Characterization of developing rice seeds under heat stress identifies OsMADS87 as a determinant of seed size sensitivity to increased temperature.
Abstract
Early seed development events are highly sensitive to increased temperature. This high sensitivity to a short-duration temperature spike reduces seed viability and seed size at maturity. The molecular basis of heat stress sensitivity during early seed development is not known. We selected rice (Oryza sativa), a highly heat-sensitive species, to explore this phenomenon. Here, we elucidate the molecular pathways that contribute to the heat sensitivity of a critical developmental window during which the endosperm transitions from syncytium to the cellularization stage in young seeds. A transcriptomic comparison of seeds exposed to moderate (35°C) and severe (39°C) heat stress with control (28°C) seeds identified a set of putative imprinted genes, which were down-regulated under severe heat stress. Several type I MADS box genes specifically expressed during the syncytial stage were differentially regulated under moderate and severe heat stress. The suppression and overaccumulation of these genes are associated with precocious and delayed cellularization under moderate and severe stress, respectively. We show that modulating the expression of OsMADS87, one of the heat-sensitive, imprinted genes associated with syncytial stage endosperm, regulates rice seed size. Transgenic seeds deficient in OsMADS87 exhibit accelerated endosperm cellularization. These seeds also have lower sensitivity to a moderate heat stress in terms of seed size reduction compared with seeds from wild-type plants and plants overexpressing OsMADS87. Our findings suggest that OsMADS87 and several other genes identified in this study could be potential targets for improving the thermal resilience of rice during reproductive development.
Global mean surface temperature is projected to continue to increase and pose a serious challenge to crop production (Shah et al., 2011; Teixeira et al., 2013). Rice (Oryza sativa), which provides more calories per capita globally than any other crop, is highly sensitive to heat stress (Peng et al., 2004; Jagadish et al., 2009; Aghamolki et al., 2014). One estimate suggests that rice yield will decrease by approximately 10% for every 1°C increase in minimum temperature (Peng et al., 2004). The grain yield of almost all crop species is highly sensitive to temperature fluctuations outside the optimal range during reproductive development (Long and Ort, 2010). Therefore, higher global temperatures coupled with increasing population pose a serious challenge for achieving food security. Importantly, the increased population pressure is disproportionately higher in the rice-growing regions of Asia and Africa.
During the rice reproductive stage, several developmental events have an impact on productivity if the plants experience a temperature spike (Shah et al., 2011). One such developmental stage is the early seed development window. The first 2 to 3 d after double fertilization are highly sensitive to environmental stresses and significantly decrease the size of the developing seeds (Folsom et al., 2014). Depending on the duration and intensity of the stress, both yield and grain quality are negatively affected (Krishnan et al., 2011; Lyman et al., 2013). For most higher plants, endosperm cellularization is the key event during early seed development (Brown et al., 1996; Hehenberger et al., 2012). After fertilization, the primary endosperm nucleus undergoes several rounds of mitotic divisions that are uncoupled from cell wall synthesis and cytokinesis to form the syncytium, a cell containing multiple free nuclei. The syncytium stage is conserved in both monocots and dicots (Olsen, 2004). In plants, the syncytium transitions from the free nuclear stage by initiating cellularization of the peripheral nucleus, which is followed by simultaneous cytokinesis and cell wall formation. After that, cell division continues to proceed in a centripetal direction until the endosperm cells occupy most of the central vacuole (Olsen, 2001). Typically, in rice, cellularization is initiated 48 h after fertilization (HAF) and the endosperm is fully cellularized by 96 HAF (Ishikawa et al., 2011; Folsom et al., 2014). Either precocious or delayed syncytium/cellularization transition in endosperm can cause abnormal rice seeds and possibly lead to seed abortion (Ishikawa et al., 2011; Sekine et al., 2013).
Several main regulators involved in early seed development have been characterized in Arabidopsis (Arabidopsis thaliana; Ohad et al., 1996; Grossniklaus et al., 1998; Kang et al., 2008). An important finding in this field was the discovery that the POLYCOMB-REPRESSIVE COMPLEX2 (PRC2), which catalyzes the trimethylation of histone H3 at Lys-27, an epigenetic mark associated with gene silencing, is essential for the transition from the syncytial stage to endosperm cellularization (Pires, 2014; Mozgova and Hennig, 2015). FERTILIZATION-INDEPENDENT SEED (FIS)-PRC2, composed of MEDEA (MEA), FIS2, and FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), functions during early seed development. The endosperms of mea, fis2, and fie mutants overproliferate and fail to cellularize, thus causing seed abortion (Ohad et al., 1996; Grossniklaus et al., 1998). Interestingly, MEA and FIS2 are imprinted and expressed preferentially from the maternal genome (Luo et al., 2000; Xiao et al., 2003). In plants, the imprinted genes have predominantly endosperm-specific expression (Köhler et al., 2012; Gehring, 2013). Several other imprinted genes, such as ARABIDOPSIS FORMIN HOMOLOG5, MATERNALLY EXPRESSED PAB C-TERMINAL, RAS-RELATED NUCLEAR PROTEIN1, and ADMETOS, also affect Arabidopsis seed development and endosperm cellularization (Tiwari et al., 2008; Fitz Gerald et al., 2009; Kradolfer et al., 2013b; Liu et al., 2014). In rice, OsFIE1 is imprinted, and its increased expression after fertilization is associated with the initiation of endosperm cellularization (Luo et al., 2009; Folsom et al., 2014). Epigenetic regulation, both DNA methylation and histone-level modifications, is important for modulating imprinting (Zhang et al., 2012). The mutants of rice METHYLTRANSFERASE1-2 (OsMET1-2) and the demethylase gene REPRESSOR OF SILENCING1a (ROS1a) also show seed developmental defects (Yamauchi et al., 2008; Ono et al., 2012; Hu et al., 2014).
In addition to the PRC2 member genes, several type I MADS box genes, some of which are regulated by PRC2, also promote endosperm proliferation or repress endosperm cellularization in Arabidopsis (Walia et al., 2009; Pires, 2014). For instance, seeds deficient in AGL62 exhibit accelerated endosperm cellularization, suggesting a role for AGL62 in suppressing the initiation of cellularization (Kang et al., 2008). Possibly, Arabidopsis type I AGLs act downstream of FIS-PRC2, because the overproliferation phenotype of the mea or fis mutant can be suppressed by reducing AGL37/PHERES1 (PHE1), AGL62, or AGL90 expression (Köhler et al., 2003; Makarevich et al., 2008; Walia et al., 2009; Hehenberger et al., 2012). Notably, several type I MADS box genes, including paternally expressed PHE1 and AGL92 and maternally expressed AGL36 and At1g59930, also are imprinted (Köhler et al., 2005; Hsieh et al., 2011; Shirzadi et al., 2011). Misregulation of one of the potential rice homologs of PHE1, OsMADS87, correlated with an altered developmental transition in interspecies rice crosses (Ishikawa et al., 2011). Besides type I MADS box genes, other subgroup members such as OsMADS29 also play a critical role in normal rice seed development (Yang et al., 2012; Yin and Xue, 2012; Nayar et al., 2013).
The expression of OsMADS87 is specific to the syncytial stage and is suppressed during endosperm cellularization (Ishikawa et al., 2011; Sekine et al., 2013). More pertinently, the expression of OsMADS87 shows temperature sensitivity and is negatively associated with an increased expression of OsFIE1, the only PRC2 complex member exhibiting thermal sensitivity in developing rice seeds (Folsom et al., 2014). Although the expression of OsMADS87 and its Arabidopsis homolog, PHE1, is associated with early endosperm development, its role in seed development, especially seed size, is not known. More importantly, we currently lack a genome-scale understanding of how increased temperature shapes the transcriptome of a rapidly growing young seed. Given the impact of heat stress on final seed size and grain quality, a better understanding of how the critical transition from the syncytial to the cellularized state of endosperm is affected by heat stress at the molecular level is essential for identifying target genes and molecular pathways for developing rice that is more resilient during this transient but highly sensitive window of reproductive development.
RESULTS
A Transient Heat Stress during Early Seed Development Impacts Final Seed Size
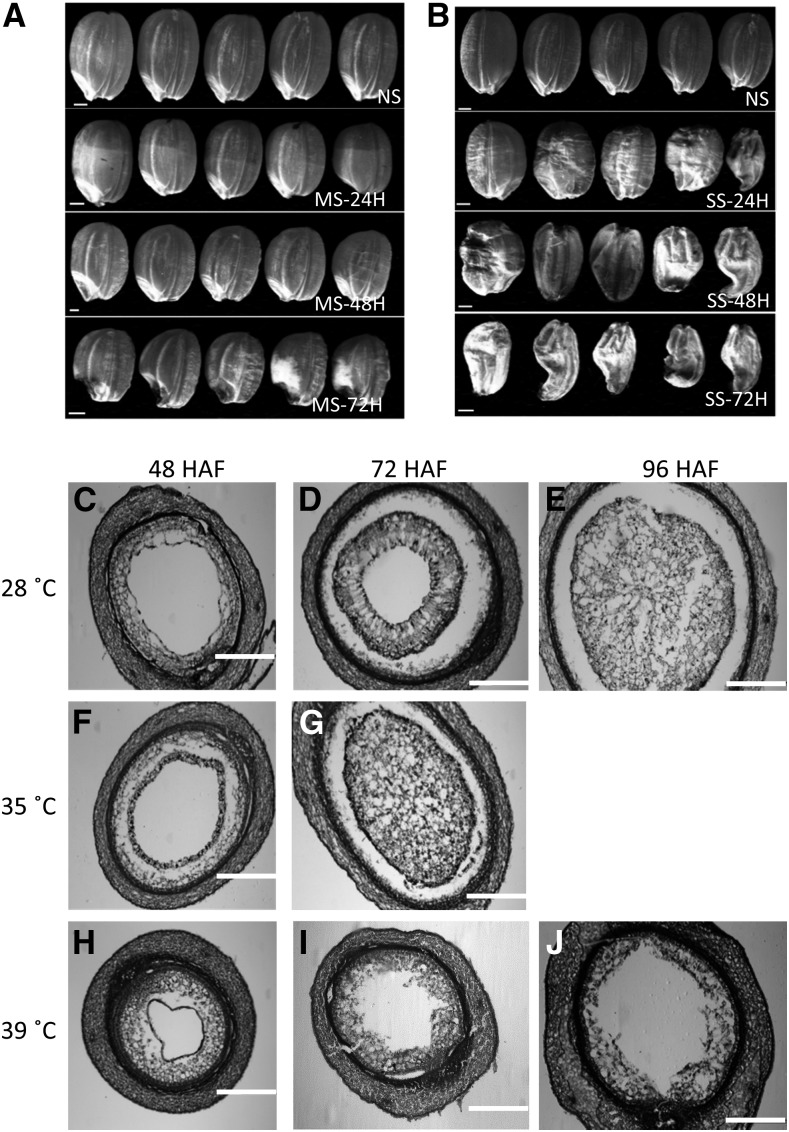
We applied moderate (day/night temperature of 35°C/30°C) and severe (39°C/34°C) heat stresses to developing rice seeds at 24 HAF and maintained them for 24, 48, or 72 h before the stress was relieved and plants were grown to maturity. A parallel set of plants was maintained under control conditions (28°C/25°C). We found that even a transitory heat stress visibly impacted the size and viability of mature grains. We observed a range of seed abnormalities (Fig. 1, A and B). Seeds exposed to severe heat stress were relatively more shriveled (Fig. 1B). For the rice variety Kitaake (ssp. japonica) used in this study, the syncytium/cellularization transition is initiated around 48 HAF but before 72 HAF under control condition (28°C; Fig. 1, C–E). Histological analysis of developing seeds showed that the transition event under moderate heat stress was accelerated, as the endosperm was completely cellularized at 72 HAF (Fig. 1, F and G). However, severe heat stress arrested the cellularization process, as the central vacuole persisted at 96 HAF (Fig. 1, H–J). These results provide developmental insights into the impact of a short-duration heat stress on mature seed size and clearly differentiate the cellular phenotypes associated with moderate and severe heat stress events. The molecular mechanisms underlying the differential developmental sensitivity of the young rice seeds remain largely unexplored.
Figure 1.
Morphological and histological analyses of control and heat-stressed seeds. A and B, Mature seed phenotypes resulting due to a short period of heat stress. The heat stress was imposed at 24 HAF for 24 to 72 h, and then plants were maintained under normal conditions (28°C) until maturity. NS, Nonstressed; MS, moderate stressed; SS, severe stressed; 24H, 48H, and 72H indicate the hours of stress treatment. Bars = 1 mm. C to J, Sections of nonstressed (C–E), moderate-stressed (F and G), and severe-stressed (H–J) young seeds at 48 HAF (C, F, and H), 72 HAF (D, G, and I), and 96 HAF (E and J). Bars = 200 μm.
Heat Stress Activates Transposable Elements
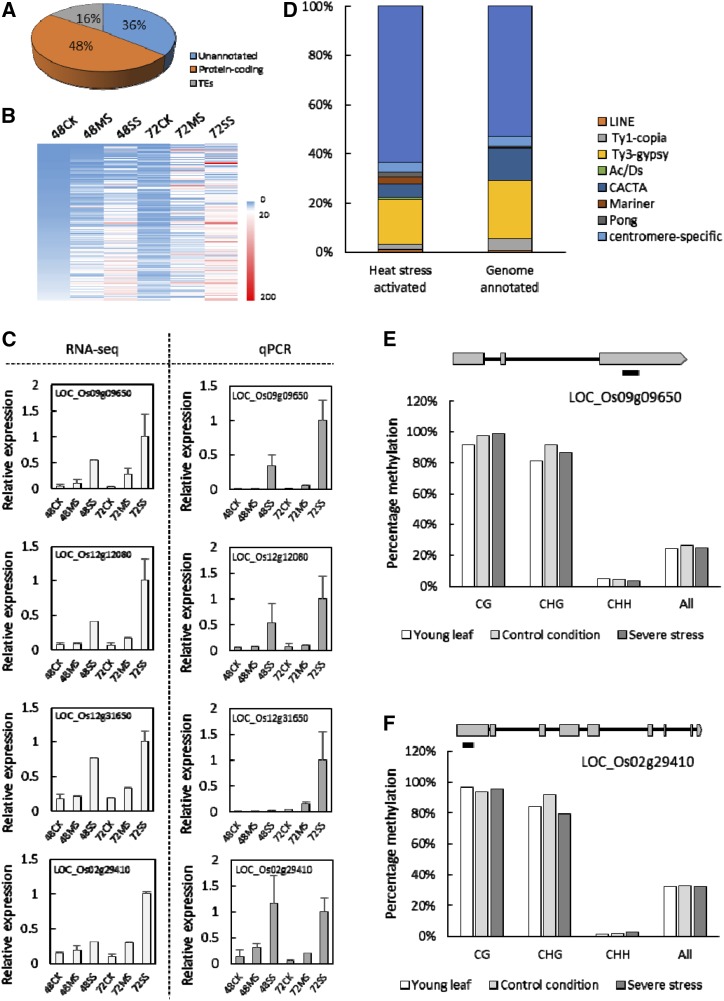
We next explored the changes in the transcriptional landscape in response to temperature-driven perturbation of endosperm developmental events. After fertilization, the rice plants were grown at 28°C/25°C (day/night) for 24 h. Then, the plants were either exposed to a moderate (35°C/30°C) or severe (39°C/34°C) heat stress or kept at 28°C/25°C (control conditions for rice). Young seeds (comprising the seed coat, embryo, and endosperm) were collected from control, moderate, and severe heat-stressed plants at 48 and 72 HAF for transcriptome analysis using the RNA sequencing (RNA-seq) approach (Supplemental Fig. S1). Of the 22,000 expressed genes detected in early developing seeds, approximately 20,706 were commonly identified in both control and heat-stressed samples (Supplemental Table S1). We found 1,265 genes to be expressed only under heat stress (Supplemental Table S2). Gene Ontology (GO) analysis of this set of heat stress-induced genes showed enrichment for secondary metabolism process, response to stimuli, oxygen binding, and transcription regulator activity (Supplemental Table S3), suggesting that these processes may be important for the heat stress response. A surprisingly high number (194 out of 1,265) of the heat-induced genes were annotated as putative transposon or retrotransposon genes (Fig. 2A; Supplemental Table S2), suggesting that temperature stress specifically activates TEs. The transcript abundance of these TEs generally increased with the severity of the stress, suggesting a temperature-sensitive quantitative response (Fig. 2B). We randomly selected two transposon- and two retrotransposon-encoding genes from the heat-induced TE set for validation using quantitative real-time PCR (qPCR) and found the data to be consistent with transcriptome analysis (Fig. 2C). Among the highly activated TEs, Pong, Ac/Ds, and Mariner subclasses of transposons are overrepresented (Fig. 2D), suggesting a higher temperature sensitivity of these TEs in rice.
Figure 2.
Transposable elements (TEs) are activated in heat-stressed young seeds. A, A significant portion of the heat stress-induced genes are TEs. B, Heat map of the expression profiles of heat-induced TEs at different time points and growth conditions. Numbers indicate hours after fertilization; CK, MS, and SS indicate nonstressed, moderate-stressed, and severe-stressed conditions, respectively. C, Gene expression validation of the four randomly selected heat-activated TEs identified from RNA-seq analysis. D, Classification of the heat-activated TEs. E and F, DNA methylation status of two selected TEs in the nonstressed leaves and seeds and severe-stressed seeds. Black bars indicate the regions investigated for methylation.
TEs are silenced by cytosine methylation in plants. To determine if the heat-induced activation of TEs is a seed-specific response resulting due to the DNA hypomethylation associated with developing rice endosperms (Zemach et al., 2010), we quantified the transcript abundance of the four previously selected TEs in heat-treated young leaves and corresponding control samples. Three of the four genes tested exhibited strong heat-induced activation in leaves, indicating that the induction of these TEs is not a seed-specific feature (Supplemental Fig. S2). To address if heat stress activation of the TE is due to an alteration in DNA methylation of the TEs, we selected two heat-induced TEs and performed targeted bisulfite sequencing of regions on these genes known to be methylated based on methylation profiles from a public database. We did not observe significant changes in cytosine methylation levels in the CG, CHG, and CHH contexts in response to heat stress (Fig. 2, E and F). Overall, our results indicate that DNA hypomethylation is unlikely to drive the temperature sensitivity of the TEs.
Identification of Putative Development-Related Genes Associated with the Initiation of Endosperm Cellularization
To identify putative rice genes whose expression is differentially regulated during the transition of endosperm from syncytium to cellularization, we compared the 48- and 72-HAF seeds developing under control conditions. We identified 1,140 differentially expressed genes, with 578 down-regulated and 562 up-regulated genes (Supplemental Table S4). These differentially expressed genes are considered as putative development-related genes (PDRGs) for normal seed development and are more likely to be involved in the transition from the syncytium to the cellularized state of the endosperm. MapMan functional category analysis indicated that cell cycle, DNA synthesis/chromatin structure, and genes associated with transcriptional regulation were significantly enriched (Supplemental Fig. S3). Among the 1,140 PDRGs, 124 genes were annotated as transcription factors (TFs). Among them, the AP2/EREBP, basic helix-loop-helix, WRKY domain, Arabidopsis response regulator, zinc finger homeodomain, and heme activator protein TF families were enriched (Supplemental Fig. S3), indicating that the endosperm undergoes active transcriptome-level reprogramming during the initiation of cellularization.
Heat Stress Impaired the Expression of Development-Related Genes in Young Seeds
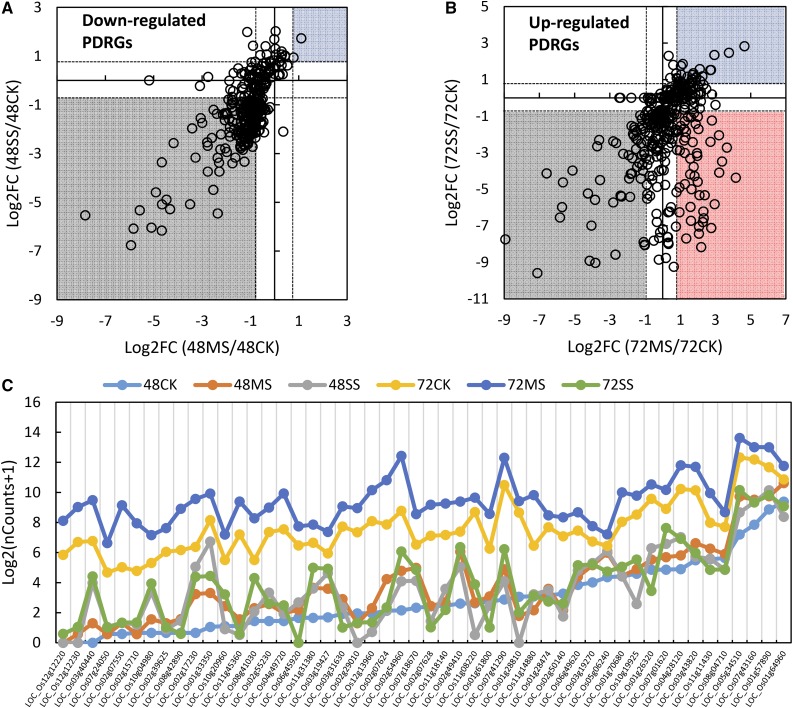
As defined above, PDRGs are a set of genes differentially regulated between 48 and 72 HAF under control conditions. We next focused on the subset of PDRGs that exhibit thermal sensitivity. This gene set is more likely to provide insights on how heat stress affects the syncytium/cellularization transition. Approximately 35% and 42% of PDRGs were differentially expressed under heat stress at 48 and 72 HAF, respectively, compared with the corresponding control conditions (Supplemental Fig. S4). Under control condition, 578 PDRGs were down-regulated at 72 HAF compared with 48 HAF. The majority of these genes were repressed at 48 HAF under heat stress (Fig. 3A). However, the up-regulated PDRGs had a more divergent response to heat stress. The expression of some PDRGs was suppressed at 72 HAF, whereas others were induced by heat stress (Fig. 3B). Notably, a group of PDRGs were up-regulated at 35°C but repressed at 39°C (Fig. 3, B and C; Supplemental Table S5). Considering that the syncytium/cellularization transition is accelerated during the 35°C treatment and delayed during the 39°C, treatment, we hypothesized that these PDRGs with contrasting expression responses to moderate and severe heat stress were associated with the accelerated and arrested syncytium/cellularization endosperm transition, respectively. We examined the expression of these transition-specific and heat-responsive PDRGs in a set of publicly available rice atlas expression data. Most of these genes are preferentially expressed in the endosperm (Supplemental Fig. S5), further supporting our hypothesis of a role for these PDRGs during early endosperm developmental events. Furthermore, this result also suggests that our analysis is enriching for endosperm-associated genes even though we sampled entire seeds for expression analysis. GO analysis indicated that the transcription factors category was significantly (P = 0.016) enriched in these moderate/severe heat stress differentially responsive PDRGs. Seven and four of the 50 PDRGs belong to the defensin (DEF) family and the ZmEBE (embryo sac/basal endosperm transfer layer/embryo-surrounding region) homologous genes, respectively (Supplemental Table S5), indicating that DEFs and ZmEBE homologs, along with additional TFs, may play critical role during the initiation of endosperm cellularization in rice.
Figure 3.
Responses of PDRGs to heat stress. A, Relative expression (indicated as log2 fold change [Log2FC]) of down-regulated PDRGs in moderate- and severe-stressed seeds at 48 HAF in comparison with nonstressed seeds. B, Relative expression of up-regulated PDRGs in moderate- and severe-stressed seeds at 72 HAF in comparison with nonstressed seeds. Gray and blue areas indicate genes that were down- and up-regulated under heat stress conditions, respectively. The red area indicates genes that were up-regulated under moderate stress but down-regulated under severe stress. C, Normalized count numbers (nCounts) of the genes specifically activated under moderate heat stress but repressed under the severe stress condition. Numbers indicate hours after fertilization; CK, MS, and SS indicate nonstressed, moderate-stressed, and severe-stressed conditions, respectively.
Phase-Specific Thermal Sensitivity of the Cell Cycle in the Endosperm
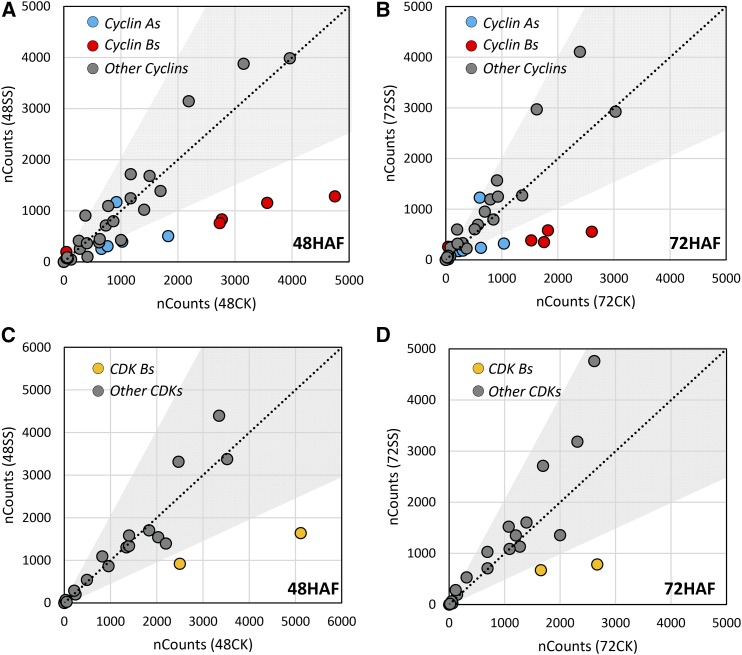
The key feature of the syncytial endosperm is the free nuclear division without cytokinesis (Olsen, 2001, 2004). Eukaryotic cell cycles are tightly regulated by several distinct protein family, such as cyclins, cyclin-dependent kinases (CDKs), cyclin-dependent kinase inhibitors, the protein kinase Wee, and E2F transcription factors (Schaller et al., 2014). Some cell cycle-related genes, such as OsCYCB1;1, OsKRP;3, and OsKRP1, mediate early endosperm development through cell cycle regulation (Barrôco et al., 2006; Guo et al., 2010; Mizutani et al., 2010). Cell cycle genes were enriched in the differential expression comparison between control and moderately heat-stressed seeds based on MapMan analysis (Supplemental Fig. S3). We categorized these genes in the context of their thermal sensitivity during the syncytial/cellularization transition phase by comparing their expression profiles at 48 and 72 HAF. Thirty-five of the 43 cyclins were expressed during early seed development in rice (Supplemental Table S6). However, only cyclin A and B group members exhibited significant transcriptional responses to heat stress. Most of these genes were down-regulated, especially under severe heat stress (Fig. 4, A and B; Supplemental Table S6). Similarly, for the CDK gene family, CDK-B group members were specifically repressed by moderate and severe heat stress (Fig. 4, C and D; Supplemental Table S6). Cyclins A and B coordinate with CDK-B to regulate the transition from the G2 phase to the M phase of cell cycle progression (De Veylder et al., 2007). The thermal sensitivity displayed by cyclins A and B and CDK-B suggests that the G2/M cell cycle phase transition is more sensitive to increased temperature compared with other cell cycle genes. An activated cyclin/CDK complex can induce the expression of the three-MYB-repeat (MYB-3R) family TFs. MYB-3Rs target the M-phase-specific activator cis-elements to activate M phase-specific genes (De Veylder et al., 2007). The rice genome has four putative MYB-3R genes, only two (LOC_Os12g13570 and LOC_Os01g12860) of which were suppressed by moderate heat stress at either 48 or 72 HAF (Supplemental Table S6). The suppression of the two heat-responsive MYB-3R genes was even stronger under severe heat stress. Collectively, our results indicate that the G2/M phase transition during the cell cycle is likely to be one of the determinants of the high heat sensitivity of young rice seeds.
Figure 4.
Expression of cyclin (A and B) and CDK (C and D) genes under control (CK) and severe stress (SS) conditions at 48 HAF (A and C) and 72 HAF (B and D). Cyclin A and B group members are labeled in blue and red, respectively, in A and B. B group members of CDKs are labeled in yellow in C and D. Dots outside of the gray areas show 2-fold changes according to the normalized count numbers (nCounts).
Heat Stress Misregulates Putative Imprinted Genes in Rice
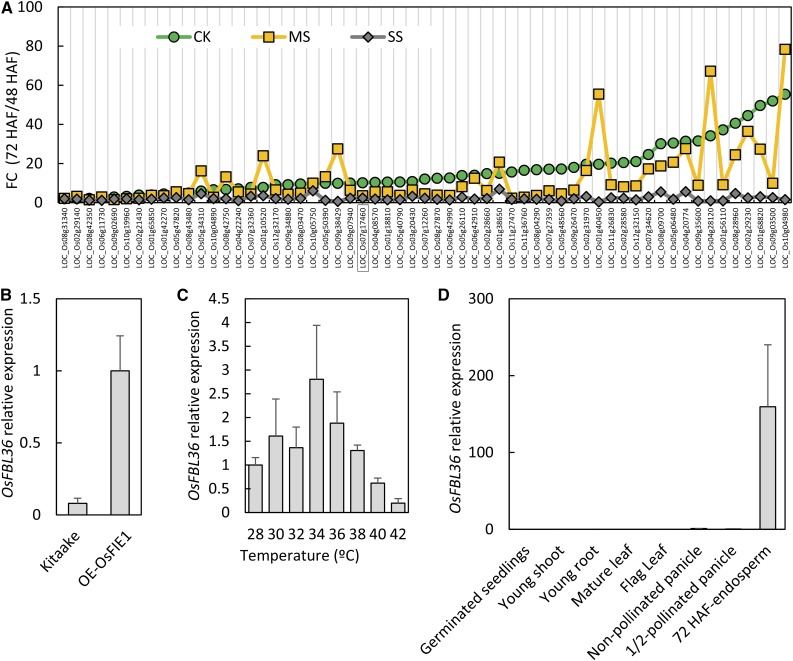
Imprinted genes of plants have been associated with endosperm development, and specifically with the cellularization process (Gehring et al., 2004; Gehring, 2013; Pires, 2014). Given the impact of heat stress on endosperm development in general and specifically on endosperm cellularization, we explored our data set to gain insights into the thermal sensitivity of putative imprinted genes in rice. A previous report had identified 168 putative imprinted genes in rice (Luo et al., 2011). Of this list, 159 genes were expressed in our developing seed samples, and more than 40% were differentially expressed (fold change > 2 and P < 0.05) between 48 and 72 HAF under the control condition. The majority (59 out of 63) of these differentially expressed imprinted genes were up-regulated at 72 HAF (Fig. 5A), suggesting that the activation of imprinted genes coincides with the initiation of endosperm cellularization. Under severe heat stress, we observed a collective lack of activation of these genes at 72 HAF. However, moderate heat stress did not maintain consistent suppression across all the putative imprinted genes (Fig. 5A). In fact, 25 of the 59 genes identified previously had higher transcript abundance at 35°C relative to the seeds developing at 28°C at 72 HAF (Fig. 5A; Supplemental Table S7). Our analysis indicates that heat stress results in mistimed release of imprinting from a subset of seed-specific genes in rice.
Figure 5.
Expression of the putative imprinted genes is interrupted by heat stress. A, Expression-level fold changes (FC) of the putative imprinted genes between 48 and 72 HAF under normal control (CK), moderate stress (MS), and severe stress (SS) conditions. B, Expression of the putative imprinted gene OsFBL36 is activated in transgenic lines overexpressing OsFIE1, a known rice imprinted gene. C, Expression-level response of OsFBL36 to 2°C increments in temperature at 48 HAF. D, Expression profile of OsFBL36 in different tissues. Error bars indicate sd.
OsFIE1 is the only functionally characterized imprinted gene in rice (Zhang et al., 2012). Exogenous expression of OsFIE1 in transgenic rice lines induces the precocious syncytium/cellularization transition of endosperm (Folsom et al., 2014). We identified a previously reported imprinted gene, OsFBL36, to be coexpressed with OsFIE1 based on rice coexpression database search and qPCR confirmation (Luo et al., 2011). Moderate heat stress up-regulated OsFBL36 as well as OsFIE1, while severe stress down-regulated both genes (Fig. 5, A and C). To further confirm the coexpression pattern of OsFIE1 and OsFBL36, we determined the transcript abundance of OsFBL36 in developing rice seeds from transgenic plants overexpressing OsFIE1. OsFBL36 was up-regulated 10-fold in OsFIE1-overexpressing seeds at 72 HAF (Fig. 5B). Previously, OsFIE1 expression was shown to respond to small increments (2°C) in temperature (Folsom et al., 2014). To further test our hypothesis of OsFBL36 coexpression with OsFIE1, we assayed OsFBL36 expression in 48-HAF seeds developing under a range of temperatures with 2°C increments. We observed that OsFBL36 transcript abundance increased until 34°C relative to the control condition seeds and decreased subsequently as the temperature exceeded 34°C (Fig. 5C). This response was similar to the OsFIE1 response, further supporting the coexpression hypothesis of OsFIE1 and OsFBL36. Additionally, similar to the seed-specific expression of OsFIE1, OsFBL36 is expressed predominantly in developing seeds (Fig. 5D).
Erasing methylation marks typically activates the expression of imprinted genes/alleles (Huh et al., 2008; Köhler et al., 2012; Gehring, 2013). Concomitant with the up-regulation of the putative imprinted genes at 72 HAF under the control condition, the expression of DNA methylase-encoding genes, including CHROMOMETHYLASE1 (OsCMT1) and OsCMT3, METHYLTRANSFERASE1 (OsMET1a and OsMET1b), and DOMAINS REARRANGED METHYLTRANSFERASE3 (OsDRM3), decreased significantly (Supplemental Table S8). Transcripts of the putative rice DNA demethylase genes ROS1a, ROS1c, ROSd, DML3a, and DML3b were detectable in developing seeds (Supplemental Table S8). Severe heat stress strongly repressed the expression of OsCMT1, OsCMT3, OsMET1a, and OsMET1b at 48 HAF. Furthermore, OsCMT3 was also down-regulated at 72 HAF by severe heat stress (Supplemental Table S8). The suppression of DNA methylase genes suggests the depletion of methylation levels. However, this result conflicts with our observation of the silencing of most imprinted genes under heat stress. This could be partially due to the fact that severe heat stress also suppressed the expression of ROS1a, the predominant demethylase gene in young rice seed (Supplemental Table S8). To determine the outcome of the temperature-induced differential regulation of known and putative methylation and demethylase genes, we ascertained the methylation status of OsFBL36 using the methylation-sensitive restriction endonuclease MspI assay. We used MspI-treated DNA from 72-HAF seeds as the template to amplify OsFBL36. Because MspI cannot digest methylated recognition sites, the yield of PCR products is indicative of the methylation status of the enzyme recognition sequence. We found that severe heat-stressed seed DNA, but not the moderate heat-stressed seed DNA, was more resistant to MspI, which was indicated as a higher yield of PCR products. This result indicated that OsFBL36 is more methylated under severe heat stress than under control conditions (Supplemental Fig. S6).
Temporal Response of Type I MADS Box Genes during Heat Stress
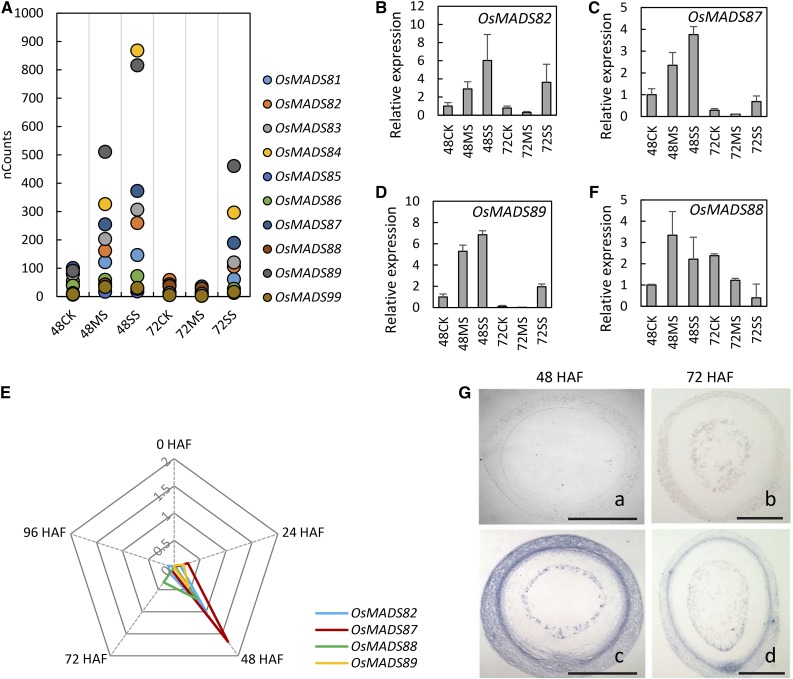
Several M-γ type I MADS box genes were repressed at 72 HAF compared with 48 HAF in rice seeds under control conditions. The repression of these MADS box genes coincided with the initiation of cellularization. At 48 HAF, these M-γ type I MADS box genes had higher transcript abundance under moderate and severe heat stress compared with control seeds (Fig. 6A). The transcript abundance of several MADS box genes was higher under severe stress compared with moderate stress at 48 HAF. At 72 HAF, however, the expression of M-γ type OsMADS genes was suppressed dramatically under moderate stress to a level even lower than in nonstressed seed (Fig. 6A). In contrast, seeds exposed to severe stress still maintained higher expression levels at 72 HAF (Fig. 6A). We selected three genes (OsMADS82, OsMADS87, and OsMADS89) for expression confirmation using qPCR (Fig. 6, B–D). A time-course experiment showed that OsMADS82, OsMADS87, and OsMADS89 are not expressed in unfertilized ovules under normal conditions (Fig. 6E). The expression was greatly increased from 24 to 48 HAF and peaked at 48 HAF. After that, genes were strongly down-regulated, with very low read counts at 72 HAF (Fig. 6A). A similar expression pattern was found for nine of the 10 M-γ type I MADS rice members. OsMADS88 was an exception, as its transcripts persisted at least until 96 HAF under control conditions (Fig. 6, E and F). This suggested that OsMADS88 may have a distinct role and/or regulation compared with other M-γ type I MADS genes in rice during early endosperm development. It is noteworthy that the expression pattern of M-γ type I OsMADS genes, which are suppressed under moderate stress but still showed relatively high levels under severe stress, matched very well with our histological observations that moderate heat stress accelerated cellularization whereas severe stress delayed it. Given the role of AGL62 (type I MADS) in regulating the cellularization timing in Arabidopsis (Kang et al., 2008), we decided to characterize OsMADS87 from our gene list for its role in endosperm development.
Figure 6.
Expression responses of M-γ type I OsMADS genes to heat stress. A, Normalized count numbers of M-γ type I OsMADS genes in the seeds grown under normal control (CK), moderate stress (MS), and severe stress (SS) conditions at 48 and 72 HAF. B to D and F, qPCR confirmation of the expression of OsMADS82 (B), OsMADS87 (C), OsMADS89 (D), and OsMADS88 (F). Error bars indicate sd. E, Time-course expression analysis of OsMADS82, OsMADS87, OsMADS88, and OsMADS89 under normal conditions. The relative expression levels of different OsMADS genes at 0, 24, 48, 72, and 96 HAF are presented. G, In situ hybridization of OsMADS87 at 48 HAF (a and c) and 72 HAF (b and d). In a and b, a sense probe was used, and in c and d, an antisense probe was used. Bars = 0.2 mm.
OsMADS87 Regulates Seed Size in Rice
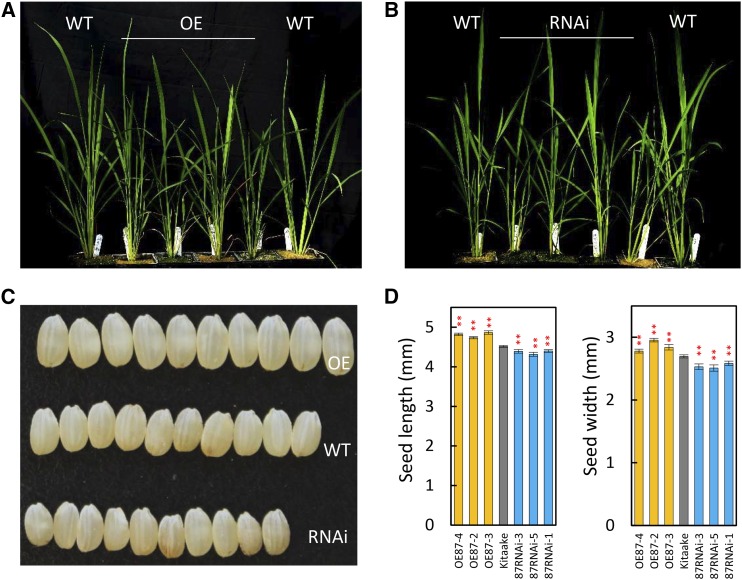
Imprinted gene OsMADS87 is one of the potential rice homologs of the Arabidopsis endosperm-specific gene PHE1 (Ishikawa et al., 2011). OsMADS87 has higher expression during the syncytial stage of rice endosperm development (Ishikawa et al., 2011; Folsom et al., 2014). However, the role of OsMADS87 during rice seed development is not known. Our in situ hybridization assay showed that the OsMADS87 gene is highly expressed in endosperm as well as in pericarp at 48 HAF, whereas gene expression is suppressed at 72 HAF (Fig. 6G). To functionally characterize OsMADS87, we generated independent rice transgenic lines either overexpressing OsMADS87 (OE) or with repressed OsMADS87 expression via RNA interference (RNAi). We found that altering OsMADS87 expression in transgenic rice lines influenced seed size. The vegetative growth of OE and RNAi lines was similar to that of the wild type, other than slightly shorter plant height (Fig. 7, A and B). However, RNAi lines produced smaller seeds and the OE lines had larger seeds at maturity compared with wild-type seeds (Fig. 7, C and D).
Figure 7.
Phenotypic and expression analyses of OsMADS87 transgenic plants. A and B, Phenotypes of OsMADS87 OE plants (A) and RNAi plants (B) at the tillering stage. C, Seed phenotypes of wild-type (WT), OE, and RNAi plants. D, Length and width of wild-type, OE, and RNAi seeds. Asterisks indicate P < 0.01. Error bars indicate sd.
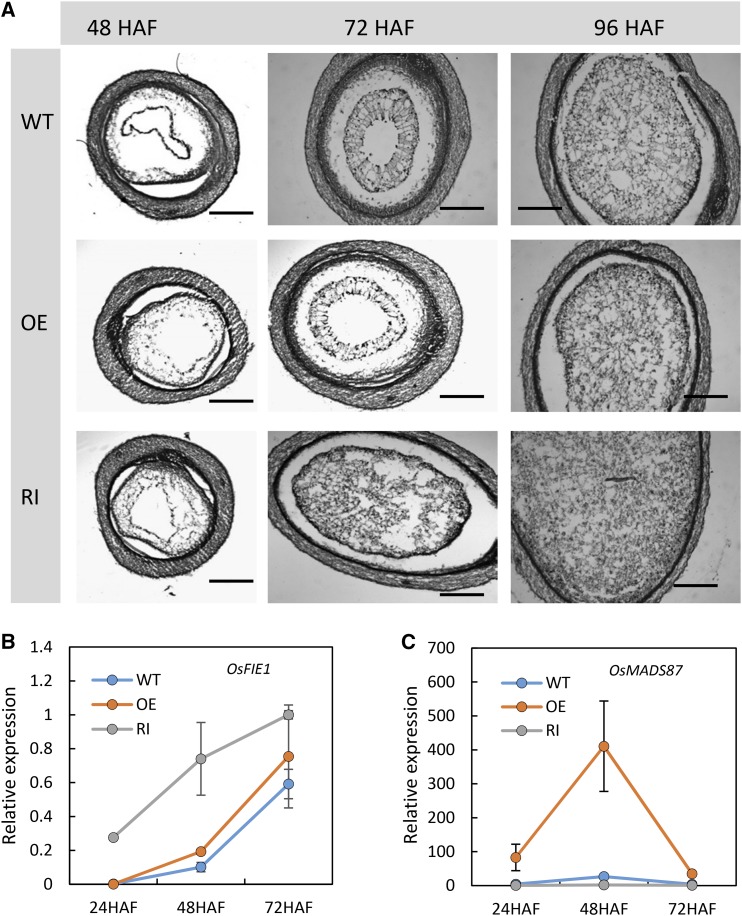
To specifically test if the alteration in seed size is due to a shift in the timing of endosperm cellularization, we examined cross sections of developing seeds from wild-type and transgenic plants at 48, 72, and 96 HAF. Reduced expression of OsMADS87 accelerated endosperm cellularization in the RNAi plants. The seeds from OsMADS87 RNAi plants were completely cellularized by 72 HAF, whereas the wild-type seeds completed cellularization at 96 HAF (Fig. 8A). However, overexpression of OsMADS87 did not exhibit any visual anomaly in the cellularization process (Fig. 8A). We next performed quantitative reverse transcription-PCR to measure the expression level of OsFIE1, a marker gene for cellularization, in developing seeds collected at 24, 48, and 72 HAF from wild-type, OE, and RNAi plants. The transcript level of OsFIE1 begins to accumulate at 48 HAF in wild-type seeds developing under control conditions and precedes the transition to cellularization (Folsom et al., 2014). The expression of OsFIE1 at 72 HAF is higher than at 48 HAF. The expression of OsFIE1 in the OsMADS87 RNAi seeds was significantly higher than in wild-type seeds at 24 and 48 HAF (Fig. 8B). This indicated that seeds deficient in OsMADS87 cellularize earlier than wild-type seeds, which typically initiate cellularization by 72 HAF. The expression of OsFIE1 was not strikingly different in OE seeds at 24, 48, and 72 HAF relative to wild-type controls (Fig. 8B). The expression patterns of OsFIE1 in different genotype plants (Fig. 7B) are consistent with the histological observation of cellularization timing (Fig. 8A). We also measured the expression of OsMADS87 in the same seed samples. Notably, the transcript abundance of OsMADS87 in OE lines is 17-, 90-, and 7-fold higher at 24, 48, and 72 HAF, respectively, relative to wild-type 24-HAF seeds. This suggests that OsMADS87 is repressed strongly at 72 HAF compared with 48 HAF even in OE lines (Fig. 8C). The expression of OsMADS87 in the RNAi lines is consistently lower at all time points (Fig. 8C).
Figure 8.
Histological and expression analyses of wild-type (WT), OE, and RNAi (RI) transgenic seeds. A, Sections of wild-type (top), OE (middle), and RNAi (bottom) seeds at 48 HAF (left), 72 HAF (middle), and 96 HAF (right). Bars = 200 μm. B and C, Relative expression of OsFIE1 (B) and OsMADS87 (C) in developing transgenic and wild-type seeds. Error bars indicate sd.
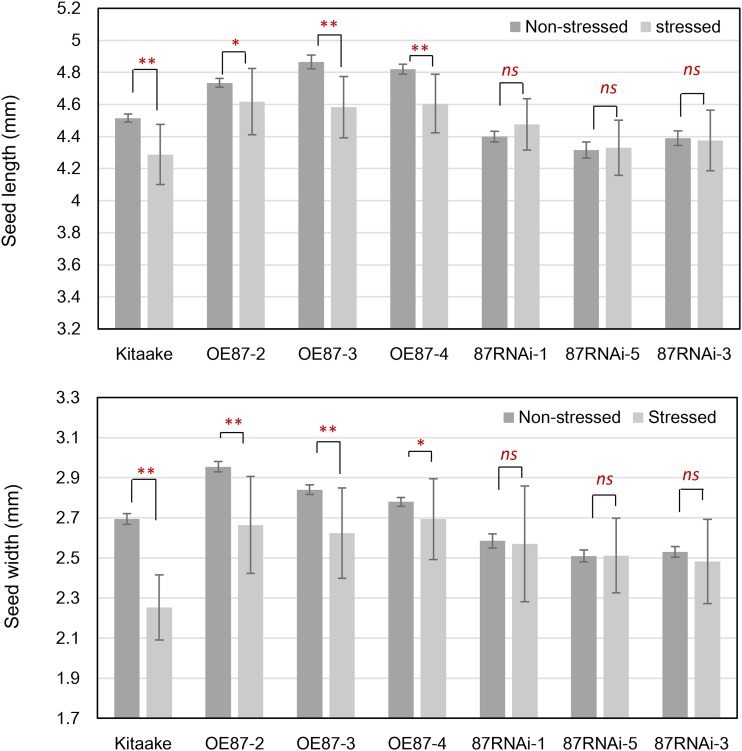
Next, we asked if altering OsMADS87 expression could directly impact the phenotypic response of seeds exposed to a short-duration heat stress. For this, OsMADS87 RNAi and OE lines and wild-type plants were moderately stressed (35°C) as described earlier. The heat stress was relieved, and the plants were allowed to reach maturity under normal conditions. We found that mature seeds from wild-type and OsMADS87 OE plants exhibited greater sensitivity in the context of reduced seed dimensions in response to heat stress compared with OsMADS87 RNAi lines (Fig. 9). Collectively, these data suggest that OsMADS87 regulates seed size and that the heat-induced deficiency of OsMADS87 during the syncytial stage accelerates endosperm cellularization.
Figure 9.
Mature seed length (top) and seed width (bottom) of wild-type, OE, and RNAi rice seeds under normal (nonstressed) and 72-h moderate stress (stressed) conditions at 24 HAF. After a short period of heat stress, the plants were moved to the normal growth condition until maturity. Error bars indicate sd; *, P < 0.05; **, P < 0.01; and ns, not significant.
DISCUSSION
Environmental stress-triggered activation of TEs has been reported in various organisms (Grandbastien, 1998; Hashida et al., 2006; Pecinka et al., 2010; Tittel-Elmer et al., 2010; Cavrak et al., 2014). Putative TEs accounted for 16% of the heat-induced genes identified in young rice seeds in our study (Fig. 2A). Changes in epigenetic modifications, such as DNA methylation and histone modifications, are considered to be the primary reason for the activation of TEs. For example, the activation of some TEs is due to reduced methylation of the Arabidopsis met1 null mutant (Mirouze et al., 2009). The suppression of TEs is temporally released in the central cell and during early stages of seed development under normal conditions (Huh et al., 2008). This process is usually accompanied by the repression of MET1 and the up-regulation of the demethylase gene, DME, in Arabidopsis (Huh et al., 2008). Rice endosperm also undergoes hypomethylation, which releases epigenetic repression from the TEs (Zemach et al., 2010). We found that TE activation was responsive to the severity of the heat stress, with higher temperature resulting in higher levels of activation (Fig. 2, B and C). The severe heat stress also strongly repressed the expression of OsCMT1, OsCMT3, OsMET1a, and OsMET1b in developing rice seeds (Supplemental Table S8). We examined the DNA methylation status of two TEs but did not find any significant changes in the DNA methylation levels of these TEs (Fig. 2, E and F). This suggested that heat stress might not be activating these TEs via DNA demethylation. Prolonged heat stress activated several repetitive elements in Arabidopsis without the loss of DNA methylation and with only minor changes of histone modification (Pecinka et al., 2010). Instead, activation of these TEs was associated with the loss of nucleosomes and heterochromatin decondensation (Pecinka et al., 2010). Whether the rice genome undergoes similar changes remains to be determined. Because we only examined the methylation status in a single region within the coding sequence of each TE, we cannot exclude the possibility of heat-induced cytosine demethylation in a region outside those assayed in our study.
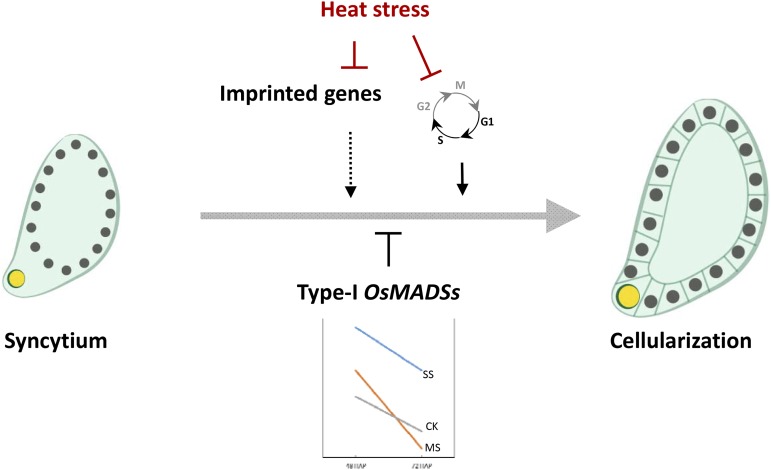
DNA methylation and/or histone modifications have been associated with a parent-of-origin expression pattern of imprinted genes in Arabidopsis (Huh et al., 2008; Köhler et al., 2012; Gehring, 2013). The DNA-demethylating glycosylase DEMETER, the DNA methytransferase MET1, and, for some cases, the core polycomb protein FIE are required to maintain the normal imprinting patterns (Huh et al., 2008). We examined the methylation status changes of a putative rice imprinted gene, OsFBL36, in response to heat stress. The result indicated that OsFBL36 had a higher methylation level under severe heat stress relative to control seeds (Supplemental Fig. S6). A large number of putative rice imprinted genes were also down-regulated in response to heat stress (Fig. 5). The down-regulation of putative imprinted genes in rice was inconsistent with our observed repression of genes encoding methylases by high temperature (Supplemental Table S8). Under control conditions, several imprinted genes were up-regulated when the endosperm transitioned from the syncytial to the cellularized stage, suggesting a transition to biparental expression (Fig. 5). Considering that severe heat stress arrests the transition from syncytium to cellularization of endosperm in rice (Fig. 1, C–J), we hypothesize that a failure to release the epigenetic repression of the silenced parental allele reduces the gene dosage under severe heat stress, thus contributing to the lack of cellularization in severe heat-stressed endosperms (Fig. 10). This potential failure to recover the expression of the imprinted alleles during cellularization could be associated with the stress-induced suppression of ROS1a, the main demethylase gene in rice (Supplemental Table S8). A similar scenario involving imprinting and genome dosage imbalance has been proposed for incompatible F1 interspecies hybrid seeds, where endosperm fails to cellularize and overproliferates, resulting in seed death (Dilkes and Comai, 2004).
Figure 10.
Proposed model incorporating the main genes and pathways that are impacted when young rice seeds are exposed to heat stress. Arrows and T bars indicate signaling promotion and inhibition, respectively. CK, MS, and SS indicate nonstressed, moderate-stressed, and severe-stressed conditions, respectively.
Our transcriptome data indicate that the regulation of imprinted genes is a contributing factor in heat stress-induced aberrations in endosperm development. Several of the genes identified from this analysis, such as OsFIE1, affect rice seed development (Folsom et al., 2014). Imprinted gene ROS1b encodes for a putative DNA demethylase. Its close homolog ROS1a also affects endosperm development via a nonequivalent maternal and paternal contribution pattern (Ono et al., 2012). Another imprinted target, CHALK5, underlies a quantitative trait locus controlling endosperm chalkiness in rice (Li et al., 2014). These findings support our hypothesis that the misregulation of imprinted genes may cause endosperm developmental abnormalities under heat stress.
AGL62, a type I MADS box gene, promotes nuclear proliferation and represses cellularization in Arabidopsis (Kang et al., 2008). Overactivation of this group of MADS box genes is associated with endosperm failure in Arabidopsis (Kang et al., 2008; Walia et al., 2009; Kradolfer et al., 2013a). However, to date, none of the rice type I MADS box genes have been functionally characterized. Rice type I MADS box genes are expressed postfertilization and largely repressed before endosperm cellularization (Fig. 6E). However, severe heat stress prevented the timely down-regulation of these genes (Fig. 6, A–D). At moderate heat stress, the expression of a subset of MADS box genes (OsMADS82, OsMADS87, and OsMADS88) was higher at 48 HAF and lower at 72 HAF relative to control seeds (Fig. 6, A–D). This suggested that suppression of the type I OsMADS genes occurs earlier than in control seeds, consistent with their syncytium-specific expression (Fig. 10). Our phenotypic characterization of OsMADS87, the rice homolog of PHE1, further substantiated the role of type I MADS box genes in regulating the developmental transition of early endosperm in rice. OsMADS87 is an imprinted gene with maternal expression (Ishikawa et al., 2011). Although the disruption of PHE1 does not have a seed size or developmental phenotype (Köhler et al., 2005), we found OsMADS87 to affect the initiation of endosperm cellularization and seed size (Fig. 7, C and D). Based on the transcript abundance of OsFIE1, which accumulates as the endosperm transitions toward cellularization (Fig. 8B), our data suggest that seeds deficient in OsMADS87 are smaller and exhibit accelerated cellularization compared with wild-type seeds. We did not observe an expected corresponding delay in the induction of OsFIE1 (Fig. 8B), indicative of cellularization initiation in OsMADS87 OE seeds, which were significantly larger at maturity. Final seed size is also determined by other factors, such as the rate of mitotic divisions after cellularization and grain-filling rate, which could possibly be disturbed due to the accumulation of OsMADS87 in cellularized endosperm. Another notable observation from our study was the relative seed size insensitivity of the OsMADS87 RNAi seeds when exposed to a short-duration moderate heat stress relative to seeds from OsMADS87 OE and wild-type plants. This suggests that, even though OsMADS87-deficient seeds are smaller than wild-type seeds under control conditions, a moderate heat stress does not further reduce the seed size to the degree observed in wild-type and OsMADS87 OE plants.
In this study, we identified a set of PDRGs differentially responding to moderate and severe heat stress. We assume that these genes are at least partially responsible for the opposite effects of moderate and severe heat stress on endosperm cellularization. Among the PDRGs, OsNF-YB1, an endosperm-specific gene, was recently reported to play a role in maintaining endosperm cell proliferation at the early endosperm developmental stage (Sun et al., 2014). Figueiredo et al. (2015) found that auxin is necessary for proper endosperm development, a process dependent on the MADS box transcription factor AGL62 (Figueiredo et al., 2015). On our PDRG list, OsIAA29 is an endosperm-specifically expressed AUXIN/INDOLE-3-ACETIC ACID family gene, which is possibly important to auxin-mediated endosperm development in rice, similar to that in Arabidopsis. In addition, a flavin monooxygenase gene involving auxin biosynthesis, OsYUCCA12, also is included in our PDRGs. Moreover, the endosperm-preferential expression pattern of the thermo differentially responsive PDRGs strongly implies that these PDRGs are very important to rice endosperm development, although we lack genetic evidence so far.
Core cell cycle-related genes, CYCLIN A and B and CDK-B group members, were largely repressed by heat stress in developing rice seeds. These genes are important for the cell cycle G2/M phase transition (Schaller et al., 2014). We hypothesize that heat stress may predominantly affect M phase of the cell cycle to alter endosperm development in young seeds (Figs. 4 and 10). This hypothesis is further supported by our observation that several activators of M phase-specific genes, the MYB-3R transcription factors, were suppressed by heat stress (Supplemental Table S6). The cell cycle genes generally showed consistent changes in response to moderate and severe stress, although the differential fold change in expression was higher during severe heat stress (Supplemental Table S6). The differential suppression of cell cycle-related genes under moderate and severe heat may have contrasting phenotypic consequences. Severe heat stress arrested seed development, as indicated by our histological observations and transcriptome analysis (Fig. 1, H–J). This could possibly be due to a repression of the cell cycle genes. However, moderate heat stress accelerated the transition toward cellularization (Fig. 1, F and G). The transition to cellularization results in the formation of cell wall around the nucleus from the syncytial stage. Concomitant with the transition to the cellularization stage, the rate of cell division decelerates, as suggested by a decrease in the expression of cell cycle genes at 72 HAF relative to 48 HAF under control conditions. Therefore, we propose that the decreased expression of cell cycle genes under moderate heat stress could be a result of precocious cellularization and the consequent decrease in the cell cycle, which typically is observed at 72 HAF under control conditions.
Our work provides novel insights into the molecular components of early rice seed development that contribute toward the thermal sensitivity of rice seeds. We highlight specific genes and gene families that are significantly affected by heat stress. Some of these genes and, perhaps, entire pathways could be potential targets for improving the climate resilience of rice in the future. As an effort in this direction, we present data for OsMADS87, which exhibited high heat sensitivity and had role in endosperm cellularization initiation and the regulation of final seed size (Fig. 7).
Given that temperature is one of the most dominant factors driving the geographical distribution of plant species, it is important to understand the molecular processes that impact the fitness and productivity of species in ecological and agricultural settings. During development, young seeds (specially the endosperm) exhibit high sensitivity to environmental and genomic perturbations, as indicated by high-temperature sensitivity and postzygotic lethality in incompatible interspecies and interploidy hybrids. Therefore, early endosperm development could possibly be conceived as a developmental bottleneck, a limitation for agricultural productivity as average temperatures continue to rise. Findings from others and the genomic overview provided by this study improve our understanding of this highly heat-sensitive developmental phase. The potential targets characterized in this study can be useful for developing more stress-resilient future crops.
MATERIALS AND METHODS
Growth Conditions and Heat Treatment
After transplantation, rice (Oryza sativa) plants were grown in a greenhouse with 14-h-light/10-h-dark diurnal conditions. The temperature was maintained between 27°C and 30°C. To apply the heat stress treatments, plants were transferred to growth chambers (14-h-day and 10-h-night settings, 28°C/25°C day/night temperature, and 50% humidity) several days before flowering. A parallel set of control plants were also grown in a growth chamber. On the day of flowering, fertilized seeds were labeled and maintained at 28°C/25°C day/night temperature for 24 h after fertilization. At 24 HAF, two different heat stress conditions were imposed on the plants. For moderate stress, the growth chamber day/night temperature conditions were set at 35°C/30°C. For the severe stress, the plants were exposed to 39°C/35°C diurnal temperatures (Supplemental Fig. S1). Corresponding control plants were maintained under a 28°C/25°C diurnal temperature regime. We selected 35°C and 39°C as the two heat stress treatments because temperatures in this range are commonly observed in tropical and subtropical rice-growing regions. Young developing seeds were collected from control, moderately stressed, and severely stressed plants at 48 and 72 HAF for RNA isolation. For morphological analysis of mature seeds, plants were moved back to the greenhouse conditions described above after 24, 48, or 72 h of stress or control treatments. Seeds were collected and analyzed at maturity after drying them for 7 d at 37°C.
DNA and RNA Extraction, Reverse Transcription-PCR, and Real-Time PCR Assay
Genomic DNA and total RNA were isolated from developing seeds at the three temperature treatments using the Qiagen DNeasy Plant Mini Kit and RNeasy Plant Mini Kit following the manual instructions. Total RNA was treated with DNaseI to remove DNA contamination. Complementary DNA was synthesized using the SuperScript VILO Kit (Invitrogen). A 5× dilution of synthesized complementary DNA was used for qPCR. IQ SYBR Green Supermix (Bio-Rad) was used for the reactions. For qPCR, we performed at least three independent biological replicates. Relative expression level was calculated by the Delta-Delta-Ct method (Livak and Schmittgen, 2001). Primers used in this study are listed in Supplemental Table S9.
RNA-seq and Differential Expression Analysis
Using Trimmomatic (Bolger et al., 2014), each 101-bp single-end RNA-seq read was trimmed to make sure the average quality score was greater than 30 and there was a minimum length of 70 bp. All trimmed short reads were mapped to the rice ssp. japonica genome (version 7.0) using TopHat (Trapnell et al., 2009), allowing up to two base mismatches per read. Reads mapped to multiple locations were discarded. Numbers of reads in genes were counted by the HTSeq-count tool using corresponding rice gene annotations, and the union resolution mode was used (Anders et al., 2015). For pairwise comparisons, differentially expressed genes were identified using DEseq (Anders and Huber, 2010) to analyze the numbers of reads aligned to genes. The thresholds for differential expression were set at fold change greater than 1.62 and adjusted P < 0.001 for the null hypothesis. The online tool agriGO (Du et al., 2010) was used for GO analysis. Functional category analysis was conducted by MapMan software (Usadel et al., 2009). The heat map of expression profiles of the genes of interest was generated by the RiceXPro database (Sato et al., 2013) according to the microarray data deposited in the database.
Seed Sectioning
Rice plants were cultured as described earlier, and plants were transferred to growth chambers for heat stress or control treatment. Developing rice seeds were harvested at different times points (48, 72, and 96 HAF) and fixed in 3.7% (w/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol, vacuum infiltrated, and stored at 4°C overnight. Samples were dehydrated through a graded ethanol series and infiltrated with xylene. Seed samples were embedded in paraffin (Fisher Scientific), sectioned at 10 µm, and stained with 0.1% Toluidine Blue. Sections were observed and photographed using a bright-field microscope (Leica DM-2500). Three independent experiments were performed to collect seeds for sectioning. The images of seed sections are representative of our observations across the replicates.
In Situ Hybridization
RNA in situ hybridization was performed according to the manual (Zachgo, 2002). Briefly, a 139-bp gene-specific fragment was amplified using primers (forward, 5′-TGAGCAACTTACCAGCCTT-3′, and reverse, 5′-CCGTGGTGTAAGGCAGTA-3′) and cloned into pGEM-T Easy vector (Promega). The sequence was confirmed and then used to generate RNA probes by in vitro transcription using the DIG RNA Labeling Kit as described in the manufacturer’s instructions (Roche). RNA transcript by T7 and SP6 polymerase was applied to slides as sense and antisense probes, respectively. Images were captured with a microscope (Leica).
Bisulfite Sequencing and MspI-PCR Assay
The same amount of genomic DNA (1 μg) was isolated from control and heat-stressed seeds and treated with sodium bisulfite using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s instructions. Three microliters of the treated DNA was used as the template for PCR. The PCR products were cloned into pGEM-T Easy vector (Promega). At least 12 colonies were selected for sequencing. For the MspI assay, the same amount of DNA was treated with MspI or water (mock) for 3 h and purified using the GeneJET gel extraction kit (Thermo Scientific). Equal amounts of digested and mock DNA were used as templates for PCR.
OsMADS87 Cloning and Transformation
The OsMADS87 (LOC_Os03g38610) full-length coding sequence was PCR amplified using specific primers. The OsMADS87 PCR product was cloned into pENTR/D-TOPO entry vector (Invitrogen) and then transferred to a final Ubi-MADS87 destination vector using LR Clonase recombination (Invitrogen). For knockdown lines, a 210-bp specific region of OsMADS87 was amplified by the use of specific primer sets and cloned into the pANDA RNAi expression vector with a Gateway cloning kit (Invitrogen). The final constructs were transformed to Agrobacterium tumefaciens strain EHA105. Rice transformation was performed as described by Cheng et al. (1997). Hygromycin was used to screen transformed resistant calli. Transgenic plants were regenerated from resistant callus. OE and RNAi T3 generation homozygous lines were used for this study.
Morphometric Measurements
Mature seeds from OsMADS87 OE and RNAi plants were used to measure seed width and length. For all measurements, seeds were obtained from 10 to 15 plants per replicate, and three replicates from three independent experiments were used for these measurements. At least 100 seeds were used for these measurements. Student’s t test was used to determine the statistical significance for seed size variation.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Scheme of the heat stress treatment for young seeds after fertilization.
Supplemental Figure S2. Relative expression of four randomly selected TEs in heat-stressed young leaves.
Supplemental Figure S3. MapMan functional category analysis of the PRDGs identified from early developed rice seeds.
Supplemental Figure S4. Venn diagrams of PDRGs and heat-responsive genes at 48 and 72 HAF.
Supplemental Figure S5. Expression profiles of the severe-stress-repressed but moderate-stress-activated PDRGs in various tissues.
Supplemental Figure S6. Methylation status of OsFBL36 detected by the methylation-sensitive restriction endonuclease MspI assay.
Supplemental Table S1. Summary of short-read quality, alignment, and number of expressed genes.
Supplemental Table S2. List of heat stress-induced genes.
Supplemental Table S3. GO enrichment of heat-induced genes.
Supplemental Table S5. Normalized count numbers of PDRGs differentially responsive to moderate and severe heat stress.
Supplemental Table S6. Normalized read number of core cell cycle genes.
Supplemental Table S7. Normalized read number of differentially expressed putative imprinted genes.
Supplemental Table S8. Normalized read number of methylase and demethylase genes.
Supplemental Table S9. Primers used in this study.
Supplementary Material
Acknowledgments
The transcriptome analysis was performed using computational resources from the Holland Computing Center at the University of Nebraska-Lincoln; we thank Dr. Paul Staswick, Dr. Sandeep Sharma, Dr. Aaron Schmitz, and Dr. Gautam Sarath for critical reading of the article.
Glossary
- HAF
hours after fertilization
- RNA-seq
RNA sequencing
- GO
Gene Ontology
- TE
transposable element
- qPCR
quantitative real-time PCR
- PDRGs
putative development-related genes
- TF
transcription factor
- RNAi
RNA interference
- OE
overexpression
Footnotes
This work was supported by the National Science Foundation (grant no. 1121648 to H.W.).
Articles can be viewed without a subscription.
References
- Aghamolki MTK, Yusop MK, Oad FC, Zakikhani H, Jaafar HZ, Kharidah SSM, Hanafi MM (2014) Response of yield and morphological characteristic of rice cultivars to heat stress at different growth stages. Int J Biol Biomol Agric Food Biotechnol Eng 8: 94–96 [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrôco RM, Peres A, Droual AM, De Veylder L, Nguyen SL, De Wolf J, Mironov V, Peerbolte R, Beemster GTS, Inzé D, et al. (2006) The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice. Plant Physiol 142: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA (1996) Development of the endosperm in rice (Oryza sativa L.): cellularization. J Plant Res 109: 301–313 [Google Scholar]
- Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O (2014) How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10: e1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Sardana RK, Altosar I (1997) Rice transformation by Agrobacterium infection: recombinant proteins from plants: production and isolation of clinically useful compounds. Methods Biotechnol 3: 1–9 [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Dilkes BP, Comai L (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C (2015) Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184. [DOI] [PubMed] [Google Scholar]
- Fitz Gerald JN, Hui PS, Berger F (2009) Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136: 3399–3404 [DOI] [PubMed] [Google Scholar]
- Folsom JJ, Begcy K, Hao X, Wang D, Walia H (2014) Rice fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol 165: 238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M. (2013) Genomic imprinting: insights from plants. Annu Rev Genet 47: 187–208 [DOI] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL (2004) Imprinting and seed development. Plant Cell (Suppl) 16: S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]; Grandbastien MA. (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3: 181–187 [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Guo J, Wang F, Song J, Sun W, Zhang XS (2010) The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231: 293–303 [DOI] [PubMed] [Google Scholar]
- Hashida SN, Uchiyama T, Martin C, Kishima Y, Sano Y, Mikami T (2006) The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 18: 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li N, Xu C, Zhong S, Lin X, Yang J, Zhou T, Yuliang A, Wu Y, Chen YR, et al. (2014) Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc Natl Acad Sci USA 111: 10642–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer RL (2008) Cellular programming of plant gene imprinting. Cell 132: 735–744 [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T (2011) Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65: 798–806 [DOI] [PubMed] [Google Scholar]
- Jagadish SVK, Cairns J, Lafitte R, Wheeler TR, Price AH, Craufurd PQ (2009) Genetic analysis of heat tolerance at anthesis in rice. Crop Sci 50: 1633–1641 [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Köhler C, Wolff P, Spillane C (2012) Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol 63: 331–352 [DOI] [PubMed] [Google Scholar]
- Kradolfer D, Hennig L, Köhler C (2013a) Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet 9: e1003163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C (2013b) An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev Cell 26: 525–535 [DOI] [PubMed] [Google Scholar]
- Krishnan P, Ramakrishnan B, Reddy KR, Reddy VR (2011) High-temperature effects on rice growth, yield, and grain quality. Adv Agron 111: 87–206 [Google Scholar]
- Li Y, Fan C, Xing Y, Yun P, Luo L, Yan B, Peng B, Xie W, Wang G, Li X, et al. (2014) Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat Genet 46: 398–404 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Liu P, Qi M, Wang Y, Chang M, Liu C, Sun M, Yang W, Ren H (2014) Arabidopsis RAN1 mediates seed development through its parental ratio by affecting the onset of endosperm cellularization. Mol Plant 7: 1316–1328 [DOI] [PubMed] [Google Scholar]
- Long SP, Ort DR (2010) More than taking the heat: crops and global change. Curr Opin Plant Biol 13: 241–248 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Platten D, Chaudhury A, Peacock WJ, Dennis ES (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2: 711–723 [DOI] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman NB, Jagadish KSV, Nalley LL, Dixon BL, Siebenmorgen T (2013) Neglecting rice milling yield and quality underestimates economic losses from high-temperature stress. PLoS ONE 8: e72157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Villar CBR, Erilova A, Köhler C (2008) Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121: 906–912 [DOI] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Naganuma T, Tsutsumi K, Saitoh Y (2010) The syncytium-specific expression of the Orysa;KRP3 CDK inhibitor: implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm. J Exp Bot 61: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Hennig L (2015) The polycomb group protein regulatory network. Annu Rev Plant Biol 66: 269–296 [DOI] [PubMed] [Google Scholar]
- Nayar S, Sharma R, Tyagi AK, Kapoor S (2013) Functional delineation of rice MADS29 reveals its role in embryo and endosperm development by affecting hormone homeostasis. J Exp Bot 64: 4239–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL (1996) A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA 93: 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 52: 233–267 [DOI] [PubMed] [Google Scholar]
- Olsen OA. (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell (Suppl) 16: S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Yamaguchi K, Fukada-Tanaka S, Terada R, Mitsui T, Iida S (2012) A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J 71: 564–574 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG (2004) Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA 101: 9971–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND. (2014) Seed evolution: parental conflicts in a multi-generational household. Biomol Concepts 5: 71–86 [DOI] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41: D1206–D1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Street IH, Kieber JJ (2014) Cytokinin and the cell cycle. Curr Opin Plant Biol 21: 7–15 [DOI] [PubMed] [Google Scholar]
- Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, Kurata N, Kinoshita T (2013) Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J 76: 792–799 [DOI] [PubMed] [Google Scholar]
- Shah F, Huang J, Cui K, Nie L, Shah T, Chen C, Wang K (2011) Impact of high-temperature stress on rice plant and its traits related to tolerance. J Agric Sci 149: 545–556 [Google Scholar]
- Shirzadi R, Andersen ED, Bjerkan KN, Gloeckle BM, Heese M, Ungru A, Winge P, Koncz C, Aalen RB, Schnittger A, et al. (2011) Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet 7: e1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ling S, Lu Z, Ouyang YD, Liu S, Yao J (2014) OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene 551: 214–221 [DOI] [PubMed] [Google Scholar]
- Teixeira EI, Fischer G, van Velthuizen H, Walter C, Ewert F (2013) Global hot-spots of heat stress on agricultural crops due to climate change. Agric Meteorol 170: 206–215 [Google Scholar]
- Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I (2010) Stress-induced activation of heterochromatic transcription. PLoS Genet 6: e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Schulz R, Ikeda Y, Dytham L, Bravo J, Mathers L, Spielman M, Guzmán P, Oakey RJ, Kinoshita T, et al. (2008) MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ 32: 1211–1229 [DOI] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL (2003) Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901 [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Moritoh S, Johzuka-Hisatomi Y, Ono A, Terada R, Nakamura I, Iida S (2008) Alternative splicing of the rice OsMET1 genes encoding maintenance DNA methyltransferase. J Plant Physiol 165: 1774–1782 [DOI] [PubMed] [Google Scholar]
- Yang X, Wu F, Lin X, Du X, Chong K, Gramzow L, Schilling S, Becker A, Theißen G, Meng Z (2012) Live and let die: the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS ONE 7: e51435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin LL, Xue HW (2012) The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell 24: 1049–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachgo S. (2002) In situ hybridization. In: Gilmartin PM, Blower C (eds) Molecular plant biology. Oxford University Press, Oxford, pp 41–63 [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cheng Z, Qin R, Qiu Y, Wang JL, Cui X, Gu L, Zhang X, Guo X, Wang D, et al. (2012) Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.