Genetic, developmental, and gene expression analyses of GhVIN1-RNAi cotton plants reveal critical roles of vacuolar invertase in floral organ development and male and female fertilities.
Abstract
Seed number and quality are key traits determining plant fitness and crop yield and rely on combined competence in male and female fertilities. Sucrose metabolism is central to reproductive success. It remains elusive, though, how individual sucrose metabolic enzymes may regulate the complex reproductive processes. Here, by silencing vacuolar invertase (VIN) genes in cotton (Gossypium hirsutum) reproductive organs, we revealed diverse roles that VIN plays in multiple reproductive processes. A set of phenotypic and genetic studies showed significant reductions of viable seeds in GhVIN1-RNAi plants, attributed to pollination failure and impaired male and female fertilities. The former was largely owing to the spatial mismatch between style and stamen and delayed pollen release from the anthers, whereas male defects came from poor pollen viability. The transgenic stamen exhibited altered expression of the genes responsible for starch metabolism and auxin and jasmonic acid signaling. Further analyses identified the reduction of GhVIN expression in the seed coat as the major cause for the reduced female fertility, which appeared to disrupt the expression of some key genes involved in trehalose and auxin metabolism and signaling, leading to programmed cell death or growth repression in the filial tissues. Together, the data provide an unprecedented example of how VIN is required to synchronize style and stamen development and the formation of male and female fertilities for seed development in a crop species, cotton.
In flowering plants, sexual reproduction involves (1) gametophytic development, producing sperm in the pollen within the anthers and eggs in the ovules embedded within the ovaries; (2) the accomplishment of specific interactions between mature pollen and the receptive stigma, followed by pollen tube elongation down to the ovules; (3) gamete fusion, known as double fertilization, resulting in a diploid embryo nucleus and a triploid endosperm nucleus; and (4) the coordinated development among seed coat, embryo, and endosperm to generate a viable seed. Accompanied by the distinctive cellular and developmental changes during these processes, complex molecular and biochemical pathways have evolved to regulate each step to ensure the success of seed production.
Sugars are important as energy source, building blocks, osmotic solutes, and signaling molecules (Ruan, 2014). As the principal product of photosynthesis, Suc is the primary carbon translocated from source leaves to nonphotosynthetic sinks, including reproductive organs. Prior to its use for metabolism and biosynthesis, Suc needs to be degraded into hexoses by either Suc synthase (Sus; EC 2.4.1.13) or invertase (INV; EC 3.2.1.26). Sus cleaves Suc in the presence of UDP into UDP-Glc and Fru and is largely involved in cell wall and starch biosynthesis in sink organs (Brill et al., 2011) and maintaining sink strength (Pozueta-Romero et al., 1999; Xu et al., 2012), especially in crop species (Ruan, 2014). INV, on the other hand, hydrolyzes Suc into Fru and Glc and plays essential roles in plant development and stress responses (Koch, 2004; Ruan, 2014). Based on their subcellular location, INV can be classified as cell wall invertase (CWIN), cytoplasmic invertase (CIN), and vacuolar invertase (VIN).
The involvement of INVs in pollen development has been observed in a wide range of species (Maddison et al., 1999; Goetz et al., 2001; Proels et al., 2006; Castro and Clément, 2007; Engelke et al., 2010; Pressman et al., 2012), especially under stress conditions such as drought (Koonjul et al., 2005) and cold (Oliver et al., 2007). Positive correlations between INV activities and seed and fruit development also have been reported in maize (Zea mays; Cheng et al., 1996; Boyer and McLaughlin, 2007), rice (Oryza sativa; Hirose et al., 2002; Wang et al., 2008), barley (Hordeum vulgare; Weschke et al., 2003), broad bean (Vicia faba; Weber et al., 1996); grape (Vitis vinifera; Davies and Robinson, 1996), and tomato (Solanum lycopersicum; Jin et al., 2009; Zanor et al., 2009).
Apart from their major roles in primary metabolism, INVs also are intimately involved in sugar signaling. For example, VIN-derived hexose signaling likely plays an indispensable role in cotton (Gossypium hirsutum) fiber (seed trichome) initiation by regulating the expression of some MYB transcription factors and auxin signaling genes (Wang et al., 2014). Furthermore, interactions between CWINs and hormone signaling pathways have been implicated in the development of wheat pollen (abscisic acid; Ji et al., 2011), maize seed (cytokinins [Rijavec et al., 2009] and auxin [LeClere et al., 2010]), rice grain (auxin; French et al., 2014), tomato fruit (ethylene; Zanor et al., 2009), and Arabidopsis (Arabidopsis thaliana) seed extrafloral nectar secretion (jasmonic acid [JA]; Millán-Cañongo et al., 2014).
Despite the progress outlined above, there is a lack of understanding of whether INVs modulate both male and female fertilities and, if so, how the regulation may be achieved at the developmental, cellular, and molecular levels. Filling this major knowledge gap is essential for better understanding the regulation of plant reproductive development and for designing better approaches to improve crop reproductive success for seed and fruit production under climate change (Ruan, 2014). Here, we provide a comprehensive analysis of the roles of VIN in reproductive development using VIN-suppressed cotton as a model. The data obtained revealed that VIN is required for both male and female fertilities and that its reduced expression in seed coat leads to programmed cell death (PCD) or growth repression in the filial tissues.
RESULTS
Silencing GhVIN1 Resulted in a High Proportion of Unviable Seeds
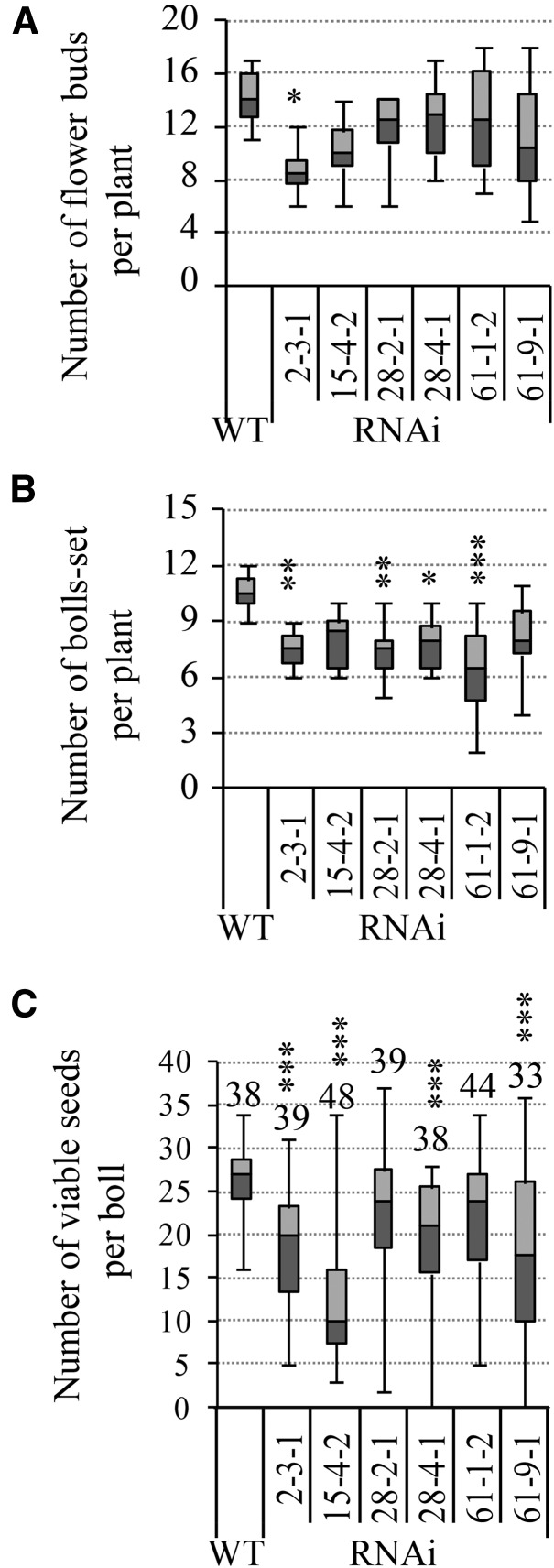
In our previous study (Wang et al., 2014), an RNA interference (RNAi) construct against the major cotton vacuolar invertase gene GhVIN1 was introduced into cotton under the control of the RD22-LIKE1 (RDL1) promoter, which is active mainly in cotton fiber and seed early in development (Wang et al., 2004). The suppression of cotton GhVIN1 resulted in a significant reduction of VIN activity in cotton seeds and, consequently, a fiberless seed phenotype (Wang et al., 2014). Apart from the blockage of fiber initiation from seed epidermis, we also observed a significant reduction of seeds in the GhVIN1-RNAi lines as compared with those of wild-type plants (Fig. 1; Supplemental Fig. S1). Detailed analyses at the T3 generation revealed that, while the number of flower buds was reduced only in one (line 2-3-1) out of six lines examined (Fig. 1A), the number of bolls that set, and viable seeds per boll, were reduced in all the transgenic lines, to 60% to 75% and 21% to 56%, respectively, of those in wild-type plants (Fig. 1, B and C). The ovule number per boll, however, was not affected significantly in the transgenic plants (Supplemental Fig. S2), excluding the possibility of reduced ovule development as a potential cause for the reduced seed production. The transgenic lines examined were homozygous for the transgenes based on PCR detection of the presence of the RDL promoter-GhVIN1 fragment in all of the tested T3 and T4 progeny for each line.
Figure 1.
GhVIN-RNAi plants exhibited reduced boll and viable seed number compared with wild-type plants (WT). Data are presented in box plots where the horizontal line within the box represents the median, while the top and bottom of the box represent the values in 75% and 25% of the population, respectively. The extent of the vertical line indicates the maximum and minimum of the data. Data in A and B were collected from eight individual plants of each T3 line at full maturity from two independent trials. For the viable seed number in C, data were collected from cotton bolls harvested from at least six individual plants of each T3 transgenic line, with the total boll number indicated above each box plot. Asterisks indicate significant differences between the wild type and a given RNAi line (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
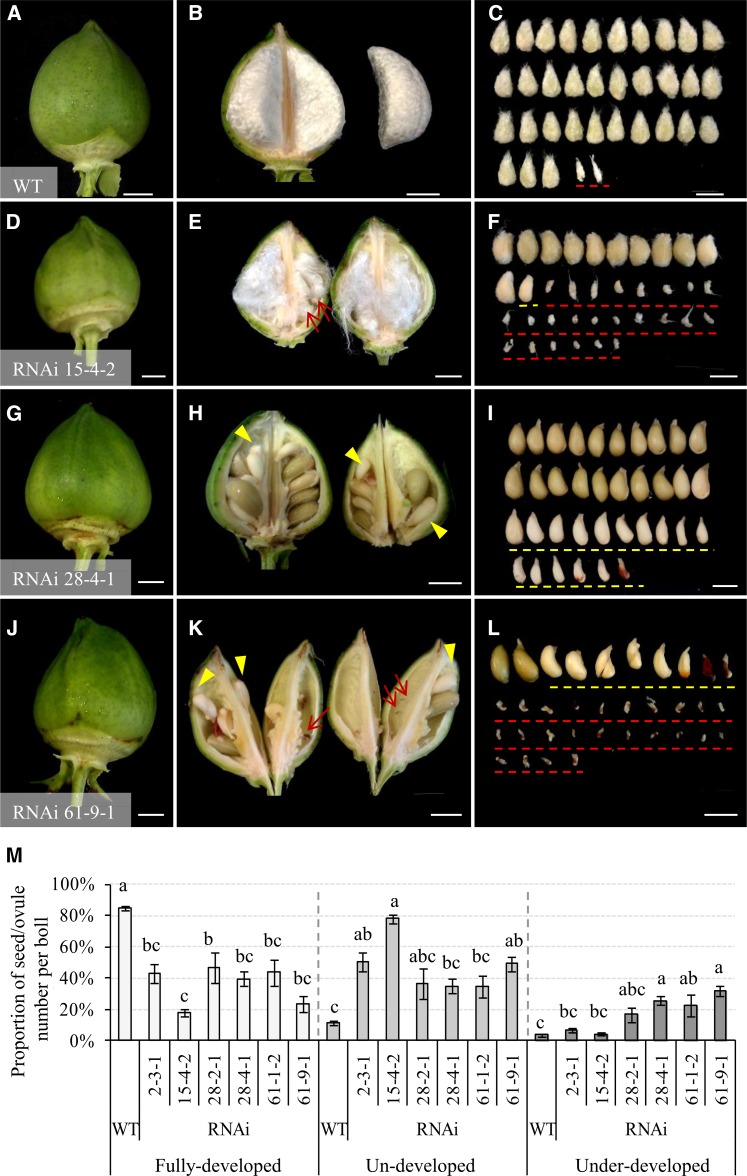
A close phenotypic analysis identified two types of unviable seeds from the transgenic lines, comprising (1) undeveloped seeds/ovules and (2) underdeveloped seeds (Fig. 2). The former became morphologically distinguishable as early as approximately 5 d after anthesis (DAA) and remained at the size of unfertilized ovules even to boll maturity (indicated by red arrows or red dashed lines in Fig. 2, E, F, K and L). The latter were expanded to some extent but were hollow or only partially filled inside the seed coat, and they could be easily sorted out from the normal seeds at about 30 DAA (yellow arrowheads or yellow dashed lines in Fig. 2, H, I, K, and L). Overall, compared with 11% of undeveloped and 3% of underdeveloped seeds observed in the wild type, an average of 50%, 78%, 36%, 35%, 34%, and 49% of total ovules failed to expand (being undeveloped) in lines 2-3-1, 15-4-2, 28-2-1, 28-4-2, 61-1-2, and 61-9-1, respectively, with approximately 7%, 4%, 17%, 26%, 23%, and 32% of the ovules becoming underdeveloped seeds in the corresponding lines (Fig. 2M). The presence of the higher proportion of these two types of unviable seeds led to a significant reduction of viable seeds across the transgenic lines examined (Fig. 2M).
Figure 2.
Unviable cotton seeds comprised undeveloped and underdeveloped seeds from the GhVIN1-RNAi lines. A to L, Representative images of 30-d cotton bolls and seeds from the wild type (WT; A–C) and lines 15-4-2 (D–F), 28-4-1 (G–I), and 61-9-1 (J–L). The undeveloped seeds are indicated by red arrows and dashed lines (E, F, K, and L), whereas the underdeveloped seeds are shown by yellow arrowheads and dashed lines (F, H, I, K, and L). Bars = 1 cm. M, Proportions of the fully developed, undeveloped, and underdeveloped seeds per boll in wild-type and T3 GhVIN1-RNAi plants. Each value is the mean ± se of at least 30 cotton bolls from eight individuals of each line in two independent trials. Different letters indicate significant differences at P < 0.05 according to one-way ANOVA.
To explore the cellular and molecular bases of the observed seed phenotypes (Figs. 1 and 2), we next selected three independent lines, 15-4-2, 28-4-1, and 61-9-1, for detailed analyses. Here, 15-4-2 had an extremely large proportion of undeveloped seeds, while the latter two lines represent those with a high ratio of underdeveloped seeds (Fig. 2). The flower bud number was not significantly affected in these three lines compared with the wild type (Fig. 1A).
GhVIN1-RNAi Plants Exhibited Mismatched Floral Structure, Delayed Anther Dehiscence, and Low Pollen Viability
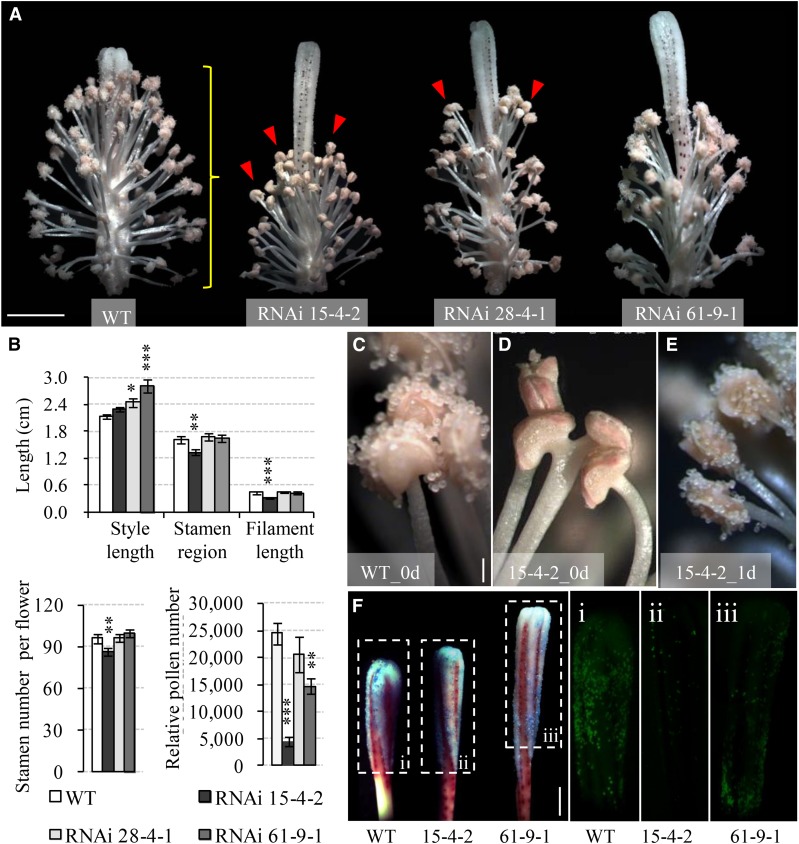
The presence of a large proportion of unviable seeds in the GhVIN1-RNAi plants was somehow unexpected, given that VIN has been typically considered to regulate cell enlargement (Wang et al., 2010). This finding prompted us to investigate the underlying developmental basis. One surprising observation was that the GhVIN1-RNAi plants had evident abnormalities in flower structure. A large proportion of the transgenic flowers displayed spatially mismatched stamen and stigma (Fig. 3A) or their malformation, or even a lack of pistil and stamen entirely (Supplemental Fig. S3). The stigma protrusion well above the stamen was owing to the increased style lengths, as in lines 28-4-1 and 61-9-1, or shortened filament length and anther coverage region, as in line 15-4-2 (Fig. 3B). Moreover, pollen number per flower was reduced to an average of 18%, 60%, and 84% of that in the wild type in lines 15-4-2, 61-9-1, and 28-4-1, respectively (Fig. 3B).
Figure 3.
Silencing GhVIN1 in cotton disrupted style and stamen development, delayed anther dehiscence, and reduced pollen number. A, Representative images of 0-d transgenic flowers with petals removed, showing the uncoordinated style protrusion and stamen development in the RNAi plants compared with the wild type (WT). The red arrowheads indicate indehiscent anthers. The yellow brace indicates the stamen region measured in B. B, GhVIN1-RNAi flowers displayed longer styles or shorter filaments, decreased stamen region, and lower stamen and pollen numbers per flower. Each value is the mean ± se, with data collected from eight flowers of four individual plants for each line. Asterisks denote significant differences (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants. C to E, Anther dehiscence occurred on the day of flowering in the wild type (C) but not in RNAi line 15-4-2 (D). The latter dehisced 1 d later (E). F, Fewer pollens were detected in transgenic styles compared with the wild type on the day of flowering. Styles were stained with Aniline Blue (left; bright field) and viewed under UV light to show the fluorescence emitted from the stained pollen grains in the boxed regions (i–iii). Bars = 1 cm in A, 200 μm in C, and 5 mm in F. The scales in D and E are the same as that in C.
We also observed a delayed dehiscence in a proportion of the transgenic flowers (indicated by arrowheads in Fig. 3A and highlighted in Fig. 3, D versus C and E). Consistently, Aniline Blue staining of 0-d styles revealed that far fewer pollen grains landed on the transgenic stigmas compared with wild-type stigmas (Fig. 3F).
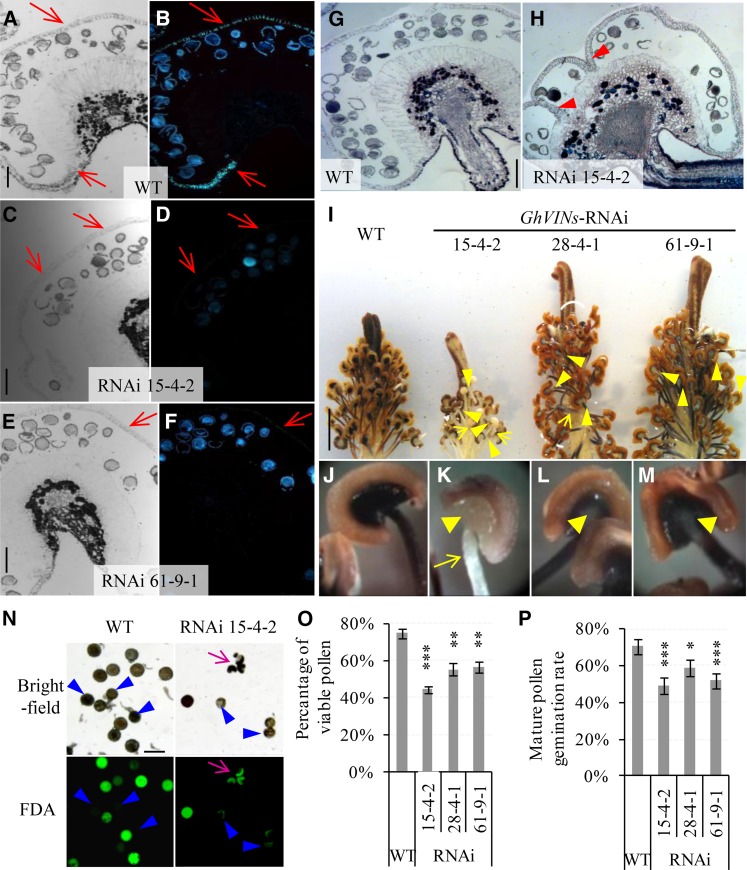
Anther opening requires cellular degeneration of septum and stomium, secondary cell wall thickening of endothecium, and water loss from anthers (Wilson et al., 2011). To this end, in contrast to the thickened wild-type anther endothecium walls that emitted callose fluorescence following Aniline Blue staining (Fig. 4, A and B), no or much reduced fluorescent signals were observed in the anther walls from lines 15-4-2 and 61-9-1 (Fig. 4, C–F), indicating compromised wall thickening in the endothecium of those anthers. Histological analyses also revealed that 50% and 31% of the −1-d anthers from lines 15-4-2 and 28-4-1 had undegenerated septum, whereas a majority of the wild-type septum had been degraded by −1 DAA (Fig. 4, H versus G). Together, the impaired septum degeneration and endothecium wall thickening are the likely cellular basis for the delayed anther dehiscence in the RNAi lines. Starch accumulation in filament and the anther-filament junction region serves as a carbon source for generating soluble sugars prior to anthesis to draw water from the anther wall osmotically, thereby contributing to anther dehiscence and opening (Keijzer, 1987; Bonner and Dickinson, 1990; Stadler et al., 1999). Staining with KI-I2 revealed that, in contrast to the strong signal of starch displayed in the wild-type stamen, the transgenic filaments, and especially their joint area with anthers, exhibited much reduced starch staining (Fig. 4, I–M). Noteworthy is that the least starch staining was observed in line 15-4-2 (Fig. 4K), which displayed the strongest phenotype of anther dehiscence delay (Fig. 3, A and D). The reduced starch accumulation in the anther-filament joint region may represent a metabolic basis for the delay of anther dehiscence in the RNAi stamen.
Figure 4.
GhVIN1-RNAi lines displayed unthickened endothecium walls, incomplete septum degradation in a certain number of −1-d anthers, reduced starch accumulation in −1-d stamens, and lowered pollen viability and pollen tube germination rates. A to F, Representative images of −1-d wild-type (WT; A and B), RNAi 15-4-2 (C and D), and RNAi 61-9-1 (E and F) anther sections stained with Aniline Blue, observed under bright field (A, C, and E) and UV light (B, D, and F). Red arrows indicate the position of the anther wall. G and H, Representative images of −1-d wild-type (G) and RNAi 15-4-2 (H) anther sections stained with Toluidine Blue. Note the remaining septum (arrowheads in H) but its absence in G. I, Representative image of wild-type and transgenic −1-d stamens and styles stained with KI-I2. J to M, Magnified views of stamen from the wild type (J), RNAi 15-4-2 (K), RNAi 28-4-1 (L), and RNAi 61-9-1 (M), respectively. Compared with the strong staining of starch in the wild-type stamen by KI-I2, indicated by the dark color, the transgenic stamen exhibited much weaker staining in filaments and especially at the anther-filament joint regions (yellow arrow and arrowheads, respectively). N, Bright-field and green fluorescence images of the same set of mature pollen grains stained with FDA from the wild type and RNAi line 15-4-2. Pink arrows indicate malformed pollen grains. Blue arrowheads point to pollen grains with lost viability. O, The proportions of viable pollen grains determined by FDA staining were reduced significantly in the transgenic lines compared with the wild type. Each value is the mean ± se from four biological replicates. P, Pollen germination rates were reduced significantly in the GhVIN1-RNAi lines compared with the wild type. Each value is the mean ± se of eight flowers from four plants for each line. Asterisks indicate significant difference between RNAi and wild-type plants based on one-way ANOVA after arcsine transformation (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). Bars = 100 μm in A, C, E, G, and N and 1 cm in I. The scales in B, D, F, and H are the same as those in A, C, E, and G, respectively.
Pollen viability staining with fluorescein diacetate (FDA) revealed significantly reduced viable pollen in the transgenic anthers (Fig. 4, N and O). Moreover, the transgenic lines also exhibited significantly reduced germination rate (Fig. 4P) and lower pollen tube elongation (Supplemental Fig. S4A). A certain number of pollen tubes, however, were able to reach the base of the ovaries in the RNAi plants (Supplemental Fig. S4, D and E).
Genetic Evidence That Both Male and Female Fertilities Were Impaired in the GhVIN1-RNAi Cotton Plants
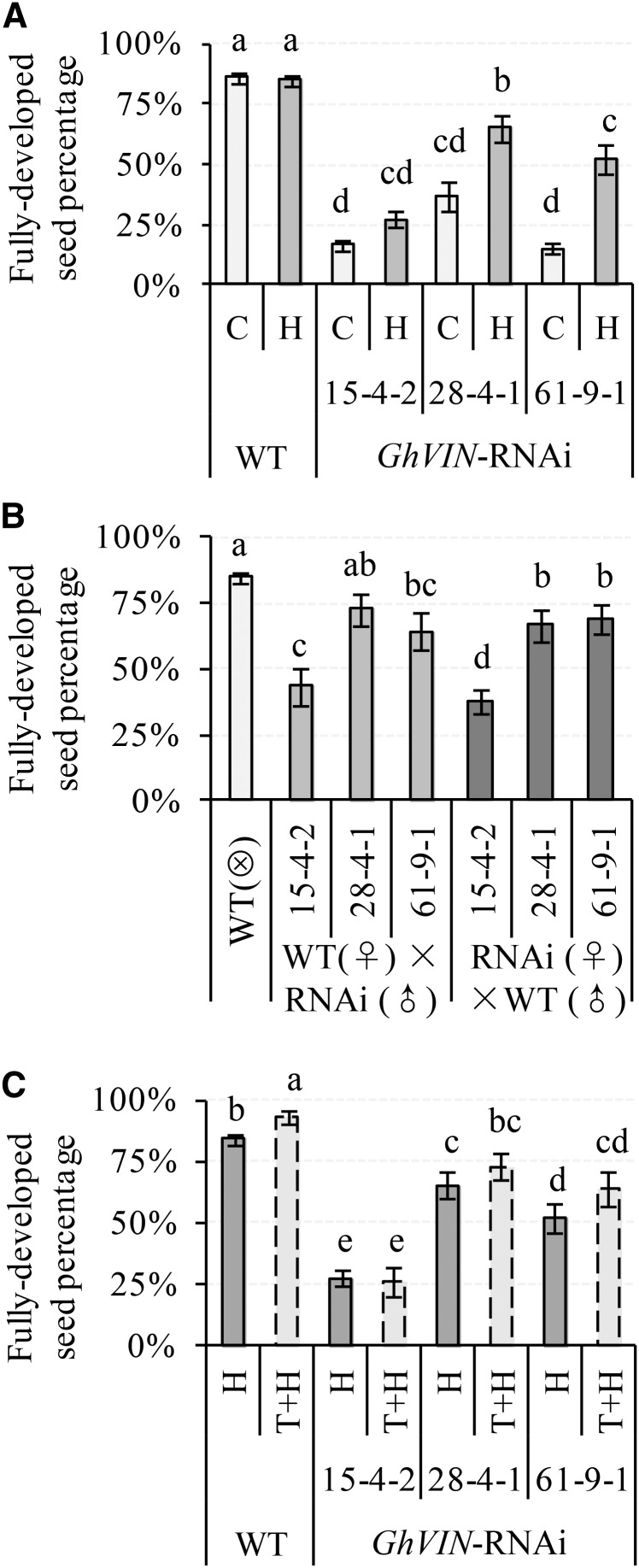
To test whether the reduced seed set in the transgenic plants was caused solely by insufficient pollen grains landed on the stigmas, we hand pollinated RNAi and wild-type cottons with their respective pollens. To analyze the proportions of the three types of seeds (normal and viable, undeveloped, and underdeveloped) between wild-type and RNAi lines, we created a generalized linear mixed model (GLMM) with a binomial error structure and logistic link function (for details, see “Materials and Methods”). GLMM has been widely used in genetics and evolution studies to analyze complex biological systems, without ignoring the random effects or violating the statistical assumptions of normal distribution and constant variances (Quinn and Keough, 2002; Jinks et al., 2006; Bolker et al., 2009).
Hand pollination did not affect seed set in the wild type, indicating sufficient pollen load under natural condition (Fig. 5A). However, it increased seed set in the transgenic lines to some extent compared with their respective controls (Fig. 5A). Accordingly, the proportion of undeveloped and underdeveloped seeds was reduced by hand pollination in some transgenic lines (Supplemental Fig. S5, A and B). It is important to note, however, that hand pollination only partially restores seed set (Fig. 5A), indicating that pollination deficiency is not the only factor accounting for low seed production in the GhVIN1-RNAi plants.
Figure 5.
Impact of hand pollination (A), reciprocal crossing (B), and bud thinning (C) on seed fertility in GhVIN1-RNAi lines. The percentages of fully developed seeds in total ovules were calculated for the nontreated control (C), hand-pollinated bolls (H), reciprocal cross hybrids, and bolls after bud thinning and hand pollination (T+H). Each value is the GLMM estimated mean ± se. Data were collected from at least 20 bolls from at least five individual plants for each line. Different letters indicate significant differences at P < 0.05 according to GLMM. WT, Wild type.
We then performed reciprocal crosses between wild-type and transgenic lines to dissect the relative paternal and maternal contributions to reduced seed production in the RNAi plants. As shown in Figure 5B, in comparison with approximately 88% of viable seed in the self-pollinated wild-type plants, pollination of wild-type stigma with pollens from RNAi lines reduced viable seed percentage by 20% to 50%. Conversely, reciprocal crosses of RNAi stigmas with wild-type pollens increased viable seed percentage to some extent, but not to the level in the wild type (Fig. 5B). The data show that both maternal and paternal defects contribute to seed infertility in GhVIN1-RNAi plants. Interestingly, compared with that of wild-type self-pollination, the proportion of underdeveloped seed remained unaffected by pollination of the wild-type flower with pollen from the RNAi lines (Supplemental Fig. S5D). Moreover, a comparable ratio of the underdeveloped seed was observed between the RNAi × wild-type hybrids and RNAi self pollination (Supplemental Fig. S5, D versus B). Hence, paternal impact seems irrelevant to this type of shrunken seeds. In other words, the underdeveloped seeds are derived mainly from maternal defects. By contrast, however, pollination of the wild-type stigma with the RNAi pollen increased the proportion of undeveloped seeds compared with that of wild-type self-pollination, while descendants from RNAi (♀) × wild-type (♂) crosses showed reduced percentages of undeveloped seeds compared with the respective self-pollinated RNAi lines (Supplemental Fig. S5, C versus A). Thus, paternal defects clearly contribute to the production of undeveloped seeds. Additionally, the proportion of undeveloped seed in RNAi 15-4-2 (♀) × wild-type (♂) hybrids is much higher than that of wild-type self-pollination but smaller than that of RNAi 15-4-2 self-pollination (Supplemental Fig. S5, C versus A), suggesting an involvement of maternal defect for the undeveloped seed.
Seed set is dependent on assimilate import and utilization in sinks (Ruan et al., 2012). To assess if the poor seed set may relate to reduced assimilate availability for individual bolls in the GhVIN1-RNAi plants, we performed thinning experiments to remove most of the flower buds to allow only four bolls to set per plant. The treatment slightly increased seed set in the wild type, but with no significant effect on the RNAi lines (Fig. 5C), indicating that the seed phenotype in the transgenic plants is not due to compromised assimilate supply.
RNAi-Mediated Suppression of GhVIN Expression Led to a Significant Decline in VIN Activity in Stamen
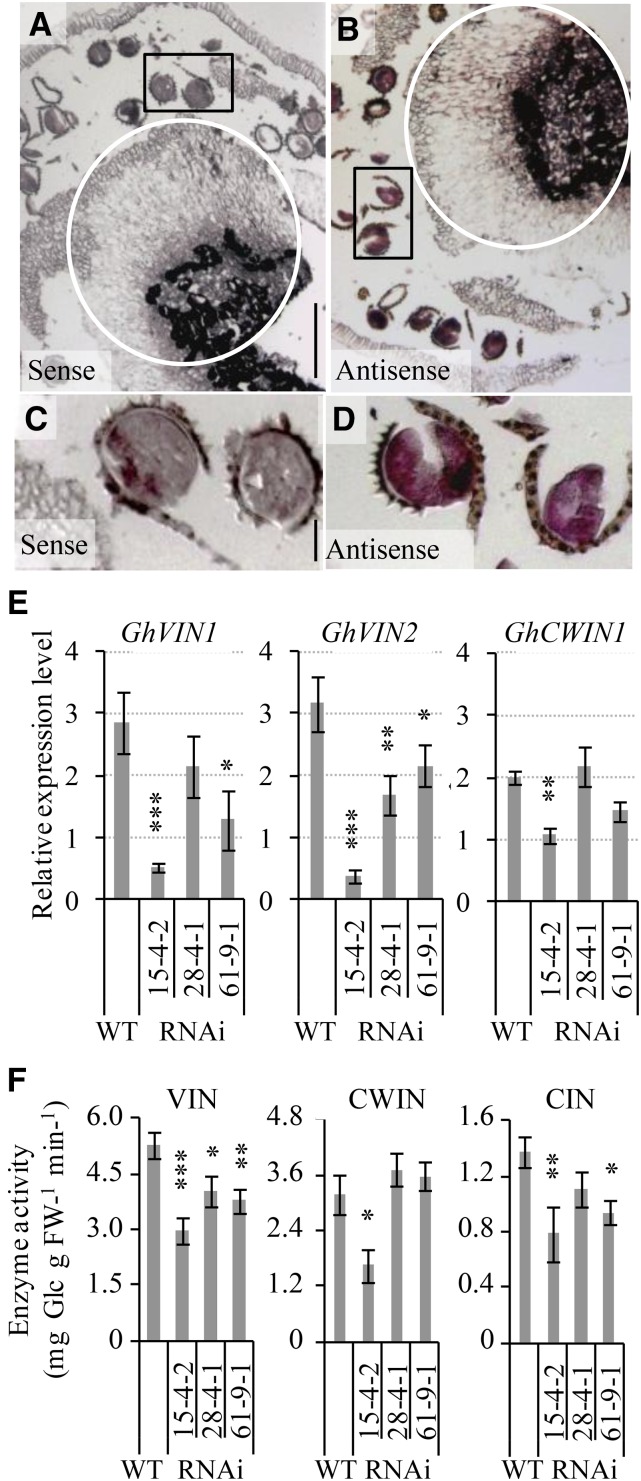
To gain insights into the roles of GhVINs in anther, we first examined its cellular expression patterns by performing in situ hybridization in wild-type anthers using an RNA antisense probe carrying 185 bp, matching the C-terminal ends of GhVIN1 and GhVIN2 mRNA sequences with 100% and 80% identity, respectively. Thus, the probe would hybridize both GhVIN1 and GhVIN2 transcripts. In comparison with the sense control (Fig. 6, A and C), GhVIN transcripts were detected abundantly in pollen grains and in the anther-filament joint area (Fig. 6, B and D). On the other hand, the RDL promoter used to drive the RNAi construct was indeed active in pollen grains and in the top part of filaments connecting anthers transformed with the RDL-GUS reporter gene (Supplemental Fig. S6, A and B). Thus, GhVIN mRNAs would be targeted by the RNAi construct for degradation in these regions. Consistently, quantitative real-time PCR (qPCR) analyses revealed that the GhVIN1 transcripts were reduced by 82%, 25%, and 55% in RNAi 15-4-2, 28-4-1, and 61-9-1 stamen (Fig. 6E). Meanwhile, the GhVIN2 mRNA levels also were reduced in the transgenic stamen (Fig. 6E), reflecting a cosilencing effect by the GhVIN1 RNAi construct. Consequently, VIN activity was decreased significantly in the stamen from all three transgenic lines compared with that in the wild type (Fig. 6F), with its degree of reduction corresponding to the level of suppression in the GhVIN1 mRNA (Fig. 6E). CWIN activity was largely unaffected in the RNAi stamen, except in transgenic line 15-4-2, which exhibited a reduction in activity and mRNA level (Fig. 6, E and F). The reduction of VIN activity was associated with a decrease in CIN activity from lines 15-4-2 and 61-9-1.
Figure 6.
GhVIN transcripts were abundant at the anther-filament joint region and in pollens of wild-type cotton stamen but were evidently reduced in GhVIN1-RNAi lines, resulting in decreased VIN activities. A to D, A longitudinal section of −1-d wild-type cotton anther hybridized with a sense (A) or an antisense (B) RNA probe for GhVINs. Note the GhVIN mRNA signals in the circled anther-filament joint area in B compared with the same region in the sense control in A. C and D are magnified views of pollens in the boxed areas of A and B, respectively, showing strong GhVIN mRNA signals in wild-type pollens. E, qPCR analyses of GhVIN1, GhVIN2, and GhCWIN1 transcripts in −1-d wild-type (WT) and transgenic stamen. Data represent means ± se (n ≥ 6). F, VIN, CWIN, and CIN activities in −1-d wild-type and RNAi stamen. Each value is the mean ± se (n = 4). Asterisks in E and F indicate significant differences (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants. FW, Fresh weight.
Collectively, the data indicate that the inhibition of GhVINs in anther and pollen (Fig. 6) impaired stamen and pollen development in the RNAi cotton plants (Figs. 3–5). These paternal defects likely resulted in pollination and fertilization failure, leading to undeveloped seeds (Figs. 1 and 2).
GhVIN1-RNAi Stamens Were Characterized by Reduced Expression of Genes for Starch Metabolism, Auxin Biosynthesis, and JA Responses and Altered Expression of Auxin Signaling Genes
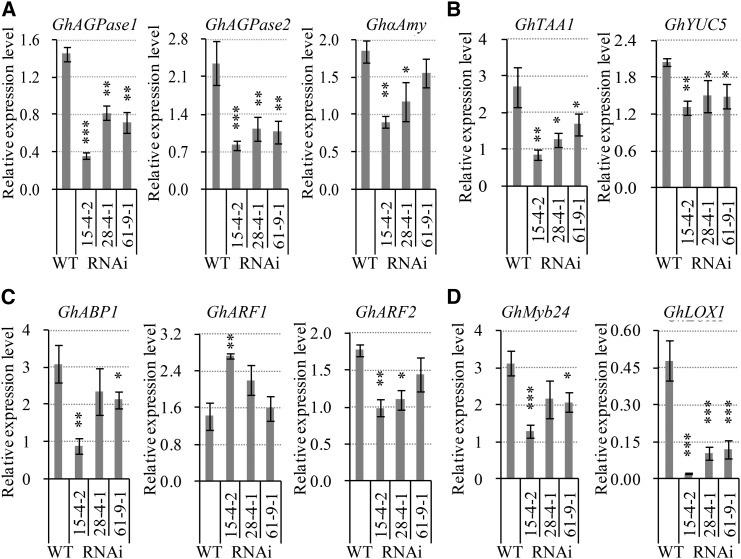
To explore the molecular basis of the VIN-mediated regulation of stamen development and anther dehiscence, we examined the potential effects of reduced VIN activity on the expression of genes responsible for carbohydrate allocation and hormonal function in −1-d stamen. The transcript levels of two ADP-Glc pyrophosphorylase genes (GhAGPase1 and GhAGPase2), encoding the rate-limiting enzyme AGPase for starch synthesis, were decreased significantly in the stamen of all three transgenic lines (Fig. 7A; Supplemental Fig. S7), consistent with the reduced starch content in the transgenic stamen (Fig. 4, I–M). Besides, down-regulated expression of an α-amylase gene, GhαAmy, also was observed in lines 15-4-2 and 28-4-1 compared with the wild type (Fig. 7A; Supplemental Fig. S7). By contrast, no detectable changes were found in mRNA levels of a cohort of anther-expressed candidate genes encoding Sus, H+/sugar transporters, and hexokinase, Glc, and Fru contents (Supplemental Fig. S7).
Figure 7.
Silencing GhVIN1 reduced the transcript levels in −1-d stamen of candidate genes for starch synthesis and degradation (A), auxin biosynthesis (B), and JA response (D) and altered the expression of some auxin signaling genes (C). Data represent means ± se (n ≥ 6), generated from the same biological replicates as in Figure 6E. Asterisks indicate significant differences (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants (WT).
Apart from starch metabolism, auxin also plays important roles in filament elongation, anther dehiscence, and pollen maturation (Feng et al., 2006; Cecchetti et al., 2008; Sundberg and Østergaard, 2009). This, together with recent progress on the roles of sugars in auxin biosynthesis and signaling (Wang and Ruan, 2013), prompted us to investigate the expression of auxin-related genes in −1-d stamen. Several stamen-expressed candidate genes involved in auxin biosynthesis, transport, and perception were chosen to measure their mRNA levels, based on previous studies (Min et al., 2014). Most notably, the transcripts of two auxin biosynthesis genes, GhTAA1 and GhYUC5, were reduced significantly in the RNAi stamen (Fig. 7B). The auxin signaling gene, GhABP1, was reduced in its transcript level in two lines, whereas GhARF1 showed increased expression in line 15-4-2, and its paralog GhARF2 exhibited a decreased mRNA level in this line as well as in line 28-4-1 (Fig. 7C). No difference was observed in the transcript levels between the transgenic and wild-type stamen for the other two auxin biosynthesis genes, GhTAR2 and GhYUC11, the auxin influx carrier GhAUX1, and two efflux transporter genes, GhPIN2 and GhPIN3 (Supplemental Fig. S8). In addition to auxin, JA is another hormone known to be a critical regulator in filament extension, anther dehiscence, and pollen viability (Ishiguro et al., 2001; Scott et al., 2004), and hexose signaling is involved in JA biosynthesis and signaling (Hamann et al., 2009). In cotton, a pollen-specific R2-R3 MYB gene, GhMYB24, and a 9-lipoxygenase gene, GhLOX1, have been identified to regulate late anther and pollen development in response to JA signaling (Marmey et al., 2007; Li et al., 2013). The GhMYB24 transcript level was reduced by 59%, 31%, and 34% of that in the wild type in the −1-d stamens of the RNAi lines 15-4-2, 28-4-1, and 61-9-1, respectively, with the GhLOX1 mRNA level reduced by approximately 90% in line 15-4-1 and by approximately 80% in the remaining two lines (Fig. 7D). These data show that suppression of GhVINs blocked the expression of these JA signaling genes in the stamen.
Female Sterility in GhVIN1-RNAi Plants Originated from Seed Rather Than Style or Ovule
In addition to the low paternal fertility, maternal defect also was found to contribute to poor seed set (Fig. 5; Supplemental Fig. S5). In broad terms, the maternal defect could derive from ovule or style in the pistil or seed coat and nucellus in the seed. To this end, the RDL1 promoter used to drive transgene expression was not active in cotton ovules (Supplemental Fig. S6, A and C; Guan et al., 2011), and the GhVIN transcripts and VIN activity also were undetectable in wild-type ovules (Wang et al., 2010, 2014). Similar to the ovules, the RNAi styles exhibited no or little RDL1-GUS signals (Supplemental Fig. S6A). Moreover, there was only a trace level of GhVIN1 mRNA detected in −1-d style tissues, with no difference between RNAi and wild-type plants (Supplemental Fig. S9). Together, both styles and ovules can be excluded as the source of maternal defects. In other words, the problem comes from the seeds.
Suppression of GhVINs in Seed Maternal Tissue Resulted in PCD or Growth Arrest in the Filial Tissue
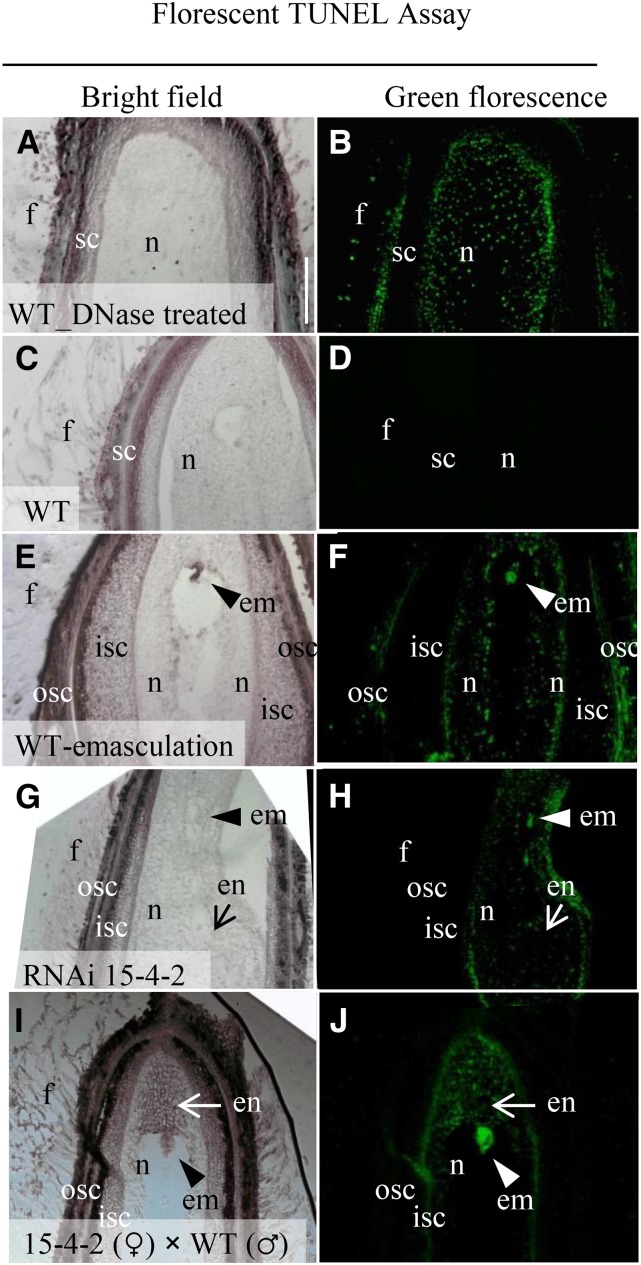
The undeveloped seeds/ovules remained at the size of ovules with no or little expansion (Fig. 2); hence, they became readily recognizable by 5 DAA. The biochemical changes underlying the growth arrest, however, must have happened beforehand. Given that inhibition of the Suc-to-Hex conversion has been shown to trigger or associate with PCD in maize ovaries (Boyer and McLaughlin, 2007) and tomato fruitlets (Li et al., 2012), we next performed a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay on 3-d cotton seeds to examine possible PCD in the GhVIN1-RNAi seeds.
As a technical positive control, seed sections were treated with DNase, which resulted in strong green fluorescent TUNEL signals throughout the entire seeds (Fig. 8, A and B). A similar PCD pattern was observed in the biological positive control (Fig. 8, E and F), in which wild-type ovules were emasculated at −1 d and harvested at 3 d. By contrast, no TUNEL-positive signals were observed in the wild-type seed derived from fertilized ovules (Fig. 8, C and D), indicating that PCD did not occur in wild-type seed at this stage. Significantly, strong PCD signals were detected in 3-d seeds from the RNAi line 15-4-2. While about half of the tested RNAi seeds exhibited PCD signals all over the seed coat and filial tissues, similar to the wild-type emasculation control (Fig. 8F), reflecting the contribution of paternal defect to the undeveloped seed, the others displayed PCD signals confined to nucellus and filial tissues but not in the seed coat (Fig. 8H). To determine if a maternal defect is involved in the cell death in RNAi seeds, a TUNEL assay also was performed on sections of hybrid seeds derived from an RNAi (♀) × wild-type (♂) cross. The analyses revealed that, among the 10 seeds tested, seven seeds displayed PCD signals strongly in the embryo and endosperm and weakly in the nucellus but not in seed coat and fiber cells (Fig. 8J). This finding indicates that the PCD in the filial tissues resulted from a maternal defect. Overall, the data matched with the early genetic analyses that the undeveloped seeds are largely due to paternal defects, with a small portion of it attributable to maternal defects (Fig. 5; Supplemental Fig. S5). The TUNEL assay was repeated using a colorimetric reaction with similar results obtained (Supplemental Fig. S10).
Figure 8.
TUNEL-positive PCD signals detected in 3-d undeveloped cotton seeds of GhVIN-RNAi line 15-4-2 and the RNAi 15-4-2 × wild-type (WT) hybrid. Fluorescent TUNEL assay was conducted on longitudinal sections of 3-d wild-type seed treated with DNase I as a technique positive control (A and B), wild-type seed (C and D), and wild-type ovule with flower bud emasculated at −1 d as a biological positive control (E and F), GhVIN-RNAi 15-4-2 seed (G and H), and RNAi 15-4-2 × wild-type hybrid seed (I and J). Note that, compared with the green fluorescent TUNEL-positive signals detected in entire seeds of the positive controls in B and F, the TUNEL signals in the RNAi 15-4-2 × wild-type hybrid seed were restricted to nucellus, embryo, and endosperm but not in seed coat and fiber cells of the RNAi sections (H and J), indicating that the PCD signal in the filial tissues was under maternal control. em, Embryo; en, endosperm; f, fiber; isc, inner seed coat; n, nucellus; osc, outer seed coat. Bar = 100 μm in A. The scales in B to J are the same as that in A.
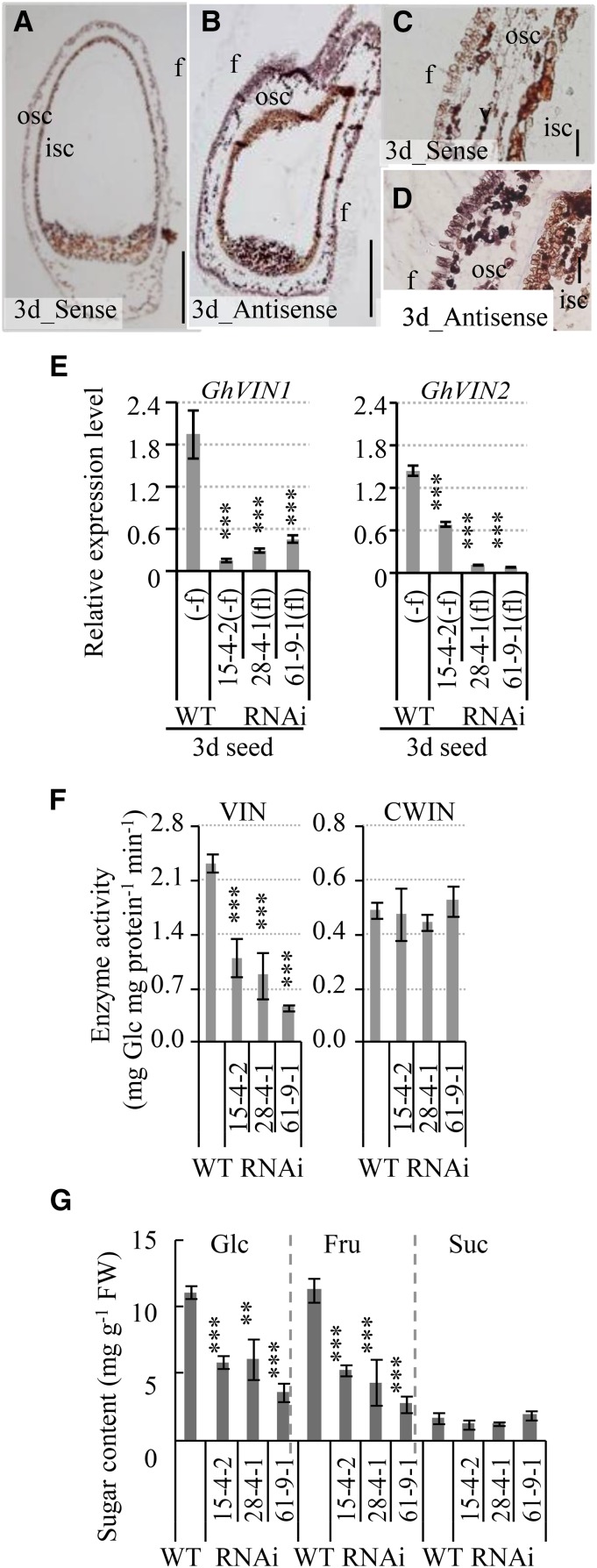
In situ hybridization analyses showed strong GhVIN mRNA signals in outer seed coat and fiber cells of 3-d wild-type seeds, with little signal detected in the filial tissues (Fig. 9, A–D). qPCR measurements revealed significant reductions of the GhVIN1 and GhVIN2 transcript levels in 3-d RNAi seeds across all three RNAi lines (Fig. 9E), leading to significant reductions of VIN activity by approximately 50% to 80% and of Glc and Fru contents by approximately 50% to 60%, with no effect on CWIN activity and Suc level (Fig. 9, F and G).
Figure 9.
GhVIN transcripts, abundant in the seed coat of 3-d wild-type cotton seeds, were reduced significantly in GhVIN-RNAi seeds, along with a reduction of VIN activity. A to D, Longitudinal section of a 3-d seed hybridized with a sense (A) and an antisense (B) RNA probe for GhVINs. C and D are magnified views of the integument region in A and B, respectively. Note the strong GhVIN mRNA signals in outer seed coat (osc) and fiber (f) cells. isc, Inner seed coat. Bars = 200 μm in A and B and 50 μm in C and D. E, Significantly reduced GhVIN1 and GhVIN2 transcripts in 3-d RNAi seeds compared with wild-type seeds (WT). For comparison with fiberless (fl) transgenic seeds from lines 28-4-1 and 61-9-1, fibers on cotton seeds of the wild type and RNAi 15-4-2 were removed (−f) to minimize the influence of GhVIN transcripts from fiber cells. Data represent means ± se (n ≥ 6). F, VIN activity, but not CWIN activity, was reduced significantly in 3-d transgenic seeds compared with wild-type seeds. Data represent means ± se (n ≥ 4). G, Sugar assays show significantly reduced Glc and Fru contents in 3-d transgenic seeds compared with wild-type seeds. Data represent means ± se (n ≥ 4). Asterisks indicate significant differences (one-way ANOVA; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants. FW, Fresh weight.
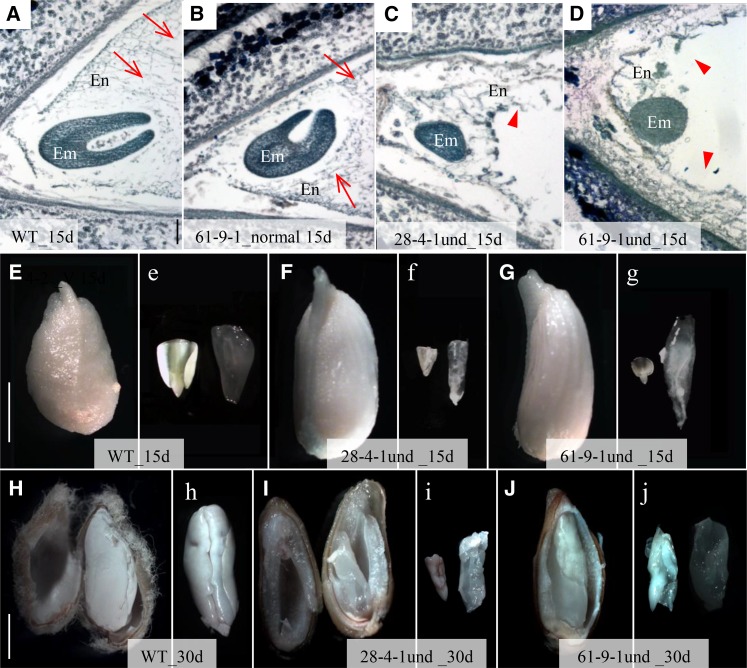
Apart from the undeveloped seeds, the transgenic cotton bolls also produced some underdeveloped seeds that were able to expand to a certain extent (Fig. 2, I and L) but were unviable as well. Histological analyses revealed that, by approximately 15 DAA, the normally developed seeds have produced torpedo embryos with cellularized endosperms (Fig. 10, A, B, E, and e), while the underdeveloped seeds were still in the globular-heart embryo stage with limited or abnormal endosperm cellularization (Fig. 10, C, D, and F–g). By 30 DAA, wild-type embryos were fully expanded with endosperm completely absorbed (Fig. 10, H and h), whereas in the underdeveloped transgenic seeds, embryo development was stunted and residual endosperm tissue remained (Fig. 10, I–j). Interestingly, many of the underdeveloped seeds had normal seed sizes (Figs. 2, I–L, and 10, I and J versus H), indicating that cell expansion in the GhVIN1-RNAi seed coat was largely unaffected and the suppression of their filial tissue growth was not due to a physical constraint imposed by the seed coat.
Figure 10.
Underdeveloped GhVIN-RNAi seeds exhibited impaired filial tissue growth. A to D, Toluidine Blue staining of longitudinal sections of 15-d fully developed seed from wild-type (WT) cotton (A) and GhVIN-RNAi 61-9-1 (B) and underdeveloped seed from RNAi 28-4-1 (C) and RNAi 61-9-1 (D). Note that the fully developed seeds had progressed to torpedo embryo stage with cellularized endosperms (arrows in A and B), whereas the underdeveloped seeds remained in the globular embryo stage with disrupted or limited endosperm cellularization (arrowheads in C and D). E to g, Compared with the 15-d wild-type seed (E and e), line 28-4-1 (F and f) and 61-9-1 (G and g) underdeveloped seeds showed slightly bigger seed size but retarded filial tissue growth. H to j, Compared with the fully developed cotyledon embryo in the wild-type seed at 30 d, where endosperm was fully absorbed (H and h), line 28-4-1 (I and i) and 61-9-1 (J and j) underdeveloped seeds exhibited stunted embryo growth with endosperm remaining. Images labeled by lowercase letters show embryo (left) and endosperm (right) tissues isolated from the seeds presented in the images labeled with the same uppercase letters. Bars = 100 μm in A and 5 mm in E and H. The scales in B to D are the same as that in A, and the scales in e to g and h to j are same as those in E and H, respectively.
Impaired Embryonic Development in GhVIN1-RNAi Seeds Was Associated with Disrupted Expression of Genes for Trehalose and Auxin Metabolism and Signaling
Finally, we examined how the suppression of GhVINs in the maternal seed tissue could lead to embryonic arrest in the underdeveloped seed by targeting seeds at 10 DAA. At this stage, the undeveloped ovule-like seeds were readily distinguishable and removed from the samples and seed coat and filial tissues could be easily separated.
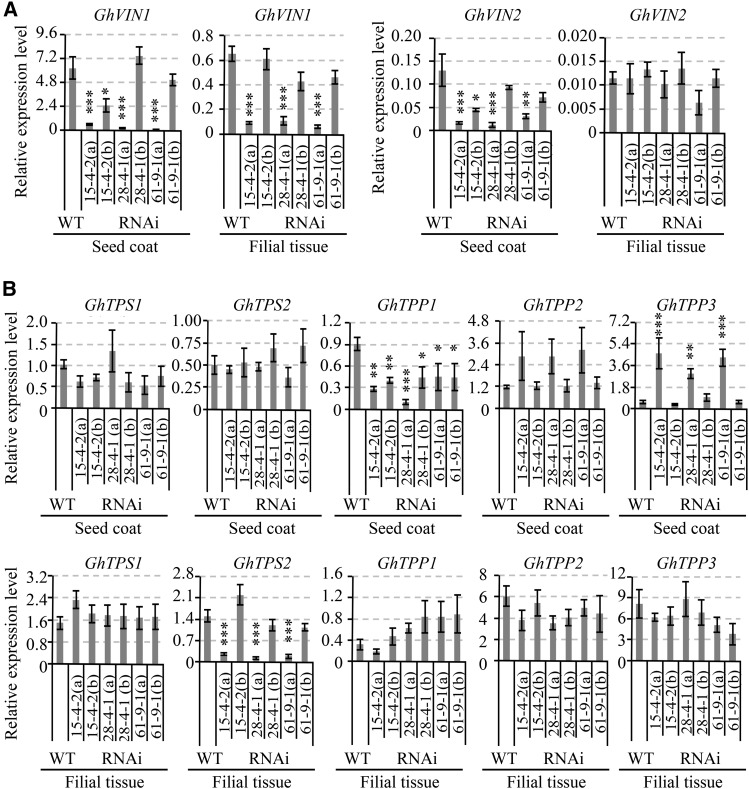
qPCR analyses revealed that, in 10-d wild-type seed, the mRNA levels of both GhVIN1 and GhVIN2 were about 10 times higher in the seed coat than in the filial tissue (Fig. 11A), consistent with in situ hybridization data from 3-d seeds (Fig. 9) and genetic evidence that the underdeveloped seed phenotype was predominantly under maternal control (Supplemental Fig. S5D). Compared with that in the wild type, GhVIN1 expression in transgenic seed coat and filial tissue was extremely low (10% or less than in the wild type) in about half of the tested samples (replicates a) but was not or only slightly reduced in the remaining seed samples (replicates b; Fig. 11A). As we used 10-d seeds from one cotton boll as one replicate, which was a mixture of normally grown transgenic seeds and underdeveloped seeds at 10 DAA, it is very likely that the cotton bolls with high proportions of underdeveloped seeds would have significantly reduced GhVIN1 transcripts (replicates a); while replicates b probably contained those with a lower percentage of underdeveloped seeds. It is worth noting that GhVIN1 is the dominant VIN gene expressed in cotton seeds (Wang et al., 2014), evidenced by its transcript levels being more than 40 times that of GhVIN2 in the seed coat and filial tissue (Fig. 11A). Compared with the wild type, the expression of GhVIN2 also was cosilenced by the GhVIN1-RNAi construct in replicates a, but not in replicates b, of the transgenic seed coat (Fig. 11A). Apart from GhVINs, the expression of CWIN and Sus genes was largely unaffected in 10-d seeds (Supplemental Fig. S11B).
Figure 11.
qPCR analysis of mRNA levels of GhVIN1 and GhVIN2 (A) and genes involved in trehalose metabolism (B) in 10-DAA wild-type (WT) and GhVIN1-RNAi cotton seed coat and filial tissue. Seeds from one cotton boll were used as one biological replicate. Note that the 10-d seeds from a given cotton boll in the transgenic line comprised two populations: underdeveloped seeds and viable seeds, which were visually indistinguishable by eye at 10 DAA. Compared with that in the wild type, GhVIN1 expression was reduced dramatically in some replicates of each transgenic line (replicates a) but was comparable or at the same order of magnitude as in the wild type in other replicates (replicates b). Data represent means ± se of at least three biological samples. Asterisks indicate significant differences (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants.
The above analyses show that the repression of GhVINs in seed coat was most likely the cause of maternal defects, but it remains intriguing how this could result in growth arrest or even cell death in the filial tissue without obvious effect on seed coat development (Figs. 8–10). In this context, trehalose-6-phosphate, an intermediate in trehalose metabolism, has emerged as a global regulator of carbon metabolism and plant growth in response to sugar availability (O’Hara et al., 2013; Lunn et al., 2014). Moreover, embryos of the Arabidopsis trehalose-6-phosphate synthase1 mutant develop more slowly than wild-type embryos and do not progress through the torpedo-to-cotyledon stage (Gόmez et al., 2005). In light of this information, we examined whether the expression of trehalose-6-phosphate metabolism-related genes was altered in GhVIN1-RNAi cotton seeds. Sequence analyses identified two trehalose-6-phosphate synthase (TPS) and three trehalose-6-phosphate phosphatase (TPP) genes from the cotton genome. Within the seed coat, while the transcript levels of two TPS genes were largely unaffected in the RNAi lines, GhTPP3 displayed increased mRNA levels in replicates a (with strong GhVIN1 suppression suggesting a high percentage of underdeveloped seeds) but not in replicates b (with weak GhVIN1 suppression and probably a low percentage of underdeveloped seeds) across all the lines examined, indicating a response to the severe GhVIN1 silencing. The GhTPP1 transcript level was reduced in both replicates a and b, which may reflect an indirect effect from the suppression of GhVIN1 (Fig. 11B). In the filial tissue, the mRNA level of GhTPS2 was reduced dramatically in replicates a across the three RNAi lines (Fig. 11B).
The formation of viable seeds requires effective communication among the maternally derived seed coat and the zygotic embryo and endosperm to ensure their coordinated development. One of the most important signaling molecules required for this communication is auxin (Locascio et al., 2014). Prompted by the regulatory roles of Glc in auxin biosynthesis (LeClere et al., 2010; Sairanen et al., 2012) and signaling (Mishra et al., 2009; Wang et al., 2014), the expression of a cohort of auxin biosynthesis and signaling genes was examined in 10-d seed. For the three tested auxin signaling genes, GhABP1, GhARF1, and GhARF2, their transcript levels were greatly reduced in seed coat, especially in those with GhVIN1 strongly suppressed samples (replicates a; Supplemental Fig. S12B). Within the filial tissue, decreased GhABP1 expression also was observed in RNAi line 28-4-1 and 61-9-1 replicates, as compared with the wild type (Supplemental Fig. S12B). Among the six highly expressed auxin biosynthesis genes, significantly reduced GhTAA1 transcript levels were observed in the filial tissues of all three lines, while the transcript levels of GhTAA1, GhYUC2, and GhYUC5 were increased in replicates a seed coat samples of lines 61-9-1, 15-4-2, and 28-4-1, respectively (Supplemental Fig. S12B). Clearly, the expression of genes related to auxin biosynthesis and signaling response was disrupted in 10-d transgenic coat seed and filial tissues.
DISCUSSION
Despite the general recognition that reproductive success relies heavily on Suc metabolism to acquire carbon nutrient, energy, and signals for the development of different reproductive tissues, it remains elusive how Suc metabolism couples with complex reproductive development at the cellular and molecular levels (Ruan et al., 2012; Nuccio et al., 2015). Here, we provide genetic and developmental evidence that VIN exerts strong control over floral development and the formation of male and female fertilities, thereby acting as a major player for seed set and subsequent development in cotton. As such, this study provides significant insights into the intimate linkage between VIN-mediated Suc metabolism and plant reproductive development and opens up new perspectives for genetic modifications to enhance crop seed development.
VIN Is Required for Floral Morphogenesis and the Formation of Male and Female Fertilities
The data obtained in this study show that the expression of cotton VIN genes is required for proper pollination, fertilization, seed set, and subsequent seed development. First, a large proportion of the flowers in the GhVIN1-RNAi cottons exhibited abnormal flower structures (Fig. 3; Supplemental Fig. S2), indicating a role of VIN in floral morphogenesis. Second, suppressing GhVIN expression in the stamen delayed anther dehiscence and, hence, pollen release (Fig. 3; Supplemental Fig. S3) as well as reduced pollen viability and pollen tube germination (Figs. 5 and 6). Third, suppression of GhVINs in seed maternal tissue resulted in PCD or growth arrest in the filial tissue (Figs. 8–10). The above reproductive defects collectively led to the production in the GhVIN1-RNAi bolls of a large number of unviable seeds classified as undeveloped seeds/ovules and underdeveloped seeds. The former resulted from (1) pollination failure, because of spatially unmatched development between stamen and pistils or delayed pollen release; (2) fertilization failure, due to poor pollen viability; and (3) seed abortion, owing to nucellus and filial tissue cell death early in seed development, caused by maternal defects. On the other hand, the underdeveloped seeds were largely attributable to female sterility, based on data from reciprocal crosses (Supplemental Fig. S5), resulting in growth arrest of the filial tissue, evident at about 10 to 15 DAA (Fig. 10). Together, these analyses identify VIN as a major regulator for diverse reproductive processes from floral development to the formation of male and female fertilities. To our knowledge, this represents an unprecedented example of the control of such diverse aspects of reproductive development by a sugar metabolic enzyme. Consistent with our finding, there have been no reports of VIN mutants in any crop species, as such a mutant is likely reproductively lethal based on what we found in this study. Biochemically, VIN activity may regulate reproductive development by modulating cytoplasmic hexose levels and sugar signaling. Flux analysis and modeling of sugar metabolism in tomato pericarp indicate that VIN-catalyzed sucrolytic activity in the vacuole induces hexose efflux from the vacuole into cytoplasm, coupled with Suc influx during cell division of fruit development (Beauvoit et al., 2014). Thus, VINs are able to regulate not only vacuolar sugar homeostasis but also cytosolic hexose levels through coupling with the activities of tonoplast sugar transporters. Altering cytosolic hexose levels could have a profound impact on gene expression through sugar signaling in parallel with its central role in sugar metabolism (Ruan, 2014). In agreement with this view is the altered gene expression for starch and trehalose metabolism and auxin and JA synthesis and signaling in the GhVIN1-RNAi stamen and seed (Figs. 7 and 11; Supplemental Fig. S12). The alterations in gene expression observed in stamen and developing seeds are likely the direct effects of decreased GhVIN gene expression, since the intervention did not appear to affect the expression of genes encoding other Suc degradation enzymes (CWIN and Sus) or sugar transporters (Fig. 9; Supplemental Figs. S7 and S11).
VIN Contributes to Male Fertility Probably by Impacting Starch Metabolism as Well as Auxin and JA Synthesis and Signaling in Stamen
The paternal defects observed in the GhVIN1-RNAi plants include delayed pollen release and reduced pollen viability (Figs. 3 and 4). The defects caused pollination failures, rendering the ovules unable to develop into seeds (Figs. 2 and 5). The delay in pollen release from the transgenic anthers may arise from a combination of (1) incomplete septum degradation and impaired endothecium secondary thickening and (2) inadequate anther wall dehydration before anthesis, due to reduced starch accumulation in filament and the anther-filament conjunction region (Fig. 4). The starch-to-sugar conversion would increase the osmotic potential in filament to facilitate water efflux from anthers, leading to anther dehiscence (Bonner and Dickinson, 1990; Stadler et al., 1999). The reduced starch in the GhVIN1-silenced stamen is likely owing to (1) decreased starch synthesis, as indicated by the reduced expression of two ADP-Glc pyrophosphorylase genes (GhAGPase1 and GhAGPase2) and (2) compromised starch hydrolysis, as suggested by the down-regulation of an α-amylase gene, GhαAmy (Fig. 7A; Supplemental Fig. S7). These data indicate that starch turnover is disrupted in the GhVIN1-RNAi stamen, which could result in not only a delay in anther dehiscence but also poor pollen viability, as starch abundance is critically required for pollen vigor (Clément et al., 1994; Goetz et al., 2001; Datta et al., 2002). Suppression of GhVINs also reduced the transcript levels of GhMYB24 and GhLOX1 in the transgenic stamens (Fig. 7). Both genes have been shown to regulate late anther and pollen development via JA signaling (Marmey et al., 2007; Li et al., 2013). Moreover, the transgenic stamen also was characterized with decreased expression of the auxin biosynthesis genes GhTAA1 and GhYUC5, the auxin signaling receptor gene GhABP1, and the auxin-responsive factor GhARF2 (Fig. 7). Auxin and JA metabolism and signaling are of importance in anther differentiation and dehiscence and pollen development (Ishiguro et al., 2001; Yang et al., 2007; Cecchetti et al., 2008; Nashilevitz et al., 2009; Li et al., 2013). Silencing GhVIN expression may alter the expression of these genes through modulating cytosolic sugar homeostasis and sugar signaling. Although the exact nature of such a regulation remains to be elucidated, our data revealed a new linkage between VIN-mediated sugar metabolism and male fertility, potentially through the VIN-mediated regulation of starch turnover and auxin and JA synthesis and signaling.
Seed Coat GhVIN Expression May Be Required for Adequate Sugar Supply and Balanced Sugar and Auxin Signaling to Support Filial Tissue Development
Apart from the paternal defects in the GhVIN1-RNAi plants, maternal sterility also was partially responsible for the generation of undeveloped seed and was largely accountable for the underdeveloped seed phenotype (Fig. 5; Supplemental Fig. S5). An intriguing finding was that, although GhVINs were predominantly expressed in the seed coat, as indicated by in situ hybridization and qPCR results (Figs. 9 and 11), the suppression of GhVINs had little phenotypic effect on the seed coat but an evident negative impact on filial tissues, characterized by their cell death at 3 DAA (Fig. 8) and growth arrest at 10 DAA (Fig. 10). Why are filial tissues more sensitive than seed coat to the down-regulation of largely maternally expressed GhVINs?
One possibility is that VIN activity in the seed coat may be essential for nutrient flow to the filial tissues. In cotton seed, Suc is unloaded symplasmically from the phloem in the outer seed coat (Ruan et al., 1997; Wang and Ruan, 2012). High VIN activities and gene expression have been observed in wild-type cotton outer seed coat during early seed development (Wang et al., 2010), along with strong Sus expression and activity in the seed epidermis (Ruan et al., 2003). These Suc-cleavage enzymes in the outer seed coat are essential for lowering local Suc concentration to facilitate phloem unloading (Ruan et al., 1996; Wang et al., 2014). Suppression of GhVIN expression could dampen the Suc-to-Glc/Fru conversion, hence reducing sink strength and phloem unloading. This is indicated by the significant decrease in Glc and Fru contents in 3-d GhVIN1-RNAi seeds (Fig. 9). Pertinently, VIN is the major enzyme hydrolyzing Suc in tomato pericarp at the cell division stage (Beauvoit et al., 2014), when unloading occurs symplasmically (Palmer et al., 2015).
For subsequent translocation into the filial tissues in cotton seed, assimilates must pass two symplasmically disconnected cellular sites: between the outer and inner seed coat, and between maternal and filial tissues (Ruan et al., 1997; Wang and Ruan, 2012). High CWIN expression at both interfaces (Wang and Ruan, 2012) may facilitate the hexose production in these sites. Thus, it is possible that hexose, derived from maternal VIN and CWIN activities, is the major carbon source transported into the filial tissues of developing cotton seed. Indeed, hexose could dominate over Suc in their import into endosperm during early seed development in Arabidopsis (Baud et al., 2005; Chen et al., 2015) and maize (Sosso et al., 2015). These findings underpin the importance of sucrolytic activities in channeling carbon from maternal to filial tissues. GhVIN1-RNAi-mediated reduction of VIN activity in the seed coat likely blocks hexose generation (Fig. 9) and its flow to filial tissues, causing carbon starvation and even PCD (Fig. 8). The correlation between low Glc and the activation of some PCD genes has been observed in maize grain under drought (Boyer and McLaughlin, 2007) and tomato fruit under heat stress (Li et al., 2012). While some seeds were blocked entirely, thus becoming ovule-like undeveloped seeds, in the GhVIN1-RNAi lines, others were able to survive the early stage in which the endosperm and embryo developed to a certain extent but became arrested at the torpedo stage (Fig. 10). The latter had comparable seed coat to fully developed seeds. After fertilization, a signal from the syncytial endosperm is considered to play a crucial role in triggering seed coat cell expansion and regulating seed size in Arabidopsis (Chaudhury et al., 2001; Garcia et al., 2003). Once seed coat cell expansion has initiated, it develops independently from endosperm and embryo (Haughn and Chaudhury, 2005). Thus, the seed coat in undeveloped seeds probably has received no or impaired initial signal for its growth, while the underdeveloped seeds may have acquired such a signal from the endosperm during nuclear division (approximately 3–5 DAA in cotton; Wang and Ruan, 2012), allowing their seed coats to be fully developed. Consistently, the endosperm in the underdeveloped seed appeared to develop until the cellularization stages at approximately 10 DAA (Fig. 10; Ruan et al., 2008), when its increased sugar demand for cell wall synthesis could become unsustainable due to the suppression of GhVIN expression in the maternal seed coat, as discussed previously. Silencing GhVINs altered the expression of two GhTPP genes in the seed coat and reduced the transcript level of GhTPS2 in the filial tissue (Fig. 11), suggesting that trehalose metabolism may have been affected in the transgenic seed, which could compromise filial tissue development. The effect of trehalose metabolism and signaling on grain set was demonstrated recently in maize (Nuccio et al., 2015). Another well-known signaling molecule involved in seed maternal-filial communication is auxin. Here, the GhVIN1-suppressed transgenic seed exhibited a disruption of gene expression in relation to auxin biosynthesis and signaling perception (Supplemental Fig. S12). Reduced gene expression for auxin biosynthesis has been observed in the maize CWIN mutant (LeClere et al., 2010). The likely compromised trehalose and auxin metabolism and signaling also may contribute to the blockage of filial development in the underdeveloped seeds.
VIN, along with CWIN, Could Act as a Gatekeeper for Reproductive Success under Abiotic Stress
Finally, much of the phenotype we observed in the GhVIN1-RNAi cotton plant resembles the symptoms of plants under abiotic stress. For example, heat-stressed cotton plants also exhibit abnormal stigma protrusion, delayed anther dehiscence, and reduced pollen viability (Brown, 2001; Snider et al., 2009; Min et al., 2014) as well as high rates of boll shedding and seed abortion (Powell, 1969; Reddy et al., 1992; Brown, 2001). Min et al. (2014) also reported that the impaired anther and pollen development under high temperature were associated with reduced expression of INV and starch synthesis genes, decreased Glc level, as well as disrupted auxin biosynthesis. Similarly, wheat male reproductive failure under water deficit was related to decreased VIN (Ivr5) and CWIN (Ivr1) gene expression in pollen (Koonjul et al., 2005). Indeed, maize soluble acid invertase (VIN gene Ivr2) was identified as an early target of drought stress during maize ovule abortion (Andersen et al., 2002). Similarly, high heat tolerance in tomato flower and young fruit correlates with strong VIN and CWIN activities (Li et al., 2012). Collectively, VINs, along with CWINs, appear to act as a common downstream gatekeeper in sustaining reproductive fertilities under abiotic stress, likely through maintaining sink strength and cytosolic sugar homeostasis and signaling.
MATERIALS AND METHODS
Plant Growth Conditions, Pollination, and Bud-Thinning Treatments
Wild-type and transgenic cotton (Gossypium hirsutum ‘Coker312’) plants were grown in a greenhouse according to Wang et al. (2010). GhVIN1-RNAi cotton lines were generated as described previously (Wang et al., 2014). Flower, stamen, ovule, or seed samples were excised from developing flower buds or cotton bolls at the specified DAA. The numbers of cotton flowers and bolls were counted throughout the growth cycle. Seed number was counted by boll maturity.
For hand pollination, flower buds were emasculated at −1 DAA, and stigmas were pollinated the next day at approximately 10 am. Each cotton stigma requires approximately 100 viable pollen grains to fully fertilize the ovules in a given cotton boll (Waller and Mamood, 1991). We collected more than 10,000 pollens from a single flower for pollination of a maximum of three stigmas to ensure adequate pollination.
For the bud-thinning experiment, all cotton buds, except four buds at nodes 6 to 9, were removed at pinhead square stage (approximately 3 weeks before flowering).
Pollen Germination and Pollen Tube Elongation
Pollen grains were collected shortly after anther dehiscence in the morning and immediately tapped onto the germination medium modified from that described by Burke et al. (2004). It contains, all in w/v, 0.05% H3BO4, 0.02% Ca(NO3)2, 0.01% KNO3, 0.02% MgSO4·0.7H2O, and 25% Suc with pH 6. Pollen grains from one flower were collected into one petri dish as one biological replicate. Pollens were incubated in the dark at 28°C for 2 h before microscopic observation (Zeiss Axiophot D-7082). A pollen grain with pollen tube length longer than or at least equal to grain diameter was considered to be germinated (Kakani et al., 2002). Germination rate was determined by dividing the number of germinated pollen grains by the total number of pollens. Pollen tube length was measured using the ImageJ program (http://rsb.info.nih.gov/ij/).
Histological Analyses
For the pollen viability test, fresh flowers were collected shortly after germination, and pollens were gently tapped into the modified pollen germination medium with the addition of 5 μg mL−1 FDA. After 5 min of incubation, pollens were examined microscopically under UV light excitation and a long-pass GFP emission filter.
To observe the number of pollens captured by the stigma, fresh flowers were collected at approximately 3 to 4 h after anther dehiscence. Stigmas were stained by 0.1% Aniline Blue in 67 mm K2HPO4-KH2PO4 buffer (pH 7.5) for 5 min. Pollen grains exhibited green fluorescence from Aniline Blue-bound callose under UV light.
For in vivo pollen tube elongation observation, stigmas were collected at various times after pollination. The tissues were fixed in cold fixation buffer (4% formaldehyde, 70% ethanol, and 10% acetic acid) overnight, rehydrated with gradient ethanol and water, softened in 1 m NaOH overnight, rinsed by 0.1 m K2HPO4-KH2PO4 buffer (pH 8.5), and then stained by 0.1% Aniline Blue for 4 h. Stigma tissue was squashed gently before observation under UV light.
For anther structure observation, −1-d anthers were fixed, dehydrated, embedded, sectioned, stained with Toluidine Blue, and examined according to Regan and Moffatt (1990). To estimate the deposition of callose and the secondary wall thickening, the sections were stained with 1% Aniline Blue in 0.1 m K2HPO4-KH2PO4 buffer, pH 8.5, for 5 min followed by visualization under UV light.
For starch localization, −1-d anther sections and −1-d flowers (with sepal and petal removed) were stained by KI-I2 (2% KI and 0.5% I2) for 10 s and 3 min, respectively.
RNA Extraction and Reverse Transcription
For RNA extraction, −1-d stamen or style from one cotton flower, and ovules, seeds, seed coats, or filial tissues from one cotton boll, were collected as one biological sample. Total RNA was isolated according to Ruan et al. (1997). About 0.5 μg of RNA was treated by RQ1 RNase-free DNase (Promega) and then reverse transcribed to complementary DNA using the SuperScript first-strand synthesis system (Invitrogen) with 50 μm oligo(dT)20 according to the manufacturer’s recommendations.
qPCR Analysis
qPCR was performed with SYBR Green and Platinum Taq DNA Polymerase (Life Technologies) on a Rotor-Gene Q instrument (Qiagen) following amplification cycles as follows: 10 min at 95°C followed by 40 rounds of 10 s at 95°C, 20 s at 60°C, and 20 s at 72°C. A product melting curve was used to confirm a single PCR product at the end of amplification. Gene-specific primers used for qPCR are listed in Supplemental Table S1, along with the GenBank accession numbers of the tested genes. Primer set efficiencies (E) were estimated for each experimental set by Rotor-Gene 6000 Series software (Qiagen).
Among the cotton reference genes, the F-box family gene GhFBX6, catalytic subunit of protein phosphatase 2A GhPP2A1, polyubiquitin gene GhUBQ14, and actin gene GhACT4 (Artico et al., 2010), a combination of GhFBX6 and GhUBQ14 displayed the most stable expression among wild-type and GhVIN1-RNAi cotton complementary DNA samples, based on analysis from the RefFinder program (http://www.leonxie.com/referencegene.php?type=reference) and geNORM software (http://medgen.ugent.be/∼jvdesomp/genorm/), and therefore were used as internal control genes in this study. All calculations of expression levels were performed on quantities (Q), which were calculated via the ΔCq (quantitation cycle) method with the formula Q = (E)ΔCq (Hellemans et al., 2007), where ΔCq equals the Cq of the sample with the lowest Cq value (highest abundance) minus the Cq of a sample. For efficiency, corrected relative amounts were calculated. The levels of target gene expression were normalized to the geometric mean of GhFBX6 and GhUBQ14 by subtracting the cycle threshold value of an internal gene set from the cycle threshold value of the target genes.
In Situ Hybridization
In situ hybridization experiments were carried out according to Wang et al. (2010).
TUNEL
Sections of paraffin-embedded cotton ovule or seed samples were dewaxed with 100% histolene, rehydrated in a graded ethanol series, and permeabilized in proteinase K. Nick-end labeling of fragmented DNA was performed using the Fluoresce In Situ Cell Death Detection Kit (Roche) or the DeadEnd Colorimetric TUNEL system (Promega), according to each manufacturer’s instructions. Slides were analyzed microscopically (Zeiss Axiophot D-7082) under bright-field (colorimetric TUNEL) or green fluorescent (fluorometric TUNEL) channel.
Invertase Enzyme Assay and Sugar Measurement
Invertase activities and sugar levels were measured enzymatically as described by Wang et al. (2010).
Statistical Analyses
Unless specified otherwise, randomization one-way ANOVA was used for the comparisons among the wild type and different RNAi lines. Means were compared using all-pairs Turky’s honestly significant difference test. Statistical calculations of ANOVA were performed using JMP 11 statistics software.
GLMM (Jinks et al., 2006; Bolker et al., 2009) was used to analyze the cotton seed numbers in the hand-pollination, cross-hybridization, and cotton bud-thinning treatments. It was a two-level hierarchical (each of three types of seed within each of three or four genotypes nested with five different treatments/control) randomized design. Statistical calculations were performed using SAS 9.2 (SAS Institute). Data sets were natural logarithm transformed because the data distribution of model residuals was not normal. P < 0.05 was considered significant.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GhVIN1-RNAi cotton plants showed reduced viable seeds at T2 generation, in comparison with that in WT.
Supplemental Figure S2. GhVIN1-RNAi cotton plants displayed ovule number per boll identical to that in WT.
Supplemental Figure S3. Suppression of GhVIN1 affected floral organ formation.
Supplemental Figure S4. In vitro and in vivo pollen tube elongation in GhVINs-RNAi and WT cotton plants.
Supplemental Figure S5. The percentages of un-developed seeds (A, C and E) and under-developed seeds (B, D, and F) after hand pollination (A–B), reciprocal crossing (C–D), and bud thinning (E–F).
Supplemental Figure S6. RDLp::GUS expression in −1 d transgenic cotton floral organ (A), stamen (B) and ovule (C), and transgenic seeds at 1 d (D), 2 d (E), 5 d (F), and 10 d (G and H).
Supplemental Figure S7. A heat map of genes involved in aspects of sugar metabolism and measured soluble sugar levels in −1 d WT and RNAi stamen.
Supplemental Figure S8. qPCR analysis of the transcript levels of auxin biosynthesis genes GhTAR2 and GhYUC11, and auxin transportation genes GhAUX1, GhPIN2, and GhPIN3 in −1 DAA stamen from WT and RNAi plants.
Supplemental Figure S9. qPCR analysis of the transcript levels of GhVIN1, GhVIN2, GhCWIN1, GhSus1 and GhSusA in −1 DAA styles from WT and RNAi plants.
Supplemental Figure S10. Colorimetric TUNEL assay on the longitudinal and cross -sections of 3d WT seed (A and C) and GhVINs-RNAi 15-4-2 seed (B and D), respectively.
Supplemental Figure S11. qPCR analyses of the expressions of GhCWIN1, GhSus1 and GhSusA in WT and RNAi 3d seeds (A) and 10d seed coat and filial tissues (B).
Supplemental Figure S12. The expressions of genes related to auxin biosynthesis (A) and signaling response (B) were disrupted in 10d GhVINs-RNAi seeds, as compare to those in WT.
Supplemental Table S1. Quantitative real-time PCR primers used in this study.
Supplementary Material
Acknowledgments
We thank Xiao-Ya Chen and Hang Lian (Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for providing the RDL-GUS transgenic cotton seeds and performing GUS staining, respectively, and Kim Colyvas (University of Newcastle, Australia) for help in statistical analyses.
Glossary
- RNAi
RNA interference
- DAA
days after anthesis
- FDA
fluorescein diacetate
- GLMM
generalized linear mixed model
- qPCR
quantitative real-time PCR
- JA
jasmonic acid
- PCD
programmed cell death
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
Footnotes
This work was supported by the Chinese National Science Foundation (grant no. 30425043) and Australian Research Council (grant no. DP110104931 to Y.-L.R.).
Articles can be viewed without a subscription.
References
- Andersen MN, Asch F, Wu Y, Jensen CR, Naested H, Mogensen VO, Koch KE (2002) Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in maize. Plant Physiol 130: 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 [DOI] [PubMed] [Google Scholar]
- Beauvoit BP, Colombié S, Monier A, Andrieu MH, Biais B, Bénard C, Chéniclet C, Dieuaide-Noubhani M, Nazaret C, Mazat JP, et al. (2014) Model-assisted analysis of sugar metabolism throughout tomato fruit development reveals enzyme and carrier properties in relation to vacuole expansion. Plant Cell 26: 3224–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White JSS (2009) Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol 24: 127–135 [DOI] [PubMed] [Google Scholar]
- Bonner LJ, Dickinson HG (1990) Anther dehiscence in Lycopersicon esculentum. 2. Water relations. New Phytol 15: 367–375 [DOI] [PubMed] [Google Scholar]
- Boyer JS, McLaughlin JE (2007) Functional reversion to identify controlling genes in multigenic responses: analysis of floral abortion. J Exp Bot 58: 267–277 [DOI] [PubMed] [Google Scholar]
- Brill E, van Thournout M, White RG, Llewellyn D, Campbell PM, Engelen S, Ruan YL, Arioli T, Furbank RT (2011) A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol 157: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PW. (2001) Heat stress and cotton yields in Arizona. In Cotton: A College of Agriculture Report, Series P-125. College of Agriculture, University of Arizona, Tucson, pp 1–7 [Google Scholar]
- Burke JJ, Velten J, Oliver MJ (2004) In vitro analysis of cotton pollen germination. Agron J 96: 359–368 [Google Scholar]
- Castro AJ, Clément C (2007) Sucrose and starch catabolism in the anther of Lilium during its development: a comparative study among the anther wall, locular fluid and microspore/pollen fractions. Planta 225: 1573–1582 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB (2015) A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1996) The miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8: 971–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément C, Chavant L, Burrus M, Audran JC (1994) Anther starch variations in Lilium during pollen development. Sex Plant Reprod 7: 347–356 [Google Scholar]
- Datta R, Chamusco KC, Chourey PS (2002) Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol 130: 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Robinson SP (1996) Sugar accumulation in grape berries: cloning of two putative vacuolar invertase cDNAs and their expression in grapevine tissues. Plant Physiol 111: 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke T, Hirsche J, Roitsch T (2010) Anther-specific carbohydrate supply and restoration of metabolically engineered male sterility. J Exp Bot 61: 2693–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XL, Ni WM, Elge S, Mueller-Roeber B, Xu ZH, Xue HW (2006) Auxin flow in anther filaments is critical for pollen grain development through regulating pollen mitosis. Plant Mol Biol 61: 215–226 [DOI] [PubMed] [Google Scholar]
- French SR, Abu-Zaitoon Y, Uddin M, Bennett K, Nonhebel HM (2014) Auxin and cell wall invertase related signaling during rice grain development. Plants 3: 95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Saingery V, Chambrier P, Mayer U, Jürgens G, Berger F (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Guivarc’h A, Kahmann U, Chriqui D, Roitsch T (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA 98: 6522–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gόmez LD, Baud S, Graham IA (2005) The role of trehalose-6-phosphate synthase in Arabidopsis embryo development. Biochem Trans Soc 33: 280–282 [DOI] [PubMed] [Google Scholar]
- Guan X, Lee JJ, Pang M, Shi X, Stelly DM, Chen ZJ (2011) Activation of Arabidopsis seed hair development by cotton fiber-related genes. PLoS ONE 6: e21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Bennett M, Mansfield J, Somerville C (2009) Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J 57: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T (2002) Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol 43: 452–459 [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dong B, Shiran B, Talbot MJ, Edlington JE, Hughes T, White RG, Gubler F, Dolferus R (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol 156: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL (2009) Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 21: 2072–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks RL, Willoughby I, Baker C (2006) Direct seeding of ash (Fraxinus excelsior L.) and sycamore (Acer pseudoplatanus L.): the effects of sowing date, pre-emergent herbicides, cultivation, and protection on seedling emergence and survival. For Ecol Manage 237: 373–386 [Google Scholar]
- Kakani VG, Prasad PVV, Craufurd PQ, Wheeler TR (2002) Response of in vitro pollen germination and pollen tube growth of groundnut (Arachis hypogaea L.) genotypes to temperature. Plant Cell Environ 25: 1651–1661 [Google Scholar]
- Keijzer CJ. (1987) The cytology of anther dehiscence and pollen dispersal. I. The opening mechanism of longitudinally dehiscing anthers. New Phytol 105: 487–498 [DOI] [PubMed] [Google Scholar]
- Koch K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Koonjul PK, Minhas JS, Nunes C, Sheoran IS, Saini HS (2005) Selective transcriptional down-regulation of anther invertases precedes the failure of pollen development in water-stressed wheat. J Exp Bot 56: 179–190 [DOI] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS (2010) Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol 153: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jiang J, Du ML, Li L, Wang XL, Li XB (2013) A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol 54: 893–906 [DOI] [PubMed] [Google Scholar]
- Li Z, Palmer WM, Martin AP, Wang R, Rainsford F, Jin Y, Patrick JW, Yang Y, Ruan YL (2012) High invertase activity in tomato reproductive organs correlates with enhanced sucrose import into, and heat tolerance of, young fruit. J Exp Bot 63: 1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Roig-Villanova I, Bernardi J, Varotto S (2014) Current perspectives on the hormonal control of seed development in Arabidopsis and maize: a focus on auxin. Front Plant Sci 5: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M (2014) Trehalose metabolism in plants. Plant J 79: 544–567 [DOI] [PubMed] [Google Scholar]
- Maddison AL, Hedley PE, Meyer RC, Aziz N, Davidson D, Machray GC (1999) Expression of tandem invertase genes associated with sexual and vegetative growth cycles in potato. Plant Mol Biol 41: 741–751 [DOI] [PubMed] [Google Scholar]
- Marmey P, Jalloul A, Alhamdia M, Assigbetse K, Cacas JL, Voloudakis AE, Champion A, Clerivet A, Montillet JL, Nicole M (2007) The 9-lipoxygenase GhLOX1 gene is associated with the hypersensitive reaction of cotton Gossypium hirsutum to Xanthomonas campestris pv malvacearum. Plant Physiol Biochem 45: 596–606 [DOI] [PubMed] [Google Scholar]
- Millán-Cañongo C, Orona-Tamayo D, Heil M (2014) Phloem sugar flux and jasmonic acid-responsive cell wall invertase control extrafloral nectar secretion in Ricinus communis. J Chem Ecol 40: 760–79 [DOI] [PubMed] [Google Scholar]
- Min L, Li Y, Hu Q, Zhu L, Gao W, Wu Y, Ding Y, Liu S, Yang X, Zhang X (2014) Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol 164: 1293–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BS, Singh M, Aggrawal P, Laxmi A (2009) Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS ONE 4: e4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Aharoni A, Kossmann J, Wolf S, Levy AA (2009) The legwd mutant uncovers the role of starch phosphorylation in pollen development and germination in tomato. Plant J 57: 1–13 [DOI] [PubMed] [Google Scholar]
- Nuccio ML, Wu J, Mowers R, Zhou HP, Meghji M, Primavesi LF, Paul MJ, Chen X, Gao Y, Haque E, et al. (2015) Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol 33: 862–869 [DOI] [PubMed] [Google Scholar]
- O’Hara LE, Paul MJ, Wingler A (2013) How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol Plant 6: 261–274 [DOI] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Palmer WM, Ru L, Jin Y, Patrick JW, Ruan YL (2015) Tomato ovary-to-fruit transition is characterized by a spatial shift of mRNAs for cell wall invertase and its inhibitor with the encoded proteins localized to sieve elements. Mol Plant 8: 315–328 [DOI] [PubMed] [Google Scholar]
- Powell RD. (1969) Effect of temperature on boll set and development of Gossypium hirsutum. Cotton Grow Rev 46: 29–36 [Google Scholar]
- Pozueta-Romero J, Perata P, Akazawa T (1999) Sucrose-starch conversion in heterotrophic tissues of plants. Crit Rev Plant Sci 18: 489–525 [Google Scholar]
- Pressman E, Shaked R, Shen S, Altahan L, Firon N (2012) Variations in carbohydrate content and sucrose-metabolizing enzymes in tomato (Solanum lycopersicum L.) stamen parts during pollen maturation. Am J Plant Sci 3: 252–260 [Google Scholar]
- Proels RK, González MC, Roitsch T (2006) Gibberellin-dependent induction of tomato extracellular invertase Lin7 is required for pollen development. Funct Plant Biol 33: 547–554 [DOI] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ (2002) Experimental Design and Data Analysis for Biologists. Cambridge University Press, Cambridge, UK [Google Scholar]
- Reddy KR, Hodges HF, McKinion JM, Wall GA (1992) Temperature effect on pima cotton growth and development. Agron J 84: 237–243 [Google Scholar]
- Regan SM, Moffatt BA (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijavec T, Kovac M, Kladnik A, Chourey PS, Dermastia M (2009) A comparative study on the role of cytokinins in caryopsis development in the maize miniature1 seed mutant and its wild type. J Integr Plant Biol 51: 840–849 [DOI] [PubMed] [Google Scholar]
- Ruan YL. (2014) Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol 65: 33–67 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Chourey PS, Delmer DP, Perez-Grau L (1997) The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol 115: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Liu Q, Xu SM, Wu LM, Wang L, Furbank RT (2008) Expression of sucrose synthase in the developing endosperm is essential for early seed development in cotton. Funct Plant Biol 35: 1–12 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Patrick JW, Bouzayen M, Osorio S, Fernie AR (2012) Molecular regulation of seed and fruit set. Trends Plant Sci 17: 656–665 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Patrick JW, Brady CJ (1996) The composition of apoplastic fluid recovered from intact developing tomato fruit. Aust J Plant Physiol 23: 9–13 [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K (2012) Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider JL, Oosterhuis DM, Skulman BW, Kawakami EM (2009) Heat stress-induced limitations to reproductive success in Gossypium hirsutum. Physiol Plant 137: 125–138 [DOI] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, et al. (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47: 1489–1493 [DOI] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N (1999) The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J 19: 269–278 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Østergaard L (2009) Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol 1: a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SM, Brill E, Llewellyn DJ, Furbank RT, Ruan YL (2012) Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol Plant 5: 430–441 [DOI] [PubMed] [Google Scholar]
- Waller GD, Mamood AN (1991) Upland and Pima cotton as pollen donors for male sterile upland seed parents. Crop Sci 31: 265–266 [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Wang L, Cook A, Patrick JW, Chen XY, Ruan YL (2014) Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J 78: 686–696 [DOI] [PubMed] [Google Scholar]
- Wang L, Li XR, Lian H, Ni DA, He YK, Chen XY, Ruan YL (2010) Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol 154: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ruan YL (2012) New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol 160: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ruan YL (2013) Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci 4: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY (2004) Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16: 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U (1996) Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 10: 823–834 [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U (2003) The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. Plant J 33: 395–411 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Song J, Taylor B, Yang C (2011) The final split: the regulation of anther dehiscence. J Exp Bot 62: 1633–1649 [DOI] [PubMed] [Google Scholar]
- Yang XY, Li JG, Pei M, Gu H, Chen ZL, Qu LJ (2007) Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep 26: 219–228 [DOI] [PubMed] [Google Scholar]
- Zanor MI, Osorio S, Nunes-Nesi A, Carrari F, Lohse M, Usadel B, Kühn C, Bleiss W, Giavalisco P, Willmitzer L, et al. (2009) RNA interference of LIN5 in Solanum lycopersicum confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol 150: 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.