The biosynthesis and spatial positioning of β-carboxysomes, the carbon fixation machinery in cyanobacteria, are modulated by light intensity and redox state of photosynthetic electron transport chain.
Abstract
Cyanobacteria have evolved effective adaptive mechanisms to improve photosynthesis and CO2 fixation. The central CO2-fixing machinery is the carboxysome, which is composed of an icosahedral proteinaceous shell encapsulating the key carbon fixation enzyme, Rubisco, in the interior. Controlled biosynthesis and ordered organization of carboxysomes are vital to the CO2-fixing activity of cyanobacterial cells. However, little is known about how carboxysome biosynthesis and spatial positioning are physiologically regulated to adjust to dynamic changes in the environment. Here, we used fluorescence tagging and live-cell confocal fluorescence imaging to explore the biosynthesis and subcellular localization of β-carboxysomes within a model cyanobacterium, Synechococcus elongatus PCC7942, in response to light variation. We demonstrated that β-carboxysome biosynthesis is accelerated in response to increasing light intensity, thereby enhancing the carbon fixation activity of the cell. Inhibition of photosynthetic electron flow impairs the accumulation of carboxysomes, indicating a close coordination between β-carboxysome biogenesis and photosynthetic electron transport. Likewise, the spatial organization of carboxysomes in the cell correlates with the redox state of photosynthetic electron transport chain. This study provides essential knowledge for us to modulate the β-carboxysome biosynthesis and function in cyanobacteria. In translational terms, the knowledge is instrumental for design and synthetic engineering of functional carboxysomes into higher plants to improve photosynthesis performance and CO2 fixation.
Compartmentalization of metabolic pathways in cells provides the fundamental basis for enhancing and modulating the cellular metabolism. Many prokaryotes have evolved specialized metabolic organelles, known as bacterial microcompartments, to sequester key metabolic pathways and thereby improve the efficiency of metabolic activities (for reviews, see Kerfeld et al., 2010; Bobik et al., 2015). Unlike eukaryotic organelles, bacterial microcompartments are assembled entirely by proteins. These organelles consist of interior enzymes that catalyze sequential metabolic reactions (Yeates et al., 2010), surrounded by a single-layer proteinaceous shell (Kerfeld et al., 2005; Tsai et al., 2007; Tanaka et al., 2008; Sutter et al., 2016). The shell facets are composed of hexameric and pentameric proteins, resulting in an overall shell architecture resembling an icosahedral viral capsid (Kinney et al., 2011; Hantke et al., 2014; Kerfeld and Erbilgin, 2015). Interactions between shell proteins are important for the self-assembly of the shell (Sutter et al., 2016). The selectively permeable shell serves to concentrate enzymes and substrates, mediate flux of metabolites, modulate the redox state, and prevent toxic intermediates from diffusing into the cytoplasm (Havemann et al., 2002; Yeates et al., 2008).
Carboxysomes were the first bacterial microcompartments to be discovered and are widely distributed among cyanobacteria and some chemoautotrophs as the central machinery for the fixation of CO2 (Shively et al., 1973). Two different types of carboxysomes have been identified (α- and β-carboxysomes), according to the types of the CO2-fixing enzyme, Rubisco (form 1A and form 1B), possessed in cyanobacteria. In most β-cyanobacteria, Rubisco is sequestered in the β-carboxysome lumen by a shell that is composed of shell and shell-associated proteins encoded by a ccmKLMNO operon (Omata et al., 2001; Long et al., 2010; Rae et al., 2012). The carboxysomal carbonic anhydrase is colocalized with Rubisco in the β-carboxysome, serving to create a CO2-rich microenvironment to favor the Rubisco activity. Some cyanobacterial species do not have the carboxysomal β-carbonic anhydrase (CcaA) homologs; instead, the N-terminal domain of CcmM functions as an active γ-carbonic anhydrase (Peña et al., 2010). The shell facets act as a selective barrier that allows the diffusion of HCO3− and retains CO2 in the interior (Dou et al., 2008). Through these mechanisms, carboxysomes elevate the CO2 concentration in the vicinity of Rubisco and thereby enhance the efficiency of carbon fixation. Supported by this nanoscale CO2-fixing machinery, cyanobacteria contribute more than 25% of global carbon fixation (Field et al., 1998; Liu et al., 1999).
The efficiency of carboxysomes in enhancing carbon fixation has attracted tremendous interest in engineering the CO2-fixing organelle in other organisms. For example, introducing β-carboxysomes into higher plants that use the ancestral C3 pathway of photosynthesis could potentially enhance photosynthetic carbon fixation and crop production (Lin et al., 2014a, 2014b). However, engineering of functional carboxysomes requires extensive understanding about the principles underlying the formation of β-carboxysomes and the physiological integration of β-carboxysomes into the cellular metabolism.
Indeed, cyanobacterial cells have evolved comprehensive systems to regulate the biosynthesis and spatial organization of carboxysomes, allowing them to modulate the capacity for photosynthetic carbon fixation. Recent studies elucidated that the β-carboxysome assembly is initiated from the packing of Rubisco enzymes, followed by the encapsulation of peripheral shell proteins (Cameron et al., 2013; Chen et al., 2013). In the model rod-shaped cyanobacterium Synechococcus elongatus PCC7942 (hereafter Synechococcus), three to four β-carboxysomes were observed to be evenly spaced along the centerline of the longitudinal axis of cells, ensuring the equal segregation of the machinery between daughter cells (Savage et al., 2010). Such specific organization of carboxysomes within cyanobacterial cells is likely to be determined by the interaction between carboxysomes and the cytoskeleton (Savage et al., 2010). Advanced understanding of the functions and assembly of β-carboxysome proteins has recently led to the construction of a chimeric protein that can functionally replace four native proteins (CcmM58, CcmM35, CcaA, and CcmN) required for carboxysome formation (Gonzalez-Esquer et al., 2015). These findings outlined the self-assembly nature and integration of carboxysomes in the cell. However, how β-carboxysome biosynthesis and organization are physiologically regulated in cyanobacteria in response to environmental changes remains poorly understood.
Here, using a combination of live-cell confocal fluorescence microscopy and biochemical and physiological approaches, we investigated the formation and spatial positioning of β-carboxysomes in Synechococcus under varying light intensities. Our study provides new insights into the regulation of β-carboxysome biosynthesis by light and the roles of photosynthetic electron flow in the carboxysome assembly. Knowledge obtained from this work is fundamental to the bioengineering and modulation of functional carboxysomes to boost photosynthetic carbon fixation in dynamic and diverse environments.
RESULTS
We chose Synechococcus as the model organism due to its superior genetic tractability and proven suitability for fluorescence imaging (Savage et al., 2010; Liu et al., 2012; Cameron et al., 2013; Cohen et al., 2014). RbcL, the large subunit of Rubisco that resides in the β-carboxysome lumen, was tagged at the C terminus with enhanced GFP (eGFP) and was visualized under confocal fluorescence microscopy to characterize the formation and positioning of carboxysomes in vivo. Homologous recombination was used to tag the genes at their native chromosomal locus under the control of their native promoters (Supplemental Fig. S1). This ensures that the fluorescently tagged proteins were expressed in context and at physiological levels.
Figure 1 represents the confocal images of RbcL:eGFP Synechococcus strain. The eGFP fluorescence (green) indicates the subcellular localization of carboxysomes, and the endogenous chlorophyll fluorescence (red) shows the organization of thylakoid membranes. In addition, the specific DNA-staining dye 4′,6-diamino-phenylindole (DAPI) was used to image chromosomes, offering the possibility to determine the cytoplasmic environment in cyanobacteria (Fig. 1A). The merged channel shows that most of the cytoplasmic volume of the Synechococcus cell is densely occupied by carboxysomes and chromosomes, and no significant fluorescence gaps were visible, implying that all carboxysomes in the RbcL:eGFP transformant are likely fluorescently visible using confocal microscopy. In the free-eGFP-expressing Synechococcus construct, the eGFP fluorescence is evenly spread across the cytoplasm. The distinct distributions of GFP fluorescence in the RbcL:eGFP and free-eGFP-expressing Synechococcus strains indicate the self-assembly of carboxysome proteins. PCR and immunoblot results indicate the RbcL:eGFP transformant could not be fully segregated; about 30% of total RbcL was fused with eGFP (Supplemental Fig. S1). The addition of GFP tag might limit the number of Rubisco proteins accommodated within the carboxysomal interior (Menon et al., 2010). Thus, there seems to be a regulation to avert full segregation and retain some unlabeled Rubisco in the carboxysome. Nevertheless, the fluorescence tagging did not affect the growth of cyanobacterial cells (Supplemental Fig. S1). Analysis of confocal images was programmed to examine statistically the number and spatial positioning of carboxysomes in the cell (Fig. 1, B and C, n = 300). On average, there are about four evenly positioned carboxysomes per cell, consistent with previous observations (Savage et al., 2010), confirming the physiological state of RbcL:eGFP cells. We also labeled the minor shell proteins in the carboxysome CcmK4 (Kerfeld et al., 2005; Savage et al., 2010; Cai et al., 2015) using eGFP. PCR results demonstrate that the ccmK4:egfp transformant was fully segregated, and the construct has similar growth rates compared to wild-type and RbcL:eGFP strains (Supplemental Fig. S1).
Figure 1.
Spatial organization of β-carboxysomes in RbcL:eGFP Synechococcus cells. A, Confocal images of a RbcL:eGFP Synechococcus cell. Green, eGFP-labeled carboxysomes; blue, DAPI-stained DNA; red, autofluorescence of the thylakoid membrane. The merged channel revealed that most of the cytoplasmic volume of the Synechococcus cell is occupied by carboxysomes and chromosomes. This subcellular organization indicated that all carboxysomes in the RbcL:eGFP cell can been visualized using confocal microscopy. The confocal image of the Synechococcus construct that expresses free eGFP illustrates that free eGFP is spread throughout the cytoplasm without specific aggregation. B, Computational programming of image analysis allows automatic identification of carboxysomes in cells in confocal images. Bar = 2 μm. C, Statistical determination of the spatial localization of carboxysomes within the cell revealed the distribution profiles of carboxysomes along both the longitudinal and short axes of the cell (n = 300). The orange squares represent the relative frequency of carboxysome localization in the cell. The developed automated analysis software routines were used in this work for analyzing the carboxysome content and positioning.
To examine whether the GFP-labeled carboxysomes can be physiologically regulated within cells, we assayed the impact of CO2 concentration on the formation of carboxysomes. Previous studies have indicated that the carboxysomes content is affected by CO2 availability (McKay et al., 1993; Harano et al., 2003; Woodger et al., 2003; Whitehead et al., 2014). Our confocal images show a striking reduction in the numbers of carboxysomes in cells aerated with 3% CO2, compared to those in cells grown in ambient air (Supplemental Fig. S2). It reveals the feasibility of using live-cell confocal imaging to monitor the in vivo regulation of carboxysome biosynthesis in response to environmental change.
Light Triggers Carboxysome Biosynthesis
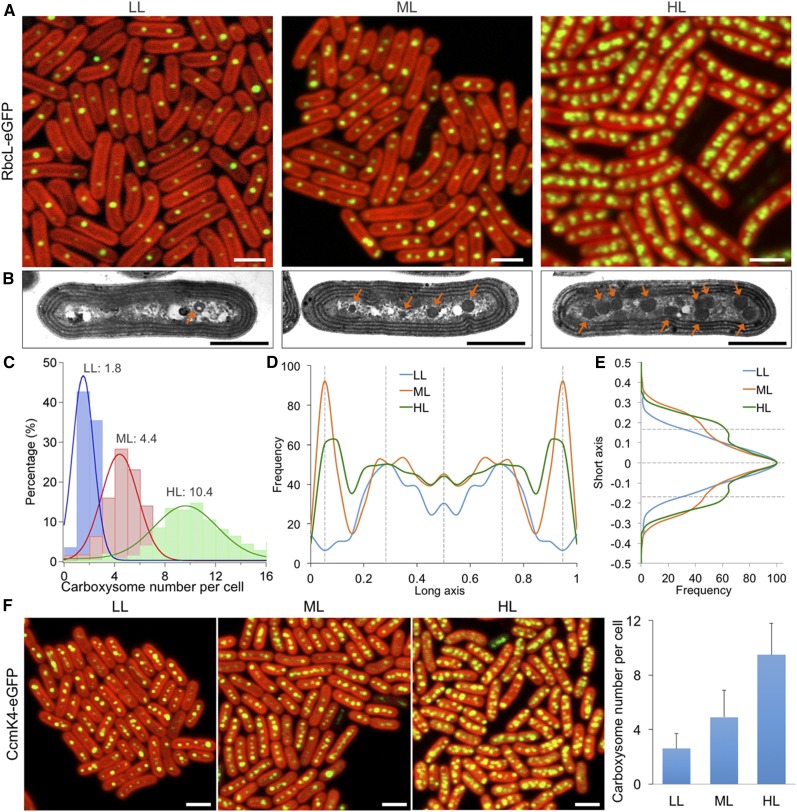
We studied the spatial distribution of carboxysomes in Synechococcus under the variation of light intensity: low light (LL; 10 μE·m−2·s−1), moderate light (ML; 50 μE·m−2·s−1), and higher light (HL; 100 μE·m−2·s−1). Confocal images of RbcL:eGFP cells show that the carboxysome abundance per cell has a strong correlation with the illumination intensity during cell growth (Fig. 2A). The number of carboxysomes per cell is higher under HL, whereas LL leads to the reduction in carboxysome numbers. The light dependence of carboxysome content was further substantiated by transmission electron microscopy results of wild-type Synechococcus cells (Fig. 2B; Supplemental Fig. S3). The numbers and positioning of carboxysomes in the cell were statistically analyzed based on the confocal images. On average, around two carboxysomes per cell (1.8 ± 1.2, n = 500) were observed under LL, whereas about four carboxysomes per cell (4.4 ± 1.9, n = 500) under ML and over 10 (10.4 ± 3.8, n = 500) under HL were detected (Fig. 2C). The data are in good agreement with the results from electron microscopy images (Supplemental Fig. S3): 1.6 ± 0.7 (LL, n = 30), 3.9 ± 0.8 (ML, n = 30), 10.2 ± 2.0 (HL, n = 30). No significant changes in cell dimensions were detected under different light intensities (Supplemental Fig. S4). These results indicate that light intensity plays an important role in determining the biosynthesis of β-carboxysomes in Synechococcus.
Figure 2.
Distinct distribution patterns of carboxysomes in Synechococcus under different light intensities. A, Confocal microscopy images of the spatial organizations of carboxysomes in the RbcL:eGFP strain grown under LL (10 μE·m−2·s−1), ML (50 μE·m−2·s−1), and HL (100 μE·m−2·s−1). Variations in the carboxysome content were observed under different light intensities. Bar = 2 μm. B, Thin-section transmission electron microscopy images of wild-type Synechococcus cells grown under LL, ML, and HL (Supplemental Fig. S3). The thylakoid membranes form regular multiple parallel layers surrounding the cytoplasm. The carboxysomes were observed as dark polyhedral particles (arrows) in the cytoplasm. Stronger light leads to the increase in carboxysome numbers in cells. Bar = 1 μm. C, Computational analysis of confocal images shows the average numbers of carboxysomes per cell under LL, ML, and HL (n = 500 for each condition). D, The positioning of carboxysomes along the normalized longitudinal axis of the Synechococcus cell under LL, ML, and HL. The relatively periodic and polar localization of carboxysomes in cells are indicated (n = 500). E, The positioning of carboxysomes along the normalized short axis of the Synechococcus cell (n = 500). HL gives rise to a wider distribution of carboxysomes from the centerline of cells. F, Confocal microscopy images of CcmK4:eGFP cells show the spatial positioning of carboxysomes grown under LL, ML, and HL. Variations in the carboxysome content were observed under different light intensities, in good agreement with those of the RbcL:eGFP strain. Error bars represent sd (n = 500 for each condition). Bar = 2 μm.
Varying light intensities could also result in different organizational patterns of carboxysomes in cells. Image analysis reveals even distribution of carboxysomes along the longitudinal axis of the cell (Fig. 2D). Carboxysomes tend to locate at approximately one-fourth position along the cell length under LL, whereas under ML and HL a polar location of carboxysomes within the cell was observed apart from the even distribution (Fig. 2D). Analysis of the positioning of carboxysomes along the cell width elucidates that increasing numbers of carboxysomes induced by stronger light present a wider distribution along the short axis of the cell, compared to the centerline positioning observed under LL (Fig. 2E). This organization likely provides a means to house more carboxysomes in a spatially crowded cytoplasm environment. Consistent with the results of the RbcL:eGFP construct, our confocal images of the CcmK4:eGFP construct also show the increase in carboxysome content triggered by stronger irradiance (Fig. 2F, n = 500). Similarly, light regulation of carboxysome content in Synechococcus was also seen in the RbcL:YFP strain (Savage et al., 2010) in which rbcL:yfp was inserted into a neutral site rather than the native locus in the genome (Supplemental Fig. S5). Together, our observations reveal a general regulation of carboxysome content and organization in Synechococcus in response to variations in light intensity.
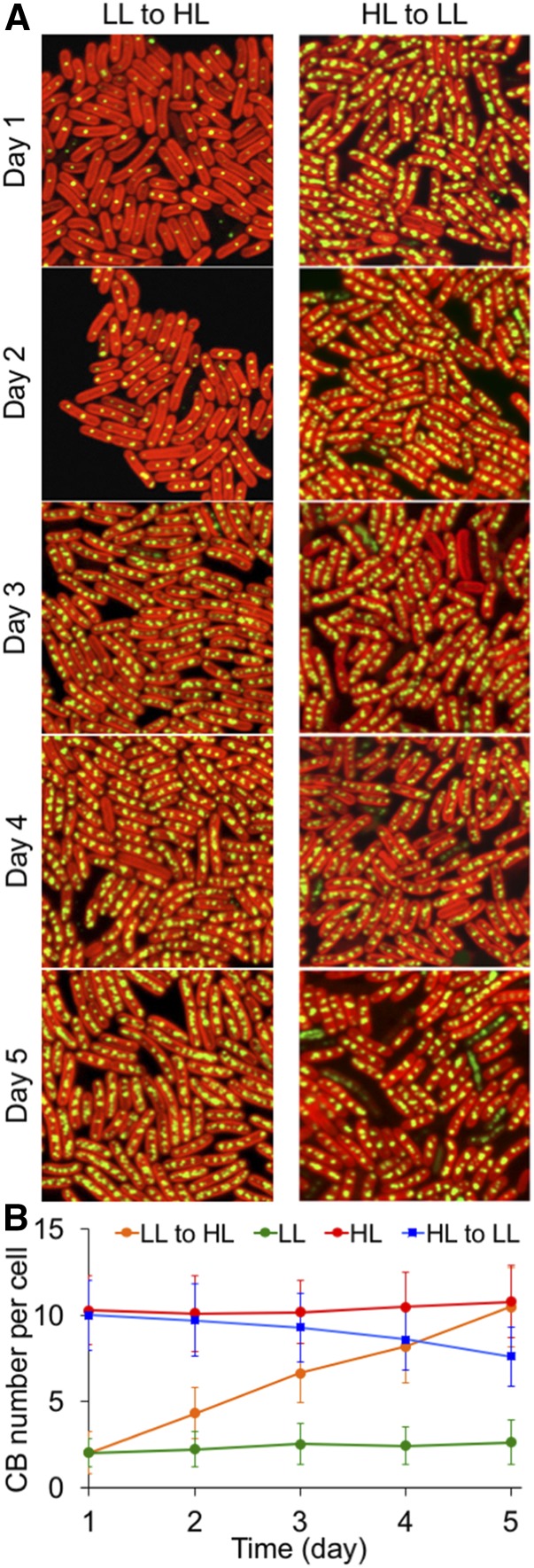
The light-regulated carboxysome biosynthesis was further characterized by time-lapse confocal imaging during cell growth (Fig. 3). HL treatment on cells that were preadapted to LL resulted in a linear increase in carboxysome content over 5 d. Reversibly, LL treatment caused a reduction in carboxysome numbers, although the rate of reduction is lower than that of the increase in carboxysome numbers. These results indicate that the light-dependent carboxysome biogenesis might function as a long-term acclimation process in cyanobacteria. On the other hand, despite potential repairing mechanism led by protein dynamics (Sutter et al., 2016), there appears to be no specific degradation pathway for carboxysomes. The stability of mature carboxysomes in vivo may be of physiological importance for the cellular metabolism (Cameron et al., 2013).
Figure 3.
Characterization of the light-regulated biosynthesis process of β-carboxysomes. A, Time-lapse confocal images of the LL-adapted RbcL:eGFP Synechococcus strain under HL treatment and the HL-adapted RbcL:eGFP strain under LL treatment. Cells from the same flasks were imaged under confocal microscopy once per day, for 5 d continuously. Changes in carboxysome content per cell were captured. B, Analysis of the average numbers of carboxysomes based on confocal images reveals an increase in carboxysome abundance induced by HL and a decline in carboxysome abundance caused by LL, compared to the numbers of carboxysomes under constant LL or HL treatments. Error bars represent sd (n = 250).
Light-Induced Carboxysome Biosynthesis Determines the Carbon Fixation Activity of Cells
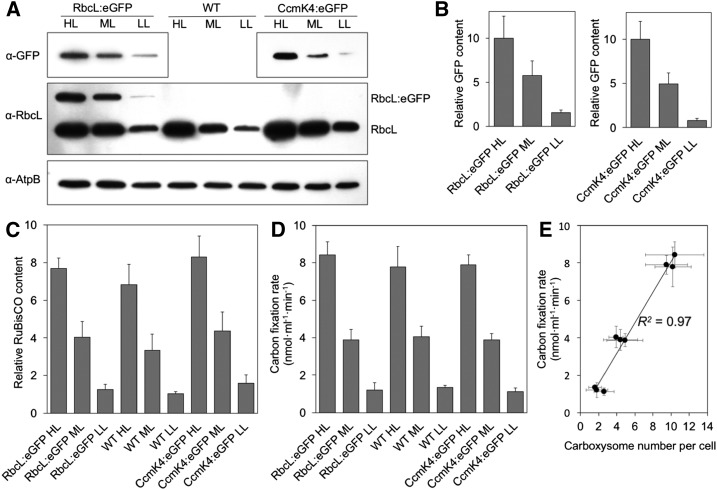
In addition to the confocal microscopy results that reveal the light-induced carboxysome content in the cell, immunoblot analysis show that the abundance of Rubisco proteins per cell, normalized using the AtpB content (Zhang et al., 2012), is also up-regulated by increasing irradiance (Fig. 4A). The Rubisco abundance under HL is about 6 and 2 times as high as those under LL and ML, respectively (Fig. 4, B and C, n = 6). Interestingly, a slight increase in Rubisco content was observed in both RbcL:eGFP and CcmK4:eGFP strains compared to wild-type cells, probably as compensation for compromised CO2-fixing activities of carboxysomes caused by fluorescence labeling. The light-induced changes in Rubisco content at the protein level was also confirmed by measuring the total fluorescence intensity of RbcL:eGFP per cell (Supplemental Fig. S6). Our results corroborate previous studies, which revealed that the transcription of carboxysome genes is stimulated in response to increasing light intensity (Watson and Tabita, 1996; Hihara et al., 2001; Gill et al., 2002; Huang et al., 2002; McGinn et al., 2004).
Figure 4.
Light modulates the biosynthesis of β-carboxysomes and cellular carbon fixation in Synechococcus. A, Immunoblot analysis using anti-RbcL and anti-GFP antibodies shows variations of the Rubisco content in RbcL:eGFP cells. HL triggers the accumulation of Rubisco. Using anti-RbcL antibody, RbcL:eGFP strains present two bands: the upper band for RbcL-eGFP and the lower band for RbcL only. AtpB was used as a loading control (Zhang et al., 2012). Gels are representative of six independent experiments. B, Immunoblot analysis implies that the GFP amount in RbcL:eGFP and CcmK4:eGFP cells varies under different light intensities (±sd, n = 6, P < 0.05). C, Densitometry of RbcL in wild-type, RbcL:eGFP, and CcmK4:eGFP cells is dependent on light intensity (±sd, n = 6, P < 0.05). D, 14C carbon fixation rates of wild-type, RbcL:eGFP, and CcmK4:eGFP cells under LL, ML, and HL at 0.5 mm RuBP (±sd, n = 6). The cell density was normalized using the AtpB content (Fig. 4A). The carbon fixation rates of cells as a function of RuBP are shown in Supplemental Fig. S7. E, The carbon fixation rate per cell is proportional to the numbers of carboxysomes within the cell (R2 = 0.97). The numbers of carboxysomes per cell were determined from electron microscopy images for the wild-type strain and confocal microscopy images for RbcL:eGFP and CcmK4:eGFP strains.
To verify the physiological coordination between carboxysome content and carbon fixation in Synechococcus, we surveyed the carbon fixation activities of cells (based on the AtpB content) under different light conditions. To examine the maximum carbon fixation rates, 0.5 mm d-ribulose 1,5-bisphosphate sodium salt hydrate (RuBP) was applied (Supplemental Fig. S7). Figure 4C depicts a strong dependence of carbon fixation rates of Synechococcus cells on light intensity. The carbon fixation rate of wild-type cells in HL (7.8 μmol·min−1·mL−1) is higher compared with those in ML (4.0 μmol·min−1·mL−1) and LL (1.3 μmol·min−1·mL−1; n = 6; Fig. 4D). Similar tendency was also observed in RbcL:eGFP and CcmK4:eGFP cells, indicating that increasing irradiance enhances the carbon fixation of Synechococcus cells (n = 6; Fig. 4D). Furthermore, there is a close correlation between the numbers of carboxysomes and carbon fixation rate of cells (Fig. 4E). Together, our results indicate explicitly that the light-intensity-regulated carboxysome biosynthesis serves as a regulatory mechanism of modulating the capacity of CO2 fixation in the cell.
Light Regulation of Carboxysome Biosynthesis Is Mediated by Photosynthetic Electron Flow
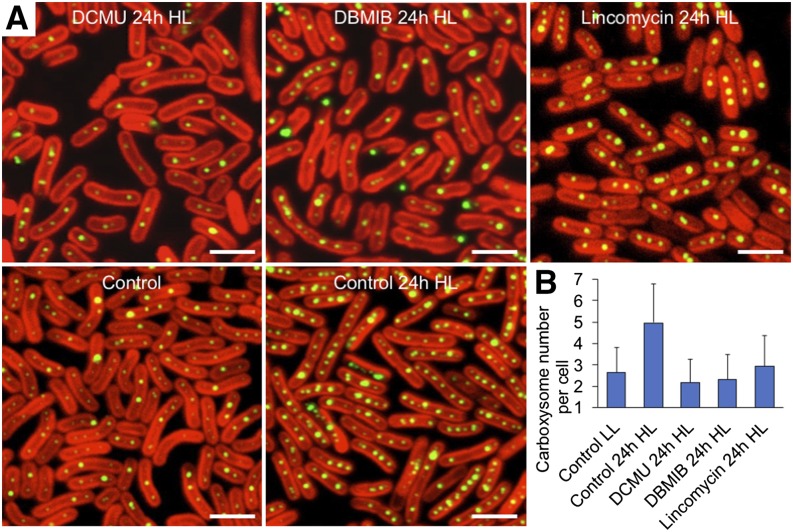
Changes in light intensity could alter electron flow and redox states of intersystem electron carriers, especially the plastoquinone (PQ) pool (Mullineaux, 2001; Liu et al., 2012). We conducted extensive studies on the carboxysome formation process in response to irradiance variations in the presence of two specific inhibitors of photosynthetic electron transport, namely, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-dibromo-3-methyl-6-isopropyl-benzoquinone (DBMIB). DCMU and DBMIB inhibit photosynthetic electron transport from the photosystem II complex to the PQ pool and from the PQ pool to the cytochrome b6f complex, respectively (Trebst, 1980). We observed that when light is switched from LL to HL for 24 h, both DCMU and DBMIB treatments suppress the O2 evolution from photosystem II of cells (Supplemental Fig. S8) and hampers the light-induced carboxysome biosynthesis (Fig. 5). It demonstrates that the inhibition of photosynthetic electron flow impairs the biogenesis of carboxysomes in cyanobacteria. The increase in carboxysome content in the cell was also inhibited by lincomycin (Fig. 5), a protein synthesis inhibitor that suppresses de novo protein synthesis (Dalla Chiesa et al., 1997). The similar effects of DCMU, DBMIB, and lincomycin on impeding the carboxysome biosynthesis suggest that the regulation of photosynthetic electron flow may affect the synthesis and assembly of carboxysome proteins to form carboxysomes.
Figure 5.
β-Carboxysome biosynthesis is regulated by photosynthetic electron flow. A, Confocal microscopy images of LL-adapted RbcL:eGFP Synechococcus cells after 24-h HL treatment, in the presence of the photosynthetic electron transport inhibitors DCMU and DBMIB or the protein synthesis inhibitor lincomycin. The control images were captured in LL-adapted Synechococcus cells grown at LL and after 24-h HL treatment without inhibitor treatments. Bar = 2 μm. B, Analysis of the average numbers of carboxysomes per cell (±sd, n = 300) based on the confocal images illustrates the suppression of carboxysome content with the treatments of DCMU and DBMIB, indicating that light intensity regulates the biosynthesis and assembly of carboxysomes through photosynthetic electron flux.
Carboxysome Localization Is Sensitive to the Redox State of Photosynthetic Electron Transport
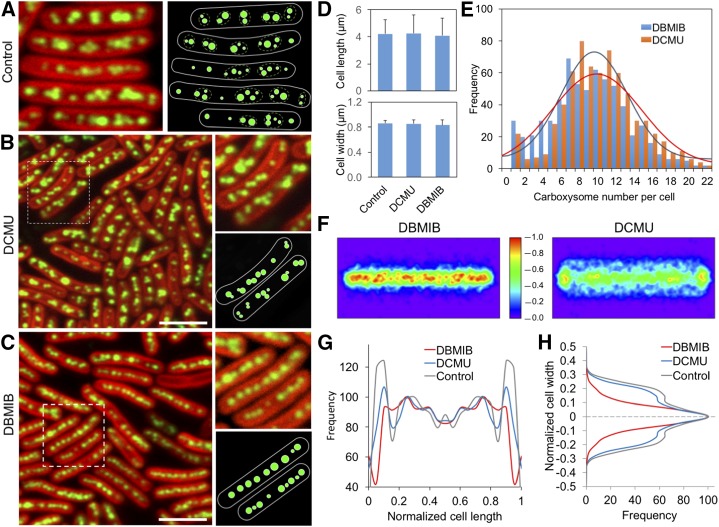
Closer inspection of the distribution of carboxysomes in HL-adapted cells illustrates that along with the dense packing of carboxysomes in the cytoplasm, a few carboxysomes aggregate preferentially and form a single large carboxysome “cluster”. Several clusters are then evenly positioned along the longitudinal axis of the cell (Fig. 6A). It was postulated that the local CO2 concentration near each carboxysome is higher (Mangan and Brenner, 2014). The carboxysome clusters may be functionally advantageous to minimizing CO2 leakage and maximizing the CO2 accumulation around all carboxysomes in the cytoplasm (Ting et al., 2007), thereby enhancing carbon fixation of cells. The equally spaced carboxysome clusters along the long axis of the cell may correlate with the positioning of chromosomes (Jain et al., 2012). It could favor the equal segregation of carboxysomes between daughter cells during cell division (Savage et al., 2010).
Figure 6.
Correlation between the spatial organization of β-carboxysomes and redox state of photosynthetic electron transport chain in Synechococcus. A, Confocal image (left) and schematic model (right) of the HL-adapted RbcL:eGFP strain show the evenly distributed carboxysome clusters (orange dotted circles), each of which contains several carboxysomes assembled together in the local cytoplasmic region. B, Confocal image of HL-adapted RbcL:eGFP cells under DCMU treatment shows the clustering distribution of carboxysomes along the centerline of the cell. Bar = 5 μm. C, Confocal image of HL-adapted Synechococcus cells under DBMIB treatment shows the linear distribution of carboxysomes along the centerline of the cell. Bar = 5 μm. D, No significant changes in the cell length and width of Synechococcus is detected during inhibitor treatments for 24 h (P > 0.05, n = 500). Error bars represent sd. E, Average numbers of carboxysomes per cell under the treatments of DCMU and DBMIB do not have remarkable changes (P > 0.05, n = 500). F, Normalized spatial distribution maps of carboxysomes in cells under the treatments of DCMU and DBMIB. The bar presents the relative frequency of carboxysome localization. G, The distributions of carboxysomes along the cell length under DCMU and DBMIB treatments (n = 500). H, DBMIB treatment results in a linear positioning of carboxysomes at the centerline of HL-adapted Synechococcus cells, whereas the DCMU treatment leads to a wider distribution of carboxysomes along the cell width (n = 500). Similar results were also obtained in LL- and ML-adapted Synechococcus cells (Supplemental Fig. S9) and CcmK4:eGFP cells (Supplemental Fig. S10).
We further surveyed the effects of DCMU and DBMIB on the spatial organization of carboxysomes in cells. In HL-adapted RbcL:eGFP cells, carboxysomes possess a dense distribution, suitable for detecting the spatial redistribution of carboxysomes. Under DCMU treatment, carboxysomes present the typical “clustering” distribution in the cytoplasm (Fig. 6B), similar to the observations in nontreated cells. By contrast, DBMIB treatment gave rise to a linear positioning of carboxysomes along the centerline of the cell (Fig. 6C). Image analysis reveals that, relative to the marked repositioning of carboxysomes, no detectable differences in the thylakoid membrane structure and cytoplasmic volume (indicated by endogenous chlorophyll fluorescence) were observed (Fig. 6D), whereas the carboxysome numbers per cell under DCMU and DBMIB treatments were comparable (Fig. 6E). The periodic distribution of carboxysomes along the long axis of DBMIB-treated cells is somewhat less significant than that of DCMU-treated cells, and the polar localization of carboxysomes seems to disappear in DBMIB-treated cells (Fig. 6, F and G). More remarkable changes were observed in carboxysome distribution along the short axis of the cell (Fig. 6, F and H). DBMIB treatment led to the relocation of carboxysomes from a wider distribution along the cell width to a narrow positioning at the centerline of the short axis of the cell, compared to the wide distribution observed in DCMU-treated and nontreated cells (Fig. 6H). DCMU and DBMIB have opposite effects on the redox state of PQ pool in photosynthetic electron transport chain: The PQ pool is oxidized by DCMU and reduced by DBMIB. The distinct effects of DCMU and DBMIB indicate that the spatial organization of β-carboxysomes in Synechococcus correlates with the redox state of photosynthetic electron transport chain. Consistent with the finding of HL-adapted cells, the reorganization of carboxysomes under DCMU and DBMIB treatments was also observed in LL- and ML-adapted RbcL:eGFP cells (Supplemental Fig. S9) and CcmK4:eGFP cells (Supplemental Fig. S10), corroborating that the redox regulation of photosynthetic electron flow could affect the spatial positioning of β-carboxysomes in Synechococcus. We further observed that changes in carboxysome localization are not clearly visible within 4-h DBMIB treatment (Supplemental Fig. S11), implying that the reorganization of carboxysomes seems to be a long-term adaptive process in response to redox regulation.
DISCUSSION
In cyanobacteria, light is fundamental to energy production, DNA replication, and the regulation of gene expression (Asayama, 2006; Ohbayashi et al., 2013). Light-dependent reactions of photosynthesis generate chemical energy, in the forms of ATP and NADPH, which is utilized to drive the Calvin-Benson-Bassham Cycle responsible for CO2 fixation into metabolizable sugars. It was found that HL could induce an increase in the transcription of Rubisco and carboxysome ccm genes (Watson and Tabita, 1996; Hihara et al., 2001; Gill et al., 2002; Huang et al., 2002). In this work, we evaluated the impact of light intensity on the regulation of β-carboxysome biosynthesis at the whole organelle and cellular levels. Our results show that increasing irradiance triggers the expression of carboxysome proteins (Fig. 4A; Supplemental Fig. S6) and formation of functional carboxysomes (Fig. 2), thereby enhancing carbon fixation of cells (Fig. 4). We further revealed the close correlation between light-regulated photosynthetic electron flow and β-carboxysome biosynthesis (Fig. 5). Given that the expression of carboxysome genes and their encoded proteins are under light-dark regulation (Watson and Tabita, 1996; Ito et al., 2009; Aryal et al., 2011), further work needs to be directed to elucidate whether de novo assembly of carboxysomes is regulated and/or potentially gated by the cyanobacterial circadian rhythm.
The spatial distribution of β-carboxysomes along the longitudinal axis of Synechococcus cells is driven by interactions with the cytoskeleton, ensuring the equal segregation of carbon fixation organelles between daughter cells (Savage et al., 2010). Synechococcus cytoplasm densely accommodates carboxysomes and chromosomes, which are interspersed with each other (Fig. 1A). It is conceivable that the organization and dynamics of carboxysomes correlate with the partitioning of chromosomes. On the other hand, the disruption of parA resulted in unequal positioning of carboxysomes, but did not interfere with chromosome organization, suggesting that the spatial partitioning of carboxysomes and chromosomes in Synechococcus is likely regulated separately (Jain et al., 2012). The detailed underlying mechanism awaits further examination.
Given the spatial constraints in the cytoplasm and the large volume of carboxysomes, the broader distribution of carboxysomes within the cell may suggest the specific associations between carboxysomes and the thylakoid membrane. Indeed, such an interaction has been deduced due to the facts that Rubisco can be found not only in the cytosol, but also near the thylakoid membranes (Agarwal et al., 2009). The structural heterogeneity and dynamics of cyanobacterial thylakoid membranes are fundamental to the physiological regulation of photosynthetic electron transport for energy conversion (Liu, 2016). It is feasible that components in the cycle have specific subcellular positioning to take advantage of the supplied energy and functionally coordinate with each other. Moreover, systems analysis suggested there might be a gradient of CO2 concentration from the cell membrane to the center of the cell cytoplasm (Mangan and Brenner, 2014). Thus, changes in the subcellular localization of carboxysomes, in particular along the short axis of Synechococcus cells, probably render a means for modulating the assimilation of CO2 within the cell.
Whether there are free Rubisco proteins that are not encapsulated within carboxysomes is an open question. Our confocal imaging did not demonstrate the existence of free Rubisco in the Synechococcus cytoplasm. It was further confirmed by our finding that no visible band of free Rubisco was determined using native-PAGE and immunoblot analysis of the soluble fraction (data not shown). However, it cannot be excluded that the amount of free Rubisco is too low to detect, given the inherent resolution limitations of confocal microscopy and the sensitivity of immunoblot analysis.
The redox state of photosynthetic electron transport chain functions as the key controller of the distribution of respiratory complexes (Liu et al., 2012), photosystem composition (Fujita et al., 1987), photosynthetic state transitions (Mullineaux and Allen, 1990), and the modulation of the circadian clock (Ivleva et al., 2006; Wood et al., 2010). In this study, we report that the redox state of the photosynthetic electron transport chain located in thylakoid membranes has an effect on the subcellular positioning of β-carboxysomes in Synechococcus. The widespread and clumping distribution of carboxysomes is determined by the oxidized state of photosynthetic electron transport chain, whereas the linear positioning of carboxysomes along the cell length is ascribed to the reduced state of photosynthetic electron transport chain (Fig. 6). Nontreated cells present similar clustering organization of carboxysomes as DCMU-treated cells, indicating that the PQ pool is oxidized upon the illumination condition, which possibly triggers the state transition to “State 1” (Mullineaux and Allen, 1990) or probably due to the high ratio of photosystem I and photosystem II in cyanobacteria (Howitt et al., 2001). Our results further reveal that the reorganization of carboxysomes appears as a long-term regulation process. It is reminiscent of the previous finding showing that the constrained diffusive dynamics of β-carboxysomes in Synechococcus (Savage et al., 2010).
It has been reported that de novo gene expression of DNA replication components in Synechococcus is dependent on the photosynthetic electron transport activity (Ohbayashi et al., 2013). Here, we show that both DCMU and DBMIB can inhibit the synthesis of carboxysome proteins and, thereby, the formation of carboxysomes (Fig. 5). Whether there are indirect effects of protein synthesis on the spatial positioning of carboxysomes needs further characterization. Our results demonstrate explicitly that DCMU and DBMIB treatments could result in distinct carboxysome positioning in the cell, whereas the carboxysome numbers, and hence probably the expression of carboxysome proteins, are comparable (Fig. 6), suggesting that the effects of electron transport inhibitors on protein synthesis seem not to correlate with the changes in carboxysome positioning.
In addition to the β-carboxysome positioning, redox regulation is also important for the β-carboxysome biosynthesis and function. Carboxysomes may preferably retain an independent redox environment from that of the cytosol, by the semipermeable shell that can selectively exclude the entry of thioredoxin and other redox equivalents into the interior (Peña et al., 2010; Rae et al., 2013). The shell encapsulation allows the establishment of an oxidizing microenvironment within the β-carboxysome (Chen et al., 2013). The thioredoxins in the cytoplasm could reduce the redox damage to carboxysome components and enhance the carboxysome biogenesis and maturation (Rae et al., 2013). In addition, the independent redox modulation of the carboxysome lumen was deduced to be vital for the activities of carboxysome enzymes. The oxidizing environment could favor the activation of CcaA (Price et al., 1992) and carbonic anhydrase function of CcmM (Peña et al., 2010).
Cyanobacterial CO2-concentrating mechanisms (CCMs) comprise carboxysomes, CO2 uptake complex NDH-1, and HCO3− transports (Price et al., 2008). To date, we have demonstrated that the in vivo distributions of carboxysomes (this work) and NDH-1 complexes (Liu et al., 2012) were both regulated by the redox state of photosynthetic electron transport chain, suggesting the potential interplay between the two components. In addition, given that the transcriptional levels of HCO3− transporters were also regulated by light through photosynthetic electron flux (McGinn et al., 2004; Burnap et al., 2013), it is likely that the organization of entire CCM pathway in cyanobacteria is modulated, in an integrated network context, by the light-mediated redox regulation of photosynthetic electron flow. Therefore, thylakoid membrane remodeling during environmental adaption might play a role in the regulation of CCM pathway in the cell, which needs to be determined experimentally in future.
CONCLUSION
In this work, we characterized extensively the biosynthesis and spatial organization of cyanobacterial carboxysomes in response to changing light levels. Our results reveal that light intensity plays an essential role in modulating the carboxysome biosynthesis and carbon fixation capacity of cyanobacterial cells. Light-regulated carboxysome biogenesis and organization in Synechococcus are coordinated with photosynthetic electron flow. This study provides essential knowledge for us to modulate the β-carboxysome biosynthesis and carbon fixation in cyanobacteria. The general principles underlying the biogenesis of carboxysomes and their functional coordination with the cellular bioenergetic network could be an important consideration when engineering functional carboxysomes and cyanobacterial CCM pathways into heterologous organisms, in specific plant chloroplasts, to boost photosynthetic performance and CO2 assimilation activity.
MATERIALS AND METHODS
Bacterial Strains, Growth Conditions, and Generation of Constructs
Synechococcus elongatus PCC7942 was grown at 30°C with constant white illumination in BG-11 medium (Rippka et al., 1979) in culture flasks with constant shaking or on BG-11 plates containing 1.5% (w/v) agar. Cultures were grown in air without additional CO2 source except for the experiment aerated with 3% CO2. For eGFP-fusion strains, BG-11 medium was supplemented with 50 µg·mL−1 apramycin. Growth of cells was monitored at OD750 using a spectrophotometer (Jenway 6300). Three biological repeats were recorded. Escherichia coli strains used in this work were DH5α and BW25113. E. coli was grown aerobically at 30 or 37°C in Luria-Broth medium. Medium supplements were used, where appropriate, at the following final concentrations: ampicillin 100 μg·mL−1, chloramphenicol 10 μg·mL−1, apramycin 50 μg·mL−1, and arabinose 100 μm.
PCR products, including ∼800-bp homologous sequence upstream and downstream of synpcc7942_1426 (rbcL) or synpcc7942_0285 (ccmK4), were cloned using the pGEM-T Easy cloning system (Promega). eGFP fusions were created by inserting the eGFP:apramycin region amplified from the plasmid pIJ786 to the C terminus of rbcL or ccmK4, using the Redirect strategy (Gust et al., 2002, 2004). Plasmids were verified by PCR and sequencing. The plasmids were then transformed into Synechococcus cells following the method described earlier (Golden, 1988). Segregation of recombinant gene/protein was checked by PCR, agarose gel electrophoresis, sequencing, and immunoblot analysis. The DNA oligonucleotides used in this work are shown in Supplemental Table S1. Free-eGFP-expressing Synechococcus strain was obtained using the pAM2991vector (a gift from Susan Golden, Addgene plasmid #40248; Ivleva et al., 2005) containing eGFP.
Light and Inhibitor Treatment
Synechococcus cultures were treated with different intensities of light illuminations (10 μE·m−2·s−1 as LL, 50 μE·m−2·s−1 as ML, and 100 μE·m−2·s−1 as HL). Cells were collected at the middle of exponential growth phase (to avoid self-shading) for the following imaging and biochemical analysis. The electron transport inhibitors DCMU (Sigma-Aldrich) and DBMIB (Sigma-Aldrich), and the protein synthesis inhibitor lincomycin were added to 20 μm, 10 μm, and 400 μg·mL−1, respectively. Cells were adapted for 24 h in the presence of DCMU, DBMIB, or lincomycin before microscopy imaging. The free-eGFP-expressing Synechococcus cells were induced with 1 mm isopropyl β-d-1-thiogalactopyranoside for 24 h prior to confocal imaging.
Confocal Microscopy and Image Analysis
Preparation of Synechococcus cells for confocal microscopy was performed as described earlier (Liu et al., 2012). Confocal laser scanning confocal microscopy used a Zeiss LSM510 or LSM710 with a 63× or 100× oil-immersion objective and excitation at 488 nm. Live-cell images were recorded from at least five different cultures. All images were captured with all pixels below saturation. Image analysis was carried out using ImageJ software (NIH Image). Results are presented as mean ± sd.
Automated analysis of cell counting, the number and positioning of carboxysomes, and the definition of thylakoid membranes per cell was programmed into the image analysis software Image SXM (http://www.imagesxm.org.uk/). The carboxysomes or thylakoid membranes were identified by processing the green or red channels of confocal images using Fourier filters to highlight the carboxysomes or the edges of thylakoid membranes. The shape and the “center of mass” of the fluorescent spots were used to determine the number and location of carboxysomes within the cell. The developed automated analysis software routines were used, in this work, for analyzing the carboxysome content and positioning.
DAPI staining was conducted as described previously (Smith and Williams, 2006). Briefly, cells were collected by centrifugation at 30°C and were then washed by phosphate buffer (pH 7.2) three times. Cells were incubated with DAPI (20 μg·mL−1) in dark at 30°C for 20 min and washed by distilled water twice before loading on a BG-11 plate for confocal imaging. The DAPI-stained chromosome was visualized under the DAPI channel (excitation = 405 nm; emission = 437–552 nm).
In Vivo Carbon Fixation Assay
Cells were harvested at exponential phase under corresponding light treatments and then resuspended in Rubisco assay buffer (100 mm EPPS, pH 8.0, and 20 mm MgCl2). Cell density was then calibrated by measuring OD750. Radiometric assay was carried out according to a previously described protocol (Price and Badger, 1989) with additional cell permeabilization treatment (Schwarz et al., 1995). Cell cultures prepared in assay buffer with the same cell density were incubated with NaH14CO3 (final concentration at 25 mm) at 30°C for 2 min and then permeabilized by mixed alkyltrimethylammonium bromide (final concentration at 0.03% [w/v]; Sigma-Aldrich). RuBP (Sigma-Aldrich) was then added with a range of concentrations (0–2.0 mm) to initialize the fixation. After 5 min, 10% formic acid was added to terminate the reaction. Samples were then dried on heat blocks at 95°C to remove unfixed NaH14CO3, and the pellets were resuspended in distilled water in the presence of scintillation cocktail (Ultima Gold XR; Perkin-Elmer). Radioactivity measurements were carried out using a scintillation counter (Tri-Carb; Perkin-Elmer). Raw readings were processed to determine the amount of fixed 14C, calibrated by blank cell samples without providing RuBP, and then converted to the total carbon fixation rates. Carbon fixation rates of cell cultures were normalized based on the AtpB quantity from immunoblot analysis. For each experiment, at least six biological repeats were prepared. Significance was assessed using a two-tailed t test.
Immunoblot Analysis
Synechococcus soluble fractions for SDS-PAGE analysis were prepared by sonication at 4°C followed by Triton treatment and centrifugation. Seventy-five micrograms of proteins was loaded on 10% (v/v) denaturing SDS-PAGE gels. Gels were electroblotted onto a PVDF membrane (Bio-Rad). Immunoblot analyses were performed using primary mouse monoclonal anti-GFP (Life Technologies), rabbit polyclonal anti-RbcL (Agrisera), anti-ATPaseB (Agrisera) antibodies, and horseradish peroxidase-conjugated goat anti-mouse igG secondary antibody (Promega) or anti-rabbit igG secondary antibody (GE Healthcare). Signals were visualized using a chemiluminescence kit (Bio-Rad). AtpB protein was used as a loading control for cell population (Zhang et al., 2012). Immunoblot protein quantification was carried out using ImageJ. For each experiment, at least three biological repeats were performed.
Transmission Electron Microscopy
Wild-type Synechococcus cells grown under different light conditions were pelleted and fixed for 1 h with 4% paraformaldehyde and 2.5% glutaraldehyde in 0.05 m sodium cacodylate buffer at pH 7.2. Cells were then postfixed with 1% osmium tetroxide for 1.5 h, dehydrated with a series of increasing alcohol concentrations (30 to 100%), and embedded in resin. Thin sections of 70 nm were cut with a diamond knife and poststained with 4% uranyl acetate and 3% lead citrate. Images were recorded using a FEI Tecnai G2 Spirit BioTWIN transmission electron microscope.
Oxygen Evolution Measurement
Oxygen evolution of cell cultures was measured at saturate light illumination at 30°C in a Clarke-type oxygen electrode (OxyLab 2; Hansatech). One milliliter of cell suspension with chlorophyll concentration of 20 μm was placed into the electrode chamber, aerated, and sealed from the atmosphere.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NC_007604.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. PCR primers.
Supplemental Figure S1. Construction and characterization of RbcL:eGFP and CcmK4:eGFP Synechococcus strains.
Supplemental Figure S2. Regulation of carboxysome biosynthesis in Synechococcus by CO2.
Supplemental Figure S3. Thin-section transmission electron microscopy images of wild-type Synechococcus cells grown under HL, ML, and LL.
Supplemental Figure S4. The sizes of Synechococcus cells remain similar under the variation of light intensity.
Supplemental Figure S5. Light regulation of carboxysome content in RbcL:YFP cells.
Supplemental Figure S6. Relative abundance of Rubisco in RbcL:eGFP Synechococcus strain under LL, ML, and HL, based on confocal image analysis.
Supplemental Figure S7. 14C carbon fixation rates of wild-type Synechococcus cells grown under LL, ML, and HL, as a function of RuBP concentration (±sd, n = 6).
Supplemental Figure S8. Oxygen evolution analysis of Synechococcus cells in the presence of DBMIB and DCMU for 24 h.
Supplemental Figure S9. Organization of carboxysomes in LL- and ML-adapted cells under the treatment of DCMU and DBMIB.
Supplemental Figure S10. The effects of DBMIB and DCMU on β-carboxysome localization in CcmK4:eGFP cells.
Supplemental Figure S11. Time-lapse confocal fluorescence imaging of RbcL:eGFP cells in the presence of DCMU and DBMIB.
Supplementary Material
Acknowledgments
We thank Dr. James Hartwell for critical comments on the manuscript. We acknowledge the Liverpool Centre for Cell Imaging for provision of confocal imaging equipment and technical assistance. We thank Prof. Ian Prior and Mrs. Alison Beckett for the support of electron microscopy.
Glossary
- DAPI
4′,6-diamino-phenylindole
- LL
low light
- ML
moderate light
- HL
higher light
- RuBP: d-ribulose 1
5-bisphosphate sodium salt hydrate
- PQ
plastoquinone
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-benzoquinone
- CCM
CO2-concentrating mechanism
References
- Agarwal R, Ortleb S, Sainis JK, Melzer M (2009) Immunoelectron microscopy for locating calvin cycle enzymes in the thylakoids of synechocystis 6803. Mol Plant 2: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal UK, Stöckel J, Krovvidi RK, Gritsenko MA, Monroe ME, Moore RJ, Koppenaal DW, Smith RD, Pakrasi HB, Jacobs JM (2011) Dynamic proteomic profiling of a unicellular cyanobacterium Cyanothece ATCC51142 across light-dark diurnal cycles. BMC Syst Biol 5: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asayama M. (2006) Regulatory system for light-responsive gene expression in photosynthesizing bacteria: cis-elements and trans-acting factors in transcription and post-transcription. Biosci Biotechnol Biochem 70: 565–573 [DOI] [PubMed] [Google Scholar]
- Bobik TA, Lehman BP, Yeates TO (2015) Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol 98: 193–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap RL, Nambudiri R, Holland S (2013) Regulation of the carbon-concentrating mechanism in the cyanobacterium Synechocystis sp. PCC6803 in response to changing light intensity and inorganic carbon availability. Photosynth Res 118: 115–124 [DOI] [PubMed] [Google Scholar]
- Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA (2015) Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol 4: 444–453 [DOI] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Chen AH, Robinson-Mosher A, Savage DF, Silver PA, Polka JK (2013) The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLoS One 8: e76127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SE, Erb ML, Selimkhanov J, Dong G, Hasty J, Pogliano J, Golden SS (2014) Dynamic localization of the cyanobacterial circadian clock proteins. Curr Biol 24: 1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Chiesa M, Friso G, Deák Z, Vass I, Barber J, Nixon PJ (1997) Reduced turnover of the D1 polypeptide and photoactivation of electron transfer in novel herbicide resistant mutants of Synechocystis sp. PCC 6803. Eur J Biochem 248: 731–740 [DOI] [PubMed] [Google Scholar]
- Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC (2008) CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem 283: 10377–10384 [DOI] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P (1998) Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281: 237–240 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Murakami A, Ohki K (1987) Regulation of photosystem composition in the cyanobacterial photosynthetic system: the regulation occurs in response to the redox state of the electron pool located between the two photosystems. Plant Cell Physiol 28: 283–292 [Google Scholar]
- Gill RT, Katsoulakis E, Schmitt W, Taroncher-Oldenburg G, Misra J, Stephanopoulos G (2002) Genome-wide dynamic transcriptional profiling of the light-to-dark transition in Synechocystis sp. strain PCC 6803. J Bacteriol 184: 3671–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SS. (1988) Mutagenesis of cyanobacteria by classical and gene-transfer-based methods. Methods Enzymol 167: 714–727 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Esquer CR, Shubitowski TB, Kerfeld CA (2015) Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell 27: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF (2004) Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54: 107–128 [DOI] [PubMed] [Google Scholar]
- Gust B, Kieser T, Chater KF (2002) REDIRECT Technology: PCR-targeting System in Streptomyces coelicolor. John Innes Centre, Norwich, UK [Google Scholar]
- Hantke MF, Hasse D, Maia FRNC, Ekeberg T, John K, Svenda M, Loh ND, Martin AV, Timneanu N, Larsson DSD, et al. (2014) High-throughput imaging of heterogeneous cell organelles with an X-ray laser. Nat Photonics 8: 943–949 [Google Scholar]
- Harano K, Ishida H, Kittaka R, Kojima K, Inoue N, Tsukamoto M, Satoh R, Himeno M, Iwaki T, Wadano A (2003) Regulation of the expression of ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) in a cyanobacterium, Synechococcus PCC7942. Photosynth Res 78: 59–65 [DOI] [PubMed] [Google Scholar]
- Havemann GD, Sampson EM, Bobik TA (2002) PduA is a shell protein of polyhedral organelles involved in coenzyme B(12)-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J Bacteriol 184: 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt CA, Cooley JW, Wiskich JT, Vermaas WF (2001) A strain of Synechocystis sp. PCC 6803 without photosynthetic oxygen evolution and respiratory oxygen consumption: implications for the study of cyclic photosynthetic electron transport. Planta 214: 46–56 [DOI] [PubMed] [Google Scholar]
- Huang L, McCluskey MP, Ni H, LaRossa RA (2002) Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J Bacteriol 184: 6845–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H (2009) Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA 106: 14168–14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS (2005) LdpA: a component of the circadian clock senses redox state of the cell. EMBO J 24: 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Gao T, LiWang AC, Golden SS (2006) Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA 103: 17468–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain IH, Vijayan V, O’Shea EK (2012) Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci USA 109: 13638–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Erbilgin O (2015) Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol 23: 22–34 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Heinhorst S, Cannon GC (2010) Bacterial microcompartments. Annu Rev Microbiol 64: 391–408 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309: 936–938 [DOI] [PubMed] [Google Scholar]
- Kinney JN, Axen SD, Kerfeld CA (2011) Comparative analysis of carboxysome shell proteins. Photosynth Res 109: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR (2014a) β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR (2014b) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Landry MR, Vaulot D, Campbell L (1999) Prochlorococcus growth rates in the central equatorial Pacific: An application of the ƒmax approach. J Geophys Res 104: 3391–3399 [Google Scholar]
- Liu LN. (2016) Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. Biochim Biophys Acta 1857: 256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LN, Bryan SJ, Huang F, Yu J, Nixon PJ, Rich PR, Mullineaux CW (2012) Control of electron transport routes through redox-regulated redistribution of respiratory complexes. Proc Natl Acad Sci USA 109: 11431–11436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Tucker L, Badger MR, Price GD (2010) Functional cyanobacterial beta-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol 153: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan N, Brenner M (2014) Systems analysis of the CO2 concentrating mechanism in cyanobacteria. eLife 3: e02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn PJ, Price GD, Badger MR (2004) High light enhances the expression of low-CO2-inducible transcripts involved in the CO2-concentrating mechanism in Synechocystis sp. PCC6803. Plant Cell Environ 27: 615–626 [Google Scholar]
- McKay RM, Gibbs S, Espie G (1993) Effect of dissolved inorganic carbon on the expression of carboxysomes, localization of Rubisco and the mode of inorganic carbon transport in cells of the cyanobacterium Synechococcus UTEX 625. Arch Microbiol 159: 21–29 [Google Scholar]
- Menon BB, Heinhorst S, Shively JM, Cannon GC (2010) The carboxysome shell is permeable to protons. J Bacteriol 192: 5881–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW. (2001) How do cyanobacteria sense and respond to light? Mol Microbiol 41: 965–971 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW, Allen JF (1990) State 1-State 2 transitions in the cyanobacterium Synechococcus 6301 are controlled by the redox state of electron carriers between photosystems I and II. Photosynth Res 23: 297–311 [DOI] [PubMed] [Google Scholar]
- Ohbayashi R, Watanabe S, Kanesaki Y, Narikawa R, Chibazakura T, Ikeuchi M, Yoshikawa H (2013) DNA replication depends on photosynthetic electron transport in cyanobacteria. FEMS Microbiol Lett 344: 138–144 [DOI] [PubMed] [Google Scholar]
- Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S (2001) Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183: 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña KL, Castel SE, de Araujo C, Espie GS, Kimber MS (2010) Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc Natl Acad Sci USA 107: 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR (1989) Isolation and characterization of High CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942: Two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol 91: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Price GD, Coleman JR, Badger MR (1992) Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol 100: 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD (2012) Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7: e43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD (2013) Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev 77: 357–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA (2010) Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327: 1258–1261 [DOI] [PubMed] [Google Scholar]
- Schwarz R, Reinhold L, Kaplan A (1995) Low activation state of ribulose-1,5-bisphosphate carboxylase/oxygenase in carboxysome-defective Synechococcus mutants. Plant Physiol 108: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE (1973) Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182: 584–586 [DOI] [PubMed] [Google Scholar]
- Smith RM, Williams SB (2006) Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA 103: 8564–8569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Faulkner M, Aussignargues C, Paasch BC, Barrett S, Kerfeld CA, Liu L-N (2016) Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy. Nano Lett 16: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO (2008) Atomic-level models of the bacterial carboxysome shell. Science 319: 1083–1086 [DOI] [PubMed] [Google Scholar]
- Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M (2007) Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J Bacteriol 189: 4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst A. (1980) Inhibitors in electron flow: Tools for the functional and structural localization of carriers and energy conservation sites. Meth Enzymol 69: 675–715 [Google Scholar]
- Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO (2007) Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol 5: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson GM, Tabita FR (1996) Regulation, unique gene organization, and unusual primary structure of carbon fixation genes from a marine phycoerythrin-containing cyanobacterium. Plant Mol Biol 32: 1103–1115 [DOI] [PubMed] [Google Scholar]
- Whitehead L, Long BM, Price GD, Badger MR (2014) Comparing the in vivo function of α-carboxysomes and β-carboxysomes in two model cyanobacteria. Plant Physiol 165: 398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TL, Bridwell-Rabb J, Kim YI, Gao T, Chang YG, LiWang A, Barondeau DP, Golden SS (2010) The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Sci USA 107: 5804–5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger FJ, Badger MR, Price GD (2003) Inorganic carbon limitation induces transcripts encoding components of the CO(2)-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol 133: 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Crowley CS, Tanaka S (2010) Bacterial microcompartment organelles: protein shell structure and evolution. Annu Rev Biophys 39: 185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (2008) Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol 6: 681–691 [DOI] [PubMed] [Google Scholar]
- Zhang P, Eisenhut M, Brandt AM, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM (2012) Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.