Phosphorylation of a subunit of cellulose synthase differentially alters the rate at which cellulose synthase moves in both directions along microtubules, thereby impacting growth and development.
Abstract
Here we report that phosphorylation status of S211 and T212 of the CESA3 component of Arabidopsis (Arabidopsis thaliana) cellulose synthase impacts the regulation of anisotropic cell expansion as well as cellulose synthesis and deposition and microtubule-dependent bidirectional mobility of CESA complexes. Mutation of S211 to Ala caused a significant decrease in the length of etiolated hypocotyls and primary roots, while root hairs were not significantly affected. By contrast, the S211E mutation stunted the growth of root hairs, but primary roots were not significantly affected. Similarly, T212E caused a decrease in the length of root hairs but not root length. However, T212E stunted the growth of etiolated hypocotyls. Live-cell imaging of fluorescently labeled CESA showed that the rate of movement of CESA particles was directionally asymmetric in etiolated hypocotyls of S211A and T212E mutants, while similar bidirectional velocities were observed with the wild-type control and S211E and T212A mutant lines. Analysis of cell wall composition and the innermost layer of cell wall suggests a role for phosphorylation of CESA3 S211 and T212 in cellulose aggregation into fibrillar bundles. These results suggest that microtubule-guided bidirectional mobility of CESA complexes is fine-tuned by phosphorylation of CESA3 S211 and T212, which may, in turn, modulate cellulose synthesis and organization, resulting in or contributing to the observed defects of anisotropic cell expansion.
Cellulose microfibrils, composed of multiple (1→4)-β-glucans, are the primary load-bearing component of plant cell walls. In diffusely growing cells, cellulose microfibrils are synthesized by plasma-membrane-localized cellulose synthase (CESA) complexes. The complexes were visualized by freeze-fracture electron microscopy as hexagonal rosettes of 25 to 30 nm in diameter (Saxena and Brown, 2005). They are thought to be composed of three structurally similar CESA proteins that are believed to be the catalytic subunits. For primary wall cellulose synthesis, CESA1 and CESA3 appear to be absolutely required, whereas CESA2, CESA5, CESA6, and CESA9 are at least partially redundant (Desprez et al., 2007; Persson et al., 2007). Cellulose synthase coaligns with and moves bidirectionally along cortical microtubules (Paredez et al., 2006). A large cellulose synthase interacting protein (CSI1) links microtubule and CESA complexes (Gu et al., 2010), and two companion proteins of cellulose synthase (CC1 and CC2) bind to microtubules and promote microtubule dynamics (Endler et al., 2015).
CESA1, CESA2, CESA3, CESA5, CESA6, and CESA9 components of cellulose synthase are phosphorylated at sites clustered in two hypervariable regions of the proteins (Nühse et al., 2004; Roitinger et al., 2015). We previously showed that phosphorylation of some CESA1 residues modulated anisotropic cell expansion and bidirectional mobility of CESA particles in a manner dependent on microtubule status (Chen et al., 2010). In diffusively growing hypocotyl cells of wild-type Arabidopsis (Arabidopsis thaliana) seedlings, CESA particles typically move with similar velocities in opposite directions along tracks of cortical microtubules (Paredez et al., 2006). However, in seedlings expressing any one of the mutated CESA1 proteins (S162E, T165E, T166A, S167E, S686A, and S688A), the complexes displayed asymmetry in the rates of bidirectional movements, and the asymmetry correlates with defects in anisotropic cell expansion (Chen et al., 2010). Chemical depolymerization of microtubules eliminates the asymmetric CESA particle movement in the mutant lines, suggesting a phosphorylation-dependent interaction between the primary wall CESA complexes and the microtubules in diffusely growing cells (Chen et al., 2010).
Phosphorylation of CESA5 modulates migration of CESA complexes containing the CESA6-related isoform, CESA5, which is coordinated by phytochrome activation (Bischoff et al., 2011). Therefore, it was of interest to examine if and how phosphorylation of CESA3 regulates interactions of CESA particles with cortical microtubules, particularly microtubule-guided bidirectional synthesis and deposition of cellulose microfibrils. In this study, we mutagenized CESA3 S211 and T212 to Ala or Glu residues to examine how elimination or mimicry of phosphorylation of the neighboring residues modulates anisotropic cell expansion and to explore molecular mechanisms underlying regulation of anisotropic cell wall expansion.
RESULTS
Effects of Phosphorylation of S211 and T212 on Diffuse Growth of Primary Roots and Etiolated Hypocotyls
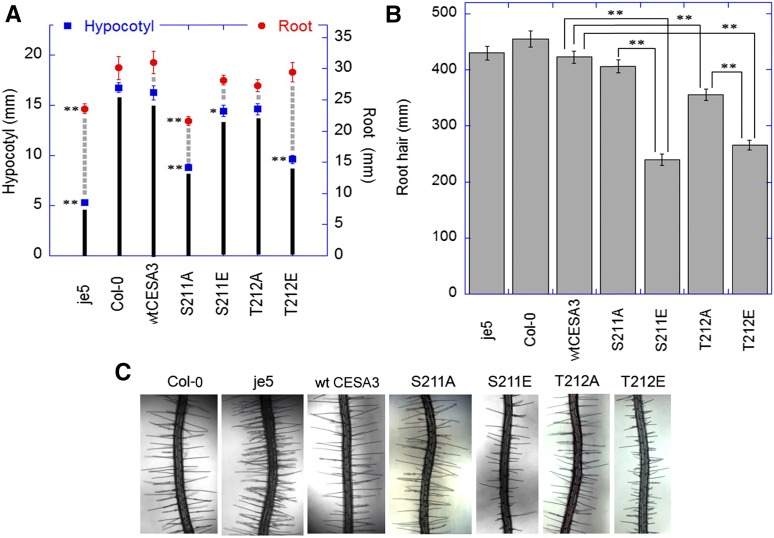
Primary roots and etiolated hypocotyls were measured to examine effects of mutations of CESA3 S211 and T212 on diffuse growth. Expression of wild-type CESA3 complemented the cesa3-1 mutation, and the elongation of primary roots and etiolated hypocotyls was restored to levels similar to those seen with ecotype Columbia (Col-0; Fig. 1A). The CESA3 gene with an Ala substitution at S211 only partially complemented cesa3-1 and led to a 28% decrease in length of primary roots and a 47% decrease in length of etiolated hypocotyls as compared with plants that expressed wild-type CESA3. Substitution of S211 with Glu had no significant effects on root elongation and only slightly reduced hypocotyl elongation (Fig. 1A). The relatively neutral effects of the S211E mutation on growth, compared with the negative effects of S211A, suggest that phosphorylation of the S211 residue results in a more active cellulose synthase. However, as noted later, S211A leads to more rapid movement of cellulose synthase complexes and a significant asymmetry in rates of movement, suggesting that rate of movement is not necessarily a reliable indicator of the quantity or quality of cellulose deposition.
Figure 1.
Lengths of primary roots and hypocotyls (A) and root hairs (B) and images of root hairs (C) of Col-0, je5, and cesa3-1 transgenic lines containing wild-type CESA3 cDNA or cesA3P mutants. A, For root measurements, seedlings were grown in continuous light on 0.5× MS plates containing 1% Suc at 22°C for 7 d. For hypocotyl measurements, seedlings were grown on 0.5× MS plates containing 1% Suc at 22°C for 5 d in darkness. *P < 0.01, **P < 0.001. Student’s t test (means ± ses, n = 20-30 seedlings). B-C, For root hair measurements, seedlings were grown in continuous light on 0.5× MS plates containing 1% Suc at 22°C for 4 d. **P < 0.001. Student’s t test (means ± ses, n = 50-100 epidermal cells from 5-10 seedlings).
By contrast to S211A, the T212A mutation did not have significant effects on the growth of etiolated hypocotyls and primary roots (Fig. 1A). However, the T212E mutation caused a 43% decrease of etiolated hypocotyl lengths, while its root lengths were not significantly altered compared with plants that expressed wild-type CESA3 (Fig. 1A). Thus, phosphorylation of T212 causes an opposite effect on the activity of CESA complexes to that of S211 in some tissues.
Effects on Tip-Growth of Root Hairs
Effects on root hair elongation were used to examine the role of phosphorylation of S211 and T212 on tip growth. While transgenic lines expressing the S211A mutation showed reduced primary root lengths (Fig. 1A), its root hair length was comparable to the wild-type control (Fig. 1B). Similarly, normal root hair elongation was observed in the CESA3 mutant je5 (Fig. 1B), while it showed reduced root lengths (Fig. 1A). By contrast, root lengths of S211E and T212E were comparable to the wild-type control (Fig. 1A), but their root hair lengths were significantly reduced (Fig. 1B). The results indicate that phosphorylation of S211 and T212 inhibits root hair elongation and that the phosphorylation events have selective effects on the growth of hypocotyls, roots, and root hairs.
Effects on Anisotropic Cell Expansion
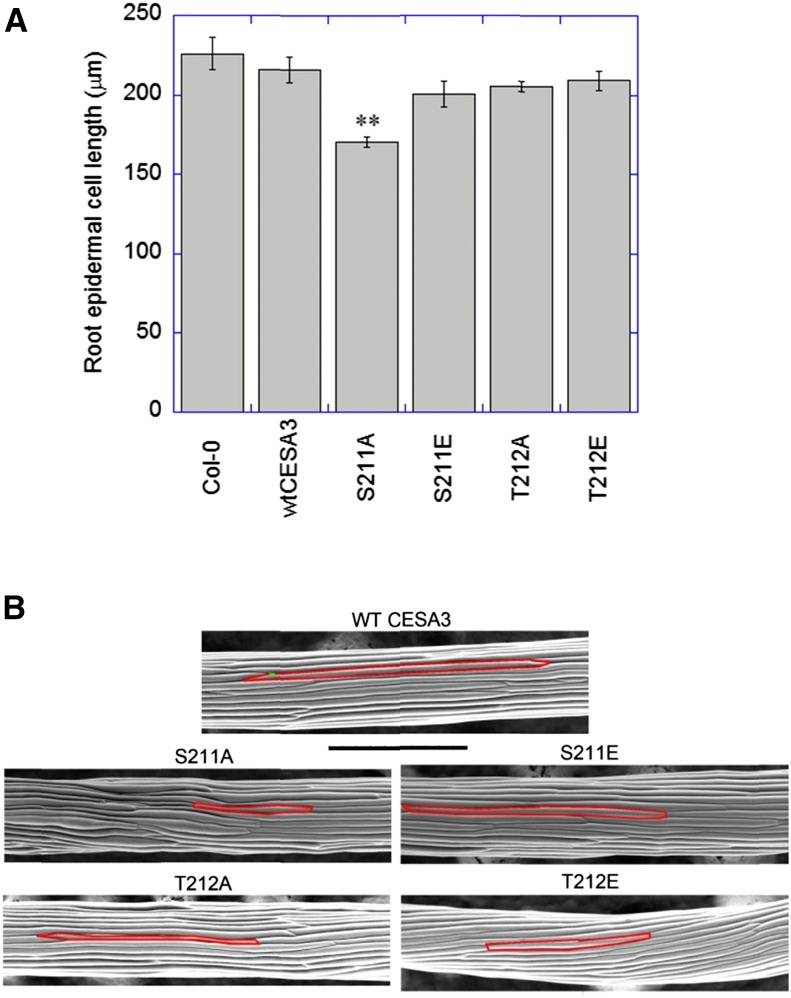
The epidermal cell lengths of primary roots of the transgenic lines expressing wild-type CESA3 were comparable to Col-0, i.e. 215 and 227 μm, respectively (Fig. 2A). The transgenic line expressing the S211A mutation that displayed reduced root lengths also exhibited reduced epidermal cell lengths, i.e. 170 μm (Fig. 2A). By contrast, the transgenic lines with relatively long roots, such as S211E, T212A, and T212E, had relatively long epidermal cells (Fig. 2A). Analysis of epidermal cell lengths of etiolated hypocotyls also demonstrated that the transgenic lines with reduced hypocotyl lengths were accompanied by reduced epidermal cell lengths, while the transgenic lines with normal hypocotyl elongation had relatively long cells (Fig. 2B). These results suggest that phosphorylation of S211 and T212 of CESA3 could lead to either promotion or inhibition of anisotropic cell expansion in rapidly expanding tissues.
Figure 2.
Root epidermal cell lengths (A) and scanning electron microscope images of etiolated hypocotyls (B). A, Seedlings grown for 7 d on 0.5× MS plates were stained with 1 μg/mL of propidium iodide in water for 4 min. Images of epidermal cells, located in the approximate middle of roots, were captured by a spinning-disk confocal microscope. **P < 0.001. Student’s t test (means ± ses, n = 60-90 cells from 4 to 6 different roots). B, Images of 5-d-old hypocotyls were obtained by using an environmental scanning electron microscope. Bar = 500 μm.
Effects on Cell Wall Composition
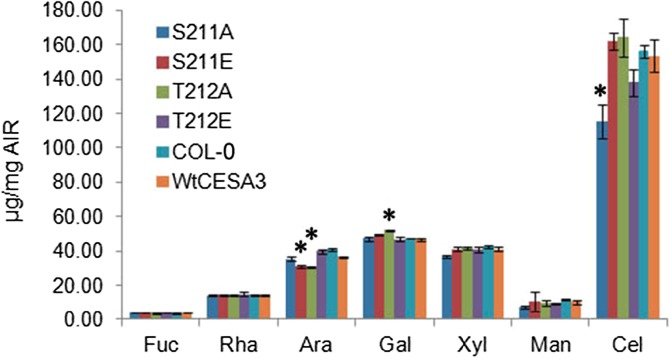
Measurements of monosaccharide composition showed that the transgenic line expressing S211A, which exhibited abnormal organ and cell expansion, had slight but significant reductions of crystalline cellulose in etiolated hypocotyls as compared with wild-type controls (Fig. 3). However, cellulose contents of T212E were not significantly reduced as compared with the wild-type controls, and expression of the S211A and T212E mutations did not cause any significant change in the contents of other sugars (Fig. 3). By contrast, arabinose contents of the mutant lines of S211E and T212A were significantly reduced, and Gal contents of T212A were significantly increased, while these mutations did not cause changes in cellulose contents (Fig. 3). These effects suggest the existence of regulatory mechanisms that adjust cell wall composition in response to as-yet-unidentified aspects of the process of cellulose deposition, or cellulose amount, or quality.
Figure 3.
Monosaccharide analysis of hypocotyls. Seedlings were grown on 0.5× MS plates containing 1% Suc at 22°C for 5 d in darkness. Cellulose content was calculated by subtracting the average amount of Glc released by 4% sulfuric acid hydrolysis from the average amount of Glc released by 72% sulfuric acid treatment. Contents of neutral sugars were measured as the amount of sugars released by 4% sulfuric acid hydrolysis. *P < 0.01, Tukey HSD test, compared to Col-0 and wild-type CESA3 (means ± sds, n = 3 biological reps).
Effects on Bidirectional Mobility of CESA Complexes
To examine directly the effects of the various mutations on cellulose synthase activity, we visualized CESA complexes in epidermal cells of dark-grown hypocotyls using a functional YFP::CESA6 fusion expressed from the native CESA6 promoter (Paredez et al., 2006). Transgenic lines were created that contained both YFP::CESA6 and CESA3 transgenes in a background that was homozygous for both prc1 (Fagard et al., 2000) and cesa3-1 (Persson et al., 2007). These lines were used to examine effects of CESA3 mutations on bidirectional mobility of CESA complexes and interactions of CESA complexes with cortical microtubules.
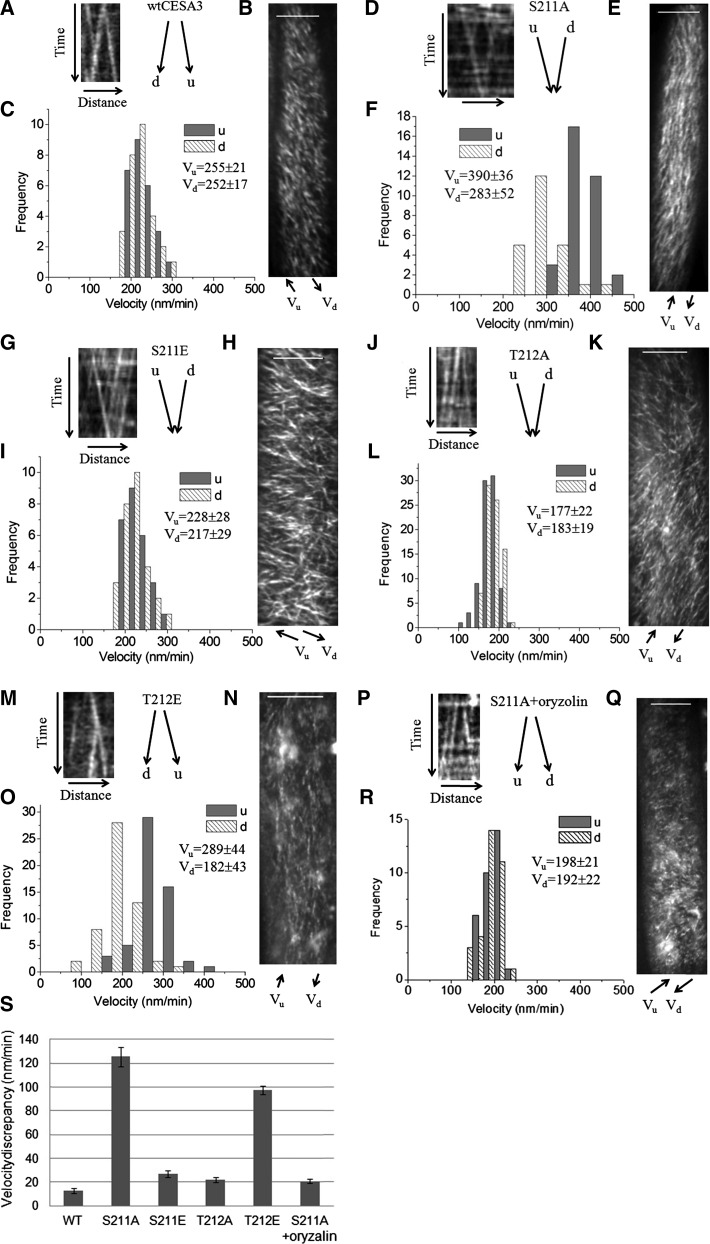
Live cell imaging showed that the transgenic lines of S211A and T212E showed a marked partitioning of particle velocities moving in opposite directions by increasing and decreasing the velocity toward one direction over the other, respectively, as reflected by bidirectional velocities derived from kymograph analysis of CESA complexes moving along individual common linear tracks (Fig. 4F, O, S; Supplemental Table S1). By contrast, the transgenic lines of S211E and T212A that displayed patterns of organ and cell growth similar to wild type showed similar bidirectional velocities along microtubule tracks by kymograph analysis (Fig. 4C, I, L, S; Supplemental Table S1). While bidirectional velocities of particles were significantly reduced by T212A compared with wild-type CESA3, the transgenic line of T212A showed normal organ and cell growth, which was also observed with the S167A mutant line of CESA1, indicating that reduced bidirectional velocities does not necessarily result in defects in anisotropic cell expansion.
Figure 4.
Kymograph analysis of bidirectional movement of CESA complexes in the wild type, S211A, S211E, T212A, and T212E transgenic lines. Time-lapse confocal images of YFP::CESA6 in hypocotyl cells of etiolated seedlings grown at 22°C for 3 d were used to measure CESA complex velocity at room temperature. Oryzalin treatment was conducted for 2 h with 20 μm of oryzalin. To analyze bidirectional movement of CESA complexes, we defined the average axis of CESA complexes movement in a given image series of a cell as the major axis and classified CESA particles as moving in either upward (u) or downward (d) direction along the major axis. A, D, G, J, M, and P, Representative kymograph displays effects of mutations on bidirectional particle translocations, derived from kymograph analysis of CESA particles moving along common linear tracks. B, E, H, K, N, and Q, Average time projections of 61 frames representing 5 min. C, F, I, L, O, and R, Histograms of particle velocities calculated from kymograph analysis of 50 to 200 particles randomly selected to represent particles from a cell. Similar results were obtained from different cells in different seedlings and their mean values were presented in (S) and Supplemental Table S1. (S) Mean values of velocity discrepancy were calculated from three to five different cells in three to five different seedlings. For each cell, 25 to 100 pairs of CESA particles moving in opposite directions along common tracks in the plasma were used to calculate the velocity discrepancy. Bar = 10 μm in B, E, H, K, N, and Q.
Oryzalin, a drug that disrupts microtubules, was applied to examine whether the observed discrepancy of bidirectional movement of CESA complexes depends upon interactions between CESA complexes and microtubules. Kymograph analysis of CESA particle movement in an S211A line indicated that, after oryzalin treatment, CESA particle velocities no longer defined a bimodal population (Fig. 4F, R, S).
All together, the results suggest that tissue or cell morphology observed with the S211A and T212E mutant lines of CESA3 correlates with bidirectional mobility of CESA particles in the plasma membrane, particularly with the velocity discrepancy of bidirectional movement of CESA complexes, and the discrepancy relates to interactions of CESA complexes with cortical microtubules, as previously observed with some mutants of CESA1 (Supplemental Table S1; Chen et al., 2010).
Effects on Cellulose Aggregation
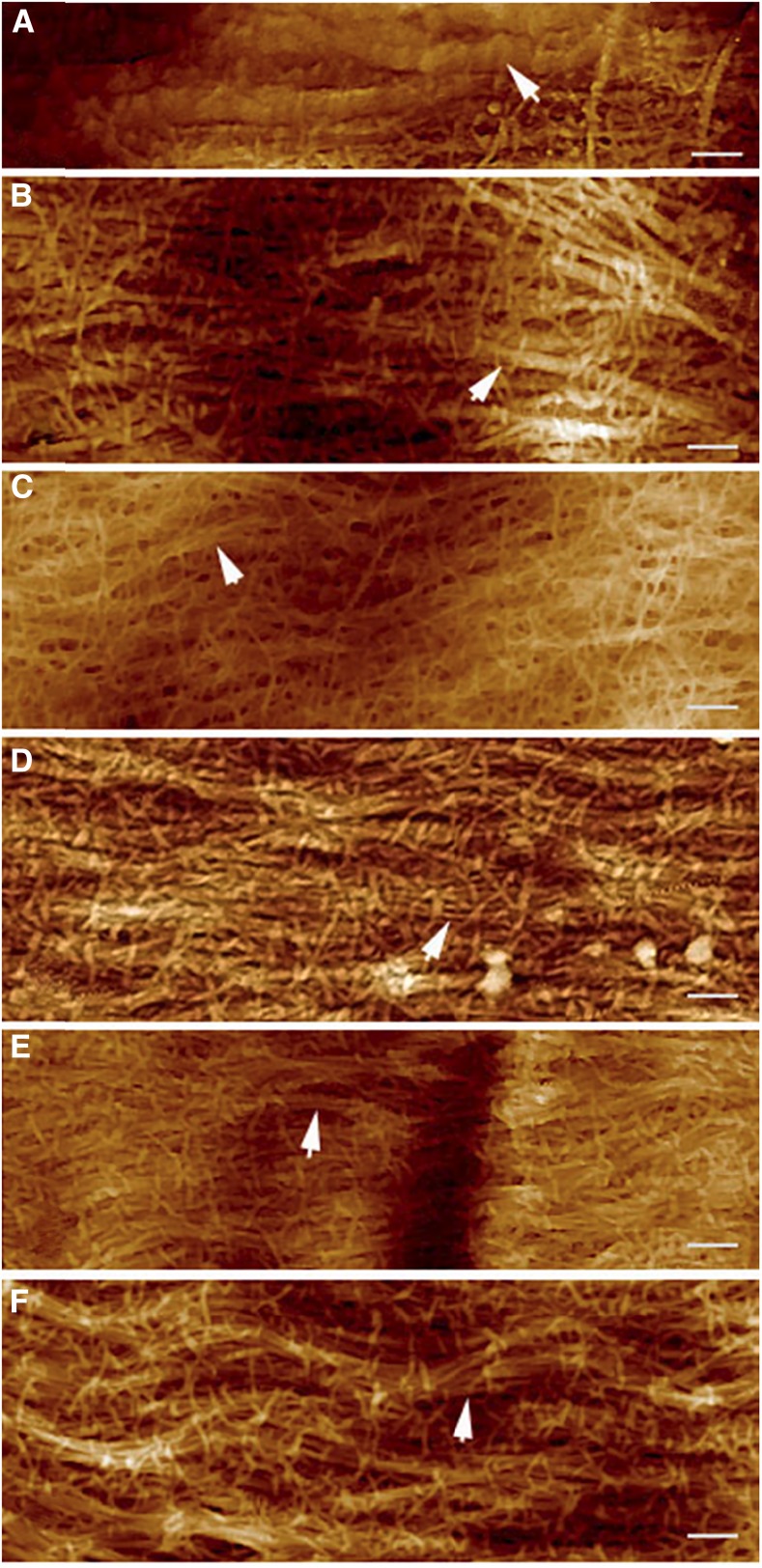
Aggregation of cellulose microfibrils to macrofibrils has been observed in the primary cell wall of maize by atomic force microscopy (Ding et al., 2014). Real-time imaging of cellulose using a fluorescence dye S4B has also shown that cellulose exists in large fibrillar bundles in the primary cell wall of Arabidopsis (Anderson et al., 2010). Thus, it has been suggested that bundles of aggregated cellulose microfibrils, not single microfibrils, might be the key structural units in primary cell walls (Thomas et al., 2013). In this study, atomic force microscopy was applied to image the primary cell wall from the upper hypocotyls of dark-grown seedlings. It was confirmed by this study that cellulose existed mainly in the form of fibrillar bundles in the cell wall of expanding hypocotyl cells (Anderson et al., 2010; Ding et al., 2014), and the diameter of the fibrillar bundles was in the range of 20 to 150 nm (Fig. 5, B-F), comparable to the size observed in the primary cell wall from maize (Ding and Himmel, 2006). Mutations of CESA3 S211A and T212E appeared to slightly affect the morphology of cell wall structure (Fig. 5, C and F), particularly the formation of fibrillar bundles, in comparison to S211E, T212A, and the wild-type control (Fig. 5B, D, and E). Fibrillar bundles of the S211A transgenic line appeared to be thinner and to be heavily cross-bridged. The T212E mutation appeared to alter the alignment of fibrillar bundles. However, we did not obtain useful quantitative measurements of the structural features on independent samples. Thus, the images are useful only inasmuch as they indicate that the mutations do not cause major obvious disruptions of cellulose structure or cross-linking.
Figure 5.
AFM height images of innermost layer of primary cell walls from hypocotyl cells just below the hook of etiolated seedlings. A, Microfibrils embedded in the matrixes of noncellulosic polymers (white arrow) with the cell wall from the wild-type CESA3 transgenic line. B, Fibrillar bundles (white arrow) observed with the cell wall from the wild-type CESA3 transgenic line. C, Altered morphology of fibrillar bundles (white arrow) of the S211A transgenic line; fibrillar bundles appeared to be thinner and to be heavily cross-bridged. D and E, Fibrillar bundles (white arrow) observed with S211E and T212A transgenic lines, respectively. F, Altered alignment of fibrillar bundles (white arrow) from the T212E transgenic line. Bar = 200 nm.
DISCUSSION
Tissue-Specific Regulation of Anisotropic Cell Expansion by Phosphorylation of S211 and T212 of CESA3
To examine the functions of phosphorylation of S211 and T212 of CESA3, these residues were mutagenized to either Ala, to prevent phosphorylation, or Glu, to mimic phosphorylation. The resulting CESA3 mutants were introduced into the null cesa3-1 mutant for further characterization. Our results indicate that altering these residues has a significant effect on the growth of etiolated hypocotyls, but a minor effect on the elongation of primary roots. The most pronounced phenotype was caused by the T212E mutation, which caused a 43% decrease in etiolated hypocotyl lengths, while its root length was not significantly altered compared with wild-type CESA3 (Fig. 1A). The apparent tissue specificity of the effects of the T212E mutation cannot be explained with currently available information. Perhaps the simplest hypothetical explanation is that the mechanism that interprets the phosphorylation status of T212 in hypocotyls is not used in root elongation. One possibility is that CESA complexes in hypocotyls are different from that in primary roots, which causes a differential response of cellulose synthase complexes to phosphorylation status of T212. Whatever the case, the phenomenon suggests the existence of cell- or organ-specific regulation of cellulose synthase.
While rapidly growing tissues, such as etiolated hypocotyls and roots, exhibit diffuse growth, root hairs elongate through tip growth. Treatment of root hairs with DCB, a cellulose synthesis inhibitor, causes growing root hairs to burst rather than expand, demonstrating the involvement of cellulose synthesis (Favery et al., 2001; DeBolt et al., 2007). The mutants of major primary-wall CESAs, including CESA3, display root hair developmental defects to varying degrees, but tip growth in these mutants was not abolished (Gu and Nielsen, 2013). Thus, it is not clear how CESA proteins are involved in the cellulose synthesis for root hair growth. Mutants of CESA3 phosphorylated residues, such as S211A, showed reduced primary root lengths, but its root hairs were relatively normal compared to wild-type controls (Fig. 1B). By contrast, the transgenic line of S211E showed relatively long primary roots, but its root hairs were relatively short (Fig. 1B), supporting the idea that mechanisms underlying cellulose synthesis for tip growth of root hairs are different from that for diffuse growth of primary roots (Park et al., 2011).
Cellulose Aggregation to Fibrillar Bundles
While root hair elongation results from tip growth, diffuse growth is achieved by anisotropic expansion of all the cell surfaces (Baskin, 2005). A typical model for diffuse growth is that while turgor pressure is isotropic, cellulose microfibrils around the cell may limit expansion to certain regions of a cell. Expansion of the cell wall then requires either widening of the spacing between microfibrils (Marga et al., 2005) or slippage between them (Cosgrove, 2005), or both. Polymer cross-bridges between microfibrils are thought to resist these deformations of the cell wall nanostructure and, thus, to control the rate of growth (Park and Cosgrove, 2015).
In this study, we confirmed the previous finding that cellulose exists in large fibrillar bundles in the primary cell walls (Fig. 5; Anderson et al., 2010; Ding et al., 2014), which might be an important mechanism for the regulation of cellulose-cellulose (Thomas et al., 2013), cellulose-pectin (Wang et al., 2015), and cellulose-xyloglucan (Park and Cosgrove, 2015) interactions. It has been observed that <8% of the cellulose surface is available for cross-bridges with noncellulosic polymers (Bootten et al., 2004; Dick-Pérez et al., 2011). Cellulose aggregation to fibrillar bundles could partially contribute to the reduced cellulose surface, as direct contact of microfibrils in the bundles as well as that between bundles might restrict the availability of cellulose surface for polymer cross-bridges (Thomas et al., 2013). As CESA particles move bidirectionally along microtubule tracks (Paredez et al., 2006; Chen et al., 2010), the observed aggregation of cellulose microfibrils could result from simultaneous synthesis and deposition of cellulose microfibrils from multiple CESA particles moving in opposite directions along microtubule tracks (Paredez et al., 2006). A recent live cell imaging study revealed that CESA complexes for the synthesis of secondary wall cellulose also move bidirectionally along microtubule tracks (Watanabe et al., 2015), indicating a common mechanism for the regulation of mechanical properties of plant cell wall.
Bidirectional Deposition of Cellulose Microfibrils and Its Potential Effects on Anisotropic Cell Expansion
It has been shown that microtubules that lay adjacent to the plasma membrane are bundled into arrays, while randomly aligned and discordant microtubules lay deeper in the cytoplasm (Barton et al., 2008). Organization of cortical microtubule arrays establishes polarity in dark-grown hypocotyl cells of Arabidopsis (Dixit et al., 2006). Common polarities were also observed in light-grown hypocotyls cells, but in groups, generating a mosaic of domains instead (Chan et al., 2007). Our studies suggest that CESA complexes can read microtubule polarity, leading to the asymmetry of bidirectional mobility of CESA particles along microtubule tracks, which can be removed upon depolymerizing microtubules with oryzalin (Fig. 4; Chen et al., 2010). It is noted that the asymmetry of bidirectional mobility of CESA complexes closely correlates with abnormal tissue and cell expansion (Supplemental Table S1). Plants carrying the S211A mutation of CESA3 showed not only asymmetrical bidirectional mobility of CESA particles but also reduced cellulose content (Fig. 3), as well as radial swelling and reduced elongation of rapidly expanding cells (Figs. 1 and 2). However, cellulose content in plants carrying the T212E mutation of CESA3 was comparable to the wild-type control (Fig. 3), and epidermal cells of etiolated hypocotyls were not obviously swollen while their lengths were significantly reduced (Figs. 1 and 2), indicating that reduced cellulose content is not solely responsible for the observed defects in cell elongation.
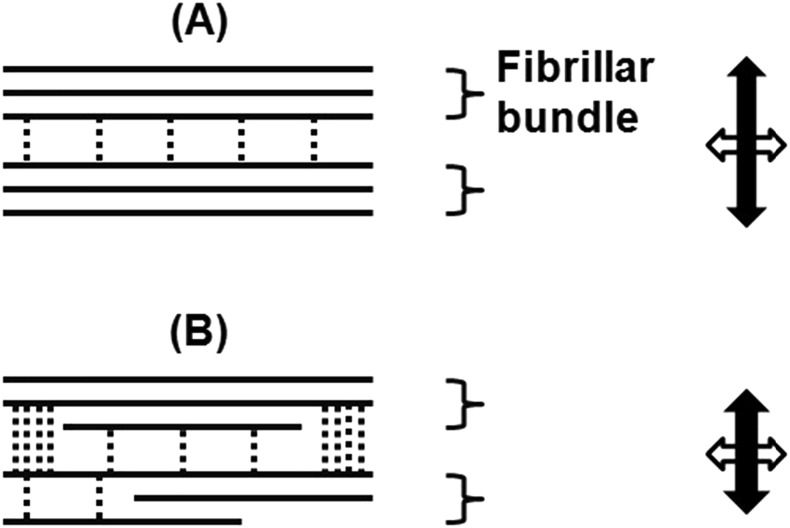
It is interesting but challenging to examine if bidirectional reading of microtubule polarity by CESA complexes affects the lifetime of the CESA complexes in the plasma membrane and, thus, microfibril synthesis and deposition. If a mismatched length of oppositely oriented microfibrils results from a bidirectional velocity discrepancy of CESA particles, it might increase the level of disorder at the interfaces between microfibrils or macrofibrils, modulating microfibril spacing, packing and alignment (Fig. 6; Park et al., 2013; Lee et al., 2014). This is consistent with the evidence that fibrillar bundles or macrofibrils in the primary cell wall are predominantly ribbon like, randomly arranged, and sometime interwoven (Fig. 5; Ding et al., 2014), and that some microfibril segments remain in direct contact with one another (Thomas et al., 2013). Thus, modulating microfibril spacing, packing, and alignment would affect not only cellulose-cellulose but also cellulose-pectin and cellulose-xyloglucan interactions (Figs. 5 and 6; Park and Cosgrove, 2015; Wang et al., 2015). Regulation of bidirectional mobility of cellulose synthase complexes by phosphorylation of CESA1 and CESA3 may provide a mechanism for spatial and temporal control of cell wall expansion.
Figure 6.
Model for regulation of anisotropic cell wall expansion by bidirectional deposition of cellulose. A, Bidirectional synthesis and deposition of microfibrils results in cellulose aggregation to fibrillar bundles or macrofibrils (Anderson et al., 2010), which are predominantly ribbon like (Ding et al., 2014), and some microfibril segments remain in direct contact with one another (Thomas et al., 2013). B, In the case of mismatched lengths of oppositely oriented microfibrils, derived from discrepancy of bidirectional mobility of cellulose synthase complexes, the spaces between one microfibril or macrofibril and the next may increase, allowing more noncellulosic polymers (dashed lines) to cross-bridge adjacent microfibrils or macrofibrils and thus retard cell elongation. It is noted that in addition to cellulose-cellulose interactions and cross-bridges between cellulose and noncellulosic polymers, other properties, such as cellulose crystallite structure as well as the packing, twisting, and ordering of microfibrils or macrofibrils, may all contribute to the overall mechanical properties of the walls (Fujita et al., 2011; Park et al., 2013).
In summary, our results suggest some aspects of the mechanisms by which phosphorylation of CESA proteins, particularly CESA1 and CESA3, plays a key role in regulating anisotropic cell expansion.
MATERIALS AND METHODS
Construction of cesA3P Mutants
pCesA3-gateway, a Ti plasmid containing the CESA3 promoter flanking recombination ATT sites, and a donor vector containing a CESA3 cDNA, were constructed and used for site-directed mutagenesis as described in a previous study (Chen et al., 2010). Sequence-verified constructs were introduced into heterozygous plants of the Arabidopsis (Arabidopsis thaliana) Col-0 cesA3 mutant (cesa3-1) by Agrobacterium-mediated transformation, then homozygous cesa3-1 mutants carrying complementing T-DNA inserts containing various mutant alleles of CESA3 were used for experiments. All lines used for phenotypic measurements were also homozygous for the T-DNA insert.
Tissue and Cell Length Measurements
Standard methods were used for measurements of tissue and cell size as described in a previous study (Chen et al., 2010). In brief, wild-type and mutant seedlings were grown on the same plates for direct comparison. Images of seedlings grown on vertically placed Murashige and Skoog (MS) plates were captured by a scanner, and the image files were analyzed by ImageJ to measure the lengths of primary roots or dark-grown hypocotyls. For measurements of root epidermal cell length, 7-d-old light-grown seedlings were stained with 1 μg/mL of propidium iodide in water for 4 min, followed by several rinses in 1× phosphate-buffered saline solution. Images of epidermal cells, located in the approximate middle of roots, were captured by a spinning-disk confocal microscope. Images of hypocotyl epidermal cells were obtained by using a Quanta 200 environmental scanning electron microscope (FEI) under a pressure of 130 Pa and a voltage of 12.5 kV with a low-vacuum detector.
Monosaccharide Analysis
Monosaccharide analysis was performed as described previously (Bauer and Ibáñez, 2014). In brief, seedlings were grown for 5 d in dark in MS liquid medium (pH 5.8) supplemented with 1% Suc. Seedling tissues were ground in liquid nitrogen, followed by one extraction with 70% ethanol and three extractions with chloroform:methanol (1:1, v/v). From each sample, two 1-mg aliquots of alcohol-insoluble residue were prepared. The first aliquot was treated with 50 µL of 72% sulfuric acid at room temperature for 1 h. Sulfuric acid was then diluted to 4% and the mixture was heated at 121°C for 1 h. The second aliquot was treated with 1.45 mL of 4% sulfuric acid at 121°C for 1 h. Released monosaccharides were analyzed using an ICS-3000 HPLC system (Thermo Fisher Scientific) equipped with a pulsed-amperometric detector. Cellulose content was calculated by subtracting the average amount of Glc released by 4% sulfuric acid hydrolysis from the average amount of Glc released by 72% sulfuric acid treatment. Contents of neutral sugars were calculated by averaging the amount of sugars released by 4% sulfuric acid hydrolysis.
Atomic Force Microscopy
Hypocotyls of 3-d-old dark-grown seedlings were bisected longitudinally and incubated in 2 m KOH at room temperature for 1 h and then in 1% Tween 20 for 30 min. After soaking, the samples were gently rinsed several times with double distilled water until the pH reached neutral. These slices were placed between glass slides under a load of 5 g for 5 min. The innermost layer of hypocotyl cells just below the hook of etiolated seedlings was examined by a multimode scanning probe atomic force microscopy with a NanoScope V controller. Contact AFM was done in air at room temperature using an SCANSYST-AIR probe with ScanAsyst imaging mode. All scans were recorded as 510- × 510-pixel images. At least three images with different scan size (i.e. 1, 2, and 5 μm) were taken in the same scan area and at least three to five different areas of the same cell were measured. The software NanoScope Analysis v1.10 was used for AFM operation and for further image processing.
Measurements of CESA Particle Velocity
Seeds were germinated on 0.5× MS agar and grown vertically in the darkness for 3 d at 22°C. Seedlings were manipulated under a red safety light to avoid the blue light response known to inhibit growth (Folta et al., 2003). Seedlings were mounted between two cover slips in water with their apexes pointing to the back of the microscope. Imaging of YFP::CESA6 particles was performed in hypocotyl cells just below the hook of etiolated seedlings. YFP was excited at 488 nm, and fluorescence was collected through a 525/50-nm band-pass filter (Chroma Technologies, Brattleboro, VT). All image processing was performed by using Metamorph (Molecular Dynamics, Sunnyvale, CA) and ImageJ (W. Rasband, National Institutes of Health, Bethesda, MD). The obtained sequential frames were averaged by using the Walking Average plug-in (three-frame window). To correct for tissue movement during imaging, subpixel frame alignment was performed using a nonlinear least squares algorithm (StackReg plug-in for ImageJ; Thévenaz et al., 1998). Since frame alignment can fail, fiducial markers in each data set were analyzed for drift, and only those data sets with <135 nm of drift over 5 min were kept for analysis. The fiducial markers used were intrinsic autofluorescent spots on cell walls.
Accession Numbers
Sequence data from this article can be found in TAIR (www.arabidopsis.org) under the following accession numbers: CESA3 (At5g05170), CESA1 (At4g32410), CESA2 (At4g39350), CESA5 (At5g09870), CESA6 (At5g64740), CESA9 (At2g21770), CC1 (At1g45688), CC2 (At5g42860), and CSI (AT2G22125).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Measurements of the rate of bidirectional movement of CESA complexes in the various cesA1P and cesA3P mutants.
Acknowledgments
We thank Jie Zhu, Ying Wang, Xuan Song, Wenting Wang, and Guixia Li for technical assistance on atomic force microscopy.
Glossary
- Col-0
ecotype Columbia
- MS
Murashige and Skoog
Footnotes
This work was supported by grants from the Energy Biosciences Institute and by the USDA National Institute of Food and Agriculture, Hatch project CA-B-PLB-0077-H and the Start-up Research Grant from the Northwest A&F University.
Articles can be viewed without a subscription.
References
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DA, Vantard M, Overall RL (2008) Analysis of cortical arrays from Tradescantia virginiana at high resolution reveals discrete microtubule subpopulations and demonstrates that confocal images of arrays can be misleading. Plant Cell 20: 982–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Bauer S, Ibáñez AB (2014) Rapid determination of cellulose. Biotechnol Bioeng 111: 2355–2357 [DOI] [PubMed] [Google Scholar]
- Bischoff V, Desprez T, Mouille G, Vernhettes S, Gonneau M, Höfte H (2011) Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr Biol 21: 1822–1827 [DOI] [PubMed] [Google Scholar]
- Bootten TJ, Harris PJ, Melton LD, Newman RH (2004) Solid-state 13C-NMR spectroscopy shows that the xyloglucans in the primary cell walls of mung bean (Vigna radiata L.) occur in different domains: a new model for xyloglucan-cellulose interactions in the cell wall. J Exp Bot 55: 571–583 [DOI] [PubMed] [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C (2007) Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat Cell Biol 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D (2007) Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA 104: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Pérez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M (2011) Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000 [DOI] [PubMed] [Google Scholar]
- Ding SY, Himmel ME (2006) The maize primary cell wall microfibril: a new model derived from direct visualization. J Agric Food Chem 54: 597–606 [DOI] [PubMed] [Google Scholar]
- Ding SY, Zhao S, Zeng YN (2014) Size, shape, and arrangement of native cellulose fibrils in maize cell walls. Cellulose 21: 863–871 [Google Scholar]
- Dixit R, Chang E, Cyr R (2006) Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol Biol Cell 17: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S (2015) A mechanism for sustained cellulose synthesis during salt stress. Cell 162: 1353–1364 [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36: 203–214 [DOI] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Hocart CH, Williamson RE, Mansfield SD, Wasteneys GO (2011) Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J 66: 915–928 [DOI] [PubMed] [Google Scholar]
- Gu F, Nielsen E (2013) Targeting and regulation of cell wall synthesis during tip growth in plants. J Integr Plant Biol 55: 835–846 [DOI] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Kafle K, Park YB, Kim SH (2014) Probing crystal structure and mesoscale assembly of cellulose microfibrils in plant cell walls, tunicate tests, and bacterial films using vibrational sum frequency generation (SFG) spectroscopy. Phys Chem Chem Phys 16: 10844–10853 [DOI] [PubMed] [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TI (2005) Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J 43: 181–190 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E (2011) A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Park YB, Lee CM, Koo BW, Park S, Cosgrove DJ, Kim SH (2013) Monitoring meso-scale ordering of cellulose in intact plant cell walls using sum frequency generation spectroscopy. Plant Physiol 163: 907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, Mechtler K (2015) Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics 14: 556–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM Jr (2005) Cellulose biosynthesis: current views and evolving concepts. Ann Bot (Lond) 96: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7: 27–41 [DOI] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Park YB, Cosgrove DJ, Hong M (2015) Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol 168: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell LM, Barkwill S, Sampathkumar A, Cartwright HN, Demura T, Ehrhardt DW, Samuels AL, Mansfield SD (2015) Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203 [DOI] [PubMed] [Google Scholar]