time point]/ [(OsWRKW104/ubq) of time point 0]. H, A working model depicts how OsABF1 modulates flowering time in response to drought stress. The transcription of OsABF1 is up-regulated under water deficit. Consequently, OsABF1 instigates the expression of OsWRKW104, which further delays flowering through inhibiting Ehd1 expression.
DISCUSSION
Drought Inhibition of Flowering, a Rice Unique Drought Avoidance Strategy
Terrestrial plants, being sessile, grow adaptively in response to limited water availability by flexible strategies, including DE (via rapid flowering and reproduction before severe water shortage leads to lethality), drought avoidance (via developmental adaptions to reduce water consumption), or drought tolerance (via adjusting osmosis, enhancing oxidative capacity, or strengthening cell walls to withstand dehydration; Chaves et al., 2003; Yue et al., 2006). For example, Arabidopsis has a short life cycle and naturally grows in xeric environments, so it relies on DE response when encountering drought stress (Supplemental Fig. S9) (Riboni et al., 2013). In contrast, lowland rice is historically grown in mesic conditions and tends to delay flowering in response to temporary drought stress (Wopereis et al., 1996; Ndjiondjop et al., 2010). Therefore, the two model plants (Arabidopsis and rice) seem to display opposite flowering time responses to drought stress. Moreover, other environmental cues also differentially affect their flowering behavior upon water scarcity. The study of Arabidopsis showed that drought-induced acceleration of flowering is dependent on LD condition (Han et al., 2013; Riboni et al., 2013). Our study in rice showed that drought delayed flowering is photoperiod independent, while other factors such as light intensity or temperature may affect the severity of drought-delayed flowering time mediated by OsABF1 (Fig. 1D). Therefore, how plants decide time of flowering seems to be ecologically diversified, and further investigations are warranted for a better understanding and discrimination of the mechanistic basis underlying the integration of environmental signals for reproductive success of different plant species.
In this report, we tested the effect of drought on rice floral transition using both indica and japonica cultivars grown in well-controlled growth chambers to avoid the effects of other environmental parameters. The result indicated that the flowering time of all tested cultivars was significantly delayed by drought regime in both LDs and SDs (Supplemental Figure S10), suggesting that drought-delayed flowering response may have already existed prior to genetic divergence of indica and japonica cultivars. Rice is more susceptible to drought sensitivity during reproductive stages (Lanceras et al., 2004), so we hypothesize that rain-fed ancestors of contemporary rice species may have postponed the transition from vegetative phase to reproductive phase as they sensed temporary water deficits and reduced water consumption. Such a drought avoidance strategy may enable rice to enhance survival by postponing blossoming until the upcoming rainy season. Although most rice cultivars tend to delay flowering under drought stress conditions, acceleration of flowering under drought conditions has also been documented in some modern rice varieties (Xu et al., 2005; Vikram et al., 2015). These examples demonstrate the complexity and diversity of rice genotype-environment interactions as a result of domestication.
OsABF1 Mediates Drought-Derived Signal to Regulate Rice Flowering Time
In this study, we showed that OsABF1 acts as a transcriptional activator to delay rice floral transition. We checked the expressions of previously known flowering associated genes and demonstrated that only Ehd1, Hd3a, and RFT1 genes, which close to the final step of the rice flowering pathway, were down-regulated by OsABF1. Several repressors of Ehd1 (including Ghd7, OsPPR37/DTH7, DTH8, OsLFL1, and OsCOL4) have been identified previously. In the Kita-ake cultivar, Ghd7 and OsPPR37/DTH7 are functionally deficient (Itoh et al., 2010; Gao et al., 2014), while the mRNA expression levels of DTH8, OsLFL1, and OsCOL4 showed no significant difference in ABF1V, ABF1E, and wild-type plants (Supplemental Figs. S2 and S3). Therefore, OsABF1 may not inhibit Ehd1 expression through up-regulation of any these repressors. By conjoint analyses of the drought-induced transcription profile and our large-scaled HTF transgenic population, we identified a direct target gene of OsABF1, named OsWRKY104, which belongs to a large WRKY TF gene family consisting of more than 100 members in rice (Berri et al., 2009). WRKY TFs have been documented to regulate a range of biological processes, including biotic or abiotic response, senescence, and development (Rushton et al., 1996; Hara et al., 2000; Johnson et al., 2002; Maré et al., 2004; Miao et al., 2004; Cai et al., 2014). Here, we showed that drought stress does enhance the expression of OsWRKY104, which may further repress floral transition. Provided that OsWRKY104 overexpression lines and EAR4-OsWRKY104 HTF transgenic lines show opposite flowering time phenotype, OsWRKY104 probably acts as a transcriptional activator in rice. Therefore, we proposed that drought stress enhances the transcription of OsABF1 and OsWRKY104, then further activates the expression of an unknown repressor of Ehd1 to delay rice heading date (Fig. 7H). The increased understanding of how rice regulates flowering time in response to water availability will benefit the development of high performance rice lines in drought-prone regions.
MATERIALS AND METHODS
Plant Materials
To generate ABF1V and ABF1E overexpression lines, OsABF1 cDNA was inserted into pBCV and pBCE expression vectors respectively (Zhao et al., 2015), using the Gateway cloning system (Invitrogen). To generate OsABF1, Ehd1 and OsWRKY104 overexpression lines, respective cDNA was inserted into the pHCF vector (Supplemental Figure S6) at PstI site using the Infusion system (Clontech). To generate OsABF1-RNAi or Ehd1-RNAi plants, a 267-bp fragment of the OsABF1 gene (from 506 to 772 bp) or a 270-bp fragment of the Ehd1 gene (from 56 to 325 bp) was inserted into the pANDA vector using the Gateway cloning system (Miki and Shimamoto, 2004). The above constructs were introduced into Agrobacterium tumefaciens strain EHA105 and transformed into wild type rice (Oryza sativa japonica cv Kita-ake) or indicated background. The T-DNA insertion mutant Osabf1-3 (rice cv Hwayoung backgroud) was identified from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/RiceGE) (Jeong et al., 2006). Homozygous line of Osabf1-3 was characterized by genotyping PCR with OsABF1 gene-specific and T-DNA specific primers.
Flowering Time Analyses
To investigate the flowering phenotypes, plants were grown under NDs in Beijing (39°54’N, 116 °23’E, nursery on May 6), China, or under LDs (14 h light, 28°C; 10 h dark, 24°C) or SDs (10 h light, 28°C; 14 h dark, 24°C) in plant growth chambers or green house. To investigate the effect of drought stress on rice flowering time, the indicated rice genotypes were sown and cultured with 1/10 Murashige and Skoog culture solution in transparent boxes for 2 weeks and then transferred to boxes containing wet soil for resurrecting. For drought treatment, the plants were not irrigated until the soil moisture content achieved approximately 30%, and then water was added to the boxes until the soil was saturated (Supplemental Figure S12). The drought regime was imposed until flowering. The control plants were grown in boxes maintaining a water level above the soil surface.
Sample Collection and Gene Expression Analyses
Seeds of wild-type rice and transgenic lines were germinated for 2 d on wet filter paper in petri dishes at 37°C. The uniformly germinated seeds were picked up and sown in bottomless 96-well plates and hydroponically grown (distiller water with 1/10 Murashige and Skoog). To test the mRNA expression of flowering-associated genes in a time course manner under LDs or SDs, plants were grown for 4 weeks and samples were collected every 4 h from the beginning of the light period. To test the mRNA level with increment of age, the latest fully expended leaves were collected just at the end of dark period from 2 to 9 weeks. To test OsABF1 or Ehd1 mRNA expression in response to PEG treatment, the roots of seedlings grown under continuous light conditions were submerged into 20% PEG4000 solution for the hours indicated. To test OsABF1 mRNA expression in response to ABA treatment, the roots were submerged in a 100-μm ABA solution and the leaves were sprayed with same solution. RNA exaction and quantitative reverse transcription PCR (qRT-PCR) were performed as previously described with three biological replicates (Meng et al., 2013).
Transactivation Activity Assay in Yeast
To test the transactivation activity, the indicated coding DNA sequence (CDS) was fused with GAL4 DNA-binding domain in the pGBKT7 vector and transformed into the yeast strain AH109. The empty vector (BD) and BD-DST vector were used as negative and positive controls, respectively. Measurement of the β-galactosidase activity was performed according to the Yeast Protocols Handbook (Clontech) using chlorophenol red-β-d-galactopyranoside (Roche Biochemical) as the substrate.
Immunoblot Analyses
The anti-OsABF1 polyclonal antibody was generated by inoculating rabbits with TF-His-OsABF1 recombination protein (Bio-med). The anti-VP16 polyclonal antibody was generated by inoculation of rabbits with a synthesized antigenic peptide consisting of hexametric repeats of VP16 (DALDDFDLDML DALDDFDLDML DALDDFDLDML DALDDFDLDML DDFDL DDFDL; Abmart). To extract the total protein for immunoblot, the young leaves were ground in liquid nitrogen and mixed with 5X SDS-PAGE loading buffer [250 mm Tris (pH 6.8), 10% (w/v) SDS, 0.5% (w/v) bromphenol blue, 50% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol], boiled for 5 min, and spun at 12,000 rpm for 5 min at room temperature. The supernatants were fractioned by 10% SDS-PAGE, and the membrane was probed with the indicated antibody.
ChIP Assay
Seedlings of 4-week-old ABF1V-1 and wild-type plants grown under continuous light were used for ChIP assays. Three g of leaves was cross-linked in 35 mL formaldehyde buffer [0.4 m Suc, 10 mm Tris-HCl (pH 8.0), 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 mm EDTA, and 1% (v/v) formaldehyde] and vacuumed twice for 15 min. To stop the reaction, 1.6 mL of 2 m Gly was added and vacuumed for 5 min. Then the samples were washed in water and ground to powder in liquid nitrogen prior to being suspended in 15 mL of Honda buffer [0.44 m Suc, 1.25% Ficoll, 2.5% Dextran T40, 20 mm HEPES KOH (pH 7.4), 10 mm MgCl2, 0.5% Triton X-100, 5 mm dithiothreitol, 1 mm PMSF, and 1 tablet/50 mL of protease inhibitor cocktail]. The nuclei were filtered through two layers of Miracloth, precipitated by centrifugation, and suspended in nuclei lysis buffer [50 mm Tris-HCl (pH 8.8), 10 mm EDTA, 1% SDS, 1 mm PMSF, and 1 tablet/50 mL of protease inhibitor cocktail]. The chromatin complexes were sonicated and incubated with anti-VP16 antibody or anti-OsABF1 antibody as described. The precipitated DNA was recovered in water for further experiments. For ChIP-qPCR assays, the DNA samples were analyzed by qPCR with SYBR Premix Ex Taq (Takara) for at least three replicates and the value was normalized to that of input DNA (% of input).
Accession Numbers
Sequence data from this article can be found in the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) databases (Kawahara et al., 2013) under the following accession numbers: OsABF1 (LOC_Os01g64730), Ehd1 (LOC_Os10g32600), Hd1 (LOC_Os06g16370), Hd3a (LOC_Os06g06320), RFT (LOC_Os06g06300), OsPHYB (LOC_Os03g19590), OsCOL4 (LOC_Os02g39710), DTH8 (LOC_Os08g07740), SE5 (LOC_Os06g40080), LFL1 (LOC_Os01g51610), OsMADS56 (LOC_Os10g39130), OsGI (LOC_Os01g08700), Ehd2 (LOC_Os10g28330), Ehd3 (LOC_Os08g01420), Ehd4 (LOC_Os03g02160), OsMADS50 (LOC_Os03g03070), OsMADS51 (LOC_Os01g69850), and OsWRKY104 (LOC_ Os11g02520).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overexpression of OsABF1 confers later flowering phenotype.
Supplemental Figure S2. Transcriptional levels of flowering-associated genes under LDs.
Supplemental Figure S3. Transcriptional levels of flowering-associated genes under SDs.
Supplemental Figure S4. Knockdown of Ehd1 suppresses the early flowering phenotype of ABF1E.
Supplemental Figure S5. Analysis of Ehd1 mRNA expression in Osabf1-3 mutant.
Supplemental Figure S6. Phylogenetic tree of OsABF1 and homologous proteins.
Supplemental Figure S7. EAR-OsWRKY104 HFT leads to early flowering phenotype.
Supplemental Figure S8. Overexpression of OsWRKY104 confers later flowering phenotype.
Supplemental Figure S9. Drought accelerates flowering in Arabidopsis.
Supplemental Figure S10. Drought delays flowering in rice.
Supplemental Figure S11. Vector map and sequence of pHCOMPARE WITH.
Supplemental Figure S12. The soil moisture content during drought regime experiment.
Supplemental Table S1. Oligonucleotide primers used in this study.
Supplementary Material
Glossary
- ABA
abscisic acid
- ABRE
ABA-responsive element
- ChIP
chromatin immunoprecipitation
- DE
drought escape
- HTF
Hybrid Transcription Factor
- LD
long day
- ND
nature-day
- PEG
polyethylene glycol
- PMSF
phenylmethylsulfonyl fluoride
- qRT-PCR
quantitative reverse transcription PCR
- RNAi
RNA interference
- SD
short day
- TF
transcription factor
Footnotes
Articles can be viewed without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro AC, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè ME, et al. (2009) Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol 9: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Chen X, Xie K, Xing Q, Wu Y, Li J, Du C, Sun Z, Guo Z (2014) Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS One 9: e102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS (2003) Understanding plant responses to drought—from genes to the whole plant. Funct Plant Biol 30: 239–264 [DOI] [PubMed] [Google Scholar]
- Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z (2012) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3: 740. [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Sarnowska E, Nagy F, Davis SJ (2010) Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana. PLoS One 5: e14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M. (2013) Plant breeding: discovery in a dry spell. Nature 501: S7–S9 [DOI] [PubMed] [Google Scholar]
- Gao H, Jin M, Zheng XM, Chen J, Yuan D, Xin Y, Wang M, Huang D, Zhang Z, Zhou K, et al. (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci USA 111: 16337–16342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang X, Wang W, Wang Y, Ming F (2013) The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8: e73541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H (2000) Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet 263: 30–37 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB (2010a) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72: 557–566 [DOI] [PubMed] [Google Scholar]
- Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB (2010b) The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167: 1512–1520 [DOI] [PubMed] [Google Scholar]
- Huang L, Zhang F, Zhang F, Wang W, Zhou Y, Fu B, Li Z (2014) Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics 15: 1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K (2011) Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics 285: 461–470 [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, b ZIPRG (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: D1182–D1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang Z, Li J, Li Z, Paek NC (2013) Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6: 1877–1888 [DOI] [PubMed] [Google Scholar]
- Kooyers NJ, Greenlee AB, Colicchio JM, Oh M, Blackman BK (2015) Replicate altitudinal clines reveal that evolutionary flexibility underlies adaptation to drought stress in annual Mimulus guttatus. New Phytol 206: 152–165 [DOI] [PubMed] [Google Scholar]
- Kwon CT, Koo BH, Kim D, Yoo SC, Paek NC (2015) Casein kinases I and 2α phosphorylate oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol Cells 38: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanceras JC, Pantuwan G, Jongdee B, Toojinda T (2004) Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol 135: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229: 605–615 [DOI] [PubMed] [Google Scholar]
- Marè C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L (2004) Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol 55: 399–416 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Urano K, Yoshiwara K, Morishita Y, Sakurai N, Suzuki H, Kojima M, Sakakibara H, Shibata D, Saito K, et al. (2014) Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol 164: 1759–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66: 603–612 [DOI] [PubMed] [Google Scholar]
- Meng Y, Li H, Wang Q, Liu B, Lin C (2013) Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 25: 4405–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Ndjiondjop M-N, Cisse F, Futakuchi K, Lorieux M, Manneh B, Bocco R, Fatondji B (2010) Effect of drought on rice (Oryza spp.) genotypes according to their drought tolerance level. In Second Africa Rice Congress, Bamako, Mali, pp 1.5.1-1.5.8 [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K (1996) The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol 112: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL (2007) Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun 360: 251–256 [DOI] [PubMed] [Google Scholar]
- Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, Lu H, Hu T, Yao N, Liu K, et al. (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45: 456–461 [DOI] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Su Z, Ma X, Guo H, Sukiran NL, Guo B, Assmann SM, Ma H (2013) Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell 25: 3785–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Chen D, Fang J, Wang P, Deng X, Chu C (2014) Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell 5: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci 6: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K (2013) Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr Opin Plant Biol 16: 228–235 [DOI] [PubMed] [Google Scholar]
- Vikram P, Swamy BP, Dixit S, Singh R, Singh BP, Miro B, Kohli A, Henry A, Singh NK, Kumar A (2015) Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci Rep 5: 14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MD, Tonsor SJ (2014) Adaptation to spring heat and drought in northeastern Spanish Arabidopsis thaliana. New Phytol 201: 323–334 [DOI] [PubMed] [Google Scholar]
- Wopereis MCS, Kropff MJ, Maligaya AR, Tuong TP (1996) Drought-stress responses of two lowland rice cultivars to soil water status. Field Crops Res 46: 21–39 [Google Scholar]
- Wu W, Zheng XM, Lu G, Zhong Z, Gao H, Chen L, Wu C, Wang HJ, Wang Q, Zhou K, et al. (2013) Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA 110: 2775–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JL, Lafitte HR, Gao YM, Fu BY, Torres R, Li ZK (2005) QTLs for drought escape and tolerance identified in a set of random introgression lines of rice. Theor Appl Genet 111: 1642–1650 [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, Liu T, Liu H, Zhang C, Zhang Z, et al. (2013) Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res 23: 969–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q (2006) Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172: 1213–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Liu J, Li HY, Lin JZ, Bian MD, Zhang CY, Zhang YX, Peng YC, Liu B, Lin C (2015) Using hybrid transcription factors to study gene function in rice. Sci China Life Sci 58: 1160–1162 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.