An effector from the potato late blight pathogen targets a host protein that negatively regulates immunity.
Abstract
Plant pathogens deliver effectors to manipulate host processes. We know little about how fungal and oomycete effectors target host proteins to promote susceptibility, yet such knowledge is vital to understand crop disease. We show that either transient expression in Nicotiana benthamiana, or stable transgenic expression in potato (Solanum tuberosum), of the Phytophthora infestans RXLR effector Pi02860 enhances leaf colonization by the pathogen. Expression of Pi02860 also attenuates cell death triggered by the P. infestans microbe-associated molecular pattern INF1, indicating that the effector suppresses pattern-triggered immunity. However, the effector does not attenuate cell death triggered by Cf4/Avr4 coexpression, showing that it does not suppress all cell death activated by cell surface receptors. Pi02860 interacts in yeast two-hybrid assays with potato NPH3/RPT2-LIKE1 (NRL1), a predicted CULLIN3-associated ubiquitin E3 ligase. Interaction of Pi02860 in planta was confirmed by coimmunoprecipitation and bimolecular fluorescence complementation assays. Virus-induced gene silencing of NRL1 in N. benthamiana resulted in reduced P. infestans colonization and accelerated INF1-mediated cell death, indicating that this host protein acts as a negative regulator of immunity. Moreover, whereas NRL1 virus-induced gene silencing had no effect on the ability of the P. infestans effector Avr3a to suppress INF1-mediated cell death, such suppression by Pi02860 was significantly attenuated, indicating that this activity of Pi02860 is mediated by NRL1. Transient overexpression of NRL1 resulted in the suppression of INF1-mediated cell death and enhanced P. infestans leaf colonization, demonstrating that NRL1 acts as a susceptibility factor to promote late blight disease.
Plant immunity is triggered by the detection of conserved microbial molecules, microbe-associated molecular patterns (MAMPs) or pathogen-associated molecular patterns (PAMPs), leading to pattern-triggered immunity (PTI), and by the detection of effectors, leading to effector-triggered immunity. Central to the successful colonization of plants by phytopathogens is the delivery of effector proteins to suppress host immunity. Secreted effectors may act outside (apoplastic effectors) or be delivered inside (intracellular or cytoplasmic effectors) host cells to attenuate PTI or effector-triggered immunity (Jones and Dangl, 2006). A broad range of host targets and activities have been elucidated for many bacterial type 3 secretion system effectors (Block and Alfano, 2011; Deslandes and Rivas, 2012; Dou and Zhou, 2012). In contrast, less is understood about the effectors from filamentous phytopathogens: the fungi and oomycetes.
The oomycete Phytophthora infestans causes the devastating late blight disease of potato (Solanum tuberosum) and tomato (Solanum lycopersicum; Kamoun et al., 2015). Among the classes of candidate virulence determinants that have been identified are the RXLR effectors (Birch et al., 2006), which are delivered inside living plant cells (Whisson et al., 2007). Following the identification of the ubiquitin E3 ligase CMPG1 as a target of Avr3a (Bos et al., 2010), the targets and/or virulence activities of a small number of other P. infestans RXLRs have been revealed. AvrBlb2 prevents the secretion of a defense protease (Bozkurt et al., 2011), while Avr2 interaction with the putative phosphatase BSL1, involved in the brassinosteroid signal transduction pathway, facilitates recognition of the effector by the resistance protein R2 (Saunders et al., 2012). Effector Pi03192 interacts with NAC transcription factors, preventing their relocalization from the endoplasmic reticulum to the host nucleus (McLellan et al., 2013). PexRD2 targets the host MAP3Kε, inhibiting signal transduction following the perception of CfAvr4 from Cladosporium fulvum by tomato resistance Cf4 (King et al., 2014), while a number of effectors act redundantly to suppress flg22-mediated mitogen-activated protein kinase (MAPK) activation and early transcriptional changes (Zheng et al., 2014), implicating this signal transduction pathway also in the response to unknown oomycete MAMPs. A K-homology class RNA-binding protein, StKRBP1, which associates with the RXLR effector Pi04089, provides, to our knowledge, the first evidence that P. infestans effectors manipulate host susceptibility factors to promote late blight disease (Wang et al., 2015). Pi04089 increases the abundance of StKRBP1, a phenomenon that also occurs during the first 24 h of P. infestans infection. Overexpression of this RNA-binding protein enhances leaf colonization by the pathogen (Wang et al., 2015). More recently, an RXLR effector from P. infestans was shown to target host PP1c isoforms. Rather than inhibiting these phosphatases, the effector forms unique holoenzymes with them to presumably dephosphorylate key substrates in the plant nucleus, leading to enhanced susceptibility (Boevink et al., 2016). Thus, the PP1c isoforms also can be regarded as susceptibility factors.
One of the key P. infestans MAMPs detected by solanaceous hosts is INF1, which elicits BAK1-dependent cell death in the model host plant Nicotiana benthamiana (Heese et al., 2007) and a range of Solanum spp. (Vleeshouwers et al., 2006). Recently, a receptor that detects INF1 and other elicitins from a broad range of oomycetes, termed ELR, was cloned from Solanum microdontum (Du et al., 2015). Overexpression of ELR in the cultivated potato enhances disease resistance to P. infestans (Du et al., 2015). INF1-mediated cell death can be suppressed by Avr3a, either by inhibition or modification of CMPG1 activity (Bos et al., 2010; Gilroy et al., 2011), and can be partially suppressed by the RXLR effector Pi18215/SFI7, which also inhibits flg22-mediated MAPK activation (Zheng et al., 2014).
In addition to CMPG1, another plant U-box (PUB) ubiquitin E3 ligase, PUB17, has been shown to positively regulate immunity (Yang et al., 2006). PUB17 functions in the host nucleus to mediate both PTI, following perception of the bacterial PAMP flg22, and cell death triggered by coexpression of Cf4/Avr4. However, it is not involved in INF1-triggered cell death (He et al., 2015). In contrast to CMPG1 and PUB17, a number of PUB E3 ligases have been shown to negatively regulate plant immunity (Duplan and Rivas, 2014). PUB12 and PUB13 work in concert to attenuate PTI by ubiquitinating the flg22 receptor FLS2, facilitating its degradation (Lu et al., 2011). PUB22, PUB23, and PUB24 also act to suppress immunity. In addition, NPR3 and NPR4 contain Broad complex, Tramtrack, and Bric-a-brac (BTB) domains that facilitate interaction with the CULLIN3 E3 ligase. NPR3 and NPR4 negatively regulate salicylic acid-mediated defenses (Fu et al., 2012). Ubiquitination is thus a posttranslational modification implicated in both positive and negative regulation of immunity.
Here, we show that either transient expression in the model host N. benthamiana, or stable transformation in potato, of the putative RXLR effector Pi02860 supports enhanced leaf colonization by P. infestans. Expression of the effector in N. benthamiana also suppresses INF1-mediated cell death, indicating that Pi02860 contributes to PTI suppression. Pi02860 localizes to the cytoplasm in N. benthamiana. Pi02860 interacts with a NONPHOTOTROPHIC HYPOCOTYL3/ROOT PHOTOTROPISM2-LIKE (NRL) protein, StNRL1, in yeast two-hybrid (Y2H) assays and in planta. Virus-induced gene silencing (VIGS) of NbNRL1 attenuates P. infestans colonization and accelerates INF1-mediated cell death. Moreover, VIGS of NbNRL1 prevents the ability of Pi02860 to suppress INF1-mediated cell death, whereas such suppression by Avr3a is unaltered, indicating that PTI suppression by Pi02860 is mediated by NRL1. In contrast, overexpression of StNRL1 alone enhances colonization and suppresses INF1-mediated cell death, indicating that StNRL1 is a negative regulator of PTI and, thus, can be regarded as a susceptibility factor.
RESULTS AND DISCUSSION
Pi02860 Promotes P. infestans Virulence and Suppresses PTI
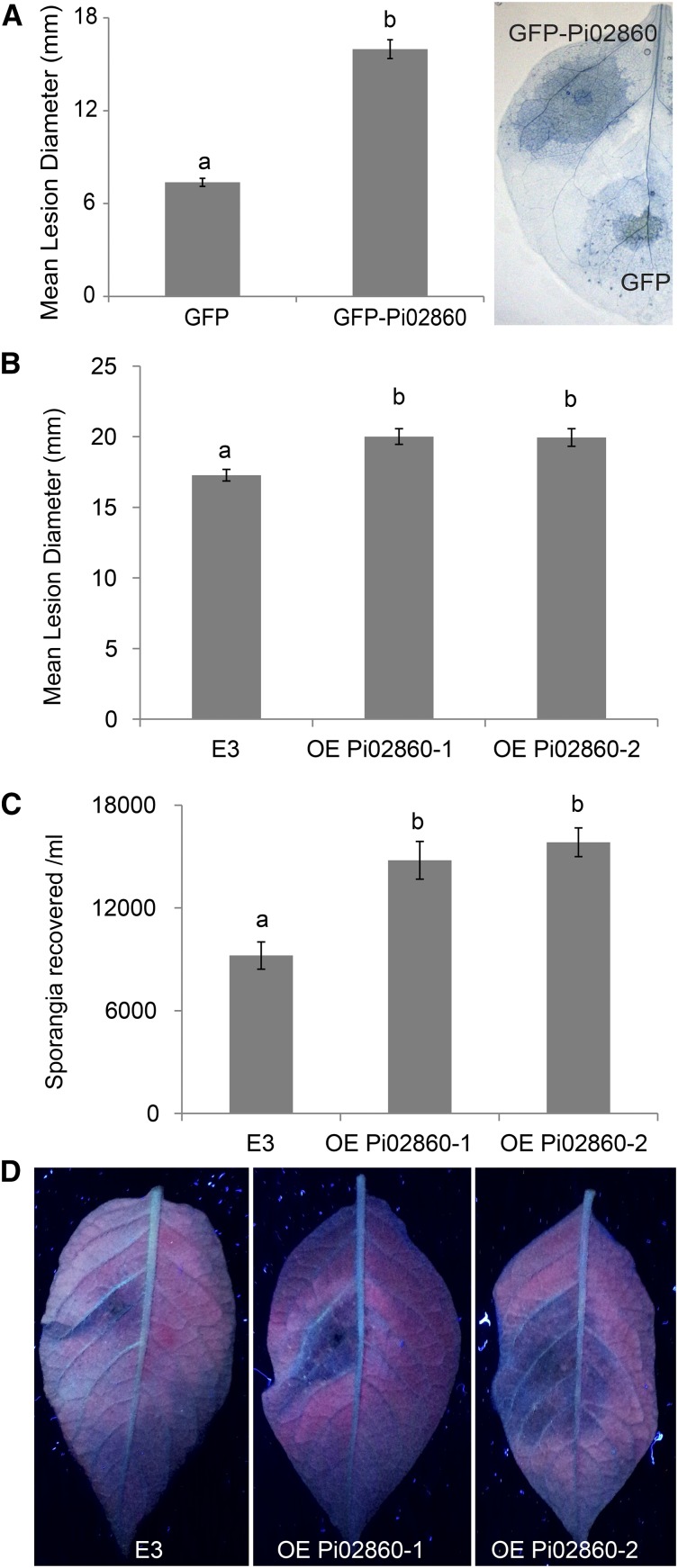
The effector Pi02860 (PITG_02860) is annotated in the P. infestans genome as a secreted RXLR-type effector protein (Haas et al., 2009). Consistent with other RXLR effectors, Pi02860 is specifically up-regulated at 2 and 3 d post inoculation (dpi) of potato plants challenged with distinct P. infestans genotypes (Haas et al., 2009; Cooke et al., 2012). As these time points correspond to the biotrophic phase of Phytophthora spp. infection (Avrova et al., 2008), this effector was cloned and tested for its ability to influence P. infestans colonization. A construct with GFP fused to the N terminus of Pi02860 in place of the signal peptide was cloned and transiently expressed in the model solanaceous P. infestans host plant N. benthamiana using Agrobacterium tumefaciens-mediated expression followed by P. infestans challenge, as performed for other RXLR effectors (McLellan et al., 2013; King et al., 2014; Zheng et al., 2014). At 6 dpi, significantly larger lesions (ANOVA, P < 0.001) were observed in areas expressing GFP-Pi02860 compared with the expression of free GFP (Fig. 1A), thus suggesting that Pi02860 confers a benefit to the pathogen consistent with effector activity. To explore this phenomenon further in the host crop plant, transgenic potato lines were made for stable expression of Pi02860, minus signal peptide-encoding sequences (Supplemental Fig. S1). These plants were subsequently challenged with P. infestans and found to support significantly larger lesions (ANOVA, P < 0.002; Fig. 1, B and D) and significantly enhanced sporulation (ANOVA, P < 0.001) of the pathogen (Fig. 1C). The enhancement of P. infestans leaf colonization promoted by Pi02860 expression inside host cells is similar to that of other recently described RXLR effectors (McLellan et al., 2013; King et al., 2014; Zheng et al., 2014; Wang et al., 2015; Boevink et al., 2016) and consistent with it modifying the host to promote susceptibility.
Figure 1.
Transient or stable overexpression of Pi02860 enhances P. infestans colonization. A, The graph shows a significant increase (P < 0.001, n = 94) in P. infestans lesion diameter following transient expression in N. benthamiana of GFP-Pi02860 compared with a free GFP control. The Trypan Blue-stained leaf image shows the extent of P. infestans colonization with GFP-Pi02860 or free GFP. B, Transgenic potato lines overexpressing Pi02860 (OE Pi02860-1 and OE Pi02860-2) allow a significant increase (ANOVA, P < 0.002, n = 83) in P. infestans lesion diameter compared with the potato cv E3 control. C, Transgenic potato lines overexpressing Pi02860 allow a significant increase (P < 0.001, n = 64) in P. infestans sporulation compared with the cv E3 control. D, Leaf images taken under UV light show an increase in P. infestans lesions in transgenic potato lines overexpressing Pi02860 compared with the cv E3 control. Lowercase letters on graphs denote statistically significant differences determined by one-way ANOVA, with pairwise comparisons performed with the Holm-Sidak method. Results shown are combinations of at least two independent biological replicates. Error bars show se.
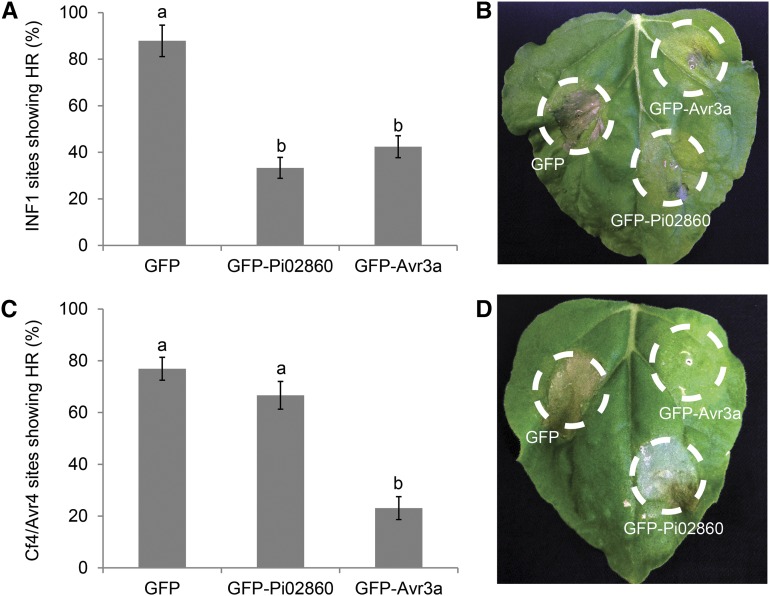
As some RXLR effectors have been demonstrated to interfere with distinct defense signaling pathways in planta (Bos et al., 2010; King et al., 2014), GFP-Pi02860 was tested to determine if it attenuated cell death signaling activated by two characterized pathways. The Phytophthora spp. PAMP INF1 triggers a hypersensitive response (HR) in some solanaceous hosts, including N. benthamiana, and this HR can be blocked by coexpressing the RXLR effector Avr3a (Bos et al., 2010). In addition, effectors Avr3a and PexRD2 are both able to suppress the HR triggered by coexpression of the Cladosporium fulvum effector Avr4 and its cognate resistance protein Cf4 by different mechanisms (Bos et al., 2010; King et al., 2014). Expression of GFP-Pi02860 was found to significantly attenuate INF1-mediated HR (ANOVA, P < 0.001) to a similar level as the GFP-Avr3a control (Fig. 2, A and B). In contrast to GFP-Avr3a, it had no significant effect on Cf4-CfAvr4 HR (P > 0.1; Fig. 2, C and D). This suggests that the function of Pi02860 may be to suppress a specific signaling pathway(s) that is triggered by the perception of P. infestans PAMPs, such as INF1, and does not extend to all cell death pathways triggered by the activation of cell surface receptors.
Figure 2.
Pi02860 suppresses INF1 but not Cf4-Avr4 HR. A, Transient overexpression of GFP-Pi02860 compared with free GFP in N. benthamiana can significantly suppress the HR (P < 0.001, n = 11) triggered by the elicitin INF1 to a similar extent to the control GFP-Avr3a. B, Representative leaf image showing INF1 HR at 6 dpi, following coexpression with the constructs indicated. C, Transient overexpression of GFP-Pi02860 or free GFP in N. benthamiana shows no significant difference (P > 0.1) in HR triggered by Cf4-Avr4, whereas GFP-Avr3a significantly suppresses this HR (P < 0.001, n = 13). D, Representative leaf image showing Cf4-Avr4 HR at 6 dpi, following coexpression with the constructs indicated. Lowercase letters on graphs denote statistically significant differences by one-way ANOVA, with pairwise comparisons performed with the Holm-Sidak method. Error bars show se.
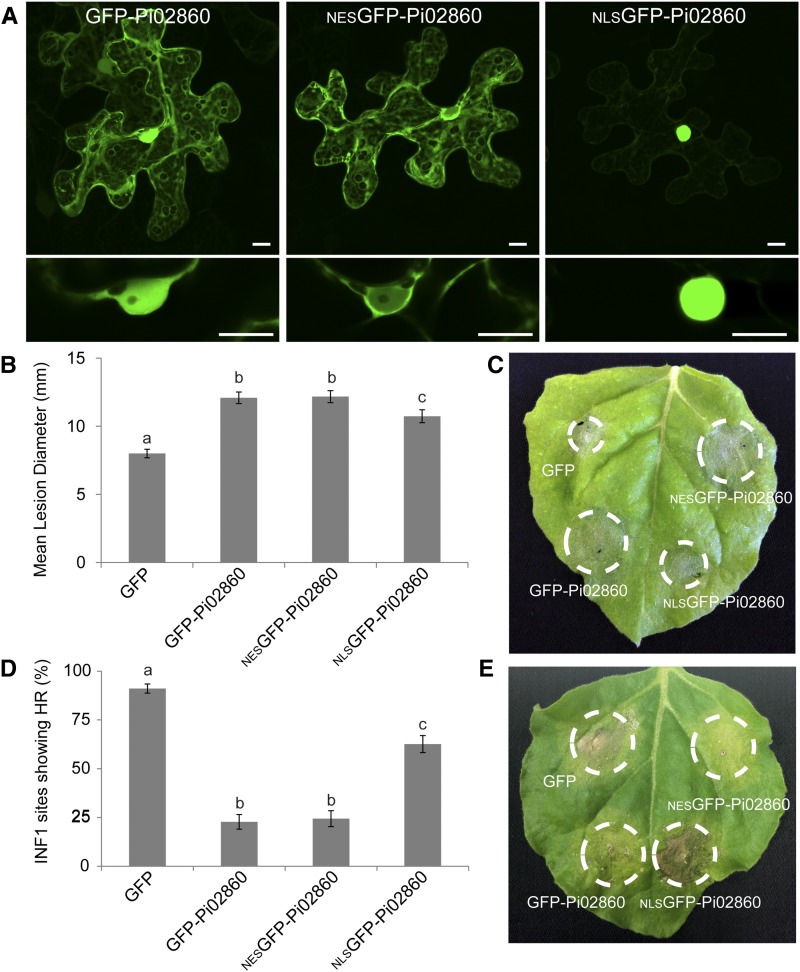
To further investigate the phenotypes associated with Pi02860 overexpression, the subcellular localization of this protein was examined in N. benthamiana using confocal microscopy. GFP-Pi02860 was found to localize throughout the plant cytoplasm and nucleoplasm (Fig. 3A). To perturb the observed localization of the effector, two additional fusion constructs were generated, to which either a nuclear export signal (NES) or a nuclear localization signal (NLS) was added to the N-terminally fused GFP, as described previously (Wang et al., 2015). Both produced intact fusion proteins when expressed in planta (Supplemental Fig. S2). On examination with confocal microscopy, NESGFP-Pi02860 was greatly reduced in the nucleoplasm but still accumulated in the cytoplasm, while NLSGFP-Pi02860 accumulated strongly in the nucleus and was largely reduced in the cytoplasm (Fig. 3A). The effects of these constructs were tested in the P. infestans virulence assay using GFP-Pi02860 and GFP as positive and negative controls, respectively. Interestingly, expression of NESGFP-Pi02860 was found to enhance leaf colonization to the same level as GFP-Pi02860, while lesion sizes following the expression of NLSGFP-Pi02860 were reduced significantly compared with GFP-Pi02860 but were still significantly larger than with free GFP (Fig. 3, B and C). A similar pattern was observed when testing these constructs for their ability to suppress INF1-mediated HR. Again, NESGFP-Pi02860 was found to suppress INF1 HR to levels similar to GFP-Pi02860. In contrast, NLSGFP-Pi02860 was significantly less able to suppress INF1-mediated HR compared with GFP-Pi02860, but the HR was nevertheless more significantly suppressed compared with that observed with free GFP expression (Fig. 3, D and E). While the NES fusion did not totally exclude GFP-Pi02860 from the nucleus and the NLS fusion still retained background levels of cytoplasmic fluorescence, these results may nevertheless indicate that the cytoplasmic localization of Pi02860 is more important for its contribution to virulence than the observed nucleoplasmic localization.
Figure 3.
Cytoplasmic localization of GFP-Pi02860 is important for P. infestans virulence and INF1 HR suppression phenotypes. A, Confocal images showing that GFP-Pi02860 is localized in the cytoplasm and nucleus, NESGFP-Pi02860 is greatly reduced in nuclear fluorescence, and NLSGFP-Pi02860 is concentrated in the nucleus and reduced in the cytoplasm. The top row shows stacked projections of single cells, while the bottom row shows single slice images of the nuclei. Bars =10 μm. B, GFP-Pi02860 and NESGFP-Pi02860 expression leads to statistically significant increases (P < 0.001, n = 68) in P. infestans lesion diameter compared with free GFP, whereas NLSGFP-Pi02860 shows an intermediate phenotype. C, Representative leaf image showing P. infestans lesions following overexpression of each construct, as indicated, in N. benthamiana. D, GFP-Pi02860 and NESGFP-Pi02860 coexpression with INF1 leads to a statistically significant decrease (P < 0.001, n = 41) in HR compared with free GFP, whereas NLSGFP-Pi02860 shows an intermediate phenotype. E, Representative leaf image showing INF1 HR following coexpression with each construct, as indicated, in N. benthamiana. Lowercase letters on graphs denote statistically significant differences by one-way ANOVA, with pairwise comparisons performed with the Holm-Sidak method. Results shown are combinations of three independent biological replicates, each with 18 infiltration zones. Error bars show se.
Pi02860 Interacts with the BTB/POZ Domain Protein StNRL1
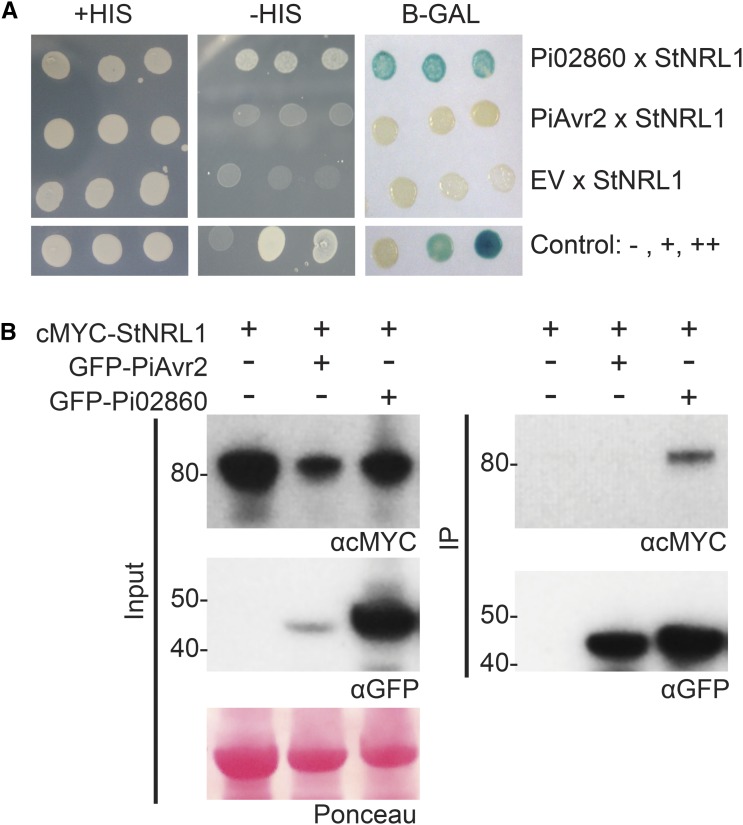
To further explore the mechanism of Pi02860 action in plants, a Y2H library made from complementary DNA (cDNA) of potato infected with P. infestans (Bos et al., 2010) was screened with a GAL4 DNA-binding domain-Pi02860 fusion (bait) construct to a depth of 0.44 × 106 yeast cotransformants. Five yeast colonies recovered from selection plates that contained GAL4 activation domain (prey) fusions yielded sequences corresponding to a potato BTB/POZ domain protein belonging to the NPH3/RPT2-like family, hereafter referred to as StNRL1. In Arabidopsis (Arabidopsis thaliana), NPH3 and RPT2 interact with phototropins, mediating blue light signaling, and are thought to be a core component of a CUL3-based ubiquitin protein ligase (E3) enzyme complex (Liscum et al., 2014). Supplemental Figure S3 shows an amino acid alignment of potato StNRL1 with its N. benthamiana equivalents NbNRL1a and NbNRLb, the Arabidopsis protein At5g67385 (AtNRL) that is a candidate ortholog (reciprocal best BLAST hit), and the characterized At5g64330 (AtNPH3) and At2g30520 (AtRPT2), indicating the conserved domains across these proteins. To confirm this interaction, a full-length StNRL1 prey construct was tested pairwise with bait constructs for Pi02860, a noninteracting RXLR control, PiAvr2, which has been shown previously to associate with the putative phosphatase BSL1 (Saunders et al., 2012), and the empty bait vector. While all transformants grew on the control plates, only yeast containing both Pi02860 and StNRL1 were able to grow on the selection plates and activate the β-galactosidase reporter (Fig. 4A).
Figure 4.
Pi02860 interacts with the potato BTB/POZ domain protein StNRL1 in Y2H and immunoprecipitation assays. A, Yeast coexpressing StNRL1 with Pi02860 grew on −His (−HIS) medium and yielded β-galactosidase (B-GAL) activity, while those coexpressed with the control PiAvr2 did not. The +HIS control shows that all yeast were able to grow in the presence of His. B, Immunoprecipitation (IP) of protein extracts from agroinfiltrated leaves using GFP-Trap confirmed that cMyc-tagged NRL1 associated specifically with GFP-Pi02860 and not with the GFP-PiAvr2 control. The expression of constructs in the leaves is indicated by +. Protein size markers are indicated in kD, and protein loading is indicated by Ponceau stain.
To confirm that this interaction also occurs in planta, a coimmunoprecipitation assay was performed by expressing cMyc-tagged StNRL1 (cMyc-StNRL1) alone or with GFP-Pi02860 or GFP-PiAvr2 and pulling down with GFP-TRAP_M beads. Figure 4B shows that, while all proteins were present in the relevant input samples, cMYC-StNRL1 was only immunoprecipitated in the presence of GFP-Pi02860 and not alone or with the GFP-PiAvr2 control.
To examine StNRL1 in more detail, GFP was fused to its N terminus to form GFP-StNRL1 and was localized in N. benthamiana using confocal microscopy. The GFP-StNRL1 fusion localized partially in the cytoplasm, but showed significant accumulation at the plasma membrane (PM), when compared with a free GFP control (Supplemental Fig. S4). Coexpression of GFP-StNRL1 with an mOrange-LTi PM marker indicated significant colocalization, which was not observed with free GFP (Fig. 5, A and B; Supplemental Fig. S4). This indicates that, while GFP-NRL1 is observed in the cytoplasm, it also strongly associates with the PM.
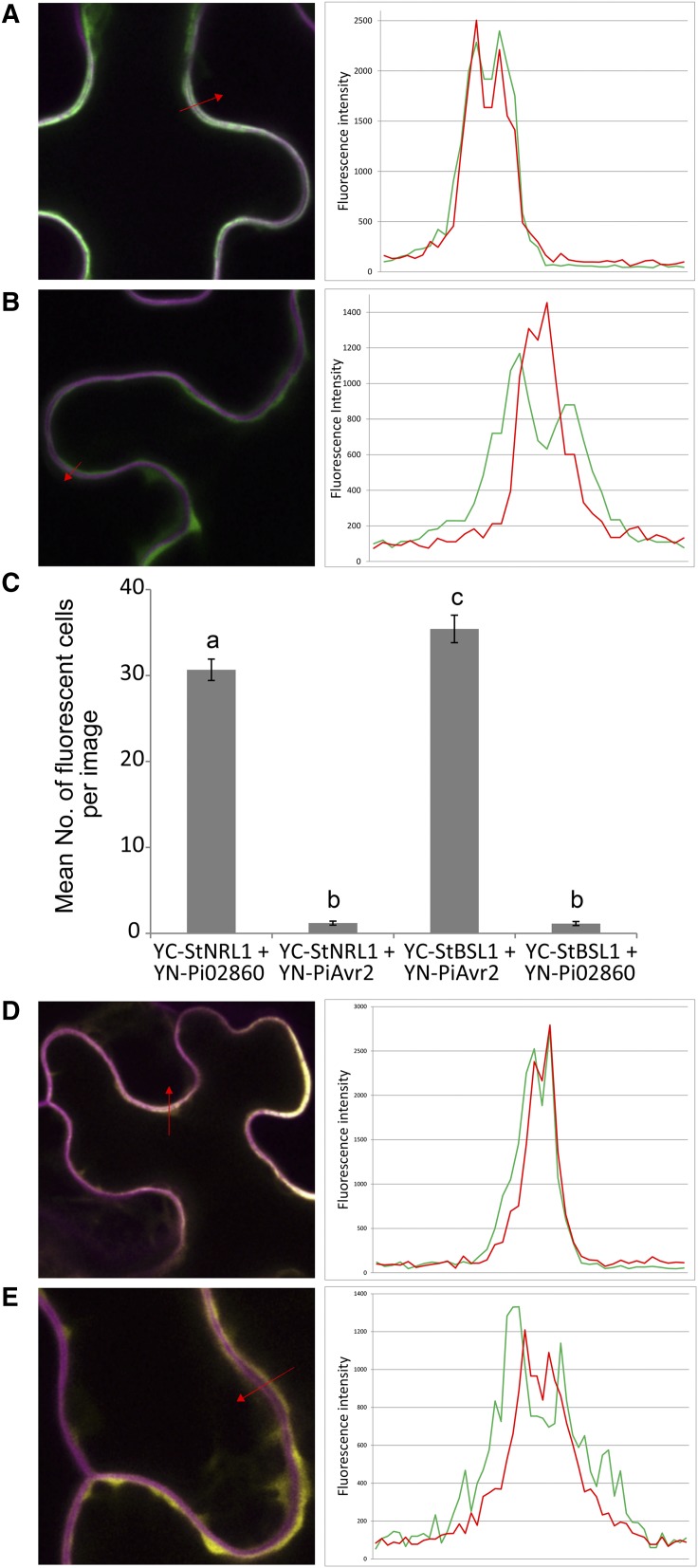
Figure 5.
GFP-tagged StNRL1 predominantly locates to the plasma membrane, and bimolecular fluorescence complementation confirms StNRL1 and Pi02860 interaction. A, Single optical slice image across PMs of two adjacent cells coexpressing GFP-StNRL1 and the mOrange-LTi PM marker, with a profile across the membranes in the location indicated by the red arrow. The plot of the profile (right) indicates that the majority of the GFP fluorescence (green line) colocates with the PM marker (red line). B, A comparable profile in a cell coexpressing the PM marker with unfused GFP, which is only present in the cytoplasm. C, Average number of fluorescent cells per image with YC-StNRL1 + YN-Pi02860 and YC-StBSL1 + YNPiAvr2 giving significantly more (P < 0.001, n = 22) reconstitution of YFP fluorescence than when noninteracting effector-interactor pairs are used. Lowercase letters on graphs denote statistically significant differences by one-way ANOVA, with pairwise comparisons performed with the Holm-Sidak method. Results shown are combinations of three independent biological replicates. Error bars show se. D, Single optical slice image across PMs of two adjacent cells coexpressing YC-StNRL1 + YN-Pi02860 and the mOrange-LTi PM marker, with a profile across the membranes in the location indicated (red arrow). The plot of the profile (right) indicates that the majority of the reconstituted YFP fluorescence (green line) colocates with the PM marker (red line). E, A comparable profile in a cell coexpressing unfused YFP in the cytoplasm with the PM marker.
A bimolecular fluorescence complementation assay (commonly referred to as split YFP) was then undertaken to establish the site of the interaction of StNRL1 and Pi02860 proteins in planta. The C-terminal fragment of yellow fluorescent protein (YFP; YC) was fused to StNRL1, while the N terminus (YN) was fused to Pi02860, to give YC-StNRL1 and YN-Pi02860, respectively. As the controls used in split YFP studies are important to rule out false-positive interactions (Boevink et al., 2014), we used YC-StBSL1 and YN-PiAvr2 as controls for a plant target and effector interacting pair, which also localize to the plant cytoplasm and PM (Saunders et al., 2012). Coexpression of either YC-StNRL1 with YN-Pi02860 or YC-StBSL1 with YN-PiAvr2 yielded fluorescence visualized by confocal microscopy, whereas there was no appreciable fluorescence when YN-Pi02860 was coexpressed with YC-BSL1 or when YN-Avr2 was coexpressed with YC-NRL1 (Supplemental Fig. S4). This was quantified by counting the number of fluorescent cells in the field of view in more than 50 low-magnification images each to show that YC-StNRL1 with YN-Pi02860 or YC-StBSL1 with YN-PiAvr2 gave significantly higher fluorescence (ANOVA, P < 0.001) than YC-StNRL1 with YN-PiAvr2 or YC-StBSL1 with YN-Pi02860 (Fig. 5C).
The YC-StNRL1 and YN-Pi02860 constructs were coexpressed in N. benthamiana with the mOrange-LTi PM marker, and YFP fluorescence was observed in the cytoplasm, but with significant accumulation at the PM, using confocal microscopy, compared with the free YFP control (Fig. 5, D and E; Supplemental Fig. S4). The presence of each of the intact fusion constructs was confirmed by immunoblotting to rule out changes in fluorescence levels being caused by construct instability (Supplemental Fig. S5).
The localization of StNRL1 at the host PM is in agreement with studies of NRL family members in Arabidopsis, all of which have been shown to interact at the PM, as part of a CUL3-RING ubiquitin ligase (CRL) complex, with phototropins, which are involved in blue light signaling (Liscum et al., 2014).
StNRL1 Silencing Retards P. infestans Colonization and Prevents Pi02860 Suppression of INF1-Mediated Cell Death
To examine a possible role for StNRL1 in plant defense against P. infestans, VIGS was used to knock down the expression of the equivalent NRL1 genes in N. benthamiana. N. benthamiana is an allotetraploid; thus, gene searches in the genome usually reveal two matching copies where the two homologous genes have not been collapsed during assembly (Bombarely et al., 2012). Consistent with that, two sequences, designated NbNRL1a and NbNRL1b, encoding proteins with 95% amino acid identity to each other, were identified in the N. benthamiana genome. The predicted NbNRL1a and NbNRL1b proteins are each 84% identical to StNRL1 (Supplemental Fig. S3). Consequently, two independent VIGS constructs, TRV-NRL V1 and TRV-NRL V2, were designed to silence both homologous copies simultaneously. Supplemental Figure S6A shows that the transcript accumulation of both NbNRL1a and NbNRL1b is reduced by 60% to 85% in plants expressing either TRV-NRL construct compared with plants expressing the TRV-GFP control. Representative images of plants expressing each TRV-NRL VIGS construct show that these plants exhibit a developmental phenotype, being stunted in growth compared with the TRV-GFP control (Supplemental Fig. S6B).
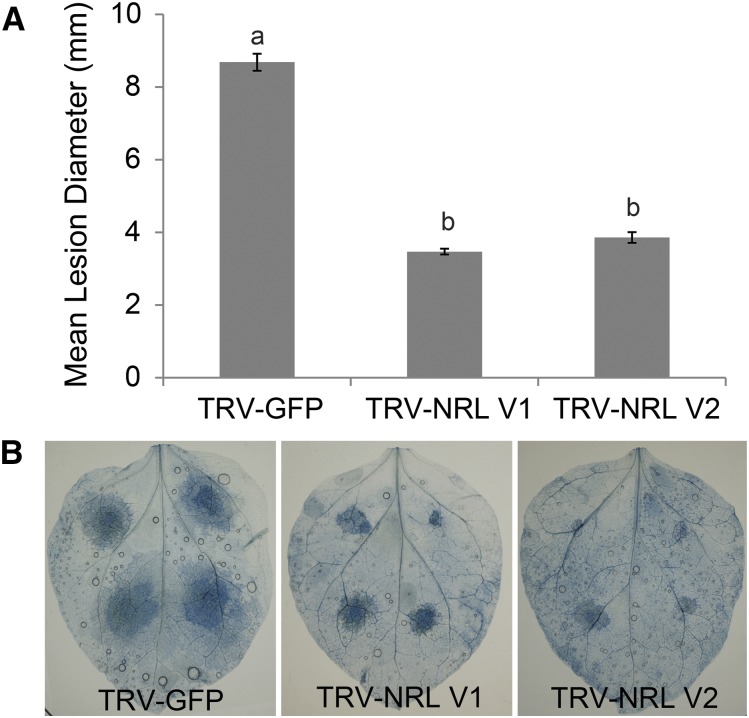
Following infection of the VIGS plants with P. infestans, it was observed that silencing of NbNRL1a and NbNRL1b led to a reduction in the ability of the pathogen to colonize these plants, with significantly smaller lesions (ANOVA, P < 0.001) developing on TRV-NRL plants compared with the TRV-GFP controls (Fig. 6). This suggests that P. infestans requires the presence of NRL1 to establish normal infections and would not support a model in which the effector Pi02860 inhibits or inactivates NRL1.
Figure 6.
Silencing of NRL1 in N. benthamiana compromises P. infestans infection. A, Silencing of NRL1 using two independent VIGS constructs (TRV-NRL V1 and TRV-NRL V2) in N. benthamiana significantly reduces (one-way ANOVA, P < 0.001, n = 464; significance denoted by lowercase letters) P. infestans lesion diameter compared with the TRV-GFP control. B, Representative leaf images stained with Trypan Blue show the extent of P. infestans leaf colonization on plants expressing each VIGS construct, as indicated.
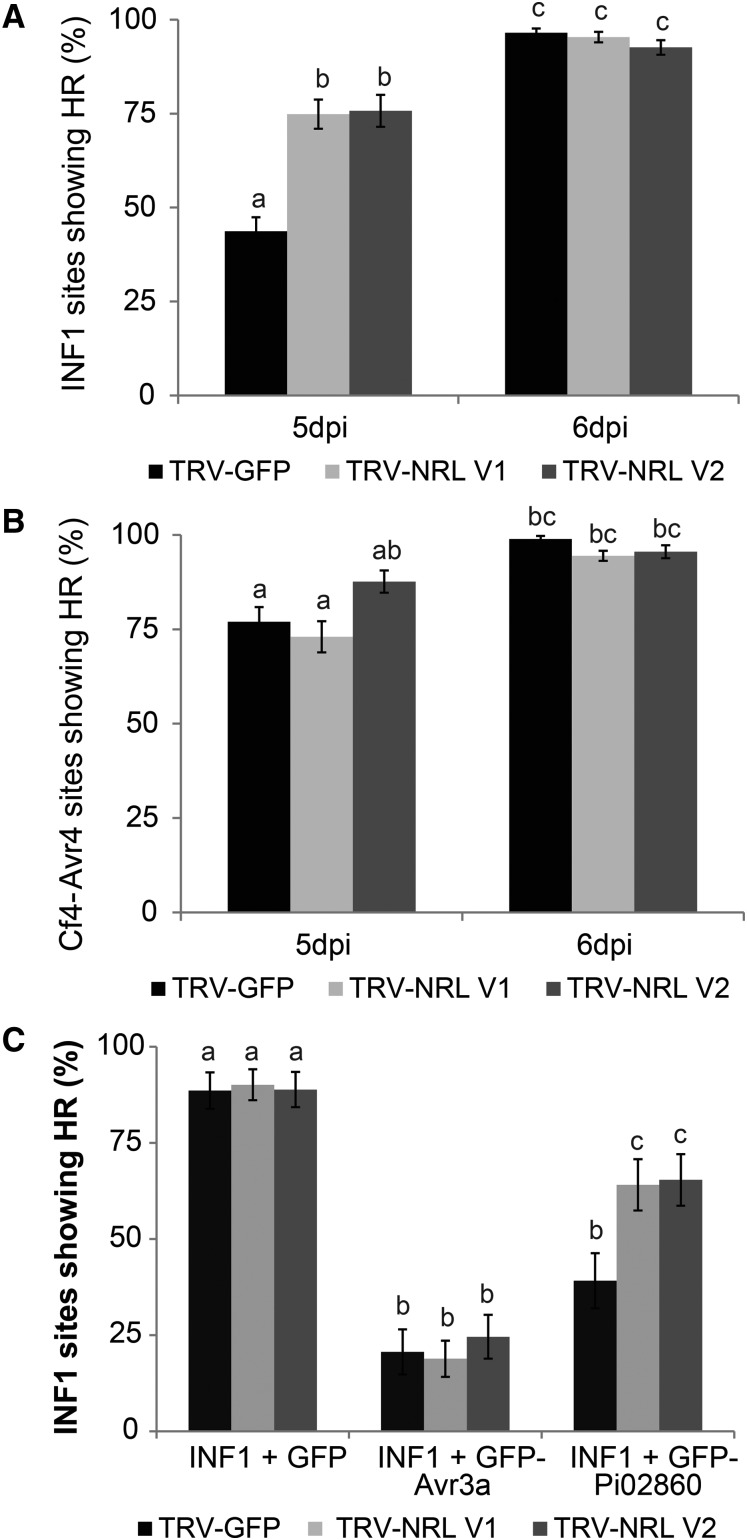
As Pi02860 also was observed to suppress INF1-mediated cell death, constructs expressing INF1 were agroinfiltrated into plants expressing each of the VIGS constructs and scored at 5 and 6 d post inoculation (dpi). At 5 dpi, a significant increase (ANOVA, P < 0.001) in INF1 HR was observed on TRV-NRL VIGS plants compared with TRV-GFP plants (Fig. 7A). This difference was not apparent by 6 dpi, indicating that the HR is accelerated by NbNRL1 silencing. The same assay was carried out to examine the Cf4-Avr4 HR, which was not affected by Pi02860 expression. As anticipated, there were no significant differences in Cf4-Avr4 HR in TRV-NRL plants compared with TRV-GFP plants at either 5 or 6 dpi (Fig. 7B). These results suggest that NRL1 acts as a negative regulator of INF1-mediated cell death.
Figure 7.
Silencing of NRL1 in N. benthamiana accelerates INF1 HR and reduces the ability of Pi02860 to attenuate INF1 HR. A, A significant increase (P < 0.001, n = 30) is seen in INF1 HR in TRV-NRL V1 and TRV-NRL V2 plants compared with the TRV-GFP control at 5 dpi but not at 6 dpi. B, No significant change (P > 0.15, n = 30) is seen in Cf4-Avr4 HR between TRV-NRL V1, TRV-NRL, and TRV-GFP at 5 or 6 dpi. C, GFP-Pi02860 expression is significantly less able to inhibit INF1 HR (P > 0.03, n = 39) in TRV-NRL V1 and TRV-NRL V2 plants compared with TRV-GFP at 6 dpi but has no significant effect on GFP-Avr3a INF1 HR suppression. Lowercase letters denote statistically significant differences by one-way ANOVA, with pairwise comparisons performed with the Holm-Sidak method. Results shown are combinations of three independent biological replicates. Error bars show se.
To investigate whether the suppression of INF1-mediated cell death by Pi02860 is dependent on the presence of NRL1, either GFP-Pi02860 or, as a control, GFP-Avr3a was coexpressed with INF1 in leaves expressing either the TRV-NRL VIGS constructs or TRV-GFP. Whereas GFP-Avr3a suppressed INF1-mediated cell death to similar amounts on all plants, the suppression of INF1-mediated cell death by GFP-Pi02860 was retained on TRV-GFP plants but was reduced significantly on plants in which NbNRL1a and NbNRL1b were silenced (Fig. 7C). Some ability to suppress INF1-mediated cell death was retained. However, this is likely due to the fact that VIGS is notoriously patchy, with some leaf areas more efficiently silenced than others, and that silencing knocked down the transcript levels of NbNRL1a and NbNRL1b by 60% to 85% (Supplemental Fig. S6), suggesting that some NRL1 protein is likely present. Nevertheless, the significant reduction in INF1 cell death suppression by GFP-Pi02860 provides a direct genetic link to indicate that this effector activity is dependent on the presence of NRL1. Taken together, these results indicate that NRL1 is a negative regulator of immunity and thus is unlikely to be inhibited by Pi02860.
StNRL1 Overexpression Suppresses INF1-Mediated Cell Death and Enhances P. infestans Colonization
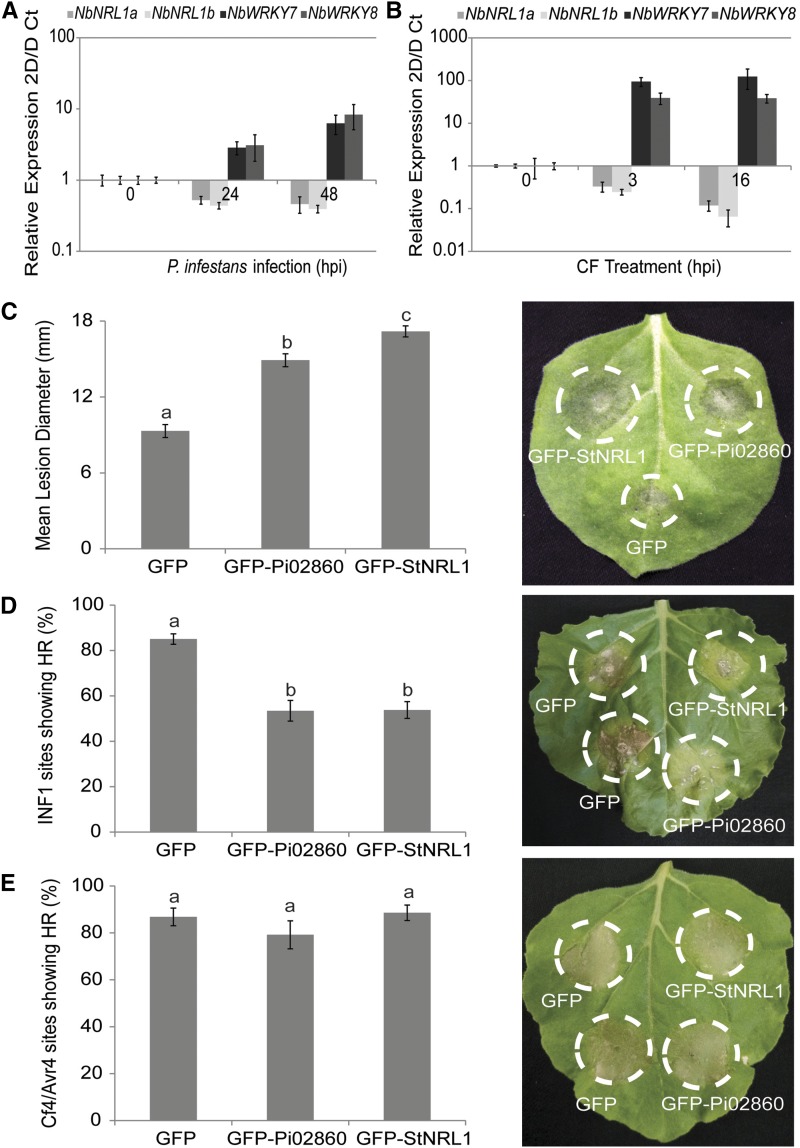
Silencing of NbNRL1a and NbNRL1b by VIGS in N. benthamiana led to accelerated INF1-triggered cell death and reduced P. infestans colonization, suggesting that NRL1 acts as a negative regulator of immunity. We thus investigated the expression of NbNRL1a and NbNRL1b during the first 48 h of P. infestans colonization of N. benthamiana, which can be regarded as the biotrophic phase of infection (Avrova et al., 2008), and at 3 and 16 h post treatment with P. infestans culture filtrate, which can be regarded as a cocktail of Phytophthora spp. PAMPs (McLellan et al., 2013). In contrast to two PTI marker genes, NbWRKY7 and NbWRKY8, which, similar to previous observations (McLellan et al., 2013), were weakly up-regulated during infection and strongly up-regulated by culture filtrate treatment, transcript accumulation of both NbNRL1a and NbNRL1b decreased weakly during infection and strongly with culture filtrate treatment (Fig. 8, A and B), indicating that they are potentially down-regulated during immune responses. This is consistent with NRL1 acting as a negative regulator of immunity. To further investigate this, we studied the effects of NRL1 overexpression.
Figure 8.
NRL1 is down-regulated by both P. infestans infection and PAMP treatment, while overexpression of NRL1 enhances P. infestans leaf colonization and suppresses INF1-triggered cell death. A, Relative expression levels of NbNRL1a, NbNRL1b, NbWRKY7, and NbWRKY8 in response to P. infestans infection. B, Relative expression levels of NbNRL1a, NbNRL1b, NbWRKY7, and NbWRKY8 in response to P. infestans culture filtrate (CF) treatment. In A and B, 2D/D Ct = log base2 delta delta cycle threshold. C, The graph shows that overexpression of GFP-Pi02860 and GFP-StNRL1 significantly increases (P < 0.001, n = 106) P. infestans lesion size compared with free GFP. The representative leaf image shows P. infestans lesions following the overexpression of each construct, as indicated, in N. benthamiana. D, The graph shows that overexpression of GFP-Pi02860 and GFP-StNRL1 significantly decreases (P < 0.001, n = 44) INF1 HR compared with free GFP. The representative leaf image shows INF1 HR with overexpression of each construct in N. benthamiana. E, The graph shows that coexpression of GFP-Pi02860 and GFP-StNRL1 has no significant effect (P = 0.325, n = 24) on Cf4-Avr4 HR compared with free GFP. The representative leaf image shows Cf4-Avr4 HR with overexpression of each construct in N. benthamiana. Lowercase letters on graphs denote statistically significant differences by one-way ANOVA, with pairwise comparisons performed with the Holm-Sidak method. Results shown are combinations of three independent biological replicates. Error bars show se.
Transient expression in N. benthamiana of either GFP-StNRL1 or GFP-Pi02860, followed by pathogen challenge, was found to result in significantly larger P. infestans lesions (P < 0.001) compared with free GFP expression, with GFP-StNRL1 overexpression having a larger infection-enhancing effect on P. infestans growth than GFP-Pi02860 (Fig. 8C). Moreover, either GFP-StNRL1 or GFP-Pi02860 expression independently suppressed INF1-mediated HR to a similarly significant level (P < 0.001) compared with free GFP expression (Fig. 8D). In contrast, the expression of either GFP-StNRL1 or GFP-Pi02860 had no significant effect on Cf4-Avr4-mediated HR (Fig. 8E).
Recently, the P. infestans effector Pi04089 was shown to interact with a K-homology class RNA-binding protein, KRBP1, which is a susceptibility factor. KRBP1 turnover is reduced in the presence of Pi04089, suggesting that the effector enhances its stability (Wang et al., 2015). We thus investigated whether such a phenomenon occurred with StNRL1 in the presence of Pi02860. However, in three independent replicates, GFP-StNRL1 protein stability was not enhanced by coexpression with cMYC-Pi02860 compared with a cMYC empty vector control (Supplemental Fig. S7).
This work shows that the P. infestans RXLR effector Pi02860, when expressed in planta, enhances pathogen colonization and suppresses cell death triggered by perception of the P. infestans PAMP INF1. We show that it does this through its interaction with the potato BTB/POZ domain family protein StNRL1, with which it interacts in the cytoplasm and at the plant plasma membrane, as silencing NRL1 compromises the ability of Pi02860 to suppress INF1-mediated cell death. In contrast, NRL1 silencing did not attenuate Avr3a suppression of INF1-triggered cell death, consistent with this effector acting through an alternative host target, the E3 ubiquitin ligase CMPG1 (Bos et al., 2010). The observation that Avr3a retains its ability to attenuate INF1-mediated cell death on NRL1 VIGS plants suggests that Avr3a acts downstream of Pi02860 in suppressing this immune response.
Recently, functional redundancy in the P. infestans effector repertoire was highlighted by the demonstration that eight out of 33 tested RXLR effectors were able to suppress early transcriptional responses to the bacterial PAMP flg22 (Zheng et al., 2014). Of these eight effectors, only three acted to suppress MAPK activation following flg22 treatment, indicating that this functional redundancy likely comprises different modes of action by these effectors, with some acting upstream and others acting downstream of MAPK activation (Zheng et al., 2014). The demonstration that Pi02860 and Avr3a (Bos et al., 2010; Gilroy et al., 2011) each suppresses INF1-mediated cell death, but through activity on different targets, further emphasizes the functional redundancy comprising diverse effector activities.
NRL1 is a member of a family of proteins that include the functionally characterized NPH3 and RPT2, which interact with phototropins at the PM to mediate blue light signaling. The BTB/POZ domain in NPH3 promotes an association with CUL3, forming a substrate adaptor in a CRL3NPH3 complex that targets the phototropin phot1 for ubiquitination. High-blue-light conditions result in either monoubiquitination/multiubiquitination or polyubiquitination, the latter of which targets phot1 for degradation by the 26S proteasome, presumably to attenuate signaling under light-sufficient conditions (for review, see Liscum et al., 2014). Under low-blue-light conditions, in contrast, only monoubiquitination/multiubiquitination of phot1 occurs, which is necessary to establish phototropic responses (Roberts et al., 2011). One of the consequences of phot1 activation by the combination of its phosphorylation and monoubiquitination/multiubiquitination is its dissociation from the PM to stimulate the relocalization of PIN proteins from endosomes to the PM, where they facilitate auxin efflux (Liscum et al., 2014). Polyubiquitination, targeting phot1 for proteasome-mediated degradation, would fail to relocalize PIN proteins and thus not stimulate auxin efflux.
Auxin is antagonistic to the defense hormone salicylic acid, and increasing cellular levels of auxin is a strategy employed by numerous pathogens to suppress immunity (Naseem and Dandekar, 2012). It is conceivable that Pi02860 could promote NRL1 activity, thus influencing phot1 levels, and therefore PIN relocalization, retaining intracellular auxin levels to antagonize immunity. However, at this stage, the function of the Arabidopsis ortholog of NRL1 is unknown, and it may not function similarly to NPH3, instead facilitating the ubiquitination and turnover of other proteins associated directly with immunity. To investigate this possibility, further work is needed to identify protein partners of StNRL1, including whether it forms a ubiquitin E3 ligase complex with CUL3.
A number of ubiquitin E3 ligases negatively regulate PTI, and these have been the subject of extensive functional studies. The E3 ligases PUB12 and PUB13 both work to attenuate PTI by ubiquitinating the flg22 receptor FLS2, facilitating its degradation (Lu et al., 2011). Recent Y2H screens have revealed potential coregulatory partners and substrates for ubiquitination by PUB13, including phosphatidylinositol-4 kinase and RABA4B, with which it complexes to negatively regulate salicylic acid-mediated defenses (Antignani et al., 2015), and the abscisic acid regulator ABI1, a PP2C family member, which is a PUB13 substrate for ubiquitination and degradation (Kong et al., 2015). In addition, PUB22, PUB23, and PUB24 also suppress immunity. PUB22 attenuates PTI by targeting the exocyst component exo70B2 for ubiquitination and degradation (Stegmann et al., 2012). A further example of E3 ligases that negatively regulate immunity are the BTB domain proteins NPR3 and NPR4, which form complexes with CUL3 to facilitate the ubiquitination and degradation of the major salicylic acid regulator NPR1 in the nucleus (Fu et al., 2012). If NRL1 forms a complex with CUL3, it may represent a further CUL3-based E3 ligase involved in the negative regulation of immunity, albeit one that is predicted to function outside of the nucleus. Identification of its substrates for ubiquitination will reveal the mechanism underlying its defense suppression.
Whereas transient silencing of NbNRL1, using VIGS, accelerated INF1 cell death and attenuated P. infestans leaf colonization, transient overexpression of StNRL1 resulted in the opposite phenotypes, indicating that, in the absence of the effector Pi02860, NRL1 is a negative regulator of immunity and, thus, can be regarded as a susceptibility factor. The term susceptibility factor has been coined to describe proteins with a wide range of activities, from cell wall alterations, to proteins that directly suppress or antagonize immunity, to those that provide metabolic changes of benefit to pathogen growth (van Schie and Takken, 2014). Many have been defined as such due to reduced pathogen colonization when they are disabled and/or increased disease development when they are overexpressed. Few such proteins have been demonstrated to be targeted by pathogen effectors. Examples include the SWEET genes that are induced by Xanthomonas spp. TAL effectors, contributing to sugar efflux to provide pathogen nutrition (Chen et al., 2010); the Pseudomonas spp. effector AvrB, which mediates the phosphorylation and activation of MPK4, a negative regulator of PTI (Cui et al., 2010); and, more recently, the P. infestans effector Pi04089, which targets and stabilizes a K-homology class RNA-binding protein, StKRBP1, the overexpression of which enhances susceptibility (Wang et al., 2015). Here, we show that the target of Pi02860, NRL1, is a susceptibility factor that directly or indirectly suppresses PTI, in the form of INF1-mediated cell death. Future work will focus on how Pi02860 supports or promotes NRL1 activity, in identifying the substrates and partner proteins of NRL1 and how it acts to enhance late blight susceptibility. Understanding how P. infestans can use endogenous host regulatory proteins and processes that may naturally undermine immunity will reveal novel means to control this pathogen. Further studies on NRL1, as a negative regulator of immunity, will indicate the mechanisms by which plants govern cross talk between biotic stress responses and other cellular processes in an attempt to balance and allocate resources.
MATERIALS AND METHODS
Vector Construction
The Phytophthora infestans putative RXLR effector gene Pi02860 was synthesized by Genscript with attL sites to generate an entry vector. To make overexpression vector PRI101-Pi02860, the effector was amplified from P. infestans cDNA with primers containing BamHI and NdeI restriction sites and ligated into PRI101 using standard molecular biology techniques. The potato (Solanum tuberosum) NPH3/RPT2-like protein StNRL1 coding sequence was amplified from potato cDNA with flanking attB sites, and PCR products were recombined into pDONR201 (Invitrogen) to generate entry clones using the Gateway (Invitrogen) primer sequences shown in Supplemental Table S1.
The effector entry clones were recombined with pDEST32 (for Y2H; Invitrogen) and pB7WGF2 (for N-terminal EGFP fusion; Karimi et al., 2002). Modified forms of pB7WGF2 were created with either an NES signal derived from PKI (amino acid sequence LALKLAGLDIN; Wen et al., 1995) or an NLS signal derived from SV40 T antigen (amino acid sequence PKKKRKV; Kalderon et al., 1984) added to the N terminus of the GFP. The effector entry clones also were recombined with pCL112 (for N-terminal YN fusion) or pCL113 (for N-terminal YC fusion) for bimolecular fluorescence complementation (Bos et al., 2010) and pGWB18 (for N-terminal tagging with the cMyc epitope; Nakagawa et al., 2007).
Potato Transformation
Agrobacterium tumefaciens-containing overexpression vector PRI101-Pi02860 was used to transform microtuber discs of the potato cv E3 (Si et al., 2003; Tian et al., 2015). Positive lines were first screened on differential medium (3% [w/v] Murashige and Skoog medium + 0.2 mg L−1 indole-3-acetic acid + 0.2 mg L−1 GA3 + 0.5 mg L−1 6-BA + 2 mg L−1 ZT + 75 mg L−1 kanamycin + 200 mg L−1 Cef, pH 5.9) and then transferred to root generation medium (3% [w/v] Murashige and Skoog medium + 50 mg L−1 kanamycin + 400 mg L−1 Cef, pH 5.9). The presence and expression level of the transgene was confirmed by semiquantitative PCR (primers are shown in Supplemental Table S1).
Plant Production and Maintenance
Nicotiana benthamiana and potato overexpression lines were grown in glasshouses in 16-h days at 22°C. Supplementary light was provided when the ambient light dropped below 200 W m−2 and shading when it was above 450 W m−2. Approximately 5-week-old N. benthamiana and 7-week-old potato plants were used.
Agroinfiltration and Infection Assays
A. tumefaciens strain AGL1 transformed with vector constructs was grown overnight in YEB medium containing selective antibiotics at 28°C, pelleted, resuspended in infiltration buffer (10 mm MES, 10 mm MgCl2, and 200 µm acetosyringone), and adjusted to the required optical density at 600 nm (OD600) before infiltration into N. benthamiana leaves (generally 0.005–0.01 for imaging purposes, 0.002 for bimolecular fluorescence complementation, 0.1 for infection assays, and 0.5 for HR assays). For coexpression, agrobacterial cultures carrying the appropriate vector constructs were mixed prior to infiltration.
P. infestans strain 88069 was used for plant infection and was cultured on rye agar at 19°C for 2 weeks. Plates were flooded with 5 mL of water and scraped with a glass rod to release sporangia. The resulting solution was collected in a falcon tube, sporangia numbers were counted using a hemocytometer and adjusted to 30,000 per mL, and 10-µL droplets were inoculated onto the abaxial side of leaves of intact N. benthamiana plants stored on moist tissue in sealed boxes. For VIGS plants, the average lesion diameter was measured and compared with that of the GFP control plants. A. tumefaciens transient assays in combination with P. infestans infection were carried out as described (McLellan et al., 2013).
Confocal Imaging
N. benthamiana cells were imaged at 2 dpi using Leica TCS SP2 AOBS, Zeiss 710, and Nikon A1R confocal microscopes with Leica HCX PL APO lbd.BL 63×/1.20 W and L 40×/0.8, Zeiss PL APO 40×/1.0, and Nikon 60×/ water-dipping objectives. GFP was excited by the 488-nm line of an argon laser, and emissions were detected between 500 and 530 nm. The pinhole was set to one airy unit for the longest wavelength fluorophore. Single optical section images and z-stacks were collected from leaf cells expressing low levels of the protein fusions to minimize the potential artifacts of ectopic protein expression. Images were projected and processed using the Leica LCS, Zen 2010, and NIS-Elements software packages. Subsequent image processing for figure generation was conducted with Adobe Photoshop CS2 and Adobe Illustrator.
Y2H and Coimmunoprecipitation
A Y2H screen with pDEST32-Pi02860 was performed in Saccharomyces cerevisiae strain MsV203 as described (McLellan et al., 2013) using the Invitrogen ProQuest system. The full-length coding sequence of the candidate interacting prey sequence, StNRL1 (accession no. Sotub02g031050.1.1), was cloned and retested with pDEST32-Pi02860 and pDEST32-PiAvr2 as a control to rule out the possibility that the observed reporter gene activation had resulted from interactions between the prey and the DNA-binding domain of the bait construct or the DNA-binding activity of the prey itself.
A. tumefaciens strain GV3101 containing the fusion protein constructs was grown overnight in YEB medium containing selective antibiotics at 28°C, pelleted, resuspended in infiltration buffer (10 mm MES, 10 mm MgCl2, and 200 µm acetosyringone), and adjusted to an OD600 of 1 before infiltration into N. benthamiana leaves. Forty-eight hours post infiltration, samples were taken and proteins were extracted. GFP-tagged Pi02860/PiAvr2 fusions were immunoprecipitated using GFP-Trap-M magnetic beads (Chromotek). The resulting samples were separated by PAGE and western blotted. Immunoprecipitated GFP fusions and coimmunoprecipitated c-Myc fusions were detected using appropriate antisera (Santa Cruz Biotechnology).
VIGS
VIGS constructs were made by cloning a 250-bp PCR fragment shared by NbNRL1a (accession no. NbS00004529g0005.1) and NbNRL1b (accession no. NbS00009404g0009.1) from N. benthamiana cDNA and cloning into pBinary tobacco rattle virus (TRV) vectors (Liu et al., 2002) between HpaI and EcoRI sites in the antisense orientation. BLAST analysis of this sequence against the P. infestans genome (http://www.broadinstitute.org/annotation/genome/phytophthora_infestans/ToolsIndex.html) did not reveal any matches that could initiate silencing in the pathogen. A TRV construct expressing GFP described previously was used as a control (McLellan et al., 2013). Primer sequences are shown in Supplemental Table S1. The two largest leaves of four-leaf-stage N. benthamiana plants were pressure infiltrated with LBA4404 A. tumefaciens strains containing a mixture of RNA1 and each NRL VIGS construct or the GFP control at OD600 = 0.5 each. Plants were used for assays or to check gene-silencing levels by quantitative reverse transcription (qRT)-PCR 2 to 3 weeks later.
Gene Expression Assay
RNA was extracted using a Qiagen RNeasy Kit with on-column DNA digestion steps according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 μg of RNA using SuperScript II RNase HReverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time qRT-PCR was performed using Power SYBR Green (Applied Biosystems) and run on a Chromo4 thermal cycler (MJ Research) using Opticon Monitor 3 software. Primer pairs were designed outside the region of cDNA targeted for silencing following the manufacturer’s guidelines. Primer sequences are given in Supplemental Table S1. Detection of real-time qRT-PCR products, calculations, and statistical analysis were performed as described previously (McLellan et al., 2013).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PITG_02860: XM_002907697.1, NRL1: XM_006339754.2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transgenic potato lines overexpress Pi02860.
Supplemental Figure S2. Immunoblots showing the stability of Pi02860 GFP fusions for relocalization experiments.
Supplemental Figure S3. Alignment of Arabidopsis, N. benthamiana, and potato NRL1 sequences.
Supplemental Figure S4. Pi02860 interaction with NRL1 largely occurs at the plant PM.
Supplemental Figure S5. Western blots showing the stability of different split YFP and GFP constructs used.
Supplemental Figure S6. Silencing levels and plant phenotypes for VIGS of NRL1 in N. benthamiana.
Supplemental Figure S7. The stability of StNRL1 is not altered by Pi02860.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank members of the Dundee Effector Consortium for helpful discussions during this work.
Glossary
- MAMP
microbe-associated molecular pattern
- PAMP
pathogen-associated molecular pattern
- PTI
pattern-triggered immunity
- VIGS
virus-induced gene silencing
- dpi
days post infection
- HR
hypersensitive response
- NES
nuclear export signal
- NLS
nuclear localization signal
- Y2H
yeast two-hybrid
- cDNA
complementary DNA
- PM
plasma membrane
- OD600
optical density at 600 nm
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the Scottish Government Rural & Environment Science & Analytical Services Division, the Biotechnology and Biological Sciences Research Council (grant nos. BB/G015244/1, BB/K018183/1, and BB/L026880/1), the China Scholarship Council (to L.Y.), and the Doctoral Fund of the Ministry of Education of China (grant no. 20110146110019 to Q.H.).
Articles can be viewed without a subscription.
References
- Antignani V, Klocko AL, Bak G, Chandrasekaran SD, Dunivin T, Nielsen E (2015) Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 27: 243–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrova AO, Boevink PC, Young V, Grenville-Briggs LJ, van West P, Birch PR, Whisson SC (2008) A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol 10: 2271–2284 [DOI] [PubMed] [Google Scholar]
- Birch PRJ, Rehmany AP, Pritchard L, Kamoun S, Beynon JL (2006) Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol 14: 8–11 [DOI] [PubMed] [Google Scholar]
- Block A, Alfano JR (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, McLellan H, Bukharova T, Engelhardt S, Birch PRJ (2014) In vivo protein–protein interaction studies with BiFC: conditions, cautions, and caveats. Methods Mol Biol 1127: 81–90 [DOI] [PubMed] [Google Scholar]
- Boevink PC, Wang X, McLellan H, He Q, Naqvi S, Armstrong MR, Zhang W, Hein I, Gilroy EM, Tian Z, et al. (2016) A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat Commun 7: 10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530 [DOI] [PubMed] [Google Scholar]
- Bos JIB, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, Oliva R, Cano LM, Jones AM, Huitema E, van der Hoorn RA, et al. (2011) Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc Natl Acad Sci USA 108: 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke DE, Cano LM, Raffaele S, Bain RA, Cooke LR, Etherington GJ, Deahl KL, Farrer RA, Gilroy EM, Goss EM, et al. (2012) Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog 8: e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM (2010) Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17: 644–655 [DOI] [PubMed] [Google Scholar]
- Dou D, Zhou JM (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Du J, Verzaux E, Chaparro-Garcia A, Bijsterbosch G, Keizer LP, Zhou J, Liebrand TWH, Xie C, Govers F, Robatzek S, et al. (2015) Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nature Plants 1: 4. [DOI] [PubMed] [Google Scholar]
- Duplan V, Rivas S (2014) E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy EM, Taylor RM, Hein I, Boevink P, Sadanandom A, Birch PRJ (2011) CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol 190: 653–666 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398 [DOI] [PubMed] [Google Scholar]
- He Q, McLellan H, Boevink PC, Sadanandom A, Xie C, Birch PRJ, Tian Z (2015) U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J Exp Bot 66: 3189–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE (1984) A short amino acid sequence able to specify nuclear location. Cell 39: 499–509 [DOI] [PubMed] [Google Scholar]
- Kamoun S, Furzer O, Jones JD, Judelson HS, Ali GS, Dalio RJ, Roy SG, Schena L, Zambounis A, Panabières F, et al. (2015) The top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol 16: 413–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- King SRF, McLellan H, Boevink PC, Armstrong MA, Hall B, Sukarta O, Bukharova T, Kamoun S, Birch PRJ, Banfield M (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signalling. Plant Cell 26: 1345–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, et al. (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Askinosie SK, Leuchtman DL, Morrow J, Willenburg KT, Coats DR (2014) Phototropism: growing towards an understanding of plant movement. Plant Cell 26: 38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, Whisson SC, Beynon JL, Birch PRJ (2013) An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Naseem M, Dandekar T (2012) The role of auxin-cytokinin antagonism in plant-pathogen interactions. PLoS Pathog 8: e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, Zheng N, Hannink M, Genschik P, Liscum E (2011) Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3NPH3. Plant Cell 23: 3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D, Breen S, Schornack S, Win J, Hein I, Bozkurt T, Champouret N, Vleeshouwers V, Birch PRJ, Gilroy EM, et al. (2012) Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24: 3420–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si HJ, Xie CH, Liu J (2003) An efficient protocol for Agrobacterium mediated transformation with microtuber and the introduction of an antisense class I patatin gene into potato. Acta Agron Sin 29: 801–805 [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Žársky V, McDowell JM, Shirasu K, Trujillo M (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, He Q, Wang H, Liu Y, Zhang Y, Shao F, Xie C (2015) The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol 56: 992–1005 [DOI] [PubMed] [Google Scholar]
- van Schie CCN, Takken FLW (2014) Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol 52: 551–581 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Driesprong JD, Kamphuis LG, Torto-Alalibo T, Van’t Slot KAE, Govers F, Visser RG, Jacobsen E, Kamoun S (2006) Agroinfection-based high-throughput screening reveals specific recognition of INF elicitins in Solanum. Mol Plant Pathol 7: 499–510 [DOI] [PubMed] [Google Scholar]
- Wang X, Boevink P, McLellan H, Armstrong M, Bukharova T, Qin Z, Birch PRJ (2015) A host KH RNA-binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol Plant 8: 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JDG, Sadanandom A (2006) The E3 ubiquitin ligase activity of Arabidopsis plant U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, McLellan H, Fraiture M, Xiaoyu L, Boevink P, Gilroy E, Ying C, Kandel K, Sessa G, Birch PRJ, et al. (2014) Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog 10: e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.