Yang et al. identify a feedback loop between gankyrin, an oncoprotein overexpressed in human hepatocellular carcinoma (HCC), and Nrf2. The positive feedback modulates a series of antioxidant enzymes that lower intracellular reactive oxygen species to confer protection from mitochondrial damage and cell death.
Abstract
Oxidative stress status has a key role in hepatocellular carcinoma (HCC) development and progression. Normally, reactive oxygen species (ROS) levels are tightly controlled by an inducible antioxidant program that responds to cellular stressors. How HCC cells respond to excessive oxidative stress remains elusive. Here, we identified a feedback loop between gankyrin, an oncoprotein overexpressed in human HCC, and Nrf2 maintaining the homeostasis in HCC cells. Mechanistically, gankyrin was found to interact with the Kelch domain of Keap1 and effectively competed with Nrf2 for Keap1 binding. Increased expression of gankyrin in HCC cells blocked the binding between Nrf2 and Keap1, inhibiting the degradation of Nrf2 by proteasome. Interestingly, accumulation and translocation of Nrf2 increased the transcription of gankyrin through binding to the ARE elements in the promoter of gankyrin. The positive feedback regulation involving gankyrin and Nrf2 modulates a series of antioxidant enzymes, thereby lowering intracellular ROS and conferring a steadier intracellular environment, which prevents mitochondrial damage and cell death induced by excessive oxidative stress. Our results indicate that gankyrin is a regulator of cellular redox homeostasis and provide a link between oxidative stress and the development of HCC.
Hepatocellular carcinoma (HCC) is a complex, heterogeneous tumor with multiple genetic aberrations. Reactive oxygen species (ROS) produce DNA oxidation and subsequent gene mutations that promote carcinogenesis (Storz, 2005). Continuous oxidative stress, which results from the generation of ROS in response to environmental factors or cellular mitochondrial dysfunction, has been associated with modification to key cellular processes, such as cell proliferation, apoptosis, and cell motility cascades, during tumor development (McCord, 2000; Fruehauf and Meyskens, 2007). However, a recent study challenged this concept by providing evidence that ROS are repressed during K-RasG12D–initiated pancreatic and lung tumorigenesis due to a MAPK pathway-mediated increase in Nrf2 transcription (DeNicola et al., 2011). Therefore, we sought to investigate the mechanism by which ROS are regulated during tumorigenesis and tumor progression. The transcription factor NF-E2–related factor 2 (Nrf2) is important for maintaining cellular homeostasis, and when cells are exposed to chemical or oxidative stress, Nrf2 regulates the antioxidant-response element (ARE)–mediated induction of cytoprotective genes (Higgins et al., 2009; Uruno and Motohashi, 2011). Nrf2 also contributes to diverse cellular functions, including differentiation, proliferation, inflammation, and lipid synthesis (Li et al., 2012). The data have increasingly shown that the aberrant expression or function of Nrf2 is associated with pathologies such as cancer, neurodegeneration, and cardiovascular disease. The disruption or alteration of the Keap1–Nrf2 interaction and the persistent activation of Nrf2 are observed in a variety of cancers, such as type-2 papillary renal cell carcinomas, lung cancer, and gallbladder cancer (Singh et al., 2006; Stacy et al., 2006; Shibata et al., 2008; Kim et al., 2010).
Gankyrin, also named 26S proteasome non-ATPase regulatory subunit 10, has been reported to be an oncoprotein that is principally overexpressed in human HCC. Gankyrin directly binds to MDM2 and accelerates the MDM2-dependent ubiquitination and degradation of p53 (Higashitsuji et al., 2005a). It has also been documented that the interaction between gankyrin and CDK4 facilitates Rb degradation (Higashitsuji et al., 2005b). Our most recent data showed that the overexpression of gankyrin accelerates HCC invasion and metastasis. Moreover, knocking down gankyrin in some HCC cells induced cell death (Li et al., 2005a). However, the roles of gankyrin in regulating oxidative stress and in maintaining cell homeostasis remain unclear.
In the present study, we investigated the role of gankyrin in regulating oxidative stress and homeostasis in HCC cells. We show that there is a positive feedback loop between gankyrin and Nrf2 that amplifies the antioxidant capacity of HCC cells, reduces oxidative stress–induced mitochondrial damage, inhibits apoptosis, and promotes the development of HCC.
RESULTS
Gankyrin expression is increased under oxidative stress conditions and participates in the elimination of ROS
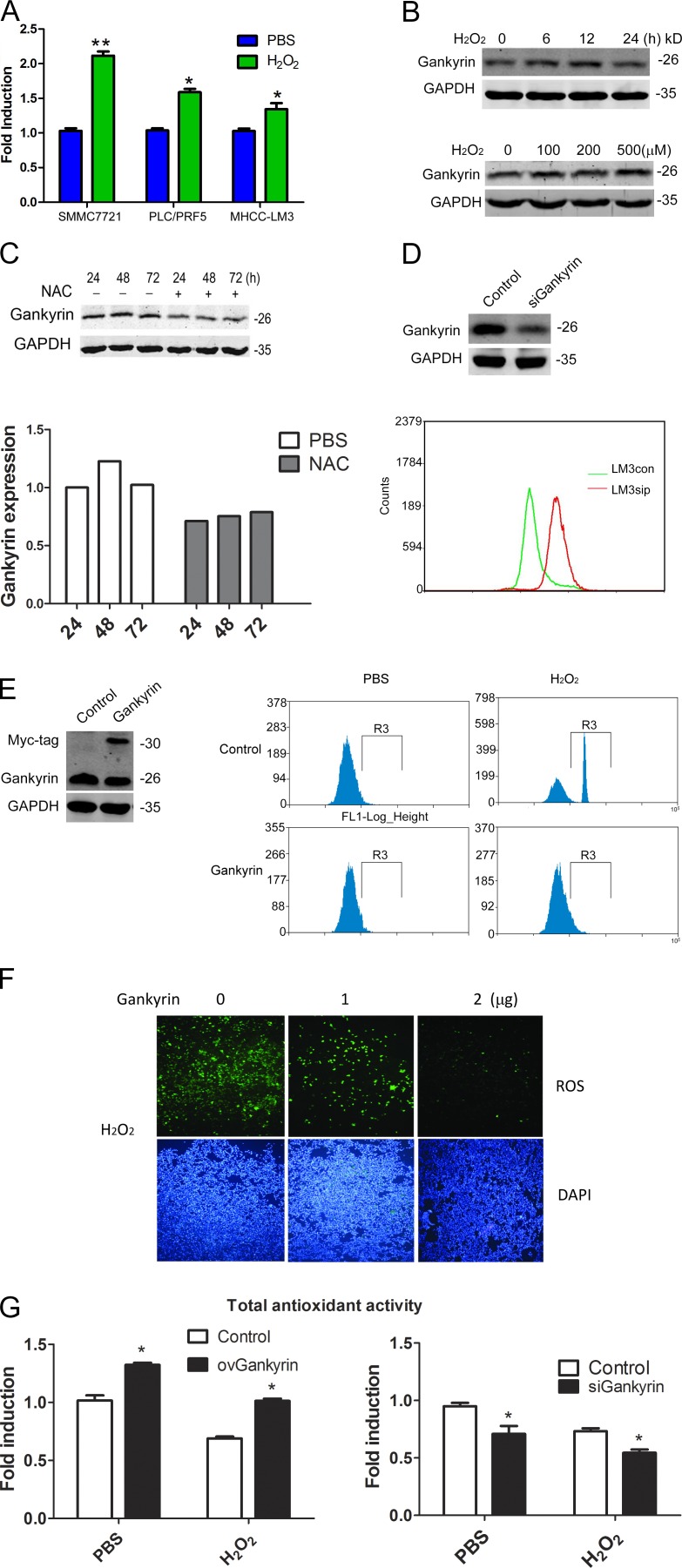
Our quantitative RT-PCR (qRT-PCR) assay revealed that hydrogen peroxide (H2O2) treatment increased the levels of gankyrin mRNA in the HCC cell lines SMMC7721, PLC/PRF/5, and MHCC-LM3 (Fig. 1 A). Western blot analysis also showed that H2O2 increased gankyrin protein levels in a time- and dose-dependent manner (Fig. 1 B). Treatment with the antioxidant N-acetyl cysteine (NAC) reduced gankyrin protein levels in MHCC-LM3 cells (Fig. 1 C). These results suggested that oxidative stress induces gankyrin expression. Next, we measured the levels of ROS in gankyrin overexpressing or depleted HCC cells. The knockdown of gankyrin markedly increased intracellular ROS in MHCC-LM3 cells (Fig. 1 D). Similarly, gankyrin overexpression significantly decreased intracellular ROS levels in SMMC7721 cells after stimulation with H2O2 (Fig. 1, E and F). In accordance with the aforementioned results, gankyrin enhanced the total antioxidant capacity of HCC cells, whereas the knockdown of gankyrin reduced this capacity (Fig. 1 G). Therefore, ROS induced the expression of gankyrin, which, via a feedback mechanism, further modulated ROS levels in HCC cells.
Figure 1.
Gankyrin expression is increased under oxidative stress and participated in elimination of ROS. (A) qRT-PCR analysis of gankyrin expression in SMMC7721, PLC/PRF/5, and MHCCLM3 cells. The data are the mean ± SEM of three independent experiments. (B) Western blot analysis of gankyrin expression at different time points or after different concentrations of H2O2 treatment in MHCCLM3 cells. (C) Western blot of gankyrin expression in MHCC-LM3 cells treated with 100 nM NAC for 24 to 72 h; the protein levels were quantified relative to the loading control. (D) ROS levels were detected in MHCCLM3 gankyrin knockdown and control cells. Cells were treated with PBS or 0.5 mM of H2O2 for 5 h, and the cells were then incubated with CM-H2DCFDA for 30 min. (E) Flow cytometry analysis to detect ROS levels in gankyrin-overexpressing and control SMMC7721 cells. (F) Fluorescence microscopy to detect ROS levels in SMMC7721 cells transiently transfected with gankyrin overexpressing plasmid. Bar, 100 µm. (G) Gankyrin regulated the total antioxidant capacity of HCC cells. SMMC7721 and MHCCLM3 cells with different gankyrin levels were treated with 0.5 mM H2O2 or PBS for 5 h, and the total antioxidant capacity was then measured with a T-AOC Assay kit. The results are the means ± SEM of three independent experiments. Data in B–F are representative of at least three experiments with similar results.*, P < 0.05.
Gankyrin regulates oxidative stress-induced cell death and mitochondrial function in HCC cells
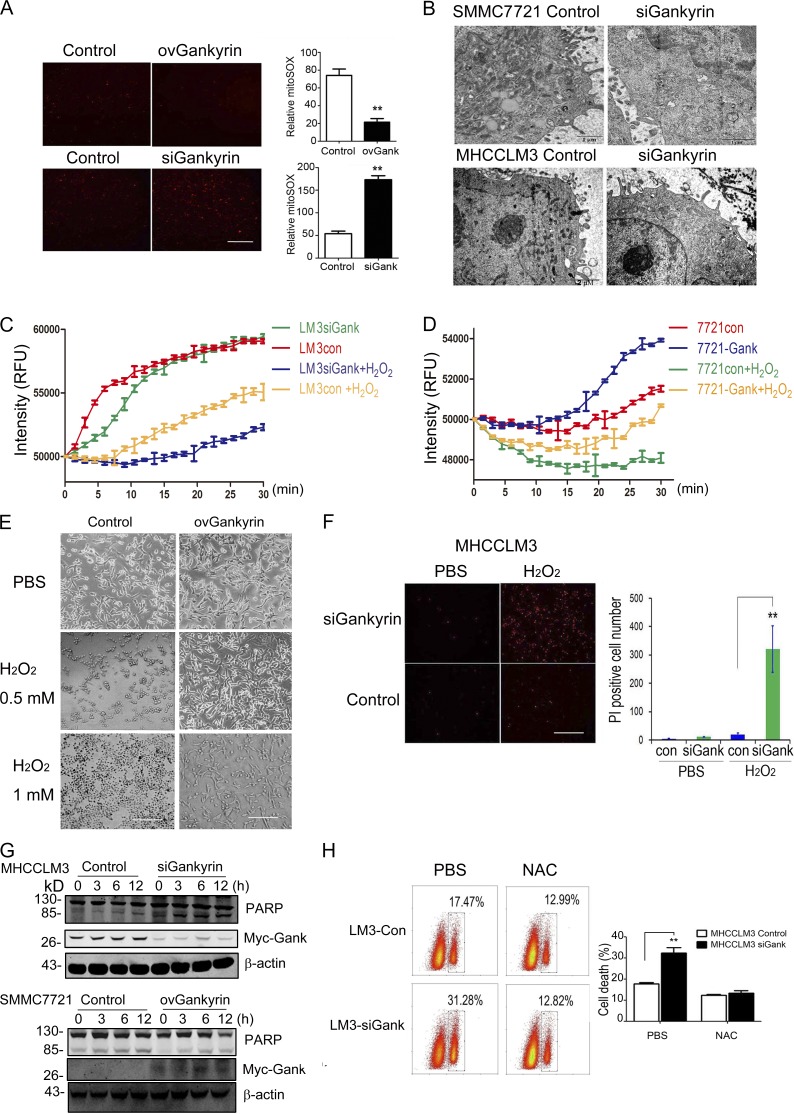
Mitochondria reportedly constitute a major cellular source of ROS, in addition to serving as the primary target of ROS-induced oxidative damage (Girish et al., 2013). In this study, we investigated the correlation between gankyrin expression and mitochondrial function. Mitochondrial-derived superoxide, a byproduct of oxidative phosphorylation, was markedly eliminated by gankyrin (Fig. 2 A). We also examined the effect of gankyrin on mitochondria using an electron microscope assay. In cells in which gankyrin was knocked down, the mitochondria appeared to be swollen and showed a loss of their internal structure and poorly defined, sparse cristae (Fig. 2 B). A mitochondrial function assay demonstrated that the depletion of gankyrin significantly reduced the consumption of O2 by the mitochondria, and that the up-regulation of gankyrin enhanced mitochondrial respiratory function with or without H2O2 stimulation (Fig. 2, C and D). Thus, gankyrin was able to protect the mitochondria from ROS-induced damage and maintain mitochondrial respiratory function.
Figure 2.
Gankyrin influenced oxidative stress–induced mitochondrial dysfunction and cell death in HCC cells. (A) Fluorescence microscopy revealed the MitoSOX levels in SMMC7721-con, SMMC7721-ovGank, MHCCLM3con, and MHCCLM3siGank cells. Cells were stained with MitoSOX Red mitochondrial superoxide indicator and the fluorescence value of MitoSOX was quantified. The data represent the mean ± SEM of three independent experiments. Bar, 100 µm. (B) Representative images of the mitochondria ultrastructure were taken by electron microscopy in SMMC7721-con, SMMC7721-siGank, MHCCLM3con, and MHCCLM3siGank cells. Representative results from three experiments are shown. Bars: 1 µm (SMMC7721); 2 µm (MHCCLM3). (C and D) Mitochondrial O2 consumption assays in MHCCLM3-con, MHCCLM3-siGank cells, SMMC7721-con and SMMC7721-ovGank cells. Cells were treated with PBS or 0.5 mM H2O2 for 5 h. Each data point represents the mean ± SEM of three wells. (E) The overexpression of gankyrin helped SMMC7721 cells gain resistance against oxidative stress–induced death. Cells were incubated with PBS (top), 0.5 mM H2O2 (middle), and 1 mM H2O2 (bottom) for 5 h, and cell death was then observed. Representative results from three experiments are shown. Bar, 100 µm. (F) Suppression of gankyrin-sensitized MHCCLM3 cells to ROS-mediated cell death. MHCCLM3-con and MHCCLM3-siGank cells were treated with H2O2 for 5 h and stained with PI for 20 min, and the number of PI-positive cells was then observed and quantified. The data represent the mean ± SEM of three experiments. Bar, 100 µm. (G) Cleaved PARP was examined in HCC cells with different gankyrin levels upon H2O2 stimulation by Western blotting analysis. Representative results from 3 experiments are shown. (H) NAC treatment attenuated the cell death resulting from gankyrin knockdown. MHCCLM3 cells were transfected with control or gankyrin-siRNA, treated with 100 nM NAC for 48 h, and stained with PI for 15 min. Flow cytometry analysis was performed to evaluate cell apoptosis. Representative results from three experiments are shown. **, P < 0.01.
Severe oxidative stress reportedly leads to progressive cell dysfunction and ultimately to cell death (Fruehauf and Meyskens, 2007). To determine whether gankyrin plays a functional role in oxidative stress-induced cell death, we manipulated gankyrin levels in HCC cells, and then treated those cells with H2O2. The overexpression of gankyrin enabled cells to gain resistance against oxidative stress–induced death (Fig. 2 E), whereas the suppression of gankyrin caused the cells to become more sensitive to ROS-mediated cell death (Fig. 2 F). These findings were further confirmed by a Western blot assay in which cleaved PARP was inhibited by H2O2 treatment in cell lines stably transfected with gankyrin (Fig. 2 G). Moreover, NAC treatment significantly inhibited the cell death that resulted from gankyrin knockdown (Fig. 2 H), suggesting that the antiapoptosis function of gankyrin might partially depend on intracellular ROS levels.
Gankyrin influences the expression of antioxidant enzymes and stabilizes the level of Nrf2 protein
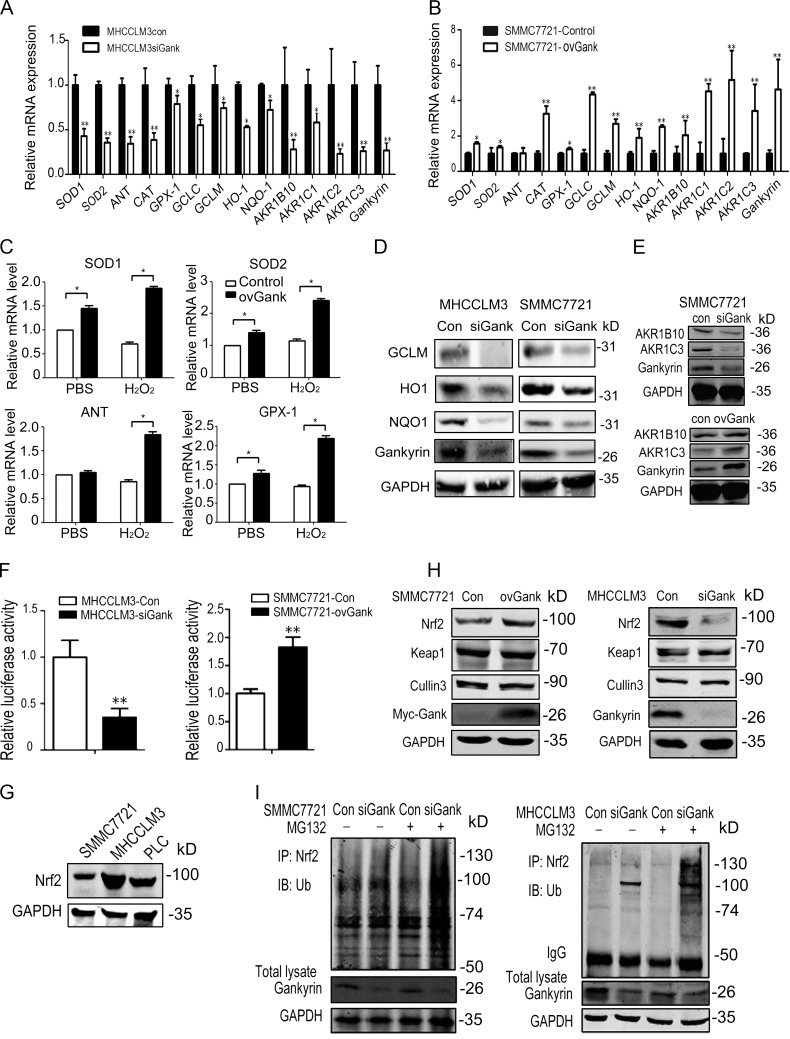
To identify how gankyrin influences intracellular ROS levels, we examined the expression of a series of antioxidant enzymes in cells in which gankyrin was either overexpressed or knocked down. The stable knockdown of gankyrin in MHCC-LM3 and SMMC7721 cells down-regulated the mRNA levels of the following: superoxide dismutase 1 (SOD1); SOD2, mitochondrial (MnSOD); glutathione peroxidase (GPx1); adenine nucleotide translocator (ANT; the key indicator of nitrative stress); catalase (CAT); glutamate-cysteine ligase, catalytic subunit (GCLC); glutamate-cysteine ligase, modifier subunit (GCLM); heme oxygenase (decycling) 1 (HO-1); NAD(P)H:quinone oxidoreductase l (NQO1); aldo-keto reductase family, member B10 (AKR1B10); aldo-keto reductase family, member C1 (AKRC1); aldo-keto reductase family, member C2 (AKR1C2); and aldo-keto reductase family, member C3 (AKR1C3; Fig. 3, A and B). The transient up-regulation of gankyrin in SMMC7721 cells increased the mRNA levels of SOD1, SOD2, GPx1, and ANT in the presence or absence of oxidation stress (Fig. 3 C). Western blot analysis also revealed that the knockdown of gankyrin decreased NQO1, GCLM, and HO-1 (Fig. 3 D). Similarly, we also observed that gankyrin affected AKR1B10 and AKR1C3 protein levels (Fig. 3 E)
Figure 3.
Gankyrin increases the expression of antioxidative enzymes. (A and B) qRT-PCR analysis was performed for SOD1, SOD2, ANT, CAT, Gpx, GCLC, GCLM, HO-1, NQO1, and AKR family members and gankyrin in MHCCLM3 and SMMC7721 control and gankyrin knockdown cells. The data are the mean ± the SEM of three independent experiments. (C) qRT-PCR analysis was performed to detect SOD1, SOD2, ANT, and Gpx expression in SMMC7721 cells that were transiently transfected with gankyrin-pcDNA3.1A vectors with or without H2O2 stimulation. The data are expressed as the mean ± SEM of three independent experiments. (D) Western blot analysis showed the levels of NQO1, HO-1, and GCLM protein in SMMC7721-con, SMMC7721-siGank, MHCCLM3-con, and MHCCLM3-siGank cells. The data shown are representative of three independent experiments. (E) Western blotting analysis showed the levels of AKR1B10 and AKR1C3 protein in gankyrin-knockdown or overexpressing SMMC7721 cells. The data shown are representative of three independent experiments. (F) Gankyrin influenced ARE luciferase reporter activity in HCC cells. MHCCLM3 and SMMC7721 cells with different gankyrin levels were transiently transfected with an ARE luciferase reporter vector or the control plasmid pRL-TK for 48 h. The cells were harvested and gankyrin reporter activities were detected. The results are means ± SEM. n = 3. *, P < 0.05; **, P < 0.01. Representative results from three experiments are shown. (G) Nrf2 protein levels were detected in three HCC cells by Western blot analysis. The data shown are representative of three independent experiments. (H) Nrf2, Keap1, and Cullin3 protein levels in SMMC7721-con, SMMC7721-ovGank cells, MHCCLM3-con, and MHCCLM3-siGank cells were detected by Western blot. The data shown are representative of three independent experiments. (I) The ubiquitination status of Nrf2 in gankyrin-knockdown and control cells. Whole-cell lysates were immunoprecipitated with an anti-Nrf2 antibody or control immunoglobulin G and analyzed by Western blot with antiubiquitin antibody. The data shown are representative of three independent experiments.
As GCLC, GCLM, NQO1, HO-1, AKR1B10, AKR1C1, AKR1C2, and AKR1C3 are transcriptional targets of Nrf2 (MacLeod et al., 2009), we hypothesized that gankyrin might influence the expression of antioxidant stress enzymes under the regulation of ARE elements. To confirm this possibility, we detected the activity of an ARE luciferase reporter in HCC cells, and we found that gankyrin significantly increased luciferase activity, whereas the knockdown of gankyrin significantly inhibited luciferase expression (Fig. 3 F).
Nrf2 protein levels were detected in three HCC cells lines, and the results showed that all had a relatively high level of Nrf2 protein (Fig. 3 G). Gankyrin overexpression markedly stabilized Nrf2 protein levels in SMMC7721 cells, whereas the silencing of endogenous gankyrin decreased Nrf2 expression in MHCC-LM3 cells (Fig. 3 H). The expression levels of Keap1 and Cullin 3, key regulators of Nrf2, were not affected. Because ubiquitination and proteasomal degradation are part of the primary degradation pathway for Nrf2, we proceeded to analyze the ubiquitination status of Nrf2 in gankyrin knockdown and control cells. Nrf2 was efficiently polyubiquitinated in cells in which gankyrin was knocked down compared with control cells (Fig. 3 I).
Gankyrin competitively binds to the Kelch domain of Keap1
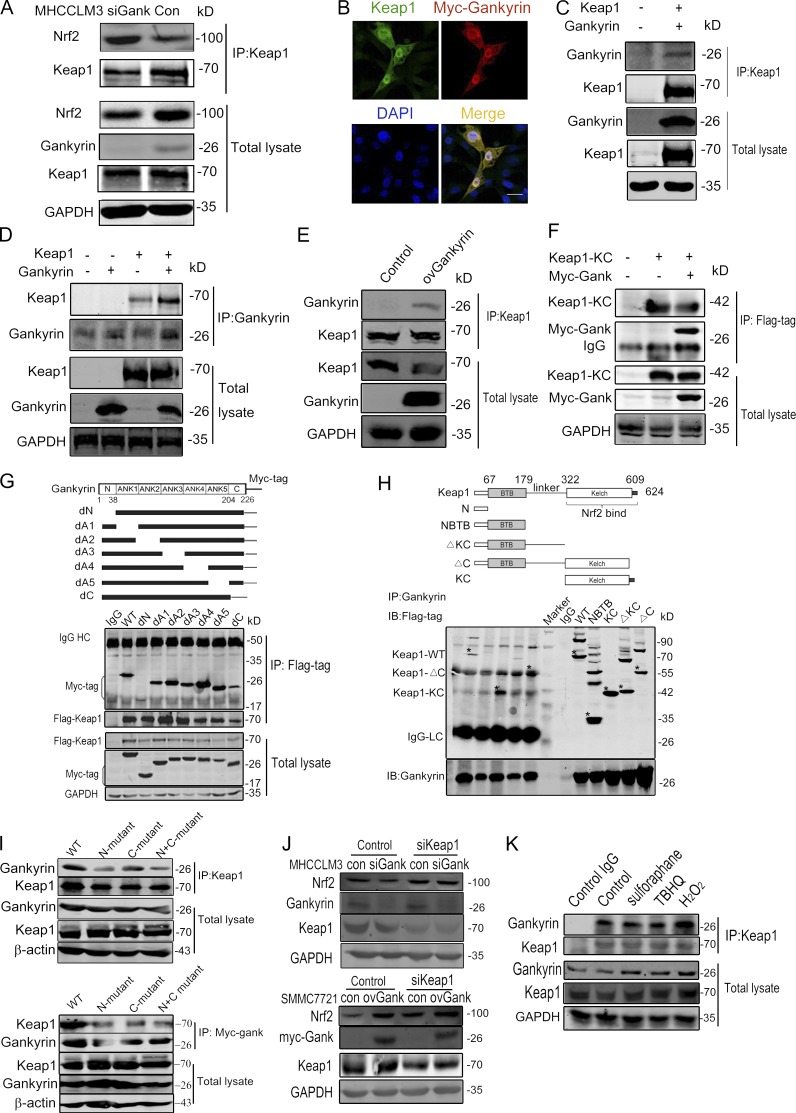
It is well known that Keap1 constitutively ubiquitinates Nrf2, resulting in the rapid degradation of Nrf2 through the proteasomal pathway. Therefore, we investigated whether gankyrin disrupted the association between Keap1 and Nrf2. Keap1 associated with more Nrf2 protein in gankyrin-knockdown cells compared with control cells (Fig. 4 A). Based on an immunofluorescence assay, Keap1 and gankyrin were co-localized throughout the cells (Fig. 4 B), and coimmunoprecipitation clearly showed the presence of gankyrin–Keap1 complexes in HEK293T cells (Fig. 4, C and D). Gankyrin–Keap1 complexes were also detected in gankyrin-overexpressing SMMC7721 cells (Fig. 4 E).
Figure 4.
Gankyrin binds to the Kelch domain of Keap1. (A) Gankyrin influenced the binding of Keap1 to Nrf2. Equal amounts of cell lysates were immunoprecipitated with an anti-Keap1 antibody. Precipitated proteins and cell lysates were blotted with anti-Nrf2, anti-gankyrin, and anti-Keap1 antibodies. (B) Confocal microscopy was performed on HEK293T cells cotransfected with Keap1 and myc-gankyrin. Bar, 10 µm. (C and D) Gankyrin and Keap1 were cotransfected into 293T cells. Whole cell lysates were immunoprecipitated with Keap1- (C) or gankyrin-specific (D) antibodies. Precipitated proteins and cell lysates were blotted with the indicated antibodies. (E) Cell lysates from SMMC7721-con and SMMC7721-ovGank cells were immunoprecipitated with anti-Keap1 antibodies, and Western blot analysis was performed with the indicated antibodies. (F) The interaction of Myc-gankyrin with Flag-tagged truncated Keap1 fragments. The top panel shows a schematic of the truncated Keap1 fragments. HEK293T cells that were cotransfected with myc-gankyrin and Flag-tagged truncated Keap1 fragments were lysed and immunoprecipitated with anti-myc antibody. Precipitates and cell lysates were blotted with anti-Flag or anti-myc antibodies. (G) The interaction of Flag-KC (Kelch domain of Keap1) with Myc-tagged gankyrin. HEK293T cells cotransfected with Flag-KC and Myc-tagged gankyrin were immunoprecipitated with anti-flag antibody and immunoblotted with anti-myc antibodies. (H) The interaction of Flag-Keap1 with Myc-tagged gankyrin mutants. The top panel shows a schematic of the gankyrin mutants. HEK293T cells cotransfected with Flag-Keap1 and myc-tagged deletion mutants of gankyrin were immunoprecipitated with anti-flag antibody. Precipitated proteins and cell lysates were blotted with anti-myc and the indicated antibodies. (I) Wild-type or ExxE motif-mutated gankyrin and Flag-Keap1 plasmids were transfected into HEK293T cells, and the cells were then lysed and immunoprecipitated with anti-myc antibody. Precipitates and cell lysates were blotted with anti-Flag or anti-myc antibody. N-mutated indicated E in aa 21-24 were mutated to A, C-mutated indicated E in aa 201-204 were mutated to A, and N+C-mutated indicated E in aa 21-24 and aa 201-204 were all mutated. (J) The knockdown of Keap1 abolished the regulatory role of gankyrin on Nrf2 protein levels. Negative control oligonucleotides or siRNA targeting Keap1 were transfected into MHCCLM3-Con, -siGank, or SMMC7721-Con, -ovGank cells. Cell lysates were blotted with anti-Nrf2 and other indicated antibodies. (K) A coimmunoprecipitation assay was used to analyze the amount of gankyrin that was associated with Keap1 after stimulation with sulforaphane, tBHQ, or H2O2. SMMC7721 cells were stimulated by sulforaphane, tBHQ, or H2O2 for 12 h, and the cells were then lysed and immunoprecipitated with an anti-Keap1 antibody. Precipitates and cell lysates were blotted with an anti-gankyrin antibody. The data are representative of at least two experiments with similar results.
To determine which region of gankyrin is required for its interaction with Keap1, we used a series of gankyrin deletion mutants. As shown in Fig. 4 H, gankyrin mutants lacking the C-terminal tail domain (aa 204–226) or N-terminal tail domain (aa 1–38) showed weaker interactions with Keap1 compared with other mutants, suggesting that these two regions may be required for binding to the Keap1 protein; the deletion of any single ankyrin repeat did not influence the interaction of gankyrin with Keap1. We further investigated which domain of Keap1 is responsible for binding to gankyrin. A series of truncated Keap1 proteins was expressed in HEK293T cells, along with myc-tagged gankyrin. Only those proteins containing the Keap1-Kelch domain were found to associate with gankyrin (Fig. 4 F), indicating that Keap1 binds to gankyrin through the Kelch domain. To further confirm the association between gankyrin and Keap1, we cotransfected the Keap1-Kelch domain along with gankyrin plasmid into cells and confirmed the interaction between the Kelch domain and gankyrin (Fig. 4 G). Given that the ETGE motif in Nrf2 is a high-affinity binding site for Keap1, it is of considerable interest that human gankyrin contains an ELKE motif between aa 21 and 24 and an ENKE motif between aa 201 and 204. Thus, the Glu residues in the ELKE and the ENKE motifs were mutated, and the interaction between Keap1 and mutated gankyrin was investigated. As shown in Fig. 4 I, Glu residues mutation in the ELKE or ENKE motifs attenuated the interaction between gankyrin and Keap1. The mutation of all Glu residues in the two motifs substantially inhibited the binding of gankyrin to Keap1.
In addition, we knocked down Keap1 to confirm that gankyrin modulates the stability of Nrf2 via Keap1. The repression of Nrf2 after gankyrin knockdown was abolished by the knockdown of Keap1; Nrf2 protein accumulated after Keap1 knockdown regardless of the level of gankyrin protein (Fig. 4 J). We performed a coimmunoprecipitation assay to analyze the amount of gankyrin that binds to Keap1 after stimulation with sulforaphane, tert-butyl hydroquinone (tBHQ; Morimitsu et al., 2002; Li et al., 2005b), or H2O2 (Fig. 4 K). The results showed that sulforaphane and tBHQ did not significantly affect gankyrin binding to Keap1, whereas stimulation with H2O2 increased the binding of Keap1 to gankyrin significantly. In conclusion, gankyrin competitively bound the Kelch domain and inhibited the interaction between Nrf2 and Keap1, which decreased the ubiquitination and subsequent degradation of Nrf2.
Nrf2 promotes gankyrin transcription
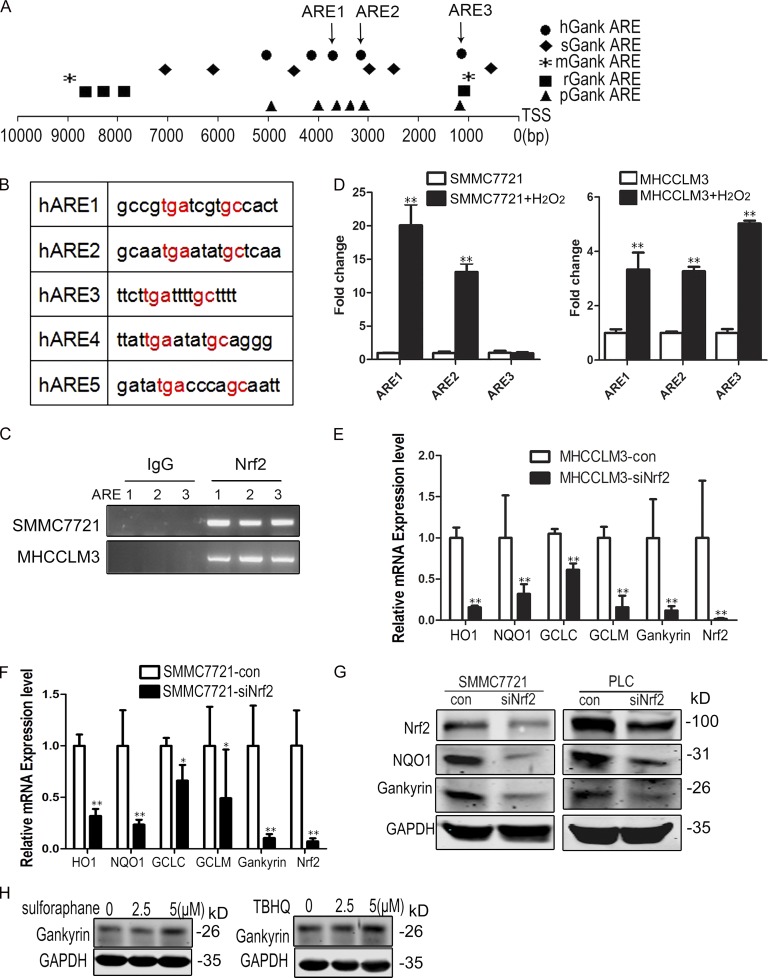
To identify potential transcriptional regulators of gankyrin under conditions of oxidative stress, we analyzed the human gankyrin gene promoter for transcription factor-binding sequences that regulate the oxidative stress response. We found that the region 10 kb upstream from the transcriptional start site contains conserved AREs (5′-RTGAYnnnGCR-3′) that were binding sites for Nrf2 (Itoh et al., 1997; Nguyen et al., 2000). We also analyzed the gankyrin promoter in different species and found that all species analyzed have AREs in the gankyrin promoter (Fig. 5, A and B).
Figure 5.
Nrf2 promotes gankyrin transcription. (A) Schematic representation of gankyrin promoters in humans (Homo species, hGank), pigs (Sus scrofa, sGank), mice (Mus musculus, mGank), rats (Rattus norvegicus, rGank), and chimpanzees (Pan troglodytes, pGank). Different symbols represent Nrf2 binding sites (ARE) in different species. TSS, transcriptional start site. (B) ARE regions and adjacent sequences in the gankyrin promoter. (C) SMMC7721 and MHCC-LM3 cells were fixed and sheered; cross-linked chromatin was prepared as described in the Materials and methods. The chromatin was precipitated using control (IgG) or Nrf2-specific antibodies (Nrf2). PCR analysis was performed using primers for ARE1, ARE2, and ARE3. The data shown are representative of three independent experiments. (D) Oxidative stress increases the binding of Nrf2 to the AREs of the gankyrin promoter. SMMC7721 and MHCCLM3 cells were treated with PBS or 0.5 mM H2O2 for 5 h, and chromatin immunoprecipitation was performed using Nrf2-specific antibodies. DNA isolated from the precipitated materials was analyzed using qPCR with the indicated primers. The ARE-specific signals from Nrf2-precipitated DNA were normalized to those from IgG-precipitated DNA. The data shown are means ± SEM of triplicate wells. (E and F) qRT-PCR analysis was performed for gankyrin and target genes of Nrf2 in PLC/RPF/5-con and PLC/RPF/5-siNrf2 or SMMC7721-con and SMMC7721-siNrf2 cells. Data represent the mean ± SEM of triplicates from an experiment that was repeated a total of three times with similar results. *, P < 0.05; **, P < 0.01. (G) Western blot analysis of gankyrin and NQO1 in SMMC7721-con, SMMC7721-siNrf2, PLC/RPF/5-con, and PLC/RPF/5-siNrf2 cells. Representative results from three experiments are shown. (H) Effects of sulforaphane and tBHQ on gankyrin expression in HCC cells. SMMC7721 cells were treated with 0 to 5 µM sulforaphane and tBHQ for 12 h, and the cells were then lysed and subjected to Western blot analysis. Representative results from three experiments are shown.
The aforementioned results suggested that Nrf2 might regulate gankyrin transcription. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) assays using IgG or Nrf2-specific antibodies in MHCC-LM3 and SMMC7721 cells after 5 h of treatment with or without H2O2, and we found that Nrf2 was able to bind to the ARE regions of gankyrin in SMMC7721 and MHCC-LM3 cells (Fig. 5 C). To determine whether oxidative stress increases the binding of Nrf2 to AREs of the gankyrin promoter, we analyzed DNA isolated from the immunoprecipitated materials by qPCR. H2O2 stimulation significantly increased the binding of Nrf2 to the ARE regions in the gankyrin promoter (Fig. 5 D).
To test whether Nrf2 modulates gankyrin expression, we knocked down Nrf2 levels in PLC/PRF/5 and SMMC7721 cells. The partial depletion of Nrf2 markedly suppressed the expression of its target genes and gankyrin (Fig. 5, E and F). Western blot analysis showed that the protein levels of gankyrin and NQO1 were decreased in Nrf2 knockdown cells (Fig. 5 G). Moreover, sulforaphane and tBHQ, both activators of Nrf2, could moderately induce the expression of gankyrin (Fig. 5 H). All of these results confirmed that gankyrin was a target gene of Nrf2.
Nrf2 and gankyrin cooperatively provide HCC cells with antioxidative stress capacity
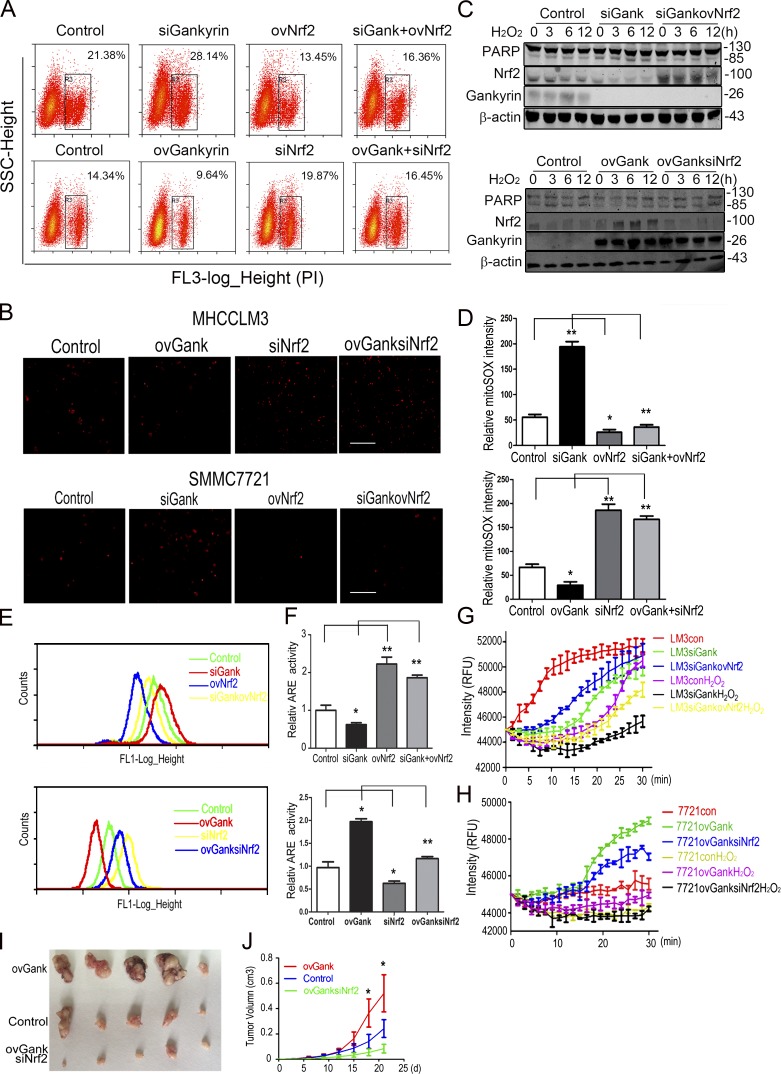
To investigate the role of Nrf2 in the gankyrin-mediated resistance to oxidative stress, we varied the levels of Nrf2 and gankyrin in HCC cells via ectopic shRNA or cDNA, and after 48 h of transfection, the cells were stimulated with 0.5 mM H2O2. Nrf2 knockdown increased the sensitivity of gankyrin-overexpressing cells to H2O2-induced toxicity, whereas the overexpression of Nrf2 helped gankyrin-knockdown cells to regain resistance to H2O2-induced toxicity (Fig. 6 A). However, gankyrin could only partially rescue the sensitivity of cells to H2O2-induced toxicity caused by Nrf2 depletion, whereas the knockdown of gankyrin only slightly reduced the protective effects of Nrf2 on cells under oxidative stress (Fig. 6, A and B). Western blot analysis showed that H2O2-induced PARP cleavage could be decreased in gankyrin-knockdown cells by the overexpression of Nrf2, whereas PARP cleavage was enhanced in gankyrin overexpressing cells in which Nrf2 was knocked down (Fig. 6 C).
Figure 6.
Nrf2 and gankyrin cooperatively provide HCC cells with increased antioxidative stress capacity. (A) MHCCLM3-con, MHCCLM3-siGank, MHCCLM3-ovNrf2, and MHCCLM3-siGankovNrf2 or SMMC7721-con, SMMC7721-ovGank, SMMC7721-siNrf2, and SMMC7721-ovGanksiNrf2 cells were treated with 0.5 mM H2O2 for 5 h and stained with PI for 15 min. Flow cytometry assays were performed to evaluate the level of apoptosis in HCC cells. Representative results from three experiments are shown. (B) Fluorescence microscopy showed the levels of cell death in MHCCLM3-con, MHCCLM3-siGank, MHCCLM3-ovNrf2, MHCCLM3-siGankovNrf2 (top), SMMC7721-con, SMMC7721-ovGank, SMMC7721-siNrf2, and SMMC7721-ovGanksiNrf2 (bottom) cells. Cells were treated with 0.5 mM H2O2 for 5 h and stained with PI for 15 min. Representative results from three experiments are shown. Bar, 100 µM. (C) Levels of cleaved PARP were evaluated in MHCCLM3-con, MHCCLM3-siGank, and MHCCLM3-siGankovNrf2 cells (top) or SMMC7721-con, SMMC7721-ovGank, and SMMC7721-ovGanksiNrf2 cells treated with 0.5 mM H2O2 for the indicated time. Representative results from three experiments are shown. (D) Quantification of the mitoSOX levels in MHCCLM3-con, MHCCLM3-siGank, MHCCLM3-ovNrf2, MHCCLM3-siGankovNrf2, SMMC7721-con, SMMC7721-ovGank, SMMC7721-siNrf2, and SMMC7721-ovGanksiNrf2 cells. The data represent the mean ± SEM of triplicates from an experiment that was repeated a total of three times with similar results. (E) Flow cytometry analyses were performed on MHCCLM3-con, MHCCLM3-siGank, MHCCLM3-ovNrf2, MHCCLM3-siGankovNrf2, and SMMC7721-con, SMMC7721-ovGank, SMMC7721-siNrf2, and SMMC7721-ovGanksiNrf2 cells to detect the levels of ROS. Representative results from three experiments are shown. (F) HCC cells were transiently transfected with an ARE luciferase reporter vector or the control plasmid pRL-TK for 48 h. The cells were then harvested, and the reporter activities were detected. The data represent the mean ± SEM of triplicates from an experiment that was repeated a total of three times with similar results. (G and H) Mitochondrial O2 consumption assays in MHCCLM3-con, MHCCLM3-siGank, MHCCLM3-siGankovNrf2, and SMMC7721-con, SMMC7721-ovGank, and SMMC7721-ovGanksiNrf2 cells. Cells were treated with PBS or 0.5 mM H2O2 for 5 h, and O2 consumption was then examined. Each data point represents the mean ± SEM of triplicates from an experiment that was repeated three times with similar results. (I and J) Tumors were excised from nude mice 25 d after subcutaneous inoculation with SMMC7721-con, SMMC7721-ovGank, or SMMC7721-ovGanksiNrf2 cells. Tumor size was measured once every 3 d, and the overall tumor volume was calculated. The data represent the mean ± SEM. n = 5. *, P < 0.05; **, P < 0.01.
Intracellular ROS and mitochondrial superoxide levels were decreased when Nrf2 was overexpressed in gankyrin-knockdown cells; in contrast, intracellular ROS and mitochondrial superoxide were both increased when the Nrf2 was knocked down in gankyrin-overexpressing cells (Fig. 6, D and E). Consistent with the aforementioned results, the ARE luciferase reporter assay also revealed that knocking down Nrf2 inhibited gankyrin-induced ARE luciferase expression and vice versa. Gankyrin had only a slight or no effect on ARE luciferase activity when Nrf2 was deleted (Fig. 6 F). However, Nrf2 could partially compensate for the effect of gankyrin on the modulation of mitochondrial respiratory function (Fig. 6, G and H).
To examine the effects of gankyrin and Nrf2 on HCC progression, we used a lentivirus to stably knockdown Nrf2 levels in gankyrin-overexpressing HCC cells. These cells were inoculated into nude mice, followed by the periodic monitoring of tumor growth. The results showed that knocking down Nrf2 significantly inhibited the tumorigenicity and subcutaneous xenograft growth of gankyrin-overexpressing cells (Fig. 6, I and J).
Gankyrin and Nrf2 overexpression in HCCs predicts a poor prognosis
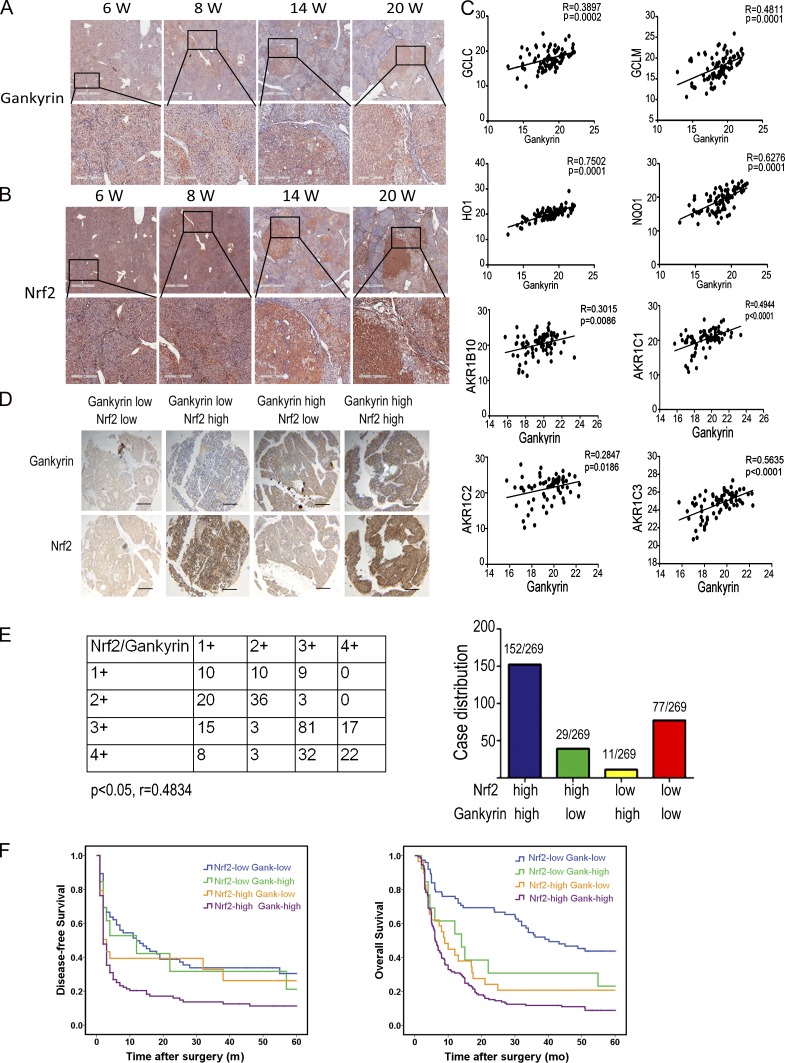
To study the role of the gankyrin-Nrf2 loop in hepatocarcinogenesis, liver samples from diethylnitrosamine (DEN)-treated Wistar rats were subjected to immunohistochemical staining. All mice developed HCC at week 20 after DEN treatment. Intriguingly, the expression of gankyrin and Nrf2 were progressively increased during the process of hepatocarcinogenesis in rats (Fig. 7, A and B). In addition, as shown in the staining results from serial sections, the increase of gankyrin expression was accompanied by an increase of Nrf2 expression in hepatocarcinogenesis. Notably, gankyrin and Nrf2 were primarily expressed in the portal area during the early stages of the DEN-induced hepatocarcinogenesis and subsequently in the carcinoma nests at the advanced stages, which suggested that gankyrin and Nrf2 might cooperatively participate in the malignant transformation from inflammation to cancer.
Figure 7.
Gankyrin and Nrf2 overexpression in HCCs predicts a poor prognosis. (A and B) Livers of Wistar rats euthanized at week 6, 8, 14, or 20 wk after DEN administration were collected and subjected to immunohistochemistry with anti-gankyrin (A) and anti-Nrf2 (B) antibodies. Bars:1 mm (top), 300 µm (bottom); n = 5 for each time point. (C) Gankyrin expression positively correlated with Nrf2 target genes in HCC specimens. The levels of gankyrin and Nrf2 target genes mRNA were detected by qRT-PCR, and the correlations between the mRNA levels of gankyrin and various antioxidative enzymes were evaluated; n = 86. (D) Immunohistochemical staining of gankyrin and Nrf2 protein levels in HCC TMA sections. Representative staining of gankyrin and Nrf2 is shown. Bar, 200 µm. (E) Graphical representation of the distribution of patients according to the staining intensities of gankyrin and Nrf2 in HCCs. (F) Kaplan-Meier curves for time to recurrence and overall survival of patients among the different groups shown in E.
In clinical HCC samples, we determined the levels of gankyrin and Nrf2 target genes mRNAs. There was a positive correlation between the level of gankyrin mRNA and GCLC, GCLM, HO-1, NQO1, and AKR family member mRNA in the HCC samples (Fig. 7 C). We further investigated a possible correlation between gankyrin and Nrf2 in 269 HCC patients using tissue microarrays (TMAs). Based on their tumoral gankyrin and Nrf2 content, patients were classified into the following four groups: group I (n = 77), low gankyrin and low Nrf2; group II (n = 29), low gankyrin and high Nrf2; group III (n = 11), high gankyrin and low Nrf2; and group IV (n = 152), high gankyrin and high Nrf2 (Fig. 7 D). In gankyrin-high HCCs, the percentage of patients with high levels of Nrf2 staining was 93.25%, which is significantly greater than that in the gankyrin-low group (P < 0.05, r = 0.4834), indicating that there is an association between gankyrin and Nrf2 expression (Fig. 7, D and E).
More importantly, differences in disease-free survival and overall survival were significant among the four groups. Patients in group IV had shorter disease-free survival (the median times of group I, II, III, and IV were 12, 12, 3, and 2 mo, respectively) and overall survival (the median times of group I, II, III, and IV were 40, 14, 9, and 6 mo, respectively) compared with the other groups (Fig. 7 F). Thus, gankyrin and Nrf2 expression is a valuable prediction factor for the prognosis of patients with HCC.
DISCUSSION
In this study, we demonstrated that there is a positive feedback regulation between gankyrin and Nrf2, which amplifies the antioxidant capability of HCC cells. Under oxidative stress, increased Nrf2 activity promotes the transcription of gankyrin and up-regulates gankyrin expression. Excess gankyrin competitively interrupted the association between Nrf2 and Keap1 and subsequently stabilized Nrf2 protein. This positive feedback regulation helps combat excessive intracellular ROS, which reduces oxidative stress-induced cell death, maintains mitochondrial stability, and further contributes to the tumorigenesis of HCC. This feedback regulation is similar to the correlation between p62/SQSTM1 and Nrf2, as Nrf2 regulates the expression of p62/SQSTM1 through ARE, and in turn p62/SQSTM1 binds to Keap1 and prevents Nrf2 degradation (Jain et al., 2010). Similar to the p62/SQSTM1 protein, the human gankyrin protein contains an ELKE motif between aa 21–24 and an ENKE motif between aa 201–204, which serves as its binding site to Keap1.
We could not directly measure the initiating event of the feedback loop between gankyrin and Nrf2 because there was not a suitable animal model available in our laboratory. Nonetheless, some studies have shown that HBV/HCV infection, alcohol, and toxic agents are able to activate Nrf2 (Burdette et al., 2010; Ivanov et al., 2011; Wang et al., 2014), whereas gankyrin is gradually increased during the hepatocarcinogenesis process in humans (Jing et al., 2014). However, it has been reported that gankyrin expression does not correlate with HBV infection (Fu et al., 2002; Guichard et al., 2012). Our results show that the expression levels of Nrf2 and gankyrin were increased in a DEN-induced rat HCC model. These data led us to hypothesize that the initiating event is the activation of Nrf2 caused by HBV/HCV infection or liver injury, which is then followed by the induction of gankyrin and other Nrf2-target genes. Increased gankyrin binds to Keap1 and prevents Keap1 from interacting with Nrf2, which results in Nrf2 accumulation. This Nrf2 accumulation then further increases the transcription of gankyrin. However, this hypothesis needs to be confirmed in clinical samples and mouse models.
The dual functions of Nrf2 in tumorigenesis have been well documented. Several studies using Nrf2 knockout mice have shown that Nrf2 protects against chemical carcinogen-induced tumor formation in the stomach, bladder, skin, and colorectum (Kensler et al., 2007; Khor et al., 2008; Marhenke et al., 2008; Jaramillo and Zhang, 2013). The mechanism by which Nrf2 protects against chemical-induced carcinogenesis might be due in part to its ability to reduce the amount of ROS and DNA damage in cells (Hirayama et al., 2003; Morito et al., 2003). Further evidence supporting the protective role of Nrf2 comes from studies of mice or humans harboring a single-nucleotide polymorphism in the promoter region of the Nrf2 gene (Cho et al., 2002; Yamamoto et al., 2004; Marzec et al., 2007). Individuals with this single-nucleotide polymorphism have significantly lower levels of Nrf2 expression and an increased risk for developing nonsmall-cell lung cancer (Suzuki et al., 2013).
Paradoxically, the role of Nrf2 in cancer promotion and in cancer resistance to therapeutic treatment has been revealed (Zhang, 2010). The elevated expression of Nrf2 target genes and the increased stability of Nrf2 in many human cancers are well documented. Mutations of Keap1 or Nrf2, modulations of the Keap1 promoters, posttranslational modulation of Keap1, competitive binding proteins to Keap1, and oncoproteins that induce Nrf2 transcription could all lead to the accumulation of Nrf2 (Zhang et al., 2010; DeNicola et al., 2011; Taguchi et al., 2011, 2012; Hanada et al., 2012; Ichimura et al., 2013). The clinical prognosis of patients with tumors expressing high levels of Nrf2 is poor (Shibata et al., 2008; Solis et al., 2010), partly due to the ability of Nrf2 to enhance cancer cell proliferation and promote chemoresistance and radioresistance. Interestingly, Satoh et al. (2013) have provided evidence that Nrf2 has two roles during carcinogenesis: one of which is preventive during tumor initiation, and a second that promotes malignant progression during lung carcinogenesis through the Kras signaling pathway. Thus, Nrf2 is a double-edged sword for cancers. In the HCC field, the somatic mutation ratios of Nrf2 and Keap1 in HCC patient have been reported to be 6.4 and 8%, respectively, which could contribute to the accumulation of Nrf2 in HCC (Guichard et al., 2012; Cleary et al., 2013). However in the present study, we found that Nrf2 was highly expressed in 67.28% of the 269 HCC tissue samples, which is far higher than the somatic mutation rate. It is reasonable to speculate that there are other factors regulating the accumulation of Nrf2 in HCC.
Gankyrin is a critical oncoprotein that is frequently overexpressed in HCC and other types of cancer (Higashitsuji et al., 2000; Fu et al., 2002). An exciting result from this study is that gankyrin is a new Nrf2 target gene, which indicates that Nrf2 can directly influence HCC tumorigenesis and progression. The region upstream of the transcriptional start site of the gankyrin gene contains several conserved AREs, which are binding sites for Nrf2. Nrf2 regulates the gene expression of a wide variety of cytoprotective phase II detoxification and antioxidant enzymes through AREs, which maintain HCC cell survival under oxidative stress and confer resistance to chemotherapeutic drugs and ionizing radiation (Venugopal and Jaiswal, 1996; Itoh et al., 1997; Homma et al., 2009; Zhang et al., 2010).
Gankyrin, also known as p28, is one of the non-ATPase subunits of PA700 (19S), a regulatory complex of the human 26S proteasome (Hori et al., 1998). Another component of the 26S proteasome with a different function, PA28, is also regulated by Nrf2 (Pickering et al., 2012). PA28 is an interferon-induced 11S complex that associates with the ends of the 20S proteasome and stimulates the in vitro breakdown of small peptide substrates, but not proteins or ubiquitin-conjugated proteins. In cells, PA28 also exists in larger complexes with the 19S particle, which allows for the ATP-dependent degradation of proteins (Davies, 2001; Pickering et al., 2010). PA28 is also induced during adaptation to oxidative stress, contributing to the overall capacity to degrade oxidized proteins and to stress resistance. Our finding that increases in gankyrin expression during adaptation to oxidative stress are largely mediated by the Nrf2 signaling transduction pathway is similar to the mechanism used by PA28. These Nrf2-dependent increases in gankyrin (19S) and PA28 (11S) are important for fully effective adaptive increases in cellular stress resistance.
In addition, some studies have reported that Nrf2 also induces the transcription of transcription factors and oncogenes. Nrf2 induces Klf9 and amplifies oxidative stress (Zucker et al., 2014). In cutaneous squamous cell carcinoma (SCC), Nrf2 directly binds to the promoter of oncogene activating transcription factor 3 (ATF3) and induces its expression, which can then promote the development of SCC (Dziunycz et al., 2014). Here, we showed that Nrf2 binds to the promoter of gankyrin and promotes its transcription, which provides a direct link between oxidative stress and HCC tumorigenesis.
In this context, our study provides a new pathway for Nrf2 accumulation and a direct link between the oxidative stress response and the development of HCC. Based on the results of this study, we propose that inhibiting the gankyrin–Keap1 interaction could be a new promising therapeutic approach against human HCC.
MATERIALS AND METHODS
Cell culture, transfection, and lentivirus infection
The human HCC cell lines SMMC7721 and MHCCLM3 were purchased from Shanghai Cell Bank of Chinese Academy of Sciences. Human embryonic kidney cell line HEK293T was obtained from ATCC. Human PLC/PRF/5 cell lines were obtained from ATCC. All cells were cultured in DMEM media supplemented with 10% fetal bovine serum (Sigma-Aldrich), and maintained at 37°C in an atmosphere of humidified air containing 5% CO2.
Plasmids pCDNA3.1A and pCDNA3.1A-Gankyrin and a series of gankyrin deletion mutants were constructed in our laboratory, as previously described (Chen et al., 2007). A series of truncated Keap1 mutants were bestowed by B. Xia (University of Medicine and Dentistry of New Jersey, New Brunswick, NJ) (Ma et al., 2012). Gankyrin overexpression/knockdown and their control lentivirus/adenovirus were generated as described previously (Li et al., 2005a; Chen et al., 2007). The sequence for knockdown gankyrin: 5′-TTTTGGCCACTGACTGAC-3′. The lentivirus or adenovirus was infected into the HCC cells with Polybrene (8 µg/ml) for 4 h. The original medium was replaced with fresh medium 12 h later.
ARE luciferase reporter vector, siRNA, and vector of Nrf2 and Keap1 were designed and purchased from GeneChem (NM 001145413). siNrf2, 5′-GGCATTTCACTAAACACAA-3′; siKeap1, 5′-CTTAATTCAGCTGAGTGTT-3′. The vectors were transfected into HCC cells with jetPEI DNA transfection reagent (Polyplus). Nrf2 cDNA was cloned from the whole cDNA synthesized by total RNA and constructed into plenti-EF3. The primers are listed as follows: Nrf2-LentiEF3, 5′-CGCGGATCCATGGATTTGATTGACATACTTTGGA-3′ and 5′-GGACTAGTCTAATGATGATGATGATGATGGTTTTTCTTAACATCTGGCTTCTT-3′.
RNA collection, cDNA synthesis, and real-time PCR analysis
Total RNA was extracted from cell lines, fresh-frozen tumor specimens, and healthy control tissues using TRIzol (Invitrogen). cDNA synthesis was performed using random hexamers (Roche) and SuperScriptII reverse transcription (Invitrogen). qRT-PCR was performed using an ABI 7900 Fast Real-Time PCR System (Applied Biosystems) and SYBR Green PCR kit (Takara Bio Inc.).The primer sequences are as follows: 18S, 5′-CGGCTACGACATCCAAGGAA-3′ and 5′-GCTGGAATTAGCGCGGCT-3′; hSOD1, 5′-AGGGCATCATCAATTTCGAGC-3′ and 5′-GCCCACCGTGTTTTCTGGA-3′; hSOD2, 5′-AACCTCAGCCCTAACGGTG-3′ and 5′-AGCAGCAATTTGTAAGTGTCCC-3′; hCatase, 5′-ACTTTGAGGTCACACATGACATT-3′ and 5′-CTGAACCCGATTCTCCAGCA-3′; hGpx-1, 5′-TGCAACCAGTTTGGGCATCA-3′ and 5′-ACCGTTCACCTCGCACTTC-3′; hAnt, 5′-TCCCCACCCAAGCTCTCAA-3′ and 5′-GTCCAGCGGGTAGACAAAGC-3′; gankyrin, 5′-GCCAAGGGTAACTTGAAGATGA-3′ and 5′-TCACAGGCTAAGTGTAGAGGAG-3′; GCLC, 5′-AGAGAAGGGGGAAAGGACAAAC-3′ and 5′-AAGTTATTGTGCAAAGAGCCTGAT-3′; GCLM, 5′-TCAGGGAGTTTCCAGATGTCTTG-3′ and 5′-TGAAGCAATGATCACAGAATCCA-3′; NQO1, 5′-GCAGTTTCTAAGAGCAGAAC-3′ and 5′-GTAGATTAGTCCTCACTCAGCCG-3′; HO-1, 5′-GGGCTAGCATGCGAAGTGAG-3′ and 5′-AGACTCCGCCCTAAGGGTTC-3′; AKR1B10, 5′-AGCAGGACGTGAGACTTCTACCTGC-3′ and 5′-TCCACCGATGGCATTACCTTTA-3′; AKR1C1, 5′-GTAAGAAACGGTTGAACTGG-3′ and 5′-AAATCCCAGGACAGGCATGA-3′; AKR1C2, 5′-TCACATGCCATTGGTTAACC-3′ and 5′-ACCCGGCTTCTATTGCCAAT-3′; AKR1C3, 5′-GTTGCCTATAGTGCTCTGGGATCT-3′ and 5′-GGACTGGGTCCTCCAAGAGG-3′.
ChIP assay
ChIP assays using an anti-Nrf2 antibody (Proteintech) were performed following a previously described procedure (Zhang et al., 2006). The primers used in the ChIP assay are described below. PCR assays were conducted in triplicate for each sample, and all experiments were repeated at least three times. Primers Used for ChIP Assays are listed as follows: gankyrinARE1, 5′-AAATTGGCTGAGTGTGTTGGTG-3′ and 5′-AGTCCACTAGGAGGGTTTCACG-3′; gankyrinARE2, 5′-CCAAACTTTCAATTGGAAGTGATTA-3′ and 5′-TGGTCAGGTATTTTGTAGAAAACCC-3′; gankyrinARE3, 5′-GTTGAAATTTGTTTTCTCTTTTGTCAT-3′ and 5′-GCCACAACTAGGTAACGATAAGAATAC-3′.
Immunoblotting, coimmunoprecipitation assays, antibodies, and chemicals
Whole-cell extracts or HCC tumor specimens were prepared in RIPA buffer and centrifuged at 12,000 g for 15 min. Protein concentrations were measured using the bicinchoninic acid assay. Immunoblotting was performed using specific primary antibodies, and immune complexes were incubated with the fluorescein-conjugated secondary antibody, and then detected using Odyssey fluorescence scanner (Li-Cor).
For coimmunoprecipitation experiments, cell lysates were prepared in RIPA buffer and protein concentrations were measured and incubated with 2 µg anti-gankyrin, anti-Keap1, anti-myc, or anti-Flag or normal mouse immunoglobulin G (Santa Cruz Biotechnology, Inc.) for 8 h at 4°C, followed by addition of Protein A/G Plus-Agarose (Santa Cruz Biotechnology, Inc.) for another 2 h. The samples from these assays were analyzed by Western blotting.
Anti-PARP and anti-Keap1 were purchased from Cell Signaling Technology. Anti-gankyrin, anti–β-actin, and anti-GAPDH were purchased from Santa Cruz Biotechnology, Inc. Anti-NQO1, anti-GCLM, and anti–HO-1 were purchase from Abclonal Inc. Anti-Nrf2 was purchased from Proteintech Inc.
Total antioxidant capacity assay
Cells were cultured in DMEM and stimulated with PBS or 0.5 mM H2O2 for 5 h. Total Antioxidant Capacity were measured with the T-AOC Assay kit (Total Antioxidant Capacity Assay kit with a Rapid ABTS method) purchased from Beyotime.
Assessment of cell death
Cells were cultured in DMEM and stimulated with PBS or 0.5 mM H2O2 for 5 h. To assess the level of cleaved PARP, the cells were harvested and analyzed by Western blotting. Cells were incubated with PI (P4170; Sigma-Aldrich) for a 30-min incubation period, washed, and resuspended in PBS. Cells were then analyzed by the flow cytometry performed with MoFlo XDP Cell Sorter (Beckman Coulter) and fluorescence microscopy (Leica Biosystems).
Transmission electron microscopy
Cells were fixed in ice-cold 2% glutaraldehyde and examined with a JEOL transmission electron microscope (JEM-1230) as described previously (Tang et al., 2009).
Mitochondrial O2 consumption assay
The assay was performed with Mito-ID O2 Extracellular Sensor kit (ENZ-51044-K; Enzo Life Sciences).
Tumor xenograft experiment
Approximately 1 × 107 SMMC7721-control, SMMC7721-ovGank, and SMMC7721-ovGanksiNrf2 cells in 0.2 ml PBS were injected subcutaneously into the right flank of the mice, which were then observed for signs of tumor development every 3 d. Once the subcutaneous tumor reached 1–1.5 cm diam, the tumor was harvested. Tumor volume was calculated as follows: V (cm3) = width2 (cm2) × length (cm)/2. All experiments were performed with at least five mice in each group, and all of the experiments were repeated three times.
Patients, specimens, and tissue microarrays
We recruited 269 patients with HCC to a training cohort, from the Eastern Hepatobiliary Surgery Hospital (Second Military Medical University, Shanghai, China) from January 2003 to January 2005. Patients following the inclusion also had available paraffin-embedded tumor tissues underwent TMA analysis: preoperative World Health Organization performance status of 0–1; Child-Pugh class A; no distant metastases, visualizable ascites, or encephalopathy; no chemotherapy or radiotherapy before surgery; curative resection; and resected lesions identified as HCC on pathological examination. Patients were excluded because of hepatic angiography after the operation indicating tumor straining and therapeutic TACE was performed. Curative resection of HCC was performed. The study was approved by the institutional ethics committee. Informed consent was obtained before surgery.
Immunohistochemistry
After screening hematoxylin and eosin-stained slides for optimal tumor content, we constructed TMA slides (Shanghai Biochip Company, Ltd.). Two cores were taken from each formalin-fixed, paraffin-embedded HCC sample and normal liver sample by using punch cores that measured 0.8 mm in greatest dimension from the center of tumor foci. Immunohistochemistry was performed as previously described (Dong et al., 2011).
Measurement of ROS levels
Cells were cultured in DMEM and stimulated with PBS or 0.5 mM H2O2 for 5 h. Then cells were incubated with 5 mM CM-H2DCFDA (Molecular Probes) or MitoSOX Red mitochondrial superoxide indicator (Invitrogen) for a 30-min incubation period. The cells were analyzed by the flow cytometry performed with MoFlo XDP Cell Sorter (Beckman Coulter) and confocal microscopy (Leica Biosystems).
Statistical analysis
The Pearson χ2 test or Fisher’s exact test was used to analyze qualitative variables. Statistical analysis was performed by the Student's t test, paired Wilcoxon signed-rank test, or Spearman rank correlation test, using the statistical software GraphPad Prism 4 (GraphPad Software). Kaplan–Meier analysis was used to determine survival. SPSS 15.0 software (SPSS Inc.) was also used for statistical analyses. The data shown represent mean values of at least three independent experiments and are expressed as mean ± SEM. Statistical significance was set at P < 0.05.
ACKNOWLEDGMENTS
We thank Dr. Bing Xia (University of Medicine and Dentistry of New Jersey, New Brunswick, NJ) for kindly providing some of the plasmids.
Research was supported by grants from the Funds for Creative Research Groups of China (81221061), the State Key Project for Liver Cancer (2013ZX10002, 2012ZX10002), Program of Shanghai Municipal Commision of Health (XBR2013090), Program of Shanghai Subject Chief Scientist (14XD1400100), and the National Natural Science Foundation of China (81370066, 81472591, and 91229205).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ANT
- adenine nucleotide translocator
- ARE
- antioxidant-response element
- GCLC
- glutamate-cysteine ligase, catalytic subunit
- GCLM
- glutamate-cysteine ligase, modifier subunit
- HCC
- hepatocellular carcinoma
- HO-1
- heme oxygenase 1
- NAC
- N-acetyl cysteine
- NQO1
- NAD(P)H:quinone oxidoreductase l
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- tBHQ
- tert-butyl hydroquinone
- TMA
- tissue microarray
References
- Burdette D., Olivarez M., and Waris G.. 2010. Activation of transcription factor Nrf2 by hepatitis C virus induces the cell-survival pathway. J. Gen. Virol. 91:681–690. 10.1099/vir.0.014340-0 [DOI] [PubMed] [Google Scholar]
- Chen Y., Li H.H., Fu J., Wang X.F., Ren Y.B., Dong L.W., Tang S.H., Liu S.Q., Wu M.C., and Wang H.Y.. 2007. Oncoprotein p28 GANK binds to RelA and retains NF-κB in the cytoplasm through nuclear export. Cell Res. 17:1020–1029. 10.1038/cr.2007.99 [DOI] [PubMed] [Google Scholar]
- Cho H.Y., Jedlicka A.E., Reddy S.P., Kensler T.W., Yamamoto M., Zhang L.Y., and Kleeberger S.R.. 2002. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26:175–182. 10.1165/ajrcmb.26.2.4501 [DOI] [PubMed] [Google Scholar]
- Cleary S.P., Jeck W.R., Zhao X., Chen K., Selitsky S.R., Savich G.L., Tan T.X., Wu M.C., Getz G., Lawrence M.S., et al. 2013. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 58:1693–1702. 10.1002/hep.26540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.J. 2001. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 83:301–310. 10.1016/S0300-9084(01)01250-0 [DOI] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. 2011. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 475:106–109. 10.1038/nature10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L.W., Hou Y.J., Tan Y.X., Tang L., Pan Y.F., Wang M., and Wang H.Y.. 2011. Prognostic significance of Beclin 1 in intrahepatic cholangiocellular carcinoma. Autophagy. 7:1222–1229. 10.4161/auto.7.10.16610 [DOI] [PubMed] [Google Scholar]
- Dziunycz P.J., Lefort K., Wu X., Freiberger S.N., Neu J., Djerbi N., Iotzowa-Weiss G., French L.E., Dotto G.P., and Hofbauer G.F.. 2014. The oncogene ATF3 is potentiated by cyclosporine A and ultraviolet light A. J. Invest. Dermatol. 134:1998–2004. 10.1038/jid.2014.77 [DOI] [PubMed] [Google Scholar]
- Fruehauf J.P., and Meyskens F.L. Jr. 2007. Reactive oxygen species: a breath of life or death? Clin. Cancer Res. 13:789–794. 10.1158/1078-0432.CCR-06-2082 [DOI] [PubMed] [Google Scholar]
- Fu X.Y., Wang H.Y., Tan L., Liu S.Q., Cao H.F., and Wu M.C.. 2002. Overexpression of p28/gankyrin in human hepatocellular carcinoma and its clinical significance. World J. Gastroenterol. 8:638–643. 10.3748/wjg.v8.i4.638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girish K.S., Paul M., Thushara R.M., Hemshekhar M., Shanmuga Sundaram M., Rangappa K.S., and Kemparaju K.. 2013. Melatonin elevates apoptosis in human platelets via ROS mediated mitochondrial damage. Biochem. Biophys. Res. Commun. 438:198–204. 10.1016/j.bbrc.2013.07.053 [DOI] [PubMed] [Google Scholar]
- Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B., Calderaro J., Bioulac-Sage P., Letexier M., Degos F., et al. 2012. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44:694–698. 10.1038/ng.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada N., Takahata T., Zhou Q., Ye X., Sun R., Itoh J., Ishiguro A., Kijima H., Mimura J., Itoh K., et al. 2012. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 12:66 10.1186/1471-2407-12-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashitsuji H., Itoh K., Nagao T., Dawson S., Nonoguchi K., Kido T., Mayer R.J., Arii S., and Fujita J.. 2000. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 6:96–99. 10.1038/71600 [DOI] [PubMed] [Google Scholar]
- Higashitsuji H., Higashitsuji H., Itoh K., Sakurai T., Nagao T., Sumitomo Y., Masuda T., Dawson S., Shimada Y., Mayer R.J., and Fujita J.. 2005a The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 8:75–87. 10.1016/j.ccr.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Higashitsuji H., Liu Y., Mayer R.J., and Fujita J.. 2005b The oncoprotein gankyrin negatively regulates both p53 and RB by enhancing proteasomal degradation. Cell Cycle. 4:1335–1337. 10.4161/cc.4.10.2107 [DOI] [PubMed] [Google Scholar]
- Higgins L.G., Kelleher M.O., Eggleston I.M., Itoh K., Yamamoto M., and Hayes J.D.. 2009. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol. Appl. Pharmacol. 237:267–280. 10.1016/j.taap.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Hirayama A., Yoh K., Nagase S., Ueda A., Itoh K., Morito N., Hirayama K., Takahashi S., Yamamoto M., and Koyama A.. 2003. EPR imaging of reducing activity in Nrf2 transcriptional factor-deficient mice. Free Radic. Biol. Med. 34:1236–1242. 10.1016/S0891-5849(03)00073-X [DOI] [PubMed] [Google Scholar]
- Homma T., Hamaoka T., Murase N., Osada T., Murakami M., Kurosawa Y., Kitahara A., Ichimura S., Yashiro K., and Katsumura T.. 2009. Low-volume muscle endurance training prevents decrease in muscle oxidative and endurance function during 21-day forearm immobilization. Acta Physiol. (Oxf.). 197:313–320. 10.1111/j.1748-1716.2009.02003.x [DOI] [PubMed] [Google Scholar]
- Hori T., Kato S., Saeki M., DeMartino G.N., Slaughter C.A., Takeuchi J., Toh-e A., and Tanaka K.. 1998. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene. 216:113–122. 10.1016/S0378-1119(98)00309-6 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., et al. 2013. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 51:618–631. 10.1016/j.molcel.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313–322. 10.1006/bbrc.1997.6943 [DOI] [PubMed] [Google Scholar]
- Ivanov A.V., Smirnova O.A., Ivanova O.N., Masalova O.V., Kochetkov S.N., and Isaguliants M.G.. 2011. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS One. 6:e24957 10.1371/journal.pone.0024957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., and Johansen T.. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285:22576–22591. 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M.C., and Zhang D.D.. 2013. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27:2179–2191. 10.1101/gad.225680.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H., Zhang G., Meng L., Meng Q., Mo H., and Tai Y.. 2014. Gradually elevated expression of Gankyrin during human hepatocarcinogenesis and its clinicopathological significance. Sci. Rep. 4:5503 10.1038/srep05503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler T.W., Wakabayashi N., and Biswal S.. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47:89–116. 10.1146/annurev.pharmtox.46.120604.141046 [DOI] [PubMed] [Google Scholar]
- Khor T.O., Huang M.T., Prawan A., Liu Y., Hao X., Yu S., Cheung W.K., Chan J.Y., Reddy B.S., Yang C.S., and Kong A.N.. 2008. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev. Res. (Phila.). 1:187–191. 10.1158/1940-6207.CAPR-08-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.R., Oh J.E., Kim M.S., Kang M.R., Park S.W., Han J.Y., Eom H.S., Yoo N.J., and Lee S.H.. 2010. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 220:446–451. 10.1002/path.2653 [DOI] [PubMed] [Google Scholar]
- Li H., Fu X., Chen Y., Hong Y., Tan Y., Cao H., Wu M., and Wang H.. 2005a Use of adenovirus-delivered siRNA to target oncoprotein p28GANK in hepatocellular carcinoma. Gastroenterology. 128:2029–2041. 10.1053/j.gastro.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Li J., Johnson D., Calkins M., Wright L., Svendsen C., and Johnson J.. 2005b Stabilization of Nrf2 by tBHQ confers protection against oxidative stress-induced cell death in human neural stem cells. Toxicol. Sci. 83:313–328. [DOI] [PubMed] [Google Scholar]
- Li Y., Paonessa J.D., and Zhang Y.. 2012. Mechanism of chemical activation of Nrf2. PLoS One. 7:e35122 10.1371/journal.pone.0035122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Cai H., Wu T., Sobhian B., Huo Y., Alcivar A., Mehta M., Cheung K.L., Ganesan S., Kong A.N., et al. 2012. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol. Cell. Biol. 32:1506–1517. 10.1128/MCB.06271-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A.K., McMahon M., Plummer S.M., Higgins L.G., Penning T.M., Igarashi K., and Hayes J.D.. 2009. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 30:1571–1580. 10.1093/carcin/bgp176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhenke S., Lamlé J., Buitrago-Molina L.E., Cañón J.M., Geffers R., Finegold M., Sporn M., Yamamoto M., Manns M.P., Grompe M., and Vogel A.. 2008. Activation of nuclear factor E2-related factor 2 in hereditary tyrosinemia type 1 and its role in survival and tumor development. Hepatology. 48:487–496. 10.1002/hep.22391 [DOI] [PubMed] [Google Scholar]
- Marzec J.M., Christie J.D., Reddy S.P., Jedlicka A.E., Vuong H., Lanken P.N., Aplenc R., Yamamoto T., Yamamoto M., Cho H.Y., and Kleeberger S.R.. 2007. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 21:2237–2246. 10.1096/fj.06-7759com [DOI] [PubMed] [Google Scholar]
- McCord J.M. 2000. The evolution of free radicals and oxidative stress. Am. J. Med. 108:652–659. 10.1016/S0002-9343(00)00412-5 [DOI] [PubMed] [Google Scholar]
- Morimitsu Y., Nakagawa Y., Hayashi K., Fujii H., Kumagai T., Nakamura Y., Osawa T., Horio F., Itoh K., Iida K., et al. 2002. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J. Biol. Chem. 277:3456–3463. 10.1074/jbc.M110244200 [DOI] [PubMed] [Google Scholar]
- Morito N., Yoh K., Itoh K., Hirayama A., Koyama A., Yamamoto M., and Takahashi S.. 2003. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 22:9275–9281. 10.1038/sj.onc.1207024 [DOI] [PubMed] [Google Scholar]
- Nguyen T., Huang H.C., and Pickett C.B.. 2000. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 275:15466–15473. 10.1074/jbc.M000361200 [DOI] [PubMed] [Google Scholar]
- Pickering A.M., Koop A.L., Teoh C.Y., Ermak G., Grune T., and Davies K.J.. 2010. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 432:585–594. 10.1042/BJ20100878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering A.M., Linder R.A., Zhang H., Forman H.J., and Davies K.J.. 2012. Nrf2-dependent induction of proteasome and Pa28αβ regulator are required for adaptation to oxidative stress. J. Biol. Chem. 287:10021–10031. 10.1074/jbc.M111.277145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H., Moriguchi T., Takai J., Ebina M., and Yamamoto M.. 2013. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res. 73:4158–4168. 10.1158/0008-5472.CAN-12-4499 [DOI] [PubMed] [Google Scholar]
- Shibata T., Kokubu A., Gotoh M., Ojima H., Ohta T., Yamamoto M., and Hirohashi S.. 2008. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 135:1358–1368: 1368.e1–1368.e4. [DOI] [PubMed] [Google Scholar]
- Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., et al. 2006. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3:e420 10.1371/journal.pmed.0030420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis L.M., Behrens C., Dong W., Suraokar M., Ozburn N.C., Moran C.A., Corvalan A.H., Biswal S., Swisher S.G., Bekele B.N., et al. 2010. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin. Cancer Res. 16:3743–3753. 10.1158/1078-0432.CCR-09-3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy D.R., Ely K., Massion P.P., Yarbrough W.G., Hallahan D.E., Sekhar K.R., and Freeman M.L.. 2006. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 28:813–818. 10.1002/hed.20430 [DOI] [PubMed] [Google Scholar]
- Storz P. 2005. Reactive oxygen species in tumor progression. Front. Biosci. 10:1881–1896. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Shibata T., Takaya K., Shiraishi K., Kohno T., Kunitoh H., Tsuta K., Furuta K., Goto K., Hosoda F., et al. 2013. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol. Cell. Biol. 33:2402–2412. 10.1128/MCB.00065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K., Motohashi H., and Yamamoto M.. 2011. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 16:123–140. 10.1111/j.1365-2443.2010.01473.x [DOI] [PubMed] [Google Scholar]
- Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., and Yamamoto M.. 2012. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. USA. 109:13561–13566. 10.1073/pnas.1121572109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Da L., Mao Y., Li Y., Li D., Xu Z., Li F., Wang Y., Tiollais P., Li T., and Zhao M.. 2009. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 49:60–71. 10.1002/hep.22581 [DOI] [PubMed] [Google Scholar]
- Uruno A., and Motohashi H.. 2011. The Keap1-Nrf2 system as an in vivo sensor for electrophiles. Nitric Oxide. 25:153–160. 10.1016/j.niox.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Venugopal R., and Jaiswal A.K.. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA. 93:14960–14965. 10.1073/pnas.93.25.14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kou Y., Wang X., Cederbaum A., and Wang R.. 2014. Multifactorial comparative proteomic study of cytochrome P450 2E1 function in chronic alcohol administration. PLoS One. 9:e92504 10.1371/journal.pone.0092504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Yoh K., Kobayashi A., Ishii Y., Kure S., Koyama A., Sakamoto T., Sekizawa K., Motohashi H., and Yamamoto M.. 2004. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem. Biophys. Res. Commun. 321:72–79. 10.1016/j.bbrc.2004.06.112 [DOI] [PubMed] [Google Scholar]
- Zhang D.D. 2010. The Nrf2-Keap1-ARE signaling pathway: The regulation and dual function of Nrf2 in cancer. Antioxid. Redox Signal. 13:1623–1626. 10.1089/ars.2010.3301 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ohta T., Maruyama A., Hosoya T., Nishikawa K., Maher J.M., Shibahara S., Itoh K., and Yamamoto M.. 2006. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol. Cell. Biol. 26:7942–7952. 10.1128/MCB.00700-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Singh A., Yegnasubramanian S., Esopi D., Kombairaju P., Bodas M., Wu H., Bova S.G., and Biswal S.. 2010. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther. 9:336–346. 10.1158/1535-7163.MCT-09-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker S.N., Fink E.E., Bagati A., Mannava S., Bianchi-Smiraglia A., Bogner P.N., Wawrzyniak J.A., Foley C., Leonova K.I., Grimm M.J., et al. 2014. Nrf2 amplifies oxidative stress via induction of Klf9. Mol. Cell. 53:916–928. 10.1016/j.molcel.2014.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]