Russell et al. show that activation of Mycobacterium tuberculosis–infected macrophages in vitro and in vivo enhances drug tolerance and renders the bacilli more refractory to drug-dependent killing.
Abstract
Successful chemotherapy against Mycobacterium tuberculosis (Mtb) must eradicate the bacterium within the context of its host cell. However, our understanding of the impact of this environment on antimycobacterial drug action remains incomplete. Intriguingly, we find that Mtb in myeloid cells isolated from the lungs of experimentally infected mice exhibit tolerance to both isoniazid and rifampin to a degree proportional to the activation status of the host cells. These data are confirmed by in vitro infections of resting versus activated macrophages where cytokine-mediated activation renders Mtb tolerant to four frontline drugs. Transcriptional analysis of intracellular Mtb exposed to drugs identified a set of genes common to all four drugs. The data imply a causal linkage between a loss of fitness caused by drug action and Mtb’s sensitivity to host-derived stresses. Interestingly, the environmental context exerts a more dominant impact on Mtb gene expression than the pressure on the drugs’ primary targets. Mtb’s stress responses to drugs resemble those mobilized after cytokine activation of the host cell. Although host-derived stresses are antimicrobial in nature, they negatively affect drug efficacy. Together, our findings demonstrate that the macrophage environment dominates Mtb’s response to drug pressure and suggest novel routes for future drug discovery programs.
Mycobacterium tuberculosis (Mtb) is one of the leading infectious disease killers worldwide, claiming nearly 1.5 million lives every year. Although frontline drugs are effective in killing Mtb in vitro, successful chemotherapy in vivo requires protracted adherence to the drug regimen. The reasons for this lengthy treatment appear to be multiple and complex and reflect the heterogeneity of both the host environment and the bacterial population.
Early studies used transcriptome analyses of Mtb’s response to drugs as a tool to facilitate the drug discovery process. Using a differential mRNA amplification method, Alland et al. (1998) delineated Mtb’s transcriptional response to isoniazid (INH) in broth culture. After this study, Wilson et al. (1999) were the first to use microarray techniques to examine gene responses in Mtb during drug treatment. This study demonstrated that the frontline drug INH induced the expression of multiple genes in the fatty acid synthase II (FAS-II) pathway, which is a known target of this drug (Vilchèze and Jacobs, 2007). After this study, Betts et al. (2003) demonstrated that transcriptional changes in response to drug treatment could be used to differentiate drugs with closely related modes of action. Boshoff et al. (2004) performed a comprehensive transcriptome study to measure the effects of 75 antimycobacterial agents and demonstrated the usefulness of gene expression profiling for determining the mechanisms of action of unknown inhibitors. In support of this theme, we recently showed that information obtained from transcriptional analysis was invaluable in the identification of the targets of inhibition of novel Mtb metabolism inhibitors identified in a large-scale chemical screen for compounds active against intracellular Mtb (VanderVen et al., 2015).
These studies have been instrumental in revealing a given drug’s mechanism of action at the whole-genome level and for providing insights into the mechanisms Mtb uses to counteract the detrimental effects of the drug. However, as they were performed in liquid medium, these studies were not designed to probe the possible impact the host environment may have on drug action. During infection, the host cell imposes multiple pressures on intracellular Mtb, such as acidic stress, restricted nutrient availability, microbicidal peptides, and toxic reactive nitrogen and oxygen species. These host-derived pressures place constraints on Mtb’s physiology and likely influence how Mtb responds to drug pressure. Several decades ago, McCune and Tompsett (1956) and McCune et al. (1956) described the persistence of drug-susceptible Mtb in animal models. Clinical studies have also linked the immune status of the human host to a reduction in the sensitivity of Mtb to frontline drugs (Wallis et al., 1999, 2012). Furthermore, the impact of host stresses is dynamic because the nature and intensity of these host-derived pressures are constantly changing and are linked to the immune status of the host during the course of infection (Aldridge et al., 2014). There is a considerable body of work that links bacterial stress response pathways to the acquisition of drug tolerance (Warner and Mizrahi, 2006; Deb et al., 2009; Baek et al., 2011), but the significance of these data within the host immune environment remains to be established.
In this study, we sought to elucidate the influence of the host immune-mediated stress on antimycobacterial chemotherapy both in vivo and in appropriate tissue culture models. The analysis of Mtb-infected myeloid cells isolated from experimental mouse infections demonstrated that the bacilli in activated cells show a greater degree of drug tolerance than those in resting cells. The macrophage is both a host cell and an effector cell through which the host’s immune response is filtered. We used transcriptional profiling to demonstrate that Mtb’s response to drug pressure inside macrophages differs significantly from that observed in broth culture and leads to an up-regulation of bacterial stress regulons common to four different frontline drugs. The data indicate that the increased stress experienced by intracellular Mtb as a consequence of exposure to drugs and host cell activation both lead to enhanced drug tolerance. Although this may be perceived initially as problematic for therapy, we believe that it identifies new opportunities for drug discovery.
RESULTS
Drug tolerance in Mtb correlates directly with the acquired immune response of the host during in vivo infection
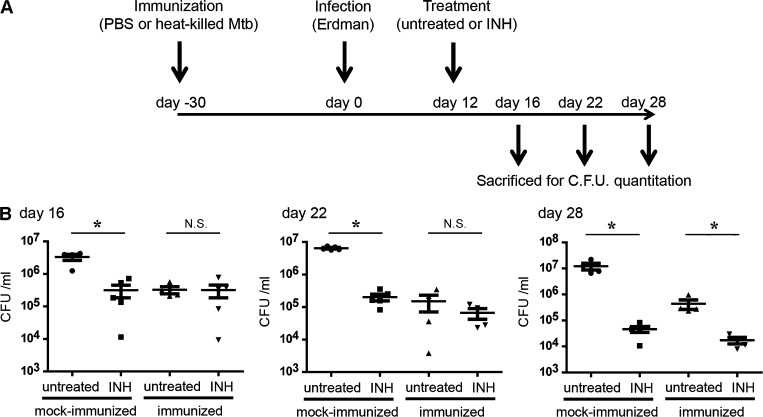
There is a growing interest in the interrelationship between host immune status and the sensitivity of Mtb to current frontline drugs. To determine the relationship between drug tolerance and immunity at the gross level, we compared the sensitivity of Mtb to INH in two groups of mice that had been injected intraperitoneally with either 106 CFUs of heat-killed Mtb (immunized) or PBS (mock immunized) 30 d before intranasal infection with 103 CFUs of viable Mtb (Fig. 1 A). This vaccination model had been developed previously, and the immunization protocol impacts bacterial burden minimally until 12 d postinfection (p.i.; Sukumar et al., 2014). For this reason, drug treatment was initiated on day 12 p.i. and maintained to 28 d p.i. By 28 d, the naive mice had also developed a robust adaptive immune response against the challenge infection (Sukumar et al., 2014). At various time points after treatment, mice were sacrificed and bacterial burden in the lungs was determined. In comparison to the control mock-immunized group, we observed lower bacterial burden in the immunized mice at all the time points examined (Fig. 1 B). After 4 d of INH treatment (day 16 p.i.), the drug treatment markedly reduced the number of viable Mtb in the mock-immunized group, whereas the drug treatment failed to further reduce the number of Mtb recovered from the immunized group (Fig. 1 B). The bacterial burden in the immunized group did decrease at later time points of drug treatment (days 22 and 28 p.i.). However, the relative impact of drug treatment was considerably less in the immunized mice than it was in the naive animals, indicating that INH treatment has reduced efficacy in an immunized host. These data show that although immunization accelerated host protective immunity and reduced bacterial burden, it also enhanced or accelerated the acquisition of drug tolerance in Mtb.
Figure 1.
In vivo mouse infection demonstrates that drug sensitivity of Mtb correlates inversely with the immune status of the host. (A) Immunization decreased bacterial burden but also reduced Mtb sensitivity to INH. C57BL/6J mice were injected with heat-killed Mtb (immunized) or PBS (mock immunized) and challenged by infection with Erdman Mtb. INH treatment was initiated 12 d p.i. and continued until 28 d. (B) At 16, 22, and 28 d p.i., mice were sacrificed and bacterial burden in the lungs was determined by CFU enumeration. *, P < 0.05 by the Mann-Whitney test. Each group contained five mice, and this experiment was repeated three times. Error bars represent SD, and horizontal lines represent the mean.
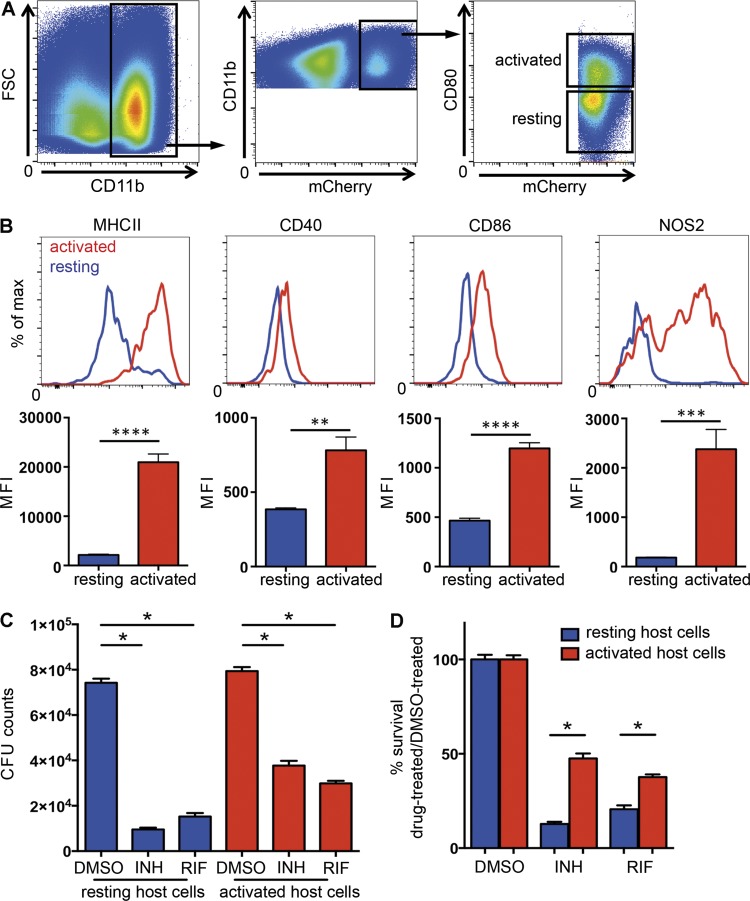
During animal infection, the majority of Mtb resides in cells of myeloid lineage, which includes macrophages, dendritic cells, and neutrophils (Srivastava et al., 2014). To probe the impact of the host cell immune status on drug sensitivity, we developed a protocol to isolate Mtb-infected myeloid cells of differing immune activation status from experimentally infected mice. To this end, we infected WT C57BL/6J mice with an Mtb strain that constitutively expresses the fluorescent protein mCherry and allowed the infection to proceed for 21 d. At that time, the lungs were harvested and digested to yield a single-cell suspension. Mtb-containing myeloid cells were recovered from the lungs and sorted according to their CD80 expression status (CD11b+ mCherry+ CD80high and CD11b+ mCherry+ CD80low) as an indicator of their activation state (Fig. 2 A). Further flow cytometric analysis of these cells revealed modest increases in surface expression of CD40 and CD86 but marked increases in the levels of expression of the classic activation markers MHCII and inducible NO synthase (iNOS) among the CD80high cell population (Fig. 2 B). These cell populations were established in culture independently, and the drug sensitivity of the bacteria was evaluated by the addition of 1 µg/ml INH or rifampin (RIF), or DMSO as a control, for 24 h before plating for CFU enumeration. The 21-d time point was selected on the basis of preliminary experiments where we found comparable numbers of activated (CD11b+ mCherry+ CD80high) and resting (CD11b+ mCherry+ CD80low) host cell populations in infected mice. Although treatment with both INH and RIF effectively reduced viable bacterial counts in both resting and activated host cell populations (Fig. 2 C), a significantly greater percentage of Mtb residing in activated host cells survived drug exposure when compared with those residing in resting cells (Fig. 2 D). The data indicate that the acquisition of a drug-tolerant phenotype in vivo correlates with the development of an acquired immune response and is mediated at the level of the host cell.
Figure 2.
Flow sorting of activated and resting Mtb-infected host cells demonstrates that the drug sensitivity of Mtb recovered from in vivo infection correlates inversely with the immune status of the host phagocyte. Mtb recovered from activated host cells in vivo were more tolerant to both INH and RIF than those recovered from resting host cells. C57BL/6J mice were infected with mCherry-expressing Erdman Mtb for 21 d, and Mtb-containing myeloid cells with different immune activation status were isolated from lung tissue using flow cytometry. (A) CD11b+ mCherry+ CD80high cells (activated population) and CD11b+ mCherry+ CD80low cells (resting population) were sorted according to the depicted gating strategies. FSC, forward scatter. (B) Flow cytometry analysis of the expression levels of MHCII, CD40, CD86, and iNOS revealed the increased expression of classical activation markers in the CD11b+ mCherry+ CD80high cells (activated population) in comparison with the CD11b+ mCherry+ CD80low cells (resting population). (C and D) Isolated cells were established in culture and subjected to treatment with 1 µg/ml INH or RIF or an equivalent volume of DMSO. After 24 h of drug treatment, bacterial survival was determined by CFU enumeration (C), and the percentage of Mtb surviving drug treatment was quantified by normalizing the bacterial load in drug-treated samples against that in DMSO-treated samples (D). Data represent the mean ± SD of duplicates from an individual experiment representative of two independent experiments. MFI, mean fluorescence intensity. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, by two-tailed Student’s t test.
Activation of the host cell in experimental in vitro tissue culture infections induces drug tolerance in Mtb
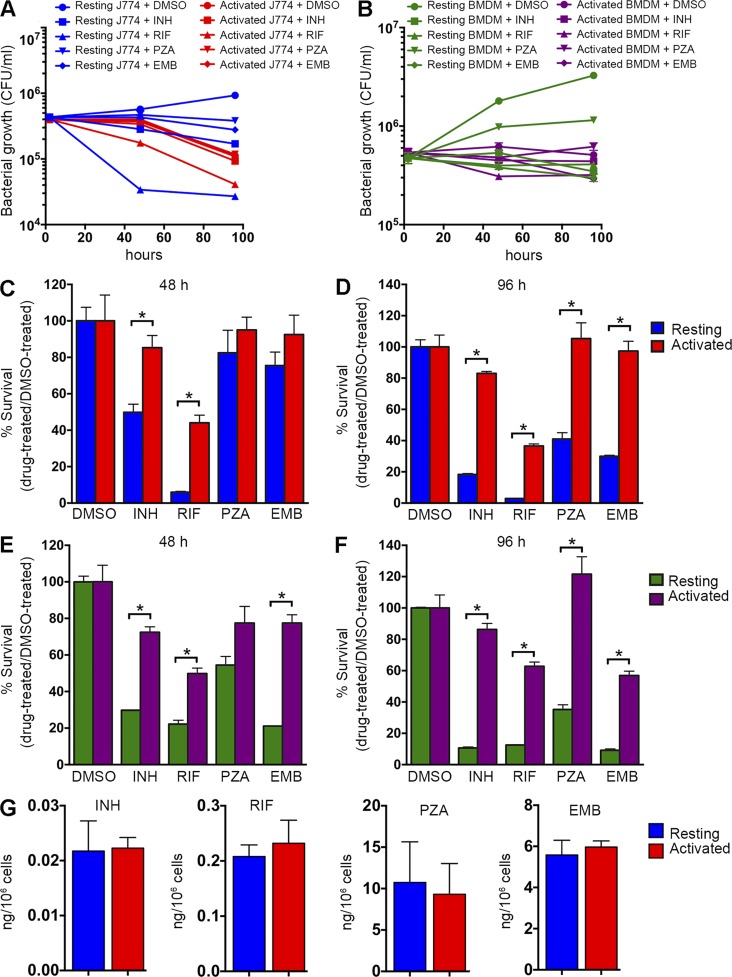
We next sought to determine whether the induction of drug tolerance in intracellular Mtb could be reproduced in both macrophage-like cell lines and primary macrophages in culture. We monitored the relative susceptibility of Mtb to frontline drugs after activation of the host cell with IFN-γ + LPS. Resting or activated J774A.1 macrophages were infected with Mtb, INH treatment was initiated 2 h p.i., and bacterial survival rate was examined after 48 and 96 h. As expected, activation of the host cell impacted bacterial survival and resulted in a decrease in bacterial counts (Fig. 3 A). Consistent with our in vivo data, Mtb residing in activated macrophages showed a significantly higher survival rate upon exposure to all four drugs (Fig. 3, A, C, and D). After 48 h of treatment with INH, Mtb in resting macrophages showed an ∼50% survival rate compared with DMSO controls, whereas bacteria in activated macrophages maintained a survival rate at ∼85% (Fig. 3, A and C). The difference in efficacy of INH was even greater at 96 h when the survival rate of Mtb was reduced to <20% in resting macrophages and the survival rate of the bacteria in activated macrophages remained at a >80% level (Fig. 3 D). We observed a similar impact of enhanced host cell pressures on drug efficacy with RIF-treated samples. The survival rate of Mtb was <10% after 48-h RIF treatment in resting macrophages, and the rate further decreased to <5% by 96 h (Fig. 3, A, C, and D). In contrast, ∼40% of Mtb survived the same dose of RIF treatment in activated macrophages at both 48- and 96-h time points. The activities of the two less potent drugs, pyrazinamide (PZA) and ethambutol (EMB), were also decreased in activated host cells. At 96 h, ∼40% of Mtb survived PZA treatment and ∼30% survived EMB treatment in resting macrophages, whereas almost 100% of Mtb survived the same dose of PZA or EMB exposure in activated macrophages (Fig. 3, A, C, and D).
Figure 3.
The drug sensitivity of Mtb in macrophages is enhanced markedly upon the activation of the host cells in vitro. Mtb in host cells activated in vitro exhibited markedly higher levels of drug tolerance than in resting host cells. (A) Resting or activated J774A.1 macrophages were infected with Mtb at an MOI of 4. 2 h later, cells were washed to remove extracellular bacteria and then treated with 0.4 µg/ml INH, 0.4 µg/ml RIF, 200 µg/ml PZA, 12 µg/ml EMB, or an equal amount of DMSO. After 48 or 96 h of treatment, macrophages were lysed and the bacterial load was determined by CFU enumeration. (B) Resting or activated primary mouse BMDMs were infected with Mtb at an MOI of 4 and treated with identical drug regimens as in A. (C–F) The bacterial survival rates in J774A.1 macrophages (C and D) and BMDMs (E and F) after 48 or 96 h of drug treatment were quantified by normalizing bacterial load in drug-treated samples against that in DMSO-treated samples. Data represent the mean ± SD of triplicates from an individual experiment and are representative of three independent experiments. *, P < 0.05 by two-tailed Student’s t test. (G) Bar graph illustrating the relative drug concentration in resting and activated J774A.1 cells treated comparably to those cells detailed in A, C, and D. The data represent the mean ± SD of triplicates from an individual experiment representative of two independent experiments. There was no statistically significant difference in the drug concentrations found in resting or activated cells in each case.
Consistent with our observations in J774A.1 macrophages, we found that Mtb developed comparable host cell–dependent antibiotic tolerance in mouse BMDMs (Fig. 3, B, E, and F). By 96 h, only ∼10% of Mtb in resting host cells survived INH treatment versus ∼80% of Mtb in activated macrophages; the survival rate in the presence of RIF is ∼15% for Mtb in resting macrophages versus ∼60% for those in activated macrophages (Fig. 3, B, E, and F). Similarly, the susceptibility of Mtb to PZA and EMB was also decreased significantly in activated macrophages. To ensure that these data were not generated artificially through differential drug sequestration in activated versus resting macrophages, we quantified the relative concentrations of the four anti-Mtb drugs in J774A.1 cells incubated under identical conditions (Fig. 3 G) and found that they were comparable under both immune states.
These data suggest that increasing host antimicrobial immune pressure induces antibiotic tolerance in Mtb. The bacterial burden is clearly reduced upon host cell activation; however, those bacilli that survive are more refractory to drug-mediated killing. The decrease of drug efficacy in activated macrophages can be explained by two non–mutually exclusive mechanisms: a preexisting drug-tolerant bacterial population was passively selected and enriched by increased host cell stresses, and/or the active adaptation to the increased host pressures coincidentally renders Mtb more tolerant to drugs. Probing the molecular mechanisms underlying this bacterial phenotype requires an understanding of the impact of drug pressure in the context of the macrophage as opposed to exposure to drugs in rich broth.
Extracellular and intracellular Mtb respond differentially to INH treatment
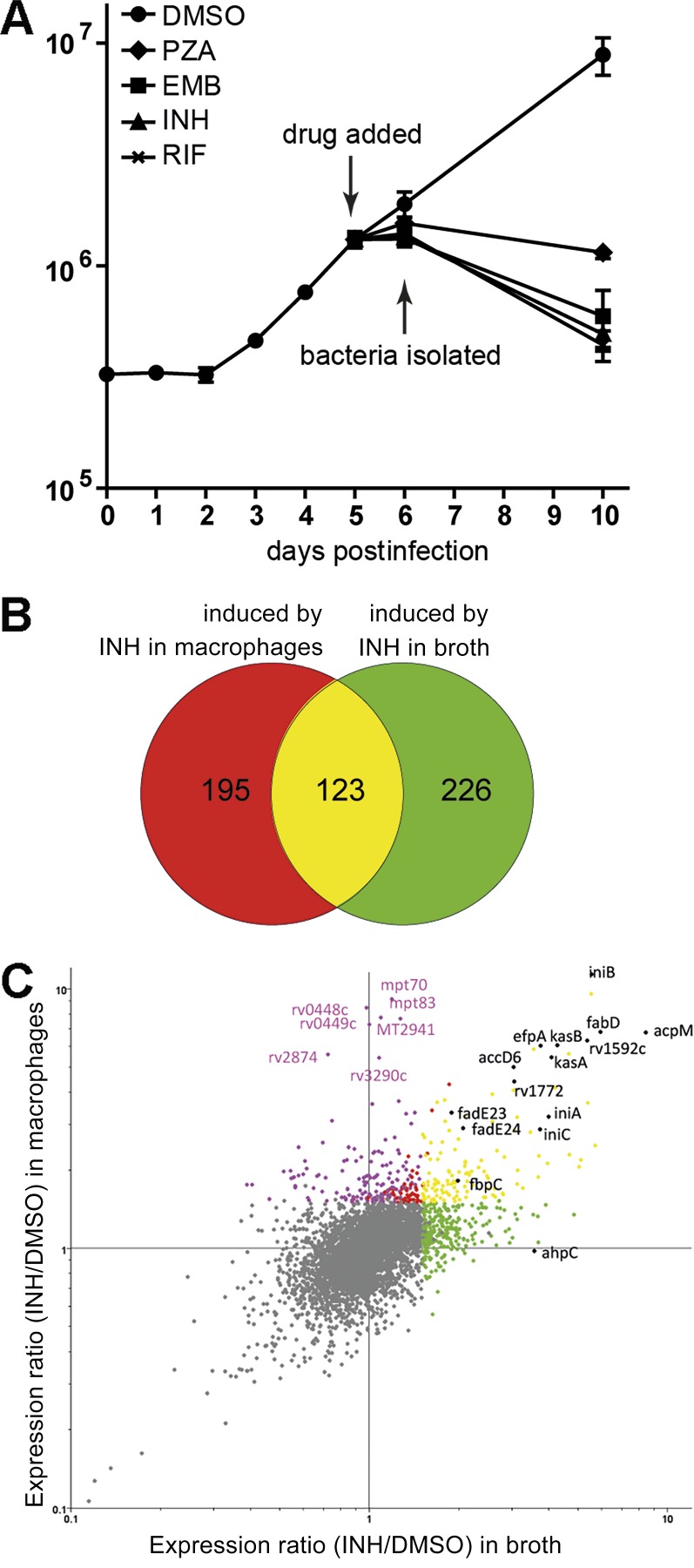
To determine the impact of the host environment on drug treatment, we used transcriptional profiling to monitor Mtb’s global gene expression in response to INH treatment during macrophage infection. Pilot experiments were performed based on published studies to establish conditions of maximal transcriptional response from Mtb under drug pressure inside a host cell (Wilson et al., 1999; Betts et al., 2003; Rohde et al., 2012). J774A.1 cells were infected for 5 d with Mtb CDC1551 before exposure to 0.2 µg/ml INH for 24 h. Under these conditions, we observed a 30% drop in CFUs at 24 h but could isolate quality RNA that generated reproducible patterns of expression (Fig. 4 A). Control RNA was isolated from parallel cultures exposed to DMSO without drugs. The gene expression profiles for Mtb in broth culture were examined under comparable conditions. The transcriptional profiles in the macrophage and in broth in both the presence and absence of drugs were compared to one another to determine those responses that were drug induced in both environments and to identify responses that were drug induced specifically within the context of the host cell.
Figure 4.
The exposure of intracellular Mtb to INH induces an environment-specific transcriptional signature. (A) Growth curve of intracellular Mtb in the presence or absence of frontline antimycobacterial drugs. 0.2 µg/ml INH, 0.4 µg/ml RIF, 200 µg/ml PZA, 12 µg/ml EMB, or an equal volume of DMSO was used for the treatment. Error bars indicate SD from the mean. (B and C) Mtb exhibited an environment-specific transcriptional response to INH. (B) Venn diagram comparing Mtb genes up-regulated upon INH exposure in broth or during macrophage infection. Data represent the mean of eight biological replicates (treatment in macrophage) or six biological replicates (treatment in broth). Genes were identified as INH induced if they were up-regulated >1.5-fold relative to DMSO-treated samples (P < 0.05 by Student’s t test). (C) Scatter plot comparison of transcriptional profiles of Mtb treated with INH in broth (x axis) or in macrophages (y axis). Genes are colored according to the Venn diagram. Purple dots indicate genes whose induction is macrophage specific. Macrophage-specific genes were defined as genes that were only induced by INH treatment in macrophages by Venn diagram analysis with the induction ratio in macrophage versus in broth being significantly different (one-way ANOVA; Benjamini and Hochberg false discovery rate; P < 0.01). Three biological replicates were included for each treatment condition. Data points are from an individual experiment conducted in triplicate and are representative of three independent experiments. Black dots indicate INH-induced genes that have been reported previously (Wilson et al., 1999).
The expression of 318 Mtb genes were up-regulated (≥1.5-fold; P < 0.05) in response to INH treatment within macrophages. Similar numbers of genes (349) were up-regulated when the bacilli were treated with INH in vitro in 7H9 broth. Although expression of many of the same genes was induced in both intra- and extracellular conditions, more than half of the genes induced were unique to their environmental conditions (Fig. 4, B and C). 123 genes were up-regulated under both conditions, comprising a list of commonly induced transcripts. INH inhibits biosynthesis of the cell wall component mycolic acid by targeting the enoyl-acpM reductase InhA in the FAS-II system (Vilchèze and Jacobs, 2007). It was therefore reassuring to note that expression of those genes related to the mycolic acid biosynthesis pathway, including FAS-II enzymes (acpM, kasA, kasB, accD6, and fabD), two acyl-CoA dehydrogenases (fadE23 and fadE24), a mycolyl transferase (fbpC), and the efflux protein gene efpA, were all highly up-regulated (Fig. 4 C). Expression of the iniBAC operon, which is responsive to cell wall damage, was also induced under both conditions (Fig. 4 C). Previous studies have documented the induction of FAS-II–related and cell wall damage genes by INH in broth culture (Wilson et al., 1999; Betts et al., 2003; Boshoff et al., 2004). Our data are consistent with these observations and confirm that mycolic acid biosynthesis is effectively targeted by INH when Mtb is inside macrophages.
Besides these commonly induced genes, however, there were significant numbers of genes that were up-regulated by INH only in Mtb inside macrophages (Fig. 4 C and Table S3). Previous transcriptional profiling of Mtb in its host cell has indicated that the bacterium experiences a range of host-dependent stresses, such as nutrient limitation, a reduction in pH, oxidative and nitrosative stress, and cell wall damage (Schnappinger et al., 2003; Rohde et al., 2007). The exposure of intracellular Mtb to INH appears to enhance the intensity of the responses to these stressors.
Although Mtb arrests the normal maturation of its phagosome, it does reside in a slightly acidic compartment with a pH of 6.4 (Sturgill-Koszycki et al., 1994; Podinovskaia et al., 2013). The two-component system PhoPR has been linked to Mtb’s response to low pH and controls many of the 291 genes up-regulated in response to acid stress (Abramovitch et al., 2011). The macrophage-specific, INH-response gene set (Table S3) contains 35 genes that are also up-regulated by in vitro acid stress. We calculated, based on the size of the pH response regulon, that the number of genes in this pathway that would overlap by random chance is eight; therefore, the 35 genes represent a significant overlap (P < 0.001; Table 1). Besides the PhoP regulon, another gene set that is significantly induced is the Rv1404 regulon, which is also responsive to acid stress (Fisher et al., 2002).
Table 1. Comparison of macrophage-specific INH-induced genes to in vitro stimuli-induced genes.
| In vitro stimulus | Reference list size | Overlap with macrophage-specific INH-induced list | Overlap by random chance | Overlap significance |
|---|---|---|---|---|
| Acid stressa | 291 | 35 | 8 | P < 0.001 |
| Nutrient starvationb | 440 | 37 | 12 | P < 0.001 |
| Oxidative stress (H2O2)c | 319 | 24 | 9 | P < 0.001 |
| NO/CO/hypoxiad | 50 | 16 | 1 | P < 0.001 |
| SDS-mediated membrane damagee | 211 | 30 | 6 | P < 0.001 |
| Triton X-100–mediated membrane damagef | 73 | 20 | 2 | P < 0.001 |
Data from Rohde et al. (2007).
Data from Betts et al. (2002) and Hampshire et al. (2004).
Data from Schnappinger et al. (2003) and Voskuil et al. (2011)
Data from Sherman et al. (2001), Ohno et al. (2003), Park et al. (2003), Voskuil et al. (2003), Kumar et al. (2008), and Shiloh et al. (2008).
Data from Manganelli et al. (2001) and He et al. (2006).
Data from He et al. (2006).
Another intriguing finding in the macrophage-specific gene list is the up-regulation of a group of genes belonging to the DosR or dormancy regulon, including seven genes residing between rv2623 and rv2630. Although induction of the DosR regulon indicates that Mtb may be experiencing additional pressures such as nitric oxide (NO), carbon monoxide (CO), or hypoxia upon INH exposure within the host cell, it also suggests that intracellular Mtb responds to the microbicidal effects of INH by entering a metabolically quiescent state.
Genes showing the highest level of induction by INH treatment within the host cell are clustered in two genomic regions, rv0445c–0449c and rv2873–2876. Two genes in the first region (rv0448c and rv0449c) and three genes in the second region (mpt83, mt2941, and mpt70) were induced more than sevenfold (Fig. 4 C). Interestingly, both clusters of genes are under the control of the extracytoplasmic function sigma factor sigK (rv0445c; Saïd-Salim et al., 2006; Rodrigue et al., 2007) and are known to be up-regulated under conditions of nutrient deprivation (Betts et al., 2002; Hampshire et al., 2004).
Other themes that emerge among those genes up-regulated upon drug pressure within the host cell include genes responsive to H2O2 (Table S4; Schnappinger et al., 2003; Voskuil et al., 2011), indicating increased exposure to oxidative stresses. The expression of genes responsive to membrane-damaging agents such as SDS or Triton X-100 was also increased, and most of those genes are regulated by the two-component system MprAB (He et al., 2006). In addition to sigK, the expression of two other alternative sigma factors (sigF and sigH) was also up-regulated. The induction of sigF has been reported after nutrient starvation in PBS (Betts et al., 2002). SigH plays a role in both nitrosative and oxidative stress responses, possibly through the regulation of thioredoxin recycling (Raman et al., 2001; Voskuil et al., 2011). In conclusion, in these initial experiments with INH, the transcriptional profiles of intracellular Mtb in response to drug pressure fell into two discrete groups of genes: genes that were routinely induced by drug treatment in both intra- and extracellular conditions, which are related predominantly to the primary target and mode of action of the drug, and genes that were only up-regulated in response to INH pressure in macrophages, which are indicative of an increased sensitivity or exposure to host-derived immune pressures (Table 1).
Exposure of intracellular Mtb to four frontline antimycobacterial agents induces a common response
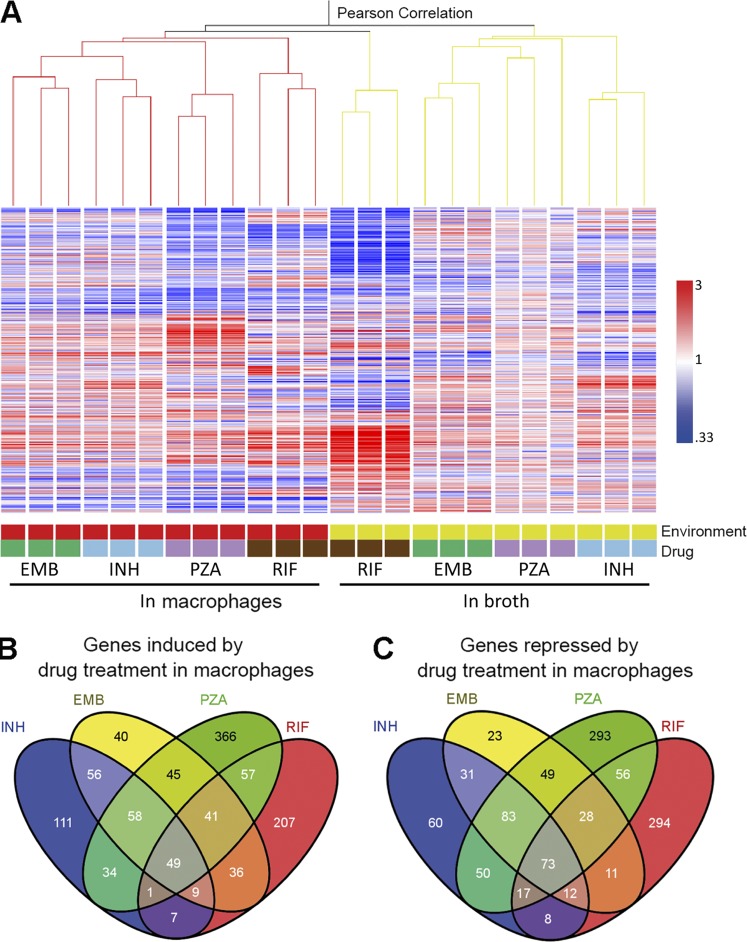
Among the approved antituberculosis agents, INH, RIF, EMB, and PZA constitute the core of treatment regimens. To gain a more holistic understanding of how the host cell environment impacts Mtb drug response, we determined the transcriptional profiles of the bacterium to EMB, RIF, and PZA during macrophage infection compared with exposure to the same drugs in broth. The correlation between the transcriptional profiles from intra- and extracellular Mtb exposed to all four drugs was examined by unsupervised condition tree clustering analysis (Fig. 5 A). Strikingly, despite the fact that all four drugs have distinct modes of action, all of the intracellular transcriptome data clustered together regardless of drug treatment (Fig. 5 A). In contrast, with the exception of RIF-treated bacteria, the profiles of Mtb exposed to drugs within macrophages segregated markedly from those treated in broth, even when the bacilli were exposed to the same drug. These results demonstrate that the environment plays a dominant role in shaping Mtb’s response to drug treatment.
Figure 5.
The exposure of intracellular Mtb to a panel of four frontline drugs induces a common transcriptional response. (A) Dominant role of the environment on Mtb’s transcriptional response to drug treatment. Condition tree analysis of Mtb transcriptional profiles in response to INH, RIF, PZA, or EMB treatment (concentrations as indicated for Fig. 4) during log phase growth in 7H9 broth or during infection of macrophages relative to DMSO-treated extracellular or intracellular Mtb. Three biological replicates were included for each treatment condition. Expression profiles were clustered using the Pearson correlation. (B and C) Shared transcriptional response across drug treatment conditions by intracellular Mtb. The four-way Venn diagrams compare genes induced or repressed by four drugs. Genes with a ≥1.5-fold change in regulation with a Student’s t test (P < 0.05) from a mean of at least three biological replicates were included for analysis.
Detailed information for the transcriptional response of intracellular Mtb to the four frontline drugs is described in Table S4. By comparing the Mtb responses to different drug treatments within the host macrophage (Fig. 5, B and C), we were able to identify a list of genes that were commonly regulated by drug pressure in the intracellular environment. This shared transcriptome consisted of 158 genes commonly induced and 213 genes commonly repressed by at least three of the four drugs (Table S5). Among them, 122 genes (49 up-regulated and 73 down-regulated) were responsive to all four drugs. Among those genes that showed increased expression, 108 genes were specifically induced within the host cell (Table S6), and the majority of them had previously been reported to respond to stresses known to be linked to the intraphagosomal environment (Table S1). Consistent with the results from INH-exposed Mtb, we observed significant overlap with genes induced by different immune pressure–mimicking stimuli. The transcription of 28 of these genes was up-regulated in acidic medium, and 27 genes were induced when nutrients were limiting. In addition, genes responsive to NO/CO/hypoxia, oxidative stress, and membrane perturbation were also identified in the list of up-regulated transcripts. Regulon analysis revealed a common set of regulons that act to direct the transcriptional remodeling of Mtb physiology in response to drug pressure within the host environment (Table S2). Among them, five regulons, DosR, MprA, PhoP, Rv1404, and Rv3058c, seem to play the major role. These five regulons accounted for more than half of the up-regulated genes in common across the different drug treatments.
In contrast, the list of transcripts of down-regulated genes (Table S5) contained many genes linked to growth, such as those involved in ribosomal production (rpl and rps genes), amino acid biosynthesis (trp, leu, ilv, met, and asp genes), respiration (atp and nuo genes), and lipid utilization (ppia, papa2, rv3553, and the rv3562–rv3563–rv3564–rv3565 gene cluster). The repression of these genes is directed by KstR2 (Kendall et al., 2010; Casabon et al., 2013), Crp (Bai et al., 2005; Rustad et al., 2014), and several other regulators (Table S2). The down-regulation of these genes reflects reduced metabolic activity and a suppression of growth and energy production in response to drug treatment.
The conclusion from the INH experiments, that drug pressure within the host macrophage increases Mtb’s sensitivity or exposure to host-dependent stresses, is also reproduced with the remaining three frontline drugs. The comparison of the transcriptional profiles induced by all four drugs indicates that when exposed to a drug inside a macrophage, irrespective of the identity of the antimycobacterial agent, the bacterium exhibits an amplified transcriptional response to a range of host-derived antimicrobial stresses.
Similarity between the transcriptional responses of intracellular Mtb to drug treatment and macrophage activation
Our transcriptome analysis revealed that, compared with untreated intracellular bacilli, Mtb exposed to drugs during macrophage infection up-regulated expression of genes linked to stresses such as acidic pH, nutrient starvation, hypoxia, and exposure to reactive oxygen and nitrogen intermediates. To clarify that this transcriptional shift was a consequence of macrophage-derived pressures, we examined the transcriptional response of Mtb within macrophages treated to up-regulate their antimicrobial activities. We reasoned that the increased hostility of activated host cells would generate pressures comparable to those experienced under drug pressure and that these studies could clarify the link between drug pressure and those host stresses that are enhanced by immune activation.
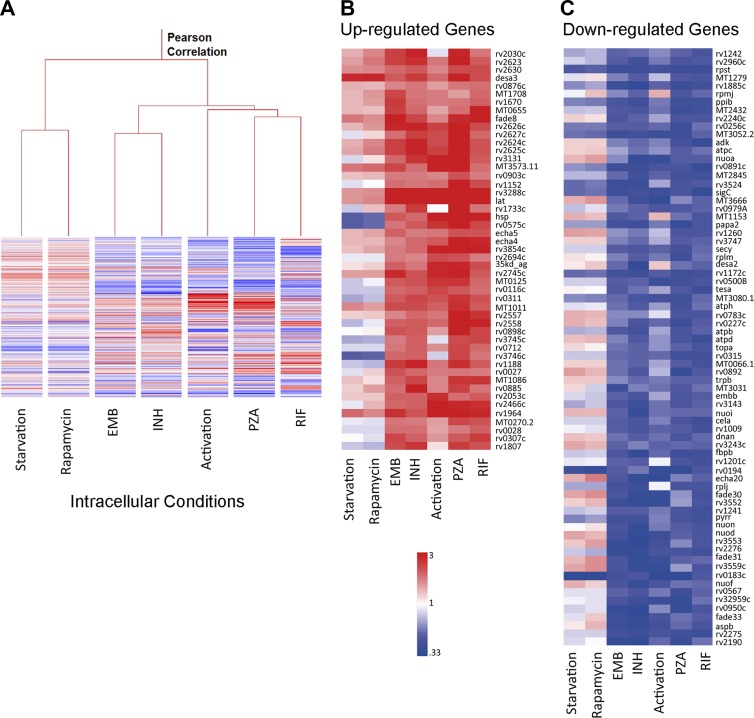
Condition tree clustering was used to compare the intracellular drug-induced expression profiles and the transcriptomes of Mtb in IFN-γ + LPS–activated macrophages or in macrophages in which autophagy was induced by either rapamycin or serum starvation (Fig. 6 A). Although both forms of treatment are known to generate enhanced antimicrobial pressures, the transcriptional responses of Mtb to cytokine-mediated activation and the induction of autophagy were different. The gene expression profiles of Mtb experiencing intracellular drug exposure clustered more closely with that of cytokine-activated macrophages in the condition tree (Fig. 6 A). A direct comparison between the Mtb transcripts up-regulated upon drug exposure or host cell activation by IFN-γ + LPS revealed a remarkable similarity (Fig. 6, B and C). Most of the genes whose expression was modulated by all four drugs were also similarly responsive in activated macrophages (Fig. 6, B and C). These data are consistent with the conclusion that the pressures encountered by intracellular Mtb during different drug exposure are derived from the host cell, and that many of these pressures are generated or augmented during macrophage activation.
Figure 6.
Activation of the host phagocyte by cytokines induces a transcriptional stress response comparable to the response induced by exposure of intracellular Mtb to four frontline drugs. (A) Condition tree analysis of intracellular Mtb expression profiles in response to different drug exposure, macrophage activation, or autophagy induced by starvation or rapamycin. Host cell activation led to an intracellular Mtb transcriptional response similar to that observed on drug exposure during macrophage infection. Data represent the mean of at least two biological replicates for each macrophage condition or at least three biological replicates for drug-treated samples. (B and C) Heat maps showing the similarity between the expression profiles of genes commonly induced (B) or repressed (C) by exposure of Mtb to the four drugs versus during infection of macrophages activated by exposure to IFN-γ + LPS before infection.
Nitrosative stress is the most potent single inducer of drug tolerance in Mtb
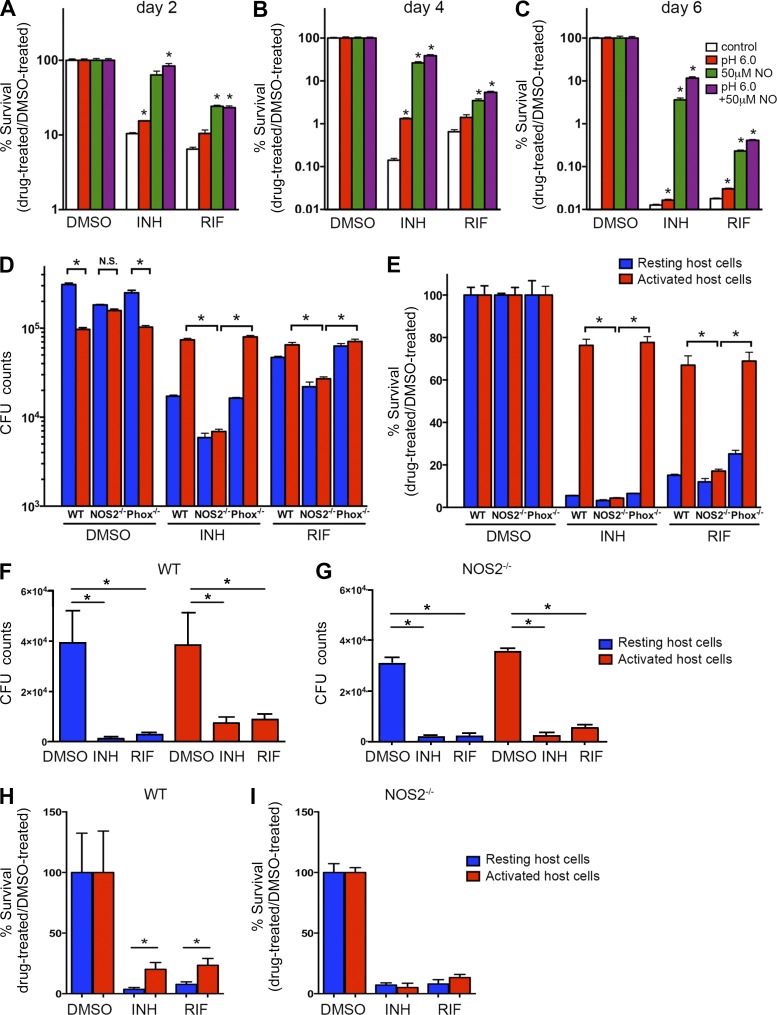
Various host-derived stresses may contribute to the increase of drug tolerance in Mtb during infection of activated macrophages. The induction of the PhoP, Rv1404, and DosR regulons indicates that acidic pH and nitrosative stress could play a role in this process. To investigate whether these stresses can directly induce drug tolerance in vitro, we tested the activities of INH and RIF against Mtb grown under acidic or nitrosative stressed conditions. In comparison to Mtb cultured in regular 7H9 broth, we consistently detected higher survival rates in Mtb cultured in 7H9 broth buffered to low pH (6.0) or in 7H9 broth containing NO through 6 d of drug treatment (Fig. 7, A–C). The impact of the two stresses on drug sensitivity was additive, as Mtb grown in low pH 7H9 broth containing NO showed the highest survival rate among all tested conditions after 6 d of drug treatment (Fig. 7 C). These data showed that at least two macrophage-related immune pressures, acidic pH and NO, are capable of inducing drug tolerance in Mtb. These results are consistent with previous studies in which the decreased susceptibility of Mtb to frontline drugs was detected under other in vitro stress conditions, such as oxygen limitation and nutrient deprivation (Lenaerts et al., 2005; Sulochana et al., 2009; Gengenbacher et al., 2010; Piccaro et al., 2013). Additionally, we observed that the survival rates were significantly higher for Mtb grown in broth containing NO than those grown in low pH broth upon drug exposure (Fig. 7, A–C), suggesting that nitrosative stress is the more potent enhancer of drug tolerance.
Figure 7.
NO is a major contributor to the induction of antibiotic tolerance in both in vitro and in vivo infection models. (A–C) Drug sensitivity of Mtb grown under acidic pH or NO stress conditions. Mtb was grown in regular 7H9 broth (control), acidified 7H9 broth (pH 6.0), 7H9 broth with 50-µM NO, and acidic 7H9 broth with NO (pH 6.0 + 50-µM NO) and exposed to 0.4 µg/ml INH, 0.4 µg/ml RIF, or an equal amount of DMSO. Bacterial survival rate was quantified after 2 (A), 4 (B), or 6 d (C) of treatment by CFU enumeration. *, P < 0.05 by two-tailed Student’s t test versus control. (D and E) Drug sensitivity of Mtb in BMDMs isolated from WT, NOS2−/−, or Phox−/− mice. Mtb survival was quantified after 96 h of drug treatment by normalizing the bacterial load in drug-treated samples against that in DMSO-treated samples. The data are shown as both CFU numbers (D) and as survival percentage relative to appropriate control conditions (E). Data represent the mean ± SD of triplicates from an individual experiment and are representative of two independent experiments. *, P < 0.05 by two-tailed Student’s t test. (F–I) C57BL/6J and NOS2−/− mice were infected with mCherry-expressing Erdman Mtb for 21 d, and Mtb-containing myeloid cells with different immune activation status were isolated from lung tissue using flow cytometry. Isolated cells were established in culture and subjected to treatment with 1 µg/ml INH or RIF or an equivalent volume of DMSO. After 24 h of drug treatment, bacterial survival was determined by CFU enumeration (F and G), and the percentage of Mtb surviving the drug treatment was quantified by normalizing the bacterial load in drug-treated samples against that in DMSO-treated samples (H and I). Data represent the mean ± SD and were pooled from two independent experiments.
Within host cells, production of NO is mainly catalyzed by iNOS (NOS2; Nathan and Shiloh, 2000). Although the significance of NO production in humans has been debated, the clear demonstration of nitrite production in human macrophages in culture (Cunningham-Bussel et al., 2013) and expression of iNOS in experimental infections in macaques (Mattila et al., 2013) resolve much of this controversy. To further test whether nitrosative stress plays a role in the induction of antibiotic tolerance during infection, we monitored drug efficacy against Mtb in macrophages lacking the NOS2 gene (NOS2−/−). For this purpose, we infected either resting or activated BMDMs derived from WT or NOS2−/− mice with Mtb and then examined bacterial load after 96 h of INH or RIF treatment, scoring both absolute bacterial CFUs (Fig. 7 D) and survival as a percentage of the drug-free controls (Fig. 7 E). In the DMSO-treated group, we observed a lower bacterial burden upon macrophage activation in WT samples. In contrast, Mtb grew similarly within resting or activated macrophages in NOS2-deficient samples. The growth differences in WT and NOS2−/− macrophages are consistent with the phenotypes reported previously (Ehrt et al., 2001). Consistent with the aforementioned observations, Mtb exhibited tolerance to both drugs when grown in activated WT macrophages. Strikingly, Mtb grown in activated NOS2−/− macrophages were significantly more sensitive to INH- and RIF-mediated killing than those grown in activated WT macrophages (Fig. 7, D and E). In fact, the survival rates were reduced to levels comparable to those detected in resting host cells (Fig. 7 E), indicating that reactive nitrogen species (RNS) plays a major role in the induction of drug tolerance. Deletion of the reactive oxygen species production gene (Phox) did not have a detectable effect on the induction of drug tolerance. This suggests that reactive oxygen species are not major contributors to the tolerance phenotype observed in activated macrophages.
To further validate the impact of NO on the induction of drug tolerance and its potential role in in vivo infections, we infected WT or NOS2−/− mice using mCherry-expressing Mtb (Fig. 7, F–I). 21 d p.i., we isolated Mtb-containing myeloid cells with different immune activation status from the lung tissues using the same flow cytometry protocols used to generate the data in Fig. 2. In agreement with the earlier experiments, the bacteria residing in activated phagocytes from WT mice were markedly less sensitive to both INH and RIF (Fig. 7, F and H) when compared with those bacteria in resting cells from the same mice. In contrast, in the NOS2−/− mice, the levels of drug susceptibility to both INH and RIF were comparable in Mtb in both resting and activated myeloid cells (Fig. 7, G and I). These data add further support to the hypothesis that nitrosative stress is a highly potent inducer of drug tolerance during Mtb infection both in tissue culture and in in vivo infections. Schnappinger et al. (2003) previously demonstrated that Mtb grown in activated NOS2−/− macrophages also exhibited decreased transcriptional responses to other types of stresses, such as oxidative stress and iron starvation. We therefore suggest that these host-derived stresses act in synergy and that the removal of one type of stress within host cells, such as NO, will have an impact that exceeds the activity of that individual stress in isolation. We believe that our data indicate that the induction of tolerance in activated macrophages is the product of the combined action of several immune-mediated stresses, among which RNS appears to play a major role.
DISCUSSION
Despite the fact that host stresses are by themselves detrimental to bacterial survival, we found that both in macrophages and in mice, enhancement of these stresses did not synergize with antibiotic treatment to eliminate the bacilli but instead induced drug tolerance, rendering the bacilli more refractory to drug-dependent killing. Our observations are in line with previous studies that reported that stress conditions, such as nutrient starvation and oxygen limitation, tended to induce drug tolerance in vitro (Lenaerts et al., 2005; Sulochana et al., 2009; Gengenbacher et al., 2010; Baek et al., 2011; Piccaro et al., 2013).
Bacterial determinants responsible for the development of drug tolerance in an activated host remain elusive, although multiple mechanisms could contribute. Previous studies have documented the presence of marked heterogeneity among the Mtb population during infection by quantitative microscopy (Abramovitch et al., 2011; Tan et al., 2013; Bhaskar et al., 2014; Sukumar et al., 2014). Using a fluorescent reporter indicative of Mtb ribosomal activity, Manina et al. (2015) showed that stress conditions and host immune pressures can amplify phenotypic heterogeneity. Therefore, it is possible that increased pressures from the host environment facilitate the selection of a slowly replicating, drug-tolerant bacterial subpopulation. Data from our transcriptional profiling support an adaptation model whereby increasing macrophage-derived stresses trigger the induction/repression of a common set of stress-response regulons, which remodel Mtb physiology to adopt a metabolically quiescent state and offer cross-protection to both environmental and drug-induced damage. This model is in line with previous findings that the induction of a DosR-regulated gene, triacylglycerol synthase (tgs1), led to reduced growth and decreased sensitivity to a range of different antituberculosis drugs (Deb et al., 2009; Baek et al., 2011). The activation of toxin–antitoxin (TA) modules mediates the formation of nonreplicating drug tolerance persisters in Escherichia coli and Salmonella (Maisonneuve et al., 2013; Helaine et al., 2014). Mtb encodes 79 TA modules in its chromosome (Sala et al., 2014), and some of them are induced upon drug treatment. Thus, it is plausible that these TA modules also contribute to the development of drug tolerance. In fact, one of the TA families, MazEF, has been shown to promote stress adaptation and drug tolerance in Mtb (Tiwari et al., 2015). The putative linkage between drug tolerance and diminished growth rate is a feature of most studies into stress-induced reduction of drug sensitivity; however, it should be noted that drug tolerance is not necessarily associated with slowed metabolism and arrested growth. It has been shown that alternative mechanisms, such as the induction of efflux pumps, can lead to decreased drug sensitivity in replicating Mycobacterium species (Adams et al., 2011). In addition, Nandakumar et al. (2014) reported recently that one of the isocitrate lyase enzymes of Mtb (icl1) can mediate broad antibiotic tolerance through a previously unknown role in antioxidant defense. In summary, Mtb appears capable of entering a drug-tolerant state through multiple pathways. Disrupting a single pathway may not block the induction of drug tolerance. In fact, we have found that the deletion of single-response regulons (DosR, PhoP, and MprA) or the treatment of the intracellular Mtb with the efflux pump inhibitor verapamil, in combination with either INH or RIF, exhibits a minimal effect on the acquisition of drug tolerance under stress (unpublished data). The phenotype is therefore multigenic with layers of redundancy between the major stress regulons of Mtb.
In conclusion, our study reveals the complex influence that the macrophage environment exerts over drug action and provides compelling arguments for drug screening strategies that incorporate the host environment. We demonstrate, both in vitro and in vivo, that immune activation promotes the induction of drug tolerance, and we suggest that persister bacilli residing in activated host cells could be part of the reason why lengthy treatment is required to eradicate Mtb that remains genetically sensitive to frontline drugs. Although both in vitro and in vivo infection data from NOS2−/− mice and macrophages indicate a dominant role for RNS in the induction of drug tolerance in Mtb, we believe that multiple stresses induced by immune activation influence this phenotypic response. Finally, there is an intriguing prediction implied by this interpretation: the identification of compounds that inhibit Mtb’s ability to respond to host stresses and mobilize a drug-tolerant phenotype could represent a novel type of drug that would show broad spectrum synergy when used in combination with existing frontline antibiotics.
MATERIALS AND METHODS
Mice
All mice were maintained under specific pathogen-free conditions at the Cornell University College of Veterinary Medicine and were handled in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee of Cornell University.
Mtb growth conditions
We used two different Mtb isolates for the tissue culture and mouse challenge experiments. CDC1551 is considerably less cytopathic than other Mtb strains and is ideally suited for tissue culture infections where the health of the host cell is a concern (Rohde et al., 2012; Podinovskaia et al., 2013). Unfortunately, it shows low virulence in mouse challenge experiments. Therefore, for the in vivo studies, we used the Erdman strain of Mtb (Sukumar et al., 2014).
Both isolates were grown in Middlebrook 7H9 broth supplemented with 10% oleic acid/albumin/dextrose/catalase (OADC), 0.2% glycerol, and 0.05% Tween 80 in standing vented tissue culture flasks. Bacterial cultures were grown from an OD600 of 0.05 to ∼0.4 before treatment with either 0.2 µg/ml INH, 0.4 µg/ml RIF, 200 µg/ml PZA, or 12 µg/ml EMB for 24 h. As a control, a parallel culture was treated with an equal volume of vehicle solvent (DMSO) for the same period of time.
Macrophage growth and infection
J774A.1 macrophages were grown in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 1 mM sodium pyruvate in a humidified incubator at 37°C and 7% CO2. For activation, macrophages were incubated in a medium containing 100 U/ml recombinant IFN-γ and 10 ng/ml LPS for 20 h. To induce autophagy, macrophages were treated with 0.5 µM rapamycin or incubated in starvation medium Earle’s balanced salts solution for 4 h as previously described (Winslow et al., 2010).
For infection, macrophages were seeded in T75 vented tissue culture flasks at a density of ∼2 × 107 per flask. The next day, midlog phase (OD600 = 0.4 to ∼0.5) Mtb cultures were harvested by centrifugation, resuspended in basal uptake buffer (PBS with 4.5 mg/ml glucose, 5 mg/ml BSA, 0.1 mg/ml CaCl2, 0.1 mg/ml MgCl2, and 1 mg/ml gelatin), passaged 10 times through a 21-gauge needle, and then used to infect macrophages at a multiplicity of infection (MOI) of ∼4. After 2 h of infection, extracellular bacteria was removed and replaced with fresh cell medium. Medium was changed daily for the duration of the infection. At 5 d p.i., different antimycobacterial drugs or an equivalent amount of DMSO were added to the infected macrophages. The same concentrations of drugs were used as in the broth treatment. All drugs were tested as endotoxin free by a ToxinSensor Chromogenic LAL Endotoxin kit (GenScript). After 24 h of treatment, medium was removed and guanidine thiocyanate buffer was added to selectively lyse host cells while leaving bacilli intact and preserving RNA transcripts as previously described (Homolka et al., 2010).
RNA isolation, linear amplification, and fluorophore labeling
Mtb RNA from extracellular and intracellular samples was isolated, amplified, and labeled as previously described (Rohde et al., 2007). In brief, Mtb in guanidine thiocyanate was pelleted, washed once with PBS supplemented with 0.1% Tween 80, and treated with 10 mg/ml lysozyme. Bacilli were then lysed in 65°C TRIzol (Invitrogen) using silicon beads in a BeadBeater. Bacterial lysate was extracted once with chloroform, and total RNA was purified from the aqueous phase using an RNeasy kit (QIAGEN). Eluted RNA samples were treated with DNase I (Ambion) for 45 min at 37°C before storage at −70°C. The MessageAmpII-Bacteria kit (Ambion) was used to generate amplified RNA (aRNA) from 100 ng of total RNA input. Amino-allyl UTP was incorporated into aRNA during in vitro transcription to allow fluorophore labeling. 10 µg of modified aRNA from each sample was then labeled with Alexa Fluors (Invitrogen). After excess dyes were quenched by hydroxylamine, the labeled aRNA was purified using a MegaClear kit (Ambion).
Microarray fabrication
Custom-designed Mtb gene expression array (design ID 025053) was fabricated by Agilent Technologies using the 8 × 15–k formatted glass slides. Each slide contains eight identical microarrays consisting of 15,744 in situ–synthesized 60-mer oligonucleotide probes. The microarray platform and all transcriptional profiles have been deposited in the Gene Expression Omnibus database (Edgar et al., 2002) under the accession numbers GPL19382 and GSE62942.
Microarray hybridization and data analysis
aRNA from drug-treated samples was labeled with Alexa Fluor 647, and aRNA from DMSO-treated samples was labeled with Alexa Fluor 555. An equal amount (400 ng) of labeled aRNA from paired samples was mixed and used for hybridization. aRNA from the DMSO-treated samples was used as a common reference to calculate relative changes in gene expression in response to drug treatments. For macrophage activation or autophagy induction arrays, aRNA generated from Mtb grown in activated or autophagic macrophages was labeled with Alexa Fluor 647, whereas aRNA from Mtb grown in resting macrophages was labeled with Alexa Fluor 555 and used as a reference.
Microarray hybridization and washes were performed using the protocols described in the Agilent Technologies Two-Color Microarray-Based Gene Expression Analysis manual. Slides were scanned by a DNA microarray scanner (G2505B; Agilent Technologies), and array data were extracted from raw image files using Feature Extraction software (version 10.7; Agilent Technologies). Data were then imported into Genespring 7.3 (Agilent Technologies) using the default Enhanced Agilent FE settings for subsequent normalization, statistical analysis, and visualization. Although we did observe batch-dependent variations, these were minor and rendered nonsignificant by the biological/technical repeats. A one-way ANOVA test was performed to identify genes with significant differences in expression between different treatment conditions. A Pearson correlation was used for gene or condition clustering analysis. Genes were identified as drug regulated if they were induced or repressed >1.5-fold with a Student’s t test p-value <0.05 from a mean of at least three biological replicates. For overlapping analyses, the numbers of genes expected to be overlapped between two gene lists by random chance and overlap significance were calculated according to Rustad et al. (2014) based on the size of each gene list in relation to the genome size. All transcriptional profiles have been deposited in the Gene Expression Omnibus database under the accession number GSE62942.
Measurement of Mtb drug sensitivity in vitro
WT, NOS2−/−, and Phox−/− mice were all on a C57BL/6J background and purchased from The Jackson Laboratory. To test drug sensitivity, either J774A.1 macrophages or mouse BMDMs were seeded in 24-well tissue culture plates at a density of ∼4 × 105 per well. For testing in activated macrophages, cells were seeded with medium containing 100 U/ml recombinant IFN-γ and 10 ng/ml LPS for 20 h before infection, and IFN-γ was kept in medium throughout the infection. The next day, midlog phase Mtb cultures were harvested and used to infect macrophages at an MOI of ∼4. After 2 h of incubation, cells were washed with medium to remove extracellular bacteria and then replaced with fresh medium containing either 0.4 µg/ml INH, 0.4 µg/ml RIF, 200 µg/ml PZA, 12 µg/ml EMB, or an equal amount of vehicle solvent DMSO. Medium supplemented with appropriate antibiotics or IFN-γ was changed every 2 d for the duration of the infection. At desired time points, macrophages were lysed with water containing 0.1% Tween 80 to release intracellular bacteria. Samples were then diluted with PBS containing 0.05% Tween 80 and plated on OADC-supplemented 7H10 agar for CFU enumeration.
To examine the drug susceptibility of Mtb under different stress conditions, midlog phase Mtb were pelleted and then resuspended in 6 ml of fresh 7H9 broth, 7H9 broth buffered to pH 6.0, 7H9 broth containing 50-µM NO donor diethylenetriamine NONOate (DETA-NONOate; Cayman Chemicals), or pH 6.0 7H9 broth containing 50 µM DETA-NONOate to a final density of OD600 = 0.1. For this test, 0.05% tyloxapol was used as the dispersal agent in 7H9 broth instead of 0.05% Tween 80 to rule out the impact of Tween 80 on Mtb survival at a low pH. After 4 h of incubation, drug treatment was initiated by adding 0.4 µg/ml INH, 0.4 µg/ml RIF, or an equal amount of vehicle solvent DMSO to culture aliquots. At desired time points, survival rate was determined by OD600 measurement as well as CFU counts.
Measurement of drug concentration in macrophages
J774 macrophages were grown in a T75 flask in DMEM supplemented with 10% FBS, 2-mM l-glutamine, and 1 mM sodium pyruvate in a tissue culture incubator at 37°C and 5% CO2. When the cells had reached ∼90% confluency, they were incubated in ice-cold PBS for 10 min and then detached gently using a cell scraper. The numbers of viable cells were counted and seeded into a 96-well plate at a density of 105 per well in 150 µl of medium. For activation, cells in a subset of wells were incubated in medium containing 100 U/ml of recombinant IFN-γ and 10 ng/ml LPS. After 20 h of incubation, both activated and resting macrophages were gently washed twice with 150 µl PBS to remove unattached and dead cells. Viable cells were quantified using PicoGreen (Invitrogen). To incubate cells with drugs, old media were removed carefully and media with drugs were added. Cells were incubated in a tissue culture incubator at 37°C and 5% CO2 for 16 h. After incubation, media were removed carefully and cells were gently washed twice with an equal volume of ice-cold PBS to remove any residual extracellular drugs. Cells were then lysed by incubation in 150 µl of deionized water for 1 h at 37°C in a tissue culture incubator. 50 µl lysate was diluted twofold and used to quantify cell numbers. The rest of the lysates were transferred to 1.5-ml centrifuge tubes and stored at −80°C before analysis by liquid chromatography coupled to tandem mass spectroscopy as described previously (Prideaux et al., 2015).
Preparation of lung tissue and cell sorting
Mice were sacrificed and lungs were aseptically removed. To prepare a single-cell suspension, lungs were minced and digested in 5% FBS/PBS solution containing 250 U/ml collagenase IV (Worthington Biochemical Corporation) and 20 U/ml DNase (Roche) for 60 min at 37°C. Lung tissues were then passed through a 70-µm cell strainer, and red blood cells were lysed with ammonium-chloride-potassium buffer.
For cell sorting, lung cell suspensions were blocked with anti–mouse CD16/32 followed by incubation with CD11b (BD) and CD80 (eBioscience) antibodies. CD11b+ mCherry+ CD80high cells (activated population) and CD11b+ mCherry+ CD80low cells (resting population) were sorted using a cell sorter (S3; Bio-Rad Laboratories) with a 100-µm nozzle. Cells were then surface stained with CD11b (BD), CD80 (eBioscience), CD40 (eBioscience), CD86 (eBioscience), and MHCII (eBioscience). For iNOS staining, surface-stained cells were fixed and permeabilized using an intracellular fixation and permeabilization buffer set (eBioscience). Cells were then incubated for 30 min with an iNOS antibody (eBioscience). Data were acquired using a flow cytometer (LSR II; BD) and analyzed with FlowJo software (Tree Star).
Measurement of drug sensitivity of Mtb in vivo
To assess the drug sensitivity of Mtb in host cells with different immune activation status, 6–8-wk-old C57BL/6J and NOS2−/− mice were inoculated intranasally with ∼2 × 103 CFUs of Erdman Mtb constitutively expressing mCherry in 25 µl PBS containing 0.05% Tween 80. 21 d p.i., 1.5 × 105 CD11b+ mCherry+ CD80high cells (activated population) and CD11b+ mCherry+ CD80low cells (resting population) were sorted as described in the previous section and treated with 1 µg/ml INH or RIF or an equal amount of DMSO. 24 h after treatment, cells were lysed and plated on 7H10 for CFU enumeration.
To directly evaluate the effects of immune activation on drug sensitivity at a whole-animal level, 6–8-wk-old C57BL/6J mice were injected intraperitoneally with either 106 CFUs of heat-killed Erdman Mtb (immunized) or PBS (mock immunized). 30 d after immunization, mice were inoculated intranasally with ∼2 × 103 CFUs of viable Erdman Mtb. 12 d p.i., INH was delivered through drinking water at a concentration of 100 µg/ml. Drug-containing water was monitored for consumption and replaced every 6 d. Mice were sacrificed after 4, 10, and 16 d of treatment. The left lung lobe together with the accessory lobe of the right lung were aseptically removed, homogenized in PBS + 0.05% Tween 80, and plated on 7H10 agar for CFU enumeration.
Online supplemental material
Table S1 shows a comparison of macrophage-specific induced genes common to all four drugs to those genes induced by in vitro stimuli. Table S2 lists regulons that are commonly regulated by different frontline drugs in macrophages. Table S3 is provided as an Excel file and lists macrophage-specific INH-induced genes. Table S4 is provided as an Excel file and lists genes differentially regulated by individual drugs in macrophages. Table S5 is provided as an Excel file and lists genes commonly regulated by different frontline drugs in macrophages. Table S6 is provided as an Excel file and lists macrophage-specific commonly induced genes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151248/DC1.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Jennifer D. Mosher at the Cornell Genomics Core Facility for microarray hybridization and scanning.
R.B. Abramovitch was supported by a National Institute of Allergy and Infectious Disease (NIAID) National Research Service Award fellowship (F32AI081482), and S. Tan was supported by an NIAID K99 award (AI114952). This work was supported by National Institutes of Health grants AI067027 and HL055936 to D.G. Russell and AI111967 to V. Dartois.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- aRNA
- amplified RNA
- EMB
- ethambutol
- INH
- isoniazid
- iNOS
- inducible NO synthase
- MOI
- multiplicity of infection
- Mtb
- Mycobacterium tuberculosis
- p.i.
- postinfection
- PZA
- pyrazinamide
- RIF
- rifampin
- RNS
- reactive nitrogen species
- TA
- toxin–antitoxin
References
- Abramovitch R.B., Rohde K.H., Hsu F.F., and Russell D.G.. 2011. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol. 80:678–694. 10.1111/j.1365-2958.2011.07601.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K.N., Takaki K., Connolly L.E., Wiedenhoft H., Winglee K., Humbert O., Edelstein P.H., Cosma C.L., and Ramakrishnan L.. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 145:39–53. 10.1016/j.cell.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge B.B., Keren I., and Fortune S.M.. 2014. The spectrum of drug susceptibility in Mycobacteria. Microbiol. Spectr. 2:MGM2-0031-2013 10.1128/microbiolspec.MGM2-0031 [DOI] [PubMed] [Google Scholar]
- Alland D., Kramnik I., Weisbrod T.R., Otsubo L., Cerny R., Miller L.P., Jacobs W.R. Jr., and Bloom B.R.. 1998. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 95:13227–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S.H., Li A.H., and Sassetti C.M.. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9:e1001065 10.1371/journal.pbio.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G., McCue L.A., and McDonough K.A.. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187:7795–7804. 10.1128/JB.187.22.7795-7804.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts J.C., Lukey P.T., Robb L.C., McAdam R.A., and Duncan K.. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731. 10.1046/j.1365-2958.2002.02779.x [DOI] [PubMed] [Google Scholar]
- Betts J.C., McLaren A., Lennon M.G., Kelly F.M., Lukey P.T., Blakemore S.J., and Duncan K.. 2003. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:2903–2913. 10.1128/AAC.47.9.2903-2913.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar A., Chawla M., Mehta M., Parikh P., Chandra P., Bhave D., Kumar D., Carroll K.S., and Singh A.. 2014. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 10:e1003902 10.1371/journal.ppat.1003902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff H.I., Myers T.G., Copp B.R., McNeil M.R., Wilson M.A., and Barry C.E. III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174–40184. 10.1074/jbc.M406796200 [DOI] [PubMed] [Google Scholar]
- Casabon I., Zhu S.H., Otani H., Liu J., Mohn W.W., and Eltis L.D.. 2013. Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol. Microbiol. 89:1201–1212. 10.1111/mmi.12340 [DOI] [PubMed] [Google Scholar]
- Cunningham-Bussel A., Zhang T., and Nathan C.F.. 2013. Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc. Natl. Acad. Sci. USA. 110:E4256–E4265. 10.1073/pnas.1316894110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb C., Lee C.M., Dubey V.S., Daniel J., Abomoelak B., Sirakova T.D., Pawar S., Rogers L., and Kolattukudy P.E.. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 4:e6077 10.1371/journal.pone.0006077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S., Schnappinger D., Bekiranov S., Drenkow J., Shi S., Gingeras T.R., Gaasterland T., Schoolnik G., and Nathan C.. 2001. Reprogramming of the macrophage transcriptome in response to interferon-γ and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123–1140. 10.1084/jem.194.8.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M.A., Plikaytis B.B., and Shinnick T.M.. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025–4032. 10.1128/JB.184.14.4025-4032.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M., Rao S.P., Pethe K., and Dick T.. 2010. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 156:81–87. 10.1099/mic.0.033084-0 [DOI] [PubMed] [Google Scholar]
- Hampshire T., Soneji S., Bacon J., James B.W., Hinds J., Laing K., Stabler R.A., Marsh P.D., and Butcher P.D.. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinb.). 84:228–238. 10.1016/j.tube.2003.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Hovey R., Kane J., Singh V., and Zahrt T.C.. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188:2134–2143. 10.1128/JB.188.6.2134-2143.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S., Cheverton A.M., Watson K.G., Faure L.M., Matthews S.A., and Holden D.W.. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 343:204–208. 10.1126/science.1244705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka S., Niemann S., Russell D.G., and Rohde K.H.. 2010. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 6:e1000988 10.1371/journal.ppat.1000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall S.L., Burgess P., Balhana R., Withers M., Ten Bokum A., Lott J.S., Gao C., Uhia-Castro I., and Stoker N.G.. 2010. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology. 156:1362–1371. 10.1099/mic.0.034538-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Deshane J.S., Crossman D.K., Bolisetty S., Yan B.S., Kramnik I., Agarwal A., and Steyn A.J.. 2008. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283:18032–18039. 10.1074/jbc.M802274200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts A.J., Gruppo V., Marietta K.S., Johnson C.M., Driscoll D.K., Tompkins N.M., Rose J.D., Reynolds R.C., and Orme I.M.. 2005. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob. Agents Chemother. 49:2294–2301. 10.1128/AAC.49.6.2294-2301.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E., Castro-Camargo M., and Gerdes K.. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 154:1140–1150. 10.1016/j.cell.2013.07.048 [DOI] [PubMed] [Google Scholar]
- Manganelli R., Voskuil M.I., Schoolnik G.K., and Smith I.. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423–437. 10.1046/j.1365-2958.2001.02525.x [DOI] [PubMed] [Google Scholar]
- Manina G., Dhar N., and McKinney J.D.. 2015. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 17:32–46. 10.1016/j.chom.2014.11.016 [DOI] [PubMed] [Google Scholar]
- Mattila J.T., Ojo O.O., Kepka-Lenhart D., Marino S., Kim J.H., Eum S.Y., Via L.E., Barry C.E. III, Klein E., Kirschner D.E., et al. 2013. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol. 191:773–784. 10.4049/jimmunol.1300113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune R.M. Jr., and Tompsett R.. 1956. Fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. I. The persistence of drug-susceptible tubercle bacilli in the tissues despite prolonged antimicrobial therapy. J. Exp. Med. 104:737–762. 10.1084/jem.104.5.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune R.M. Jr., McDermott W., and Tompsett R.. 1956. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J. Exp. Med. 104:763–802. 10.1084/jem.104.5.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar M., Nathan C., and Rhee K.Y.. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 5:4306 10.1038/ncomms5306 [DOI] [PubMed] [Google Scholar]
- Nathan C., and Shiloh M.U.. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA. 97:8841–8848. 10.1073/pnas.97.16.8841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Zhu G., Mohan V.P., Chu D., Kohno S., Jacobs W.R. Jr., and Chan J.. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell. Microbiol. 5:637–648. 10.1046/j.1462-5822.2003.00307.x [DOI] [PubMed] [Google Scholar]
- Park H.D., Guinn K.M., Harrell M.I., Liao R., Voskuil M.I., Tompa M., Schoolnik G.K., and Sherman D.R.. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843. 10.1046/j.1365-2958.2003.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccaro G., Giannoni F., Filippini P., Mustazzolu A., and Fattorini L.. 2013. Activities of drug combinations against Mycobacterium tuberculosis grown in aerobic and hypoxic acidic conditions. Antimicrob. Agents Chemother. 57:1428–1433. 10.1128/AAC.02154-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podinovskaia M., Lee W., Caldwell S., and Russell D.G.. 2013. Infection of macrophages with Mycobacterium tuberculosis induces global modifications to phagosomal function. Cell. Microbiol. 15:843–859. 10.1111/cmi.12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prideaux B., Via L.E., Zimmerman M.D., Eum S., Sarathy J., O’Brien P., Chen C., Kaya F., Weiner D.M., Chen P.Y., et al. 2015. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat. Med. 21:1223–1227. 10.1038/nm.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Song T., Puyang X., Bardarov S., Jacobs W.R. Jr., and Husson R.N.. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119–6125. 10.1128/JB.183.20.6119-6125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue S., Brodeur J., Jacques P.E., Gervais A.L., Brzezinski R., and Gaudreau L.. 2007. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189:1505–1513. 10.1128/JB.01371-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K.H., Abramovitch R.B., and Russell D.G.. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2:352–364. 10.1016/j.chom.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Rohde K.H., Veiga D.F., Caldwell S., Balázsi G., and Russell D.G.. 2012. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 8:e1002769 10.1371/journal.ppat.1002769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad T.R., Minch K.J., Ma S., Winkler J.K., Hobbs S., Hickey M., Brabant W., Turkarslan S., Price N.D., Baliga N.S., and Sherman D.R.. 2014. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 15:502 10.1186/s13059-014-0502-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saïd-Salim B., Mostowy S., Kristof A.S., and Behr M.A.. 2006. Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis. Mol. Microbiol. 62:1251–1263. 10.1111/j.1365-2958.2006.05455.x [DOI] [PubMed] [Google Scholar]
- Sala A., Bordes P., and Genevaux P.. 2014. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel). 6:1002–1020. 10.3390/toxins6031002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D., Ehrt S., Voskuil M.I., Liu Y., Mangan J.A., Monahan I.M., Dolganov G., Efron B., Butcher P.D., Nathan C., and Schoolnik G.K.. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704. 10.1084/jem.20030846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman D.R., Voskuil M., Schnappinger D., Liao R., Harrell M.I., and Schoolnik G.K.. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA. 98:7534–7539. 10.1073/pnas.121172498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh M.U., Manzanillo P., and Cox J.S.. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 3:323–330. 10.1016/j.chom.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Ernst J.D., and Desvignes L.. 2014. Beyond macrophages: the diversity of mononuclear cells in tuberculosis. Immunol. Rev. 262:179–192. 10.1111/imr.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki S., Schlesinger P.H., Chakraborty P., Haddix P.L., Collins H.L., Fok A.K., Allen R.D., Gluck S.L., Heuser J., and Russell D.G.. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 263:678–681. 10.1126/science.8303277 [DOI] [PubMed] [Google Scholar]
- Sukumar N., Tan S., Aldridge B.B., and Russell D.G.. 2014. Exploitation of Mycobacterium tuberculosis reporter strains to probe the impact of vaccination at sites of infection. PLoS Pathog. 10:e1004394 10.1371/journal.ppat.1004394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulochana S., Mitchison D.A., Kubendiren G., Venkatesan P., and Paramasivan C.N.. 2009. Bactericidal activity of moxifloxacin on exponential and stationary phase cultures of Mycobacterium tuberculosis. J. Chemother. 21:127–134. 10.1179/joc.2009.21.2.127 [DOI] [PubMed] [Google Scholar]
- Tan S., Sukumar N., Abramovitch R.B., Parish T., and Russell D.G.. 2013. Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog. 9:e1003282 10.1371/journal.ppat.1003282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P., Arora G., Singh M., Kidwai S., Narayan O.P., and Singh R.. 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6:6059 10.1038/ncomms7059 [DOI] [PubMed] [Google Scholar]
- VanderVen B.C., Fahey R.J., Lee W., Liu Y., Abramovitch R.B., Memmott C., Crowe A.M., Eltis L.D., Perola E., Deininger D.D., et al. 2015. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog. 11:e1004679 10.1371/journal.ppat.1004679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchèze C., and Jacobs W.R. Jr. 2007. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu. Rev. Microbiol. 61:35–50. 10.1146/annurev.micro.61.111606.122346 [DOI] [PubMed] [Google Scholar]
- Voskuil M.I., Schnappinger D., Visconti K.C., Harrell M.I., Dolganov G.M., Sherman D.R., and Schoolnik G.K.. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713. 10.1084/jem.20030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil M.I., Bartek I.L., Visconti K., and Schoolnik G.K.. 2011. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2:105 10.3389/fmicb.2011.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis R.S., Patil S., Cheon S.H., Edmonds K., Phillips M., Perkins M.D., Joloba M., Namale A., Johnson J.L., Teixeira L., et al. 1999. Drug tolerance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:2600–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis R.S., Jakubiec W., Mitton-Fry M., Ladutko L., Campbell S., Paige D., Silvia A., and Miller P.F.. 2012. Rapid evaluation in whole blood culture of regimens for XDR-TB containing PNU-100480 (sutezolid), TMC207, PA-824, SQ109, and pyrazinamide. PLoS One. 7:e30479 10.1371/journal.pone.0030479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner D.F., and Mizrahi V.. 2006. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin. Microbiol. Rev. 19:558–570. 10.1128/CMR.00060-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M., DeRisi J., Kristensen H.H., Imboden P., Rane S., Brown P.O., and Schoolnik G.K.. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA. 96:12833–12838. 10.1073/pnas.96.22.12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow A.R., Chen C.W., Corrochano S., Acevedo-Arozena A., Gordon D.E., Peden A.A., Lichtenberg M., Menzies F.M., Ravikumar B., Imarisio S., et al. 2010. α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J. Cell Biol. 190:1023–1037. 10.1083/jcb.201003122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.