Xylan biosynthesis in the Golgi apparatus is affected by IRX9, IRX10, and IRX14A in Asparagus.
Abstract
Heteroxylans are abundant components of plant cell walls and provide important raw materials for the food, pharmaceutical, and biofuel industries. A number of studies in Arabidopsis (Arabidopsis thaliana) have suggested that the IRREGULAR XYLEM9 (IRX9), IRX10, and IRX14 proteins, as well as their homologs, are involved in xylan synthesis via a Golgi-localized complex termed the xylan synthase complex (XSC). However, both the biochemical and cell biological research lags the genetic and molecular evidence. In this study, we characterized garden asparagus (Asparagus officinalis) stem xylan biosynthesis genes (AoIRX9, AoIRX9L, AoIRX10, AoIRX14A, and AoIRX14B) by heterologous expression in Nicotiana benthamiana. We reconstituted and partially purified an active XSC and showed that three proteins, AoIRX9, AoIRX10, and AoIRX14A, are necessary for xylan xylosyltranferase activity in planta. To better understand the XSC structure and its composition, we carried out coimmunoprecipitation and bimolecular fluorescence complementation analysis to show the molecular interactions between these three IRX proteins. Using a site-directed mutagenesis approach, we showed that the DxD motifs of AoIRX10 and AoIRX14A are crucial for the catalytic activity. These data provide, to our knowledge, the first lines of biochemical and cell biological evidence that AoIRX9, AoIRX10, and AoIRX14A are core components of a Golgi-localized XSC, each with distinct roles for effective heteroxylan biosynthesis.
Heteroxylans are abundant noncellulosic matrix polysaccharides of plant cell walls. They are the main component of the primary wall of commelinid monocots (including grasses) and also the secondary wall of most woody dicot species (Faik, 2010; Rennie and Scheller, 2014). Heteroxylans have a linear backbone of β-1,4-linked d-xylopyranosyl residues (Xylp) with backbone decorations that vary between cell types and tissues, developmental stages, and species. The dominant decorations are α-1,2- and/or α-1,3-linked l-arabinofuranosyl residues (Araf) in grass primary walls and α-1,2-linked (methyl)glucuronopyranosyl residues in woody tissues (Bastawde, 1992; Ebringerova et al., 2005). In commelinid monocots, the Araf residues can be further decorated with a ferulic acid residue that, through dimerization, can cross-link heteroxylans into the wall (Ishii 1997). The α-1,2-GlcA residues can be further modified with α-1,2-linked galactopyranosyl residues (Galp) found in heteroxylans of Eucalyptus globulus (Shatalov et al., 1999) and Arabidopsis (Arabidopsis thaliana; Zhong et al., 2014). Recently, an atypical xylan was found with a 1,2-pentosyl residue (not determined if it was Ara or Xyl) linked to the α-1,2-GlcA side chain of the xylan backbone in the primary wall from Arabidopsis (Chong et al., 2015; Mortimer et al., 2015). Additionally, the reducing end (RE) sequence of heteroxylans from dicots and gymnosperms differs from that of grasses. In the dicots/gymnosperms, it is a characteristic tetrasaccharide glycosyl sequence, β-d-Xylp-(1→3)-α-l-Rhap-(1→2)-α-d-GalpA-(1→4)-d-Xylp, that is absent in the grasses (for review, see Hao and Mohnen, 2014). Recently, Ratnayake et al. (2014) reported that the RE glycosyl sequence of wheat (Triticum aestivum) endosperm arabinoxylan constitutes a linear (1→4)-β-d-Xylp backbone that may be monosubstituted either with an α-l-Araf residue at the RE β-d-Xylp residue and/or the penultimate RE β-d-Xyl residue or with an α-d-GlcpA residue at the RE β-d-Xylp residue. These studies, and many others, highlight the diversity of heteroxylan structures and, therefore, the complex nature of heteroxylan biosynthesis.
Most noncellulosic plant cell wall polysaccharides are predicted to be synthesized in the Golgi apparatus (GA) by glycosyltransferase (GT) enzyme complexes (Oikawa et al., 2013), with the exception of cellulose, callose, and (1,3;1,4)-β-glucans, which are assembled at the plasma membrane (Doblin et al., 2010; Wilson et al., 2015). Some examples of Golgi-located polysaccharide biosynthesis complexes include GALACTURONOSYLTRANSFERASE1 (GAUT1)/GAUT7 involved in pectin biosynthesis (Atmodjo et al., 2011), CSLC4/XylT (for xylan xylosyltranferase) involved in xyloglucan biosynthesis (Cocuron et al., 2007; Chou et al., 2012), and ARAD1/ARAD2 involved in arabinan biosynthesis (Harholt et al., 2012). Genetic studies in Arabidopsis have identified several GTs that are predicted to be involved in a xylan backbone biosynthesis complex, namely Arabidopsis IRREGULAR XYLEM9 (AtIRX9), AtIRX10, AtIRX14, AtPARVUS, and AtIRX8, as well as their homologs (Zhong et al., 2005; Brown et al., 2007, 2009; Lee et al., 2007a, 2007b; Peña et al., 2007; Wu et al., 2009, 2010; Keppler and Showalter 2010). Among these, AtIRX9 and AtIRX14 (both GT43 family members) and AtIRX10 (GT47), as well as their functionally redundant homologs, are believed to be involved directly in xylan backbone biosynthesis and to form a complex in the GA (Rennie and Scheller 2014). Orthologs of the AtIRX proteins have been identified in many other species, including wheat (Zeng et al., 2010), Populus trichocarpa (Lee et al., 2012b), Physcomitrella patens (Hörnblad et al., 2013), Plantago ovata (Jensen et al., 2013), rice (Oryza sativa; Chen et al., 2013; Chiniquy et al., 2013), Gossypium hirsutum (Li et al., 2014), Neolamarckia cadamba (Zhao et al., 2014), and garden asparagus (Asparagus officinalis; Song et al., 2015). Enzymes involved in xylan backbone biosynthesis as well as side chain decorations, such as the arabinosyltransferases, glucuronosyltransferases, and acetylation enzymes, were recently reviewed by Rennie and Scheller (2014) and Hao and Mohnen (2014).
Several biochemical studies have contributed toward unraveling the core partners involved in xylan biosynthesis, but the issue is complex and the full suite of proteins has not yet been elucidated (Rennie and Scheller, 2014). Nevertheless, some progress has been made. Microsomes extracted from tobacco (Nicotiana tabacum) BY2 cells coexpressing AtIRX9 and AtIRX14 have been shown to have xylan synthase activity in vitro (Lee et al., 2012a). Furthermore, two independent studies have demonstrated using a heterologous expression approach that AtIRX10L (Urbanowicz et al., 2014) and IRX10 from P. ovata and P. patens (Jensen et al., 2014) also have distributive xylan XylT activity. Interestingly, Ren et al. (2014), using site-directed mutagenesis (SDM) and genetic approaches, showed that AtIRX9 is not involved directly in catalytic activity, because mutant variants of potential catalytic domains of AtIRX9 were still able to complement Arabidopsis irx9 mutants. Taken together, these observations are consistent with the hypothesis that AtIRX9, AtIRX10, and AtIRX14 form a xylan synthase complex (XSC; Rennie and Scheller, 2014). However, unequivocal biochemical (or cell biological) proof of the nature of their interaction(s)/stoichiometry in planta is still lacking, predominantly because of the low abundance of these Golgi-localized proteins, which makes the purification and characterization of the enzyme complexes challenging.
In order to define the biochemical activity of these Golgi-localized GTs, different heterologous expression systems are routinely used for functional characterization and, therefore, overcome the inherent difficulties of purifying these low-abundance membrane-bound proteins. Nicotiana benthamiana is widely used as an efficient and high-level expression system for functional characterization of candidate genes (Voinnet et al., 2003). It has been utilized to examine the biochemical activities of several plant cell wall polysaccharide biosynthetic GTs, including xylogalacturonan (pectin) xylosyltransferase (Jensen et al., 2008), xylan glucuronosyltransferase (Rennie et al., 2012), β-(1,4)-galactan synthase (Liwanag et al., 2012), arabinogalactan-protein galactosyltransferase (Geshi et al., 2013), and (1,3;1,4)-β-glucan CSLF glucan synthase (Wilson et al., 2015). Other heterologous expression systems (e.g. either the yeast Pichia pastoris or mammalian cell lines) also have been used to characterize the functions of GTs such as xyloglucan xylosyltransferase (Faik et al., 2002) and pectin homogalacturonan galacturonosyltransferase (Sterling et al., 2006).
We previously demonstrated a high level of xylan XylT activity (around 10-fold higher than any other native in vitro system, including Arabidopsis) in vegetative spears of asparagus, a noncommelinid monocot species, and also identified five putative xylan backbone biosynthesis genes (AoIRX9, AoIRX9L, AoIRX10, AoIRX14A, and AoIRX14B; Song et al., 2015). Furthermore, we also showed that the xylan content, the xylan XylT activity, and the expression of the backbone biosynthesis genes changed along the developmental gradient of the spear as it transitioned from an elongation zone to a maturation zone (Song et al., 2015). To characterize the precise biochemical function(s) of these genes, we now show that three of these proteins (AoIRX9, AoIRX10, and AoIRX14A) must be heterologously coexpressed for xylan XylT activity equivalent to that observed using asparagus microsomal membranes (MMs). We also show that all three proteins are required for GA retention. Using SDM, we demonstrated that the DxD motifs of AoIRX10 and AoIRX14A are essential for catalytic activity. Bimolecular fluorescence complementation (BiFC) analysis indicated that AoIRX14A interacts directly with AoIRX9, while AoIRX10 likely interacts indirectly with AoIRX9 and/or AoIRX14A. These data provide, to our knowledge, the first lines of direct evidence that AoIRX9, AoIRX10, and AoIRX14A are part of a core XSC together with potentially unknown partner(s) in the GA.
RESULTS
Expression of Asparagus Xylan Biosynthesis Genes in N. benthamiana
We previously demonstrated that garden asparagus spears have strikingly high XylT enzyme activity and identified five predicted xylan biosynthesis genes (AoIRX9, AoIRX9L, AoIRX10, AoIRX14A, and AoIRX14B; Song et al., 2015). In this study we sought to test which of these genes are essential for the observed native activity by expressing candidates heterologously in N. benthamiana, since an efficient transformation procedure for asparagus is not available. The full-length open reading frames of AoIRX9, AoIRX9L, AoIRX14A, AoIRX14B, and AoIRX10 were cloned into a binary vector under the control of the strong, constitutively active cauliflower mosaic virus 35S promoter and expressed either singly or in combinations in N. benthamiana leaves.
To verify the expression of these asparagus IRX proteins, we developed antibodies against AoIRX9, AoIRX10, and AoIRX14A for protein detection and pull-down assays. Using western-blot analysis, we demonstrated that the antibodies detect the protein that they were raised against (Fig. 1). The bands detected by the AoIRX10- and AoIRX14A-directed antibodies matched the predicted molecular mass (47.4 and 57.4 kD, respectively). A weak band above 100 kD also was detected with anti-AoIRX14A, possibly indicating the existence of the homodimer (Fig. 1). However, other bands also were visible, suggesting that this antibody is less specific than that of AoIRX10. Interestingly, a band at approximately 55 kD was detected with anti-AoIRX9, which is higher than the predicted molecular mass of 40.8 kD for this protein (Fig. 1), suggesting that there may be posttranslational modifications occurring. The AoIRX9 antibody detected a single band in MMs from N. benthamiana expressing either AoIRX9 or the triple combination of AoIRX9/10/14A, but no signal was detected in MMs expressing AoIRX9L, AoIRX10, and/or AoIRX14A (Fig. 1). Similarly, AoIRX10 and separately AoIRX14A antibodies were only able to detect signal in MMs that expressed their cognate protein (Fig. 1). The expression levels of each AoIRX protein are not significantly affected when coexpressed with other proteins, since the western-blot signal intensities of singly expressed and coexpressed AoIRX proteins are similar (Fig. 1).
Figure 1.
Western-blot analysis of MMs prepared from N. benthamiana leaves infiltrated with asparagus xylan biosynthesis gene constructs. MMs (20 µg of protein) from N. benthamiana leaves infiltrated with either single AoIRX9/10/14 proteins or the triple combination were extracted and analyzed via western blotting using antibodies against AoIRX9 (A), AoIRX10 (B), and AoIRX14A (C). The predicted molecular masses for native AoIRX9, AoIRX10, and AoIRX14A are 40.8, 47.4, and 57.4 kD, respectively.
Xylan Synthase Activity in MMs Prepared from N. benthamiana Leaves Coexpressing Predicted Xylan Biosynthesis Genes from Asparagus
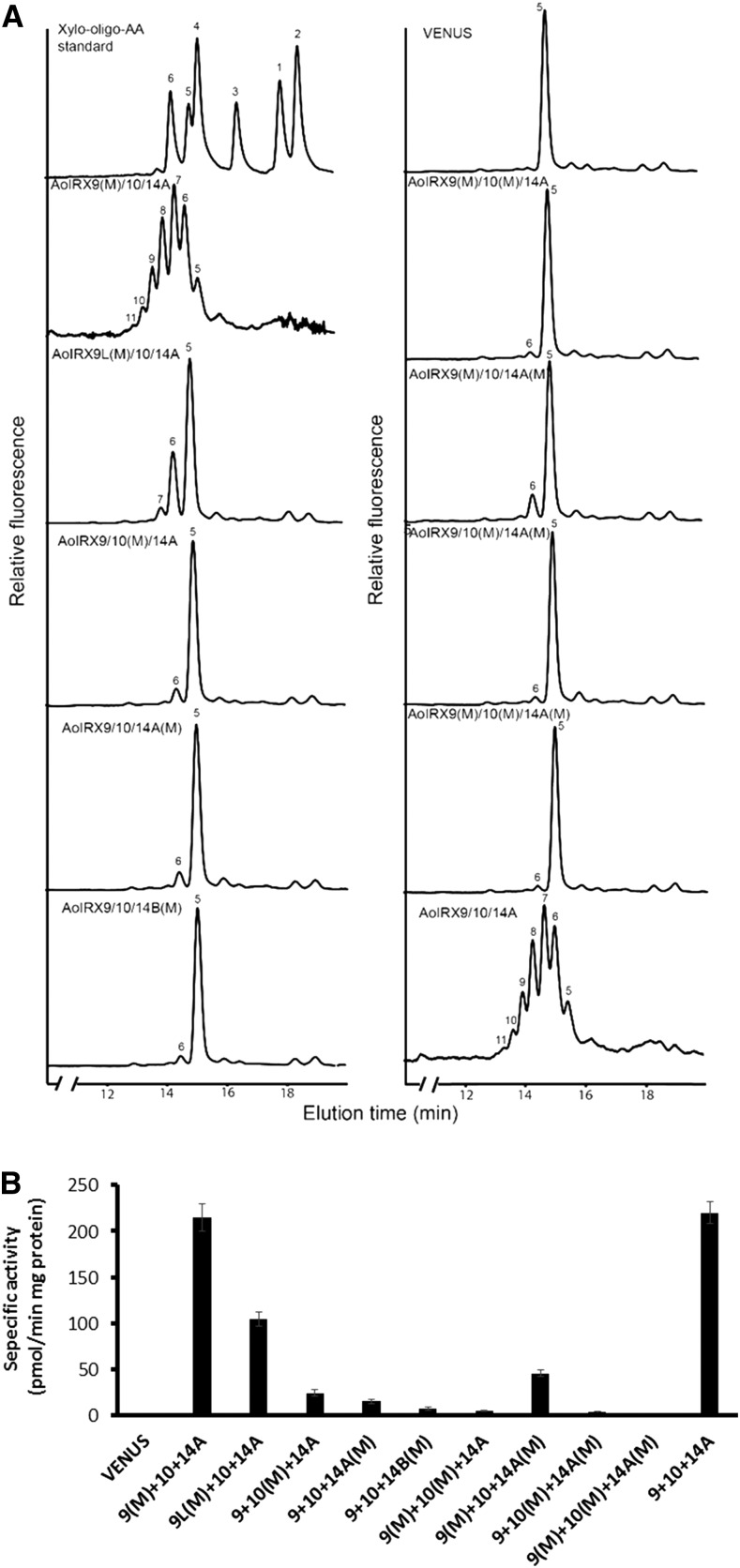
As heteroxylans are synthesized in the GA, xylan XylT activity was assayed using MMs prepared from N. benthamiana leaves expressing either single or combinations of asparagus IRX proteins. Asparagus spear MMs were used as a positive control for the XylT activity. As demonstrated previously by Song et al. (2015), asparagus MMs are capable of incorporating up to six Xyl residues onto the Xyl5-AA fluorescent acceptor within 1 h under standard assay conditions, producing β-1,4-xylooligosaccharides up to a degree of polymerization of 11 (Fig. 2). MMs prepared from N. benthamiana leaves infiltrated with a VENUS control contained only the Xyl5-AA acceptor, indicating that there is no detectable endogenous XylT activity in N. benthamiana leaf MMs under these assay conditions. Individual expression of each of the five AoIRXs showed no significant incorporation of Xyl onto the Xyl5-AA acceptor except for AoIRX10 and AoIRX14A, which produced a small peak of Xyl6-AA (Fig. 2). To quantify the XylT activity, we used UDP-[14C]Xyl as a donor to monitor Xyl incorporation (Fig. 2). Consistent with the RP-HPLC results, single expression of either AoIRX9 or AoIRX9L leads to negligible Xyl incorporation. The expression of AoIRX10, AoIRX14A, or AoIRX14B individually results in a very low level of incorporation, equivalent to less than 7% of the XylT activity of native asparagus MMs.
Figure 2.
Xylan XylT activity of MMs from N. benthamiana leaves expressing combinations of IRX9, IRX10, and IRX14 genes from asparagus. Asparagus xylan synthase proteins (AoIRX9, AoIRX9L, AoIRX10, AoIRX14A, and AoIRX14B) were expressed in N. benthamiana leaves alone and/or in various combinations, and the MMs were isolated and assayed for xylan XylT activity. N. benthamiana leaves expressing VENUS and native asparagus MMs were used as negative and positive controls, respectively. A, XylT products were fractionated by reverse-phase (RP)-HPLC and detected with a fluorescence detector. Xylooligosaccharides labeled with AA (Xyl1–6-AA) were used as standards. The numbers on the plots indicate the degree of polymerization of xylooligosaccharides. B, Xylan XylT was measured using UDP-[14C]Xyl as the donor and Xyl5-AA as the acceptor. Values represent means ± se of three biological replicates.
Previous studies have proposed that AtIRX9, AtIRX10, and AtIRX14 are part of an active xylan XylT complex in the GA for xylan backbone biosynthesis during Arabidopsis secondary wall development (Rennie and Scheller, 2014). To test this hypothesis, we coinfiltrated combinations of two or more asparagus IRX genes and assayed MMs for xylan XylT activity as before. When AoIRX10 was coexpressed with either AoIRX9 or AoIRX14A, two Xyl additions onto the Xyl5-AA acceptor were detected, although the activity was approximately 75% lower than that observed using native asparagus MMs (Fig. 2). In contrast, coexpression of AoIRX9 and AoIRX14A did not show additional Xyl incorporation compared with AoIRX14A alone (Fig. 2). However, when all three genes (AoIRX9, AoIRX10, and AoIRX14A) were coexpressed, the MMs incorporated up to six additional Xyl residues onto the Xyl5-AA acceptor, with a similar degree of polymerization profile and activity level (approximately 97%) to the native asparagus MMs in assays with the same amount of total protein (Fig. 2). This indicates that there is cooperative xylan XylT activity when AoIRX9, AoIRX10, and AoIRX14A are coexpressed and that they likely form an XSC. Interestingly, combinations including AoIRX9/10/14B, AoIRX9L/10/14A, and AoIRX9L/10/14B showed lower activities compared with the AoIRX9/10/14A triple combination, suggesting that AoIRX9L and AoIRX14B are less active compared with AoIRX9 and AoIRX14A, respectively.
Given the increase in xylan XylT activity with coinfiltration of all three asparagus IRX9, IRX10, and IRX14A, we tested if the additional expression of AoIRX9L and AoIRX14B with the triple combination could further boost xylan XylT activity. Interestingly, coexpression of all five genes led to Xyl incorporation up to degree of polymerization 9 and an approximately 24% reduction of xylan XylT activity compared with AoIRX9/10/14A coexpression (Fig. 2). This suggests that AoIRX9 and AoIRX9L, and separately AoIRX14A and AoIRX14B, may be competing with each other in the XSC. MMs from N. benthamiana coexpressing AoIRX9/10/14A and VENUS had a similar XylT activity compared with the AoIRX9/10/14A triple combination, providing further evidence that AoIRX9L and AoIRX14B are competing with AoIRX9 and AoIRX14A, respectively, rather than acting as a dilution factor.
From these data, we concluded that AoIRX9, AoIRX10, and AoIRX14A in combination are part of a core XSC in the N. benthamiana heterologous expression system.
The DxD Motif of AoIRX10 and AoIRX14A Is Important for Catalytic Activity
Most GTs with a GT-A fold have a conserved DxD motif (where x stands for any amino acid except for Pro and x is optional) shown to be the nucleotide sugar (donor) binding site through the coordination of a divalent cation and, hence, essential for catalytic activity (Breton et al., 2006). To test if this motif is critical for catalytic activity in our proteins of interest, we used an SDM approach to alter the predicted DxD motif by substituting the conserved Asp residues (D) in the IRX sequences with Asn (N) residues. Interestingly, a DxD motif could not be identified in AoIRX9, a GT43 family protein predicted to have a GT-A fold, although alignments with IRX9 and IRX9L proteins from other species identified a single, highly conserved Asp residue (Asp-298 in AoIRX9 and Asp-364 in AoIRX9L) and a DDD motif in AoIRX9L (DDD287–289; Supplemental Fig. S1). To test if these conserved residues were part of a catalytic binding site, we targeted these residues for SDM (D298N for AoIRX9 and D287N/D289N for AoIRX9L). Coexpression of either AoIRX9 (D298N) or AoIRX9L (D287N/D289N) with AoIRX10 and AoIRX14A (Fig. 3) did not significantly alter XylT activity compared with the triple expression of either wild-type AoIRX9 or AoIRX9L with AoIRX10 and AoIRX14A (compare Figs. 2 and 3). This indicated that the conserved Asp residue of AoIRX9 was not critical for xylan synthesis, and neither was the conserved DxD motif of AoIRX9L.
Figure 3.
Xylan XylT activity of MMs from N. benthamiana leaves coexpressing combinations of wild-type and mutant versions of AoIRX genes from asparagus. Combinations of three wild-type (AoIRX9 or AoIRX9L, AoIRX10, and AoIRX14A or AoIRX14B) and mutated (designated with M) genes were coexpressed in N. benthamiana leaves, and the MMs were extracted for xylan XylT activity analysis. N. benthamiana leaves (co)expressing VENUS and AoIRX9/10/14A were used as negative and positive controls, respectively. A, Xylan XylT products were fractionated by RP-HPLC and detected with a fluorescence detector. The numbers on the plots indicate the degree of polymerization of xylooligosaccharides. B, Xylan XylT was measured using UDP-[14C]Xyl as the donor and Xyl5-AA as the acceptor. Values represent means ± se of three biological replicates.
Interspecies sequence alignments of IRX14 proteins have revealed that the DxD motif at amino acid residues 276 to 277 in AoIRX14A and 247 to 248 in AoIRX14B are highly conserved (Supplemental Fig. S2). Xylan XylT activity assays of N. benthamiana leaves expressing either AoIRX14A (D276N/D277N) or AoIRX14B (D247N/D278N) with AoIRX9 and AoIRX10 showed that there was a noticeable impact on xylan XylT activity compared with their wild-type controls (Fig. 3). This suggests that residues 276 to 277 in AoIRX14A and 247 to 248 in AoIRX14B are likely to be catalytically important for xylan XylT activity.
Although GT47 enzymes are predicted to have a GT-B-type fold and to be metal ion independent, AoIRX10 was found to have a highly conserved DxD motif near the C terminus at amino acid residues 310 to 311 (Supplemental Fig. S3). Coexpression of AoIRX10 (D310N/D311N) with AoIRX9 and AoIRX14A also led to a significant loss of xylan XylT (Fig. 3), suggesting that this motif is essential for xylan XylT activity.
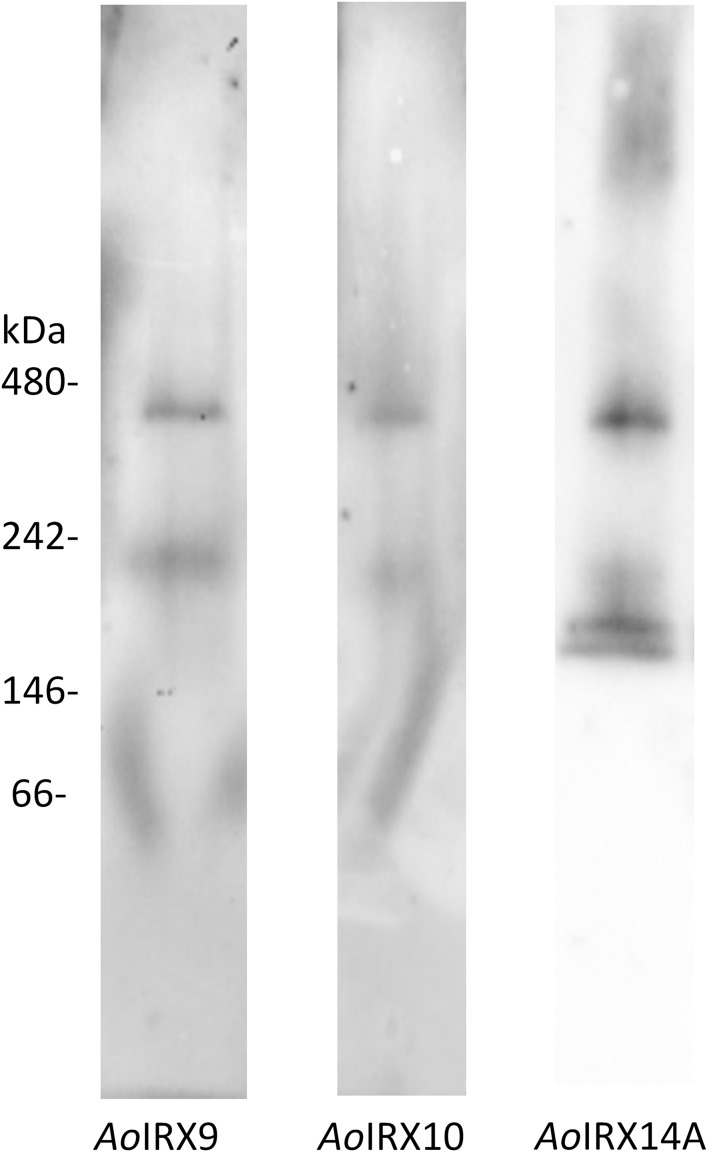
MMs from N. benthamiana leaf coexpressing AoIRX9/10/14A were solubilized with 1% (w/v) n-dodecyl β-d-maltoside (DDM), separated on a native protein gel, and then immunoblotted with AoIRX9, AoIRX10, and AoIRX14A antibodies. Intriguingly, the AoIRX9, AoIRX10, and AoIRX14A antibodies all recognized bands at similar positions just below the 242- and 480-kD markers (Fig. 4), suggesting that these three proteins interact to form heterotrimeric complexes. Interestingly, no monomers of the individual AoIRXs were detected.
Figure 4.
Western blot of a native gel containing solubilized MMs of N. benthamiana leaves coexpressing AoIRX9, AoIRX10, and AoIRX14A. MMs extracted from N. benthamiana leaves coexpressing AoIRX9, AoIRX10, and AoIRX14A were solubilized using 1% (w/v) DDM, separated on a single native gel in three adjacent lanes, and then transferred to a single membrane that was marked for subsequent realignment. The membrane of each lane was then cut and probed with different antibodies against AoIRX9 (left), AoIRX10 (middle), and AoIRX14A (right). The membrane was realigned and subsequently imaged. No monomer bands of AoIRX9, AoIRX10, and AoIRX14A were detected; however, there were some bands detected for AoIRX9 and AoIRX14A between 146 and 242 kD, suggesting the presence of lower oligomers of the protein complex.
To further examine the XSC, detergent (DDM)-solubilized MMs from N. benthamiana coexpressing AoIRX9/10/14A were coimmunoprecipitated with the AoIRX9 antibody. Tryptic peptides originating from all three AoIRX proteins could be identified via proteomic analysis (Supplemental Fig. S4), further indicating that AoIRX9, AoIRX10, and AoIRX14A interact either directly or indirectly and form a multiprotein complex.
AoIRX9, AoIRX10, and AoIRX14A Are Detected in a Protein Complex Coimmunoprecipitated with a GFP-Trap Approach
In an attempt to partially purify the XSC in the heterologous expression system, we used the fluorescent translational fusion constructs (VENUS) of each of the three AoIRX proteins (AoIRX9, AoIRX10, and AoIRX14A) and coexpressed them in N. benthamiana leaves. Western-blot analysis demonstrated that each of the translational fusions was of the expected size and that the VENUS-conjugated AoIRX9, AoIRX10, and AoIRX14A proteins were expressed at similar levels (Supplemental Fig. S5). We subsequently enriched for the XSC using a GFP-Trap approach. Xylan XylT activity assays using the enriched products showed a similar product profile to that of asparagus MMs (Fig. 5). The GFP-Trap-enriched XSC fraction was analyzed by tandem mass spectrometry, and 32 proteins with more than two peptides were identified (Supplemental Table S1). AoIRX9, AoIRX10, and AoIRX14A as well as VENUS were the top four matches according to the exponentially modified protein abundance index (Ishihama et al., 2005), with multiple peptides (eight, seven, six, and six peptides, respectively) assigned, indicating that a successful enrichment was achieved with the GFP-Trap procedure. Twenty-eight N. benthamiana proteins also were identified in the same fraction (Supplemental Table S1); however, no other GTs or proteins previously implicated in xylan biosynthesis were found.
Figure 5.
Xylan XylT activity of the GFP-Trap-purified proteins from MMs of N. benthamiana leaves coexpressing asparagus IRX9-VENUS, IRX10-VENUS, and IRX14A-VENUS. The activity of untransformed, unpurified native asparagus microsomal membranes (top) was used as a positive control. Three asparagus xylan XylTs (AoIRX9, AoIRX10, and AoIRX14A) fused at the C terminus to VENUS were coexpressed in N. benthamiana leaves, the MMs were extracted and detergent solubilized, and the resulting proteins were enriched by coimmunoprecipitation with a GFP-Trap (bottom). Xylan XylT products were analyzed by RP-HPLC. Numbers above the peaks designate the degree of polymerization of xylooligosaccharides. Xyl5-AA was the acceptor.
Targeting to the GA Requires Coexpression of AoIRX9, AoIRX10, and AoIRX14A
Biochemical enzyme assays and antixylan epitope labeling of subcellular compartments indicate that xylan biosynthesis occurs in the GA (Porchia et al., 2002; Rennie and Scheller, 2014). The xylan XylT activity assay on MMs expressing these reporter constructs confirmed that the VENUS tag of the AoIRX proteins does not interfere significantly with xylan XylT activity (Fig. 5). To confirm that the subcellular location of the AoIRX9, AoIRX10, and AoIRX14A proteins is in the GA, we tested C-terminal AoIRX translational fusion constructs with VENUS in N. benthamiana (Figs. 6–8).
Figure 6.
Subcellular location of asparagus IRX9-VENUS in N. benthamiana leaves expressed with or without asparagus IRX10/IRX14A. Fluorescence images show IRX9-VENUS (yellow) expressed in N. benthamiana leaves alone (A), with IRX10 (F), with IRX14A (K), or with both IRX10 and IRX14A (P). B, G, L, and Q show coexpression of the endoplasmic reticulum (ER) marker SP-AtWAK2-CFP-HDEL (cyan), while C, H, M, and R show coexpression of the GA marker GmMan1(1-49)-RFP (magenta). D, I, N, and S show the merged images, and E, J, O, and T show the merged images overlaid with the transmitted light images. IRX9-VENUS signal is detected in a reticulate pattern that largely overlaps that of the ER marker when it is expressed alone or in combination with IRX10 (compare A and F with B and G, respectively). When IRX9 is coexpressed with IRX14A, some punctate structures are observed that overlap that of the GA marker (arrowheads; compare K and M; composite image N). Strikingly, when AoIRX9-VENUS was coexpressed with AoIRX10 and AoIRX14A, its signal was not observed in a reticulate pattern reminiscent of ER labeling (compare P and Q) but rather appeared as discrete punctate bodies that overlapped that of the Golgi marker (arrowheads; compare P and R; composite image S). Overlapping signal is artificially colored white.
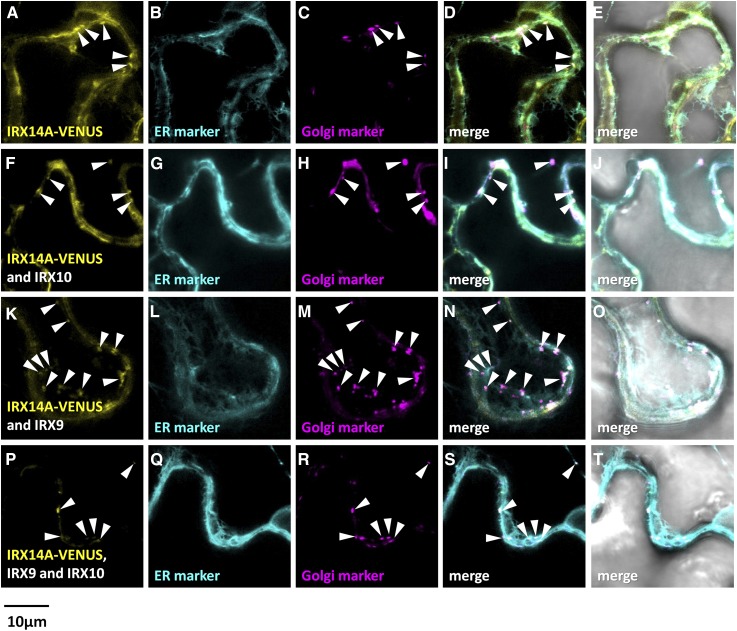
Figure 8.
Fluorescent localization of asparagus IRX14A-VENUS proteins expressed in N. benthamiana leaves with or without IRX9/IRX10. Fluorescence images show IRX14-VENUS (yellow) expressed in N. benthamiana leaves alone (A), with IRX10 (F), with IRX9 (K), or with both IRX10 and IRX9 (P). B, G, L, and Q show the ER marker SP-AtWAK2-CFP-HDEL (cyan). C, H, M, and R show the Golgi marker GmMan1(1-49)-RFP (magenta). D, I, N, and S show the merged images, and E, J, O, and T show the merged images overlaid with the transmitted light images. Golgi localization of IRX14-VENUS is marked with white arrowheads. Overlapping signal is artificially colored white.
When viewed on the confocal microscope, N. benthamiana leaves infiltrated with AoIRX9-VENUS showed a reticulate-like pattern that overlapped that of the ER marker AtWAK2-CFP-HDEL to a greater extent than the GmMAN1(1-49)-RFP Golgi marker (Fig. 6). When AoIRX9-VENUS was coexpressed with unlabeled AoIRX10, a similar ER-predominant localization pattern was observed (Fig. 6), while coexpression of AoIRX9-VENUS and unlabeled AoIRX14A showed labeling that overlapped with the ER marker and also somewhat with the Golgi marker. Strikingly, coexpression of AoIRX9-VENUS with unlabeled AoIRX10 and AoIRX14A resulted in a subcellular location that coincides almost exclusively with the Golgi marker (Fig. 6).
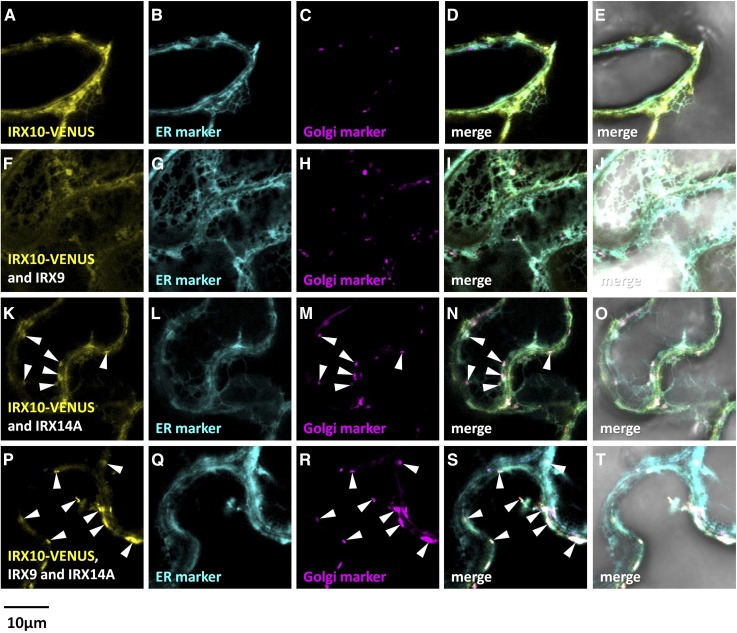
Analysis of the reciprocal combinations of AoIRX10-VENUS and AoIRX14A-VENUS translational fusions gave similar results (Figs. 7 and 8). Expression of AoIRX10-VENUS alone generated an expression pattern that predominantly overlapped with that of the ER marker (Fig. 7). Coexpression of AoIRX10-VENUS with either of the untagged partner proteins, AoIRX9 or AoIRX14A, led to an ER-like localization with partial overlap with the Golgi marker (Fig. 7). Expression of AoIRX10-VENUS with both untagged AoIRX9 and AoIRX14A resulted in fluorescence predominantly in punctate bodies that colocalized with the Golgi marker, although some reticulate ER patterns still could be observed (Fig. 7). Golgi targeting of AoIRX14A-VENUS was enhanced by the coexpression of all three proteins (Fig. 8).
Figure 7.
Fluorescent localization of asparagus IRX10-VENUS proteins expressed in N. benthamiana leaves with or without IRX9/IRX14A. Fluorescence images show IRX10-VENUS (yellow) expressed in N. benthamiana leaves alone (A), with IRX9 (F), with IRX14A (K), or with both IRX9 and IRX14A (P). B, G, L, and Q show the ER marker SP-AtWAK2-CFP-HDEL (cyan). C, H, M, and R show the Golgi marker GmMan1(1-49)-RFP (magenta). D, I, N, and S show the merged images, and E, J, O, and T show the merged images overlaid with the transmitted light images. Golgi localization of IRX10-VENUS is marked with white arrowheads. Overlapping signal is artificially colored white.
AoIRX9, AoIRX10, and AoIRX14A Form a Xylan Biosynthesis Complex
To further dissect the interactions between AoIRX9, AoIRX10, and AoIRX14A, a BiFC approach was adopted (Kerppola, 2006). Since all three proteins are required for colocation to the GA, all tests were conducted in the presence of their unlabeled partners. Infiltration combinations and the results of this experiment are summarized in Supplemental Table S2.
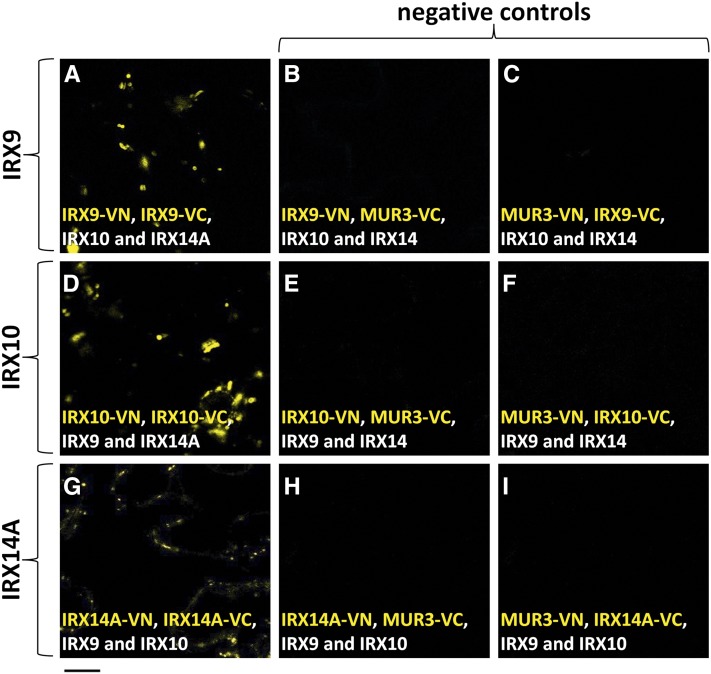
BiFC requires many controls to demonstrate that the signal detected in this assay reflects specific protein-protein interactions. As a positive control, we first established that MUR3 (GenBank accession no. AB844168.1) is able to form a homodimer, as described previously (Wilson et al., 2015), by coexpressing MUR3-VN and MUR3-VC (Supplemental Fig. S6). MUR3 is known to be involved in xyloglucan biosynthesis (Madson et al., 2003) and is not predicted to interact with proteins involved in xylan biosynthesis. A fluorescent signal was detected in the GA (Supplemental Fig. S6), confirming that the system was working appropriately. Fluorescence was also detected when AoIRX9-VN and AoIRX9-VC, AoIRX10-VN and AoIRX10-VC, or AoIRX14A-VN and AoIRX14A-VC (Fig. 9) were coexpressed in the presence of the two other untagged partner proteins, indicating that all three proteins are able to form homodimers. The fluorescence detected in these three tests was considered to signify a specific protein-protein interaction, as the negative controls consisting of combinations of either AoIRX-VN or AoIRX-VC with either MUR3-VC or MUR3-VN, respectively, had no detectable signal under the same conditions (Supplemental Figs. S6–S8). The ER marker MANI(49)-RFP was used as an internal control for all BiFC experiments and confirmed that the lack of fluorescence observed in these negative controls was not due to experimental conditions disrupting efficient transformation.
Figure 9.
IRX9, IRX10, and IRX14A can homodimerize in planta. BiFC was used to test if IRX9, IRX10, and IRX14A can homodimerize in planta. The N- and C-terminal halves of VENUS fused to IRX9 (A), IRX10 (D), and IRX14A (G) were coexpressed in N. benthamiana leaves in the presence of the two other untagged IRX proteins. Fluorescent signal indicates an interaction between the two tagged proteins. A strong punctate signal was observed in all three cases, indicating that IRX9, IRX10, and IRX14A are able to homodimerize. When either IRX9/10/14A-VN (B, E, and H, respectively) or IRX9/10/14A-VC (C, F, and I, respectively) were independently expressed with a noninteracting partner (MUR3-VC/VN; negative controls), no signal was detectable, as expected. Expanded results and positive and negative controls are shown in Supplemental Figures S6 to S8.
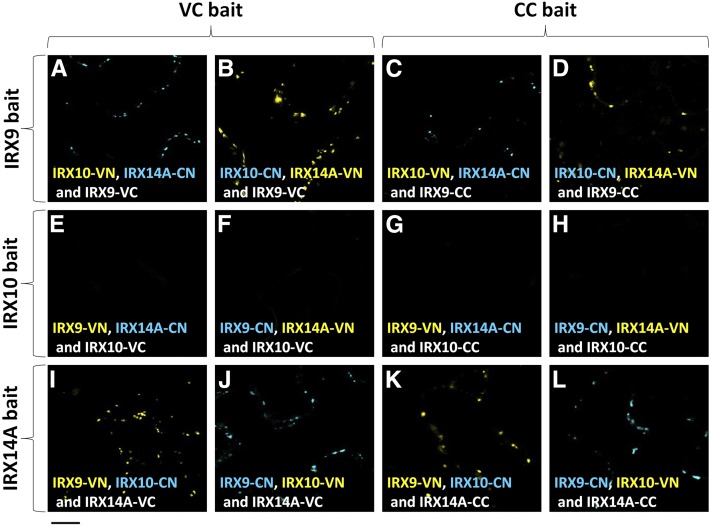
Since our previous results suggest that AoIRX9, AoIRX10, and AoIRX14A form a multiprotein XSC, we used a multicolor bimolecular fluorescence complementation (mcBiFC) approach (Lee et al., 2008) to test whether heterodimerization could occur between the AoIRX proteins in planta. mcBiFC experiments enable the comparison of the subcellular distributions of protein complexes formed with different interaction partners and the analysis of competition between mutually exclusive interaction partners for binding a shared partner present at limiting concentration. The results from this experiment are summarized in Figure 10 and Supplemental Table S2. Twelve independent tests indicate that AoIRX9 and AoIRX14A are able to interact with each other but not with AoIRX10 in planta (Fig. 10). This suggests that an additional unknown protein is part of the XSC, which bridges AoIRX10 with AoIRX9 and/or AoIRX14A. Expanded figures showing individual channels for each experiment are provided in Supplemental Figures S9 to S14. No fluorescence was detected in any channel on the negative control combinations including either MUR3-VN or MUR3-VC.
Figure 10.
Using mcBiFC to elucidate protein-protein interactions in the XSC. mcBiFC was used to test if IRX9, IRX10, and IRX14A can heterodimerize in planta by expressing split VENUS and split CFP constructs in N. benthamiana. The C-terminal half of either VENUS (VC) or CFP (CC) is fused to a protein of interest and expressed in the presence of two other proteins carrying either the N-terminal half of VENUS (VN) or CFP (CN). The fluorescence color is determined predominantly by the N-terminal half of the reconstituted protein. Signal in either the yellow or blue spectrum indicates an interaction between the respective proteins. A to D, First, we tested which proteins can interact with IRX9 by using it as a bait by fusing IRX9 to either VC (A and B) or CC (C and D) and expressing it in the presence of IRX10 and IRX14A. A signal was observed in all four tests, indicating an interaction between IRX9 and IRX14A only. No interaction was detected between IRX9 and IRX10. E to H, We then tested which proteins can interact with IRX10 by using it as a bait by fusing IRX10 to either VC (E and F) or CC (G and H) and expressing it in the presence of IRX9 and IRX14A. No signal was detected between any of the proteins, indicating that IRX10 does not interact directly with either IRX9 or IRX14A. I to L, Finally, we tested which proteins can interact with IRX14A by using it as a bait and fusing IRX14A to either VC (I and J) or CC (K and L) and expressing it in the presence of IRX9 and IRX10. A signal was observed in all four combinations, indicating an interaction between IRX14A and IRX9 only. No interaction was detected between IRX14 and IRX10. Twelve independent tests indicate that IRX9 and IRX14A are able to form heterodimers with each other but not with IRX10. Expanded results and negative controls are shown in Supplemental Figures S9 to S14.
The XSC Is Located in the Lumen of the GA
In order for mcBiFC to be informative, the split fluorescent halves must be located in the same compartment. One possible explanation for the lack of detectable signal in combinations including AoIRX10-VN and AoIRX10-VC is that the C termini of all three proteins are not located in the GA lumen, as predicted. Therefore, to validate the topology for these proteins, we used the previously described Golgi protein membrane topology (GO-PROMTO) assay (Søgaard et al., 2012) and transiently expressed combinations of constructs with either cytosolic (VN/VC-TMD) or lumenal (TMD-VN/VC) fluorescent GO-PROMTO reporters in N. benthamiana leaves. For all three genes, each of the four possible pairs of constructs was tested, and the results are shown in Figure 11. Fluorescence complementation was never observed using the cytosolic reporters, but signal was detected consistently upon coexpression with the GA lumenal reporter, indicating that the C terminus of all three proteins is located in the Golgi lumen. These data confirm that the XSC is located in Golgi membranes with the catalytic site facing the lumen.
Figure 11.
GO-PROMTO analysis of AoIRX9, AoIRX10, and AoIRX14 in N. benthamiana leaves 3 d postinfiltration. Coexpression of either cytosolic (VN/VC-TMD; A) or lumenal (TMD-VN/VC; B) fluorescent GO-PROMTO reporters in N. benthamiana leaves yielded strong fluorescent signals. C-terminal VN or VC fusions of AoIRX9 (C–F), AoIRX10 (G–J), and AoIRX14 (K–N) were coexpressed with the cytosolic reporters VN-TMD (C, G, and K) or VC-TMD (D, H, and L) or the lumenal reporters TMD-VN (E, I, and M) or TMD-VC (F, J, and N), respectively. Only combinations including either of the luminal reporters had detectable fluorescent signal, indicating that the C terminus of these proteins is located in the lumen of the GA.
DISCUSSION
AoIRX9, AoIRX10, and AoIRX10A Must Be in a Complex to Form an Active XSC in Planta for Both Optimal Activity and Appropriate Subcellular Targeting
Many plant cell wall polysaccharides are synthesized by an enzyme complex located at either the GA or the plasma membrane (Oikawa et al., 2013). Previous studies have indicated that this is indeed likely to be the case for xylan biosynthesis, with key proteins predicted to be involved in a core XSC (IRX9, IRX10, and/or IRX14; Zeng et al., 2010; Ren et al., 2014; Rennie and Scheller 2014; Jiang et al., 2016). Here, we show that asparagus AoIRX9, AoIRX10, and AoIRX14A are all required to form an active XSC, as only coexpression of all three proteins led to xylan XylT activity at levels similar to that observed using native endogenous asparagus MMs (Fig. 2). At first glance, this observation may appear contradictory with previously published results from other groups. For instance, Lee et al. (2012a) found that MMs from tobacco BY2 suspension-cultured cells coexpressing AtIRX9 and AtIRX14 have active XylT activity. We have shown that the coexpression of AoIRX9 and AoIRX14 in N. benthamiana leaves also resulted in XylT activity. However, the levels of activity were significantly lower than when all three proteins were present. Indeed, it is possible that the expression of AtIRX9 and AtIRX14 in BY2 cells may recruit the endogenous IRX10 to form an XSC. This is supported by our data showing that AoIRX9 appears to drive XSC assembly to/in Golgi membranes (Figs. 6–8), and it is the relocation from the ER to the GA of all three proteins that causes the increase in XylT activity (Fig. 2). Our observations also are consistent with recent findings by Jiang et al. (2016), who showed that wheat TaGT43 plays a critical role in initiating XSC assembly in the ER prior to the transport of the XSC to the GA. Heterologous expression studies have shown that IRX10 proteins from multiple species have distributive xylan XylT activity (Jensen et al., 2014; Urbanowicz et al., 2014), while our observation is that AoIRX10 heterologous expression alone in N. benthamiana leaves was only capable of adding a single Xyl residue to the Xyl5-AA acceptor (Fig. 2). This may be due to IRX10 functional variations between plant species and/or differences in the heterologous expression systems (for review, see Hao and Mohnen, 2014). Similarly, expression of AoIRX14A also was capable of adding a single Xyl residue to the Xyl5-AA acceptor. Given that N. benthamiana likely has the genes required for xylan XylT activity, it may well be that expression of either AoIRX10 or AoIRX14A in N. benthamiana activates a latent activity in their MMs, rather than the heterologously expressed proteins necessarily having xylan XylT activity.
Heterologous expression of one and/or two of the asparagus IRX9/10/14 proteins was able to produce some XylT activity in N. benthamiana MMs. However, the coexpression of all three proteins, AoIRX9, AoIRX10, and AoIRX14A, was sufficient to replicate native asparagus activity. Furthermore, for appropriate subcellular localization of these asparagus proteins to the GA, all three must be coexpressed, supporting the hypothesis that these proteins form a functional XSC core.
Interactions between IRX Proteins in Different Species
Since the structure of heteroxylans varies between different species and tissue types, it is conceivable that the biosynthetic machinery, both the composition and function of individual proteins in the XSC, for each type of heteroxylan/species may be different. For example, P. ovata mucilage is rich in heteroxylan, but the expression level of GT43 genes (PoIRX9 and PoIRX14) in mucilage-secreting cells is very low (Jensen et al., 2013), suggesting that the multiple PoIRX10 genes (GT47) identified in this system play a predominant role. Similarly, Zeng et al. (2010) identified TaIRX10 and TaIRX14 proteins in a wheat glucuronoarabinoxylan synthase complex isolated from etiolated seedlings but not TaIRX9. These studies suggested that the components of the XSC have diversified in different species and/or tissue types through evolution. The regulation of stem secondary wall biosynthesis is an elegant and complex process (Zhong and Ye, 2015) that involves a number of players, including upstream transcription factors. AtIRX9 may play an important regulatory role in xylan biosynthesis in stem secondary wall formation.
Different laboratory-based experimental techniques have been utilized to study protein-protein interactions, including yeast two-hybrid approaches (Jones et al., 2014), the reversible Renilla luciferase protein complementation assay (Stefan et al., 2007), BiFC (Bracha-Drori et al., 2004; Wilson et al., 2015; Jiang et al., 2016), and affinity purification followed by mass spectrometry-based proteomics (Van Leene et al., 2008). A comparison of the pros and cons of each strategy can be found in a review by Zhang et al. (2010). Lund et al. (2015) studied the protein-protein interactions of Arabidopsis IRX9, IRX9L, IRX10, IRX10L, IRX14, and IRX14L via Renilla luciferase protein complementation assay and showed that all the interactions between each protein pair (homodimer or heterodimer) were negative. One explanation for their observations is that the interactions between these proteins is weak (Ren et al., 2014) and, therefore, could not be detected via this assay method. Another equally plausible explanation in light of our observations is that all three proteins need to be coexpressed simultaneously to form an active complex in the GA. When the asparagus IRX proteins were expressed either alone or in pairs, they were not effectively targeted to the GA. It is possible that other non-GT proteins, such as AtIRX15 (Brown et al., 2011; Jensen et al., 2011), are part of the XSC, although our preliminary proteomic analysis did not identify any obvious candidates.
Asparagus IRX9, IRX10, and IRX14A Have Distinct Roles in the XSC
Plant polysaccharide synthase complexes often contain multiple GTs with distinct physiological functions. One example is the pectin homogalacturonan synthase complex, containing two GAUT1 catalytic proteins and one GAUT7 protein that acts as a membrane anchor protein (Atmodjo et al., 2011). The exact stoichiometry of the structure of the XSC and the precise roles that each of the different GTs play within the complex are still unknown. AoIRX9 does not have a conserved DxD motif (Supplemental Fig. S1), which is thought to be involved in the binding of UDP-sugar substrates (Chang et al., 2011). This suggests that AoIRX9 is not catalytically active in the asparagus XSC. SDM of a conserved aspartic residue (D) in AoIRX9 and a DxD motif in AoIRX9L did not significantly affect XylT activity of the XSC when coexpressed with AoIRX10 and AoIRX14A (Fig. 3), indicating that neither of these Asp residue motifs is catalytically important. However, AoIRX10 and AoIRX14 coexpressed without AoIRX9 results in a significant diminution of activity, showing that AoIRX9 is essential for maximal XSC activity by playing a structural, rather than a catalytic, role in the complex. Interestingly, proteomic analysis identified that the Asn-177 residue of AoIRX9 is N-glycosylated (Ford et al., 2015), although this site is conserved in P. patens, P. trichocarpa, and ryegrass (Lolium perenne) but not in Arabidopsis or rice (Supplemental Fig. S1). The physiological role of this posttranslational modification on AoIRX9 is being investigated.
In contrast, the SDM of AoIRX14A, AoIRX14B, and AoIRX10 significantly affected the xylan XylT activity (Fig. 3), suggesting that both AoIRX10 and AoIRX14 are catalytically important for the asparagus XSC. This raises the question of whether AoIRX10 and AoIRX14A play slightly different roles within the complex. AoIRX14A is a GT43 family protein, members of which are predicted to have a GT-A fold, while AoIRX10 belongs to the GT47 family, classified as having a GT-B fold (Lairson et al., 2008). GT-B fold enzymes are metal ion independent, and indeed, AtIRX10 does not require metal ions for its xylan XylT synthase catalytic activity (Urbanowicz et al., 2014). It is possible that the DxD motif is important for protein dimerization and/or protein folding (Wiggins and Munro, 1998). Thus, the exact role of the AoIRX10 DxD motif in the XSC remains elusive.
Ren et al. (2014) found that SDM of the AtIRX14 DxD motif results in a nonfunctional GT and speculated that Arabidopsis IRX14 functions in binding UDP-Xyl and transferring the donor to another unknown protein instead of binding it directly for Xyl incorporation. IRX10 appears to be the most active of the three XSC candidates, as it has been shown to have xylan XylT activity when expressed in nonplant heterologous systems (Jensen et al., 2014; Urbanowicz et al., 2014). When AoIRX10 was expressed alone in N. benthamiana leaves, a single Xyl residue was incorporated onto the acceptor (Fig. 2), similar to the activity of Arabidopsis IRX10 expressed in P. pastoris (Jensen et al., 2014). Expression of AoIRX14 alone did not give as high XylT activity as AoIRX10 (Fig. 2). Combining the xylan XylT activity and XSC subcellular localization data, we propose that AoIRX9 may act as a structural protein anchoring the complex to the Golgi membrane to initiate the recruitment of the other XSC members. AoIRX10 does not have a predicted transmembrane domain (Supplemental Fig. S3) but has been localized within the Golgi lumen (Søgaard et al., 2012) and, therefore, would have to be affiliated with the XSC as a peripheral protein. We have shown through BiFC that AoIRX9, AoIRX10, and AoIRX14A all homodimerize (Fig. 9) and that AoIRX9 interacts directly with AoIRX14A but not AoIRX10 (Fig. 10). We postulate that AoIRX9 recruits AoIRX14A, which in turn recruits another yet to be identified protein that bridges AoIRX10 to AoIRX9 and AoIRX14. We suggest that AoIRX14 performs the initial priming reaction, thereby facilitating subsequent xylan XylT activity by AoIRX10. Our proposed model of the core XSC is shown schematically in Figure 12. In spite of numerous in vitro studies demonstrating xylan XylT activity in the absence of a primer, models of xylan backbone initiation and extension in planta must accommodate the presence of a unique tetrasaccharide RE sequence in dicot/gymnosperm heteroxylans (York and O’Neill, 2008; Hao and Mohnen, 2014). Thus, how this rudimentary XSC model (Fig. 12) synthesizes the xylooligosaccharide structures primed from the Xyl5-AA acceptor in our in vitro MM assays relates to the in planta assembly mechanism using a potential RE primer sequence remains to be determined.
Figure 12.
Proposed model for the core asparagus XSC in the GA. The asparagus XSC contains homodimers of IRX9, IRX10, and IRX14A with potential interaction partner(s). AoIRX9 and AoIRX14 form heterodimers that are anchored to the GA membrane, with their C termini facing the Golgi lumen. AoIRX10 is located in the Golgi lumen and lacks a transmembrane domain. The shapes with question marks indicate other interacting proteins that may act to bridge IRX10 to IRX9 and IRX14A. We postulate that perhaps IRX14 performs the initial priming reaction, thereby facilitating subsequent xylan XylT activity by IRX10 within the Golgi lumen.
Asparagus spears provide an ideal system for heteroxylan biosynthesis, where significant quantities of both primary and secondary wall heteroxylan exist in the top and bottom portions of the spear, respectively (Song et al., 2015). AoIRX9L is highly expressed in the top of the asparagus spear, where the expression of AoIRX9 is very low (Song et al., 2015). Many questions still remain, including how plants delicately regulate the transition from primary wall to secondary wall biosynthesis. Mortimer et al. (2015) proposed that the Arabidopsis primary wall xylan biosynthetic machinery likely consists of AtGUX3, AtIRX9L, AtIRX10L, and AtIRX14. It is possible that two distinct sets of XSCs are responsible for primary and secondary wall xylan biosynthesis, as proposed previously by Ratke et al. (2015). This would be similar to the cellulose synthases (CESAs) for primary wall (CESA1, CESA3, and CESA6) and secondary wall (CESA4, CESA7, and CESA8) in Arabidopsis (Carroll et al., 2012). Purification of the XSC from native systems such as asparagus with high xylan XylT activity and subsequent proteomics analysis to help identify the complete XSC protein complement is one of a number of approaches that will lead to a better understanding of the heteroxylan biosynthesis mechanism(s).
MATERIALS AND METHODS
Plant Materials
Nicotiana benthamiana plants were grown in soil in a glasshouse with continuous cool-white fluorescent light and natural daylight at 20°C to 26°C as described previously (Wilson et al., 2015). Asparagus (Asparagus officinalis) spears were generously provided by Vizzarri Farm in Victoria, Australia, and processed within 2 h of harvest as described previously (Song et al., 2015).
Molecular Biology
RNA was isolated from emerging asparagus spears, and first-strand complementary DNA synthesis was carried out using the Tetro cDNA Synthesis Kit (Bioline) with 1 µg of total RNA following the manufacturer’s instructions. The sequences of primers used in this study are shown in Supplemental Table S3. To generate C-terminal fluorescently labeled fusions of AoIRX9, AoIRX10, and AoIRX14A, primers (pFUERTE)UFP_Fwd and UFP_Rev were used to PCR amplify VENUS, while primers (pFUERTE)AoIRX_Fwd and Rev were used to amplify the respective asparagus IRX coding sequences. Phusion High-Fidelity Master Mix (New England Biolabs) was used for amplification. PCR products were cloned using the Gibson assembly method (New England Biolabs) into the binary vector pFUERTE, which contains the cauliflower mosaic virus 35S promoter, a multiple cloning site (MCS), and the 3′ octopine synthase gene terminator sequence (Wilson et al., 2015). SDM was carried out using the Gibson assembly method, where changes to the DNA sequence were incorporated in the PCR primers used for amplification of the two separate SDM AoIRX coding fragments and cloned into pFUERTE vector.
BiFC constructs were generated by amplifying PCR products using primers (BiFC)AoIRX Fwd and AoIRX(BiFC) Rev and cloning them into SfoI- and KpnI-linearized pURIL (binary vector containing the first 155 amino acids of VENUS; VN155) and pDOX (C-terminal version containing the last 84 amino acids of VENUS; VC84) using the Gibson assembly method (Wilson et al., 2015; Lampugnani et al., 2016). For mcBiFC experiments, new reporters were generated using the CFP mCERULEAN (Rizzo et al., 2004) split at amino acid 155, such that the N-terminal version contained the first 155 amino acids (CN155) and the C-terminal version contained the last 84 amino acids (CC84). CN155 and CC84 were synthesized downstream of a 20-amino acid Gly linker and cloned into pFUERTE as described previously (Wilson et al., 2015; Lampugnani et al., 2016). The resulting vectors 35S:MCS‐gLINKER‐CN and 35S:MCS‐gLINKER‐CC were named pGANTHET and pWALKER, respectively. mcBiFC analysis involves expressing a bait protein translationally fused to the C-terminal half of either VENUS (VC) or CFP (CC) in the presence of two prey proteins carrying the N-terminal half of either VENUS (VN) or CFP (CN), respectively. The fluorescence color is determined by the N-terminal half of the reconstituted protein. Signal in either the yellow or blue spectrum indicates an interaction between the bait protein and one or more prey proteins. All mcBiFC constructs used in this study were generated using the primers described previously for BiFC and cloned into SfoI- and KpnI-linearized pGANTHET or pWALKER.
All generated constructs were sequence verified and transformed into Agrobacterium tumefaciens strain AGL1 by electroporation as described previously (Lampugnani et al., 2012) with the helper plasmid pSOUP (Hellens et al., 2000).
Transient Expression in N. benthamiana Leaves
A. tumefaciens strain AGL1 carrying the constructs described earlier and another strain generating the P19 protein to suppress gene silencing (Voinnet et al., 2003) were grown in 2YT liquid medium containing kanamycin (50 µg mL−1), ampicillin (100 µg mL−1), and tetracycline (10 µg mL−1) to an OD 600 nm = 1 at 28°C. Cells were then pelleted, subsequently resuspended in infiltration medium (10 mm MgCl2 and 0.6 mm acetosyringone, with the addition of 10 mm MES, pH 5.7, for enzymatic studies), and adjusted to a final optical density at 600 nm = 1.2. After 2 h of incubation at room temperature, the cells were infiltrated into the abaxial surface of 4-week-old N. benthamiana leaves using a 1-mL needleless syringe. After 3 d, leaf sectors were excised, mounted in water, and processed as described below (“Microscopy”). Each inoculation was performed on triplicate leaves, and all transformations were performed on at least two separate occasions.
Assay of Xylan XylT Activity and Enzyme Purification
MMs were isolated from N. benthamiana leaves 3 d after A. tumefaciens infiltration following the method of Song et al. (2015). Xylan XylT assays using either RP-HPLC (unlabeled donors) or radiolabeling (labeled donors) also follow Song et al. (2015) with minor modifications. RP-HPLC buffer B was changed to 50 mm ammonium acetate (pH 4.3) instead of sodium acetate for better separation of the fluorescently labeled xylooligosaccharide products.
C-terminal VENUS-conjugated AoIRX9, AoIRX10, and AoIRX14A were coexpressed in N. benthamiana leaves. For coimmunoprecipitation, MMs (around 500 µg of protein) were solubilized with 1% (w/v) DDM for 20 min on ice and centrifuged at 100,000g for 60 min at 4°C. The supernatant was mixed with 20 µL of GFP-Trap_A beads (Chromotek), incubated at 4°C for 3 h, and washed three times with HEPES-KOH extraction buffer (Song et al., 2015) with 150 mm NaCl. Protein-bound agarose beads were added directly to the XylT assay and assayed using the standard conditions outlined above.
Western Blotting
Polyclonal antibodies were generated and affinity purified by GenScript. The antigen regions of AoIRX9, AoIRX10, and AoIRX14A were chosen using the OptimumAntigen design tool (GenScript). The AoIRX9 antibody was directed toward the peptide sequence LTASPSSSQSKNRR, the AoIRX10 antibody toward SSPVRTLNPEEA, and the AoIRXa14A antibody toward NSEPLVQNEKKSEE. To confirm the expression of the asparagus IRX proteins and to test the specificity of these antibodies, MMs of N. benthamiana leaves expressing asparagus IRX proteins were extracted and used in western blotting. SDS-PAGE and western blotting were performed according to Wilson et al. (2015) unless otherwise specified. Briefly, SDS-PAGE and protein transfer was performed using the XCell SureLock Mini-Cell system (Life Technologies). Proteins were transferred to a 0.45-µm polyvinylidene difluoride membrane (Thermo Scientific) using NuPAGE transfer buffer (Life Technologies) at 30 V for 1 h. Membranes were blocked in 5% (w/v) skim milk powder in Tris-buffered saline buffer containing 0.1% (v/v) Tween 20 (TBST) for 1 h at room temperature and incubated with a primary antibody (1:1,000 dilution) in TBST containing 5% (w/v) skim milk powder for 1 h at room temperature with gentle shaking. After washing with TBST for 10 min (three times), the membranes were incubated with the secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit [Thermo Scientific]; 1:5,000 dilution) in TBST containing 5% (w/v) skim milk powder for 1 h at room temperature with shaking. Subsequently, the membranes were washed with TBST for 10 min (three times) before WesternBright ECL HRP substrate (Advansta) was added to the membrane. Signals were detected using a ChemiDoc MP imaging system and processed via Image Lab software (Bio-Rad). In the case of the western blot of native gels containing solubilized MMs of N. benthamiana leaves coexpressing AoIRX9, AoIRX10, and AoIRX14A, the MMs were solubilized using 1% (w/v) DDM, separated on a single native gel in three adjacent lanes, and then transferred to a single membrane that was marked for subsequent realignment. The membrane for each lane was then cut and probed separately with the different antibodies against AoIRX9, AoIRX10, and AoIRX14A. The membrane was then realigned and subsequently imaged.
Proteomic Analysis
Purified protein samples were digested with trypsin (Promega), and the peptides were analyzed by liquid chromatography-tandem mass spectrometry using an LTQ Orbitrap Elite (Thermo Scientific) with an EASY nano electrospray interface coupled to an Ultimate 3000 RSLC nanosystem (Dionex). The nanoLC system was equipped with an Acclaim Pepmap nano-trap column (C18; 100 Å, 75 μm × 2 cm; Dionex) and a Thermo EASY-Spray column (Pepmap RSLC C18; 2 μm, 100 Å, 75 μm × 25 cm). Sample was loaded onto the enrichment (trap) column at an isocratic flow of 4 μL min−1 of 3% (v/v) acetonitrile containing 0.1% (v/v) formic acid for 5 min before the enrichment column was switched in line with the analytical column. The eluents used for liquid chromatography were 0.1% (v/v) formic acid (solvent A) and 0.1% (v/v) formic acid in acetonitrile (solvent B). The following gradient of solvents was used: 3% B increasing to 12% B over 1 min, 12% B increasing to 35% B over 20 min, followed by 35% B increasing to 80% B over 2 min. The LTQ Orbitrap Elite mass spectrometer was operated in the data-dependent mode with nano-electrospray ionization spray voltage of +2 kV, capillary temperature of 250°C, and S-lens RF value of 55%. Spectra were acquired in positive mode with full-scan range from mass-to-charge ratio 300 to 1,650, with resolution of 100,000 FWHM and a target value of 1 × 106 ions. Each sample was analyzed twice. The analysis used higher collision dissociation for fragmentation for the 10 most intense ions detected in the full-scan mode with charge state n ≥ 2, isolated, and fragmented using normalized collision energy of 35 and activation time of 100 ms.
The higher collision dissociation spectra were analyzed using Mascot (Perkins et al., 1999) to search against an N. benthamiana database (in addition to the heterologously expressed proteins; Bombarely et al., 2012) with the following parameters: enzyme, trypsin; fixed modifications, carbamidomethyl; mass spectrometry peptide tolerance, 5 ppm; tandem mass spectrometry tolerance, 0.1 D; number of missed cleavages, up to one. A protein was determined significant based on a false discovery rate of 1% using a randomized concatenated decoy database.
Microscopy
Imaging of fluorescent constructs was carried out on an inverted Leica SP5 confocal microscope using a 63× PL Apo BL oil objective (numerical aperture of 1.4). A 458-nm laser line, attenuated to 40%, was used to excite mCERULEAN; a 514-nm laser line, attenuated to 20%, was used to excite mVENUS; and a 561-nm laser line, attenuated to 20%, was used to excite mCHERRY. Emissions were detected from 465 to 505 nm, 525 to 555 nm, and 572 to 614 nm, and photodetectors were set at 800, offset by −5. Images were collected using an average of eight optical slices. GA and ER markers used in this study have been described previously (Lee et al., 2014; Wilson et al., 2015).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. IRX9 protein sequence alignments.
Supplemental Figure S2. IRX14 protein sequence alignment.
Supplemental Figure S3. IRX10 protein sequences alignment.
Supplemental Figure S4. Proteomic analysis of GFP-trap-enriched IRX proteins.
Supplemental Figure S5. Western blot of VENUS-conjugated IRX proteins.
Supplemental Figure S6. Expanded images from Figure 9, A to C.
Supplemental Figure S7. Expanded images from Figure 9, D to F.
Supplemental Figure S8. Expanded images from Figure 9, G to I.
Supplemental Figure S9. Negative controls for IRX9 in Figure 10.
Supplemental Figure S10. Negative controls for IRX10 in Figure 10.
Supplemental Figure S11. Negative controls for IRX14A in Figure 10.
Supplemental Figure S12. Expanded images from Figure 10, A to D.
Supplemental Figure S13. Expanded images from Figure 10, E to H.
Supplemental Figure S14. Expanded images from Figure 10, I I to L.
Supplemental Table S1. Protein list of the GFP-trap-enriched XSC.
Supplemental Table S2. Summary of (mc)BiFC assays.
Supplemental Table S3. Primer sequences.
Supplementary Material
Acknowledgments
We thank the Biological Optical Microscopy Platform at the University of Melbourne for access to equipment and Ed Newbigin, Terrance Quon, Joshua Heazlewood, and Staffan Persson for critical comments on the work.
Glossary
- RE
reducing end
- GA
Golgi apparatus
- SDM
site-directed mutagenesis
- XSC
xylan synthase complex
- MM
microsomal membrane
- BiFC
bimolecular fluorescence complementation
- RP
reverse-phase
- DDM
n-dodecyl β-d-maltoside
- ER
endoplasmic reticulum
- mcBiFC
multicolor bimolecular fluorescence complementation
- GO-PROMTO
Golgi protein membrane topology
- TBST
Tris-buffered saline buffer containing 0.1% (v/v) Tween 20
Footnotes
This work was supported by Australian Research Council Centre of Excellence in Plant Cell Walls (grant no. CE110001007 to W.Z., E.R.L., K.L.P., J.Z., I.M.F., K.F., M.S.D., and A.B.), the China National Natural Science Foundation (grant no. 31170165 to A.-M.W.), and the Zhejiang Provincial Natural Science Foundation of China (grant no. LR15C160001 to L.S.).
Articles can be viewed without a subscription.
References
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA III, Orlando R, Scheller HV, Mohnen D (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastawde KB. (1992) Xylan structure, microbial xylanases, and their mode of action. World J Microbiol Biotechnol 8: 353–368 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A (2006) Structures and mechanisms of glycosyltransferases. Glycobiology 16: 29R–37R [DOI] [PubMed] [Google Scholar]
- Brown D, Wightman R, Zhang Z, Gomez LD, Atanassov I, Bukowski JP, Tryfona T, McQueen-Mason SJ, Dupree P, Turner S (2011) Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J 66: 401–413 [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Carroll A, Mansoori N, Li S, Lei L, Vernhettes S, Visser RG, Somerville C, Gu Y, Trindade LM (2012) Complexes with mixed primary and secondary cellulose synthases are functional in Arabidopsis plants. Plant Physiol 160: 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Singh S, Phillips GN Jr, Thorson JS (2011) Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr Opin Biotechnol 22: 800–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vega-Sánchez ME, Verhertbruggen Y, Chiniquy D, Canlas PE, Fagerström A, Prak L, Christensen U, Oikawa A, Chern M, et al. (2013) Inactivation of OsIRX10 leads to decreased xylan content in rice culm cell walls and improved biomass saccharification. Mol Plant 6: 570–573 [DOI] [PubMed] [Google Scholar]
- Chiniquy D, Varanasi P, Oh T, Harholt J, Katnelson J, Singh S, Auer M, Simmons B, Adams PD, Scheller HV, et al. (2013) Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front Plant Sci 4: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SL, Koutaniemi S, Juvonen M, Derba-Maceluch M, Mellerowicz EJ, Tenkanen M (2015) Glucuronic acid in Arabidopsis thaliana xylans carries a novel pentose substituent. Int J Biol Macromol 79: 807–812 [DOI] [PubMed] [Google Scholar]
- Chou YH, Pogorelko G, Zabotina OA (2012) Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 159: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG (2007) A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Pettolino F, Bacic A (2010) Plant cell walls: the skeleton of the plant world. Funct Plant Biol 37: 357–381 [Google Scholar]
- Ebringerova A, Hromadkova Z, Heinze T (2005) Hemicellulose. Adv Polm Sci 186: 1–67 [Google Scholar]
- Faik A. (2010) Xylan biosynthesis: news from the grass. Plant Physiol 153: 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford KL, Zeng W, Heazlewood JL, Bacic A (2015) Characterization of protein N-glycosylation by tandem mass spectrometry using complementary fragmentation techniques. Front Plant Sci 6: 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T, et al. (2013) A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J 76: 128–137 [DOI] [PubMed] [Google Scholar]
- Hao Z, Mohnen D (2014) A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit Rev Biochem Mol Biol 49: 212–241 [DOI] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A, et al. (2012) ARAD proteins associated with pectic arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236: 115–128 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hörnblad E, Ulfstedt M, Ronne H, Marchant A (2013) Partial functional conservation of IRX10 homologs in Physcomitrella patens and Arabidopsis thaliana indicates an evolutionary step contributing to vascular formation in land plants. BMC Plant Biol 13: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Ishii T. (1997) Structure and functions of feruloylated polysaccharides. Plant Sci 127: 111–127 [Google Scholar]
- Jensen JK, Johnson N, Wilkerson CG (2013) Discovery of diversity in xylan biosynthetic genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Front Plant Sci 4: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson NR, Wilkerson CG (2014) Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J 80: 207–215 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron JC, Orler R, Ralph J, Wilkerson CG (2011) The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J 66: 387–400 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, et al. (2008) Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20: 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Wiemels RE, Soya A, Whitley R, Held M, Faik A (2016) Composition, assembly, and trafficking of a wheat xylan synthase complex (XSC). Plant Physiol (in press) 10.1104/pp.15.01777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Xuan Y, Xu M, Wang RS, Ho CH, Lalonde S, You CH, Sardi MI, Parsa SA, Smith-Valle E, et al. (2014) Border control: a membrane-linked interactome of Arabidopsis. Science 344: 711–716 [DOI] [PubMed] [Google Scholar]
- Keppler BD, Showalter AM (2010) IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Mol Plant 3: 834–841 [DOI] [PubMed] [Google Scholar]
- Kerppola TK. (2006) Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc 1: 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77: 521–555 [DOI] [PubMed] [Google Scholar]
- Lampugnani ER, Ho YY, Moller IE, Koh PL, Golz JF, Bacic T, Newbigin E (2016) A glycosyltransferase from Nicotiana alata pollen mediates synthesis of a linear (1,5)-α-l-arabinan when expressed in Arabidopsis. Plant Physiol (in press) 10.1104/pp.15.02005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Kilinc A, Smyth DR (2012) PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J 71: 724–735 [DOI] [PubMed] [Google Scholar]
- Lee C, O’Neill MA, Tsumuraya Y, Darvill AG, Ye ZH (2007a) The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol 48: 1624–1634 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH (2007b) The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol 48: 1659–1672 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Ye ZH (2012a) Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol 53: 135–143 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Ye ZH (2012b) Biochemical characterization of xylan xylosyltransferases involved in wood formation in poplar. Plant Signal Behav 7: 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lampugnani ER, Bacic A, Golz JF (2014) SEUSS and SEUSS-LIKE 2 coordinate auxin distribution and KNOXI activity during embryogenesis. Plant J 80: 122–135 [DOI] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Huang J, Qin L, Huang Y, Zeng W, Rao Y, Li J, Li X, Xu W (2014) Two cotton fiber-associated glycosyltransferases, GhGT43A1 and GhGT43C1, function in hemicellulose glucuronoxylan biosynthesis during plant development. Physiol Plant 152: 367–379 [DOI] [PubMed] [Google Scholar]
- Liwanag AJ, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MC, Clausen MH, Scheller HV (2012) Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell 24: 5024–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund CH, Bromley JR, Stenbæk A, Rasmussen RE, Scheller HV, Sakuragi Y (2015) A reversible Renilla luciferase protein complementation assay for rapid identification of protein-protein interactions reveals the existence of an interaction network involved in xyloglucan biosynthesis in the plant Golgi apparatus. J Exp Bot 66: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer JC, Faria-Blanc N, Yu X, Tryfona T, Sorieul M, Ng YZ, Zhang Z, Stott K, Anders N, Dupree P (2015) An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14. Plant J 83: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa A, Lund CH, Sakuragi Y, Scheller HV (2013) Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci 18: 49–58 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Porchia AC, Sørensen SO, Scheller HV (2002) Arabinoxylan biosynthesis in wheat: characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol 130: 432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratke C, Pawar PM, Balasubramanian VK, Naumann M, Duncranz ML, Derba-Maceluch M, Gorzsás A, Endo S, Ezcurra I, Mellerowicz EJ (2015) Populus GT43 family members group into distinct sets required for primary and secondary wall xylan biosynthesis and include useful promoters for wood modification. Plant Biotechnol J 13: 26–37 [DOI] [PubMed] [Google Scholar]
- Ratnayake S, Beahan CT, Callahan DL, Bacic A (2014) The reducing end sequence of wheat endosperm cell wall arabinoxylans. Carbohydr Res 386: 23–32 [DOI] [PubMed] [Google Scholar]
- Ren Y, Hansen SF, Ebert B, Lau J, Scheller HV (2014) Site-directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. PLoS ONE 9: e105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Hansen SF, Baidoo EE, Hadi MZ, Keasling JD, Scheller HV (2012) Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol 159: 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Scheller HV (2014) Xylan biosynthesis. Curr Opin Biotechnol 26: 100–107 [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW (2004) An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22: 445–449 [DOI] [PubMed] [Google Scholar]
- Shatalov AA, Evtuguin DV, Pascoal Neto C (1999) (2-O-Alpha-D-galactopyranosyl-4-O-methyl-alpha-D-glucurono)-D-xylan from Eucalyptus globulus Labill. Carbohydr Res 320: 93–99 [DOI] [PubMed] [Google Scholar]
- Søgaard C, Stenbæk A, Bernard S, Hadi M, Driouich A, Scheller HV, Sakuragi Y (2012) GO-PROMTO illuminates protein membrane topologies of glycan biosynthetic enzymes in the Golgi apparatus of living tissues. PLoS ONE 7: e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zeng W, Wu A, Picard K, Lampugnani ER, Cheetamun R, Beahan C, Cassin A, Lonsdale A, Doblin MS, et al. (2015) Asparagus spears as a model to study heteroxylan biosynthesis during secondary wall development. PLoS ONE 10: e0123878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan E, Aquin S, Berger N, Landry CR, Nyfeler B, Bouvier M, Michnick SW (2007) Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc Natl Acad Sci USA 104: 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Witters E, Inzé D, De Jaeger G (2008) Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wiggins CAR, Munro S (1998) Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA 95: 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Ho YY, Lampugnani ER, Van de Meene AM, Bain MP, Bacic A, Doblin MS (2015) Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3;1,4)-β-D-glucan in grasses. Plant Cell 27: 754–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57: 718–731 [DOI] [PubMed] [Google Scholar]
- York WS, O’Neill MA (2008) Biochemical control of xylan biosynthesis: which end is up? Curr Opin Plant Biol 11: 258–265 [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A (2010) A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 154: 78–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao P, Yuan JS (2010) Plant protein-protein interaction network and interactome. Curr Genomics 11: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Ouyang K, Gan S, Zeng W, Song L, Zhao S, Li J, Doblin MS, Bacic A, Chen XY, et al. (2014) Biochemical and molecular changes associated with heteroxylan biosynthesis in Neolamarckia cadamba (Rubiaceae) during xylogenesis. Front Plant Sci 5: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Peña MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH III, Darvill AG, York WS, Ye ZH (2005) Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Teng Q, Lee C, Ye ZH (2014) Identification of a disaccharide side chain 2-O-α-D-galactopyranosyl-α-D-glucuronic acid in Arabidopsis xylan. Plant Signal Behav 9: e27933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2015) Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56: 195–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.