Ploegh et al. raised an alpaca single-domain antibody (VHH) against the inflammasome adaptor ASC. VHHASC blocks inflammasome activation in vitro and in living cells, and demonstrates a role of the ASC CARD domain in cross-linking ASC Pyrin domain filaments.
Abstract
Myeloid cells assemble inflammasomes in response to infection or cell damage; cytosolic sensors activate pro–caspase-1, indirectly for the most part, via the adaptors ASC and NLRC4. This leads to secretion of proinflammatory cytokines and pyroptosis. To explore complex formation under physiological conditions, we generated an alpaca single domain antibody, VHHASC, which specifically recognizes the CARD of human ASC via its type II interface. VHHASC not only impairs ASCCARD interactions in vitro, but also inhibits inflammasome activation in response to NLRP3, AIM2, and NAIP triggers when expressed in living cells, highlighting a role of ASC in all three types of inflammasomes. VHHASC leaves the Pyrin domain of ASC functional and stabilizes a filamentous intermediate of inflammasome activation. Incorporation of VHHASC-EGFP into these structures allowed the visualization of endogenous ASCPYD filaments for the first time. These data revealed that cross-linking of ASCPYD filaments via ASCCARD mediates the assembly of ASC foci.
The innate immune system employs a diverse set of genetically encoded sensors to detect evidence of infection or cell damage in the different compartments of the cell. Cytosolic sensors and adaptors in myeloid cells integrate information to initiate a strong inflammatory response through the assembly of macromolecular protein complexes called inflammasomes. Activation of inflammasomes culminates in the activation of caspase-1, which enables the maturation and release of proinflammatory cytokines, such as IL-1β and -18, as well as cell death by pyroptosis (Vanaja et al., 2015).
The sensor involved determines the specificity of the inflammasome and is typically a member of two conserved protein families: NLRs (nucleotide-binding domain [NBD]– and leucine-rich repeat [LRR]–containing proteins), and ALRs (AIM2-like receptors). These sensors recruit caspase recruitment domain (CARD)–containing pro–caspase-1 indirectly via the interposition of CARD-containing ASC or NLRC4. A diverse array of cell damage signals, including potassium efflux, activates NLRP3 inflammasomes through an unknown mechanism, which then recruits ASC via interactions between Pyrin domains (PYDs). AIM2 directly binds to DNA with its HIN domain and also engages ASC via PYD–PYD interactions. Human NAIP/NLRC4 inflammasomes contain two NLRs: NAIP, which senses components of bacterial type III secretion systems or flagellin, and NLRC4, which, once activated by NAIP, can recruit pro–caspase-1, although ASC was required for efficient cytokine secretion in mice (Broz et al., 2010b; Yang et al., 2013; Kortmann et al., 2015; Vance, 2015). Recent cryo-electron microscopy experiments showed that a single NAIP family member primes the self-propagated incorporation of 9–11 NLRC4 monomers into a wheel-like structure (Hu et al., 2015; Zhang et al., 2015).
Local polymerization of PYD and CARD domains, both members of the death domain family, determines activation thresholds and amplifies the signal (Cai et al., 2014; Lu et al., 2014; Sborgi et al., 2015). In their active conformation, the PYD of NLRP3 and AIM2 nucleate the formation of ASCPYD filaments, whereas locally concentrated ASCCARD induces the polymerization of pro–caspase-1 CARD. These structures have thus far been shown only in vitro, or in cells that overexpress single domains of ASC fused to fluorescent proteins. Their presence therefore demands verification at physiological protein levels in the relevant cell type (Lechtenberg et al., 2014). Local concentration of pro–caspase-1 autocatalytically activates caspase-1, which in turn catalyzes the conversion of pro-cytokines into mature IL-1β and -18.
Although we understand some of the molecular triggers and consequences of inflammasome activation, the underlying cell biology and the molecular interactions involved require further study. Reconstitution of defined steps of inflammasome activation in vitro or in unrelated control cells has been quite informative, but—short of their deletion, mutation, or overexpression—inflammasome components in their physiological context are challenging targets for molecular perturbations. Many inflammasome components are prone to oligomerization, or self-activate when artificially overexpressed (Fernandes-Alnemri et al., 2007; Shenoy et al., 2012), emphasizing the need for functional studies in relevant cell types with endogenous expression levels of inflammasome components.
Antibodies are valuable tools to perturb protein function in vitro, or when microinjected into living cells (Doxsey et al., 1987; Antonin et al., 2000). Nonetheless, application of full-sized antibodies has been limited to a few select cases, mostly because of technical challenges. These include the time required to generate and produce antibodies, their bulk, inefficient delivery methods, and sensitivity to the reducing environment of the cytosol.
The description of heavy chain-only antibodies in camelids was a landmark discovery (Hamers-Casterman et al., 1993; Helma et al., 2015) that provided a means to overcome many of these challenges. Heavy chain-only antibodies are comprised of two identical heavy chains and recognize their antigen by means of a single, heavy chain variable domain (VHH). These VHH segments can be expressed as stand-alone fragments and exhibit affinities comparable to conventional antibodies. VHHs are stable in the absence of disulfide bonds or glycosylation and can thus be easily produced in bacteria or expressed in the cytosol of eukaryotic cells (Muyldermans, 2013). VHHs have been used as crystallization chaperones owing to their ability to stabilize protein structures (Pardon et al., 2014; Burg et al., 2015). VHHs can induce or selectively bind distinct conformations, mask epitopes required for protein–protein interactions, or inhibit enzymatic activities (Kirchhofer et al., 2010; Paalanen et al., 2011; Irannejad et al., 2013). VHHs directed against distinct inflammasome components would thus constitute excellent tools to probe and molecularly analyze specific steps of inflammasome activation.
We generated a VHH against the adaptor protein ASC to elucidate the molecular mechanism that allows ASC to greatly amplify the signaling response to many inflammasome triggers. We show that VHHASC specifically blocks CARD–CARD interactions of ASCCARD, and thus inhibits inflammasome activation by agents that trigger NLRP3, AIM2, and NAIP in human macrophage-like cells. By stabilizing an intermediate step of inflammasome activation, VHHASC confirmed the formation of ASCPYD filaments at endogenous ASC levels, and revealed that ASC CARD mediates the coalescence of multiple ASC filaments into a single macromolecular ASC focus per cell.
RESULTS
Identification of an alpaca VHH specific for human ASC
To study molecular interactions in the course of inflammasome activation, we generated alpaca VHHs that bind to the adaptor protein ASC. We immunized an alpaca with purified ASC, produced as a fusion with maltose-binding protein (MBP). After five booster immunizations, we isolated peripheral blood lymphocytes from the immunized alpaca, extracted RNA, prepared cDNA, and amplified VHH-coding sequences by PCR. The amplified VHH cDNAs were cloned into a phagemid vector to yield a plasmid library that encodes the VHH repertoire contained in the collected lymphocytes.
To identify VHHs that bind human ASC, we immobilized MBP-ASC to beads and selected ASC-binding phage by two rounds of phage display. Because we observed a strong immune response to MBP, we depleted phages displaying MBP-binding VHHs before each round of phage display and panned for ASC-specific VHHs in the presence of excess soluble MBP. 96 VHH clones obtained by this procedure were sequenced and tested by ELISA for binding to the fusion protein MBP-ASC and MBP alone. Of these 96 clones, 43 bound to MBP-ASC, but not to MBP, and all encoded the same VHH, VHHASC.
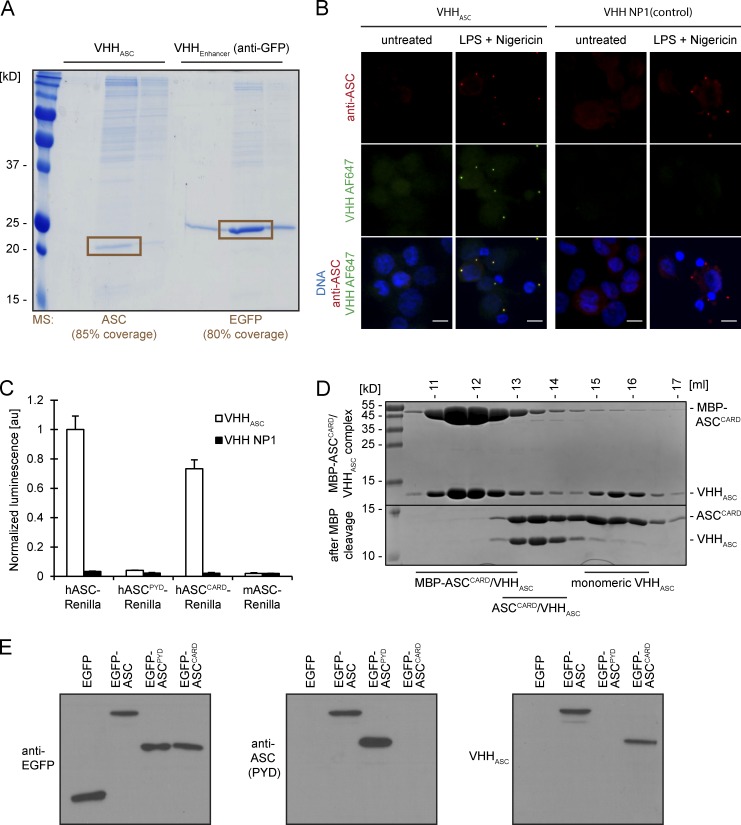
To test its specificity, we expressed VHHASC in Escherichia coli and coupled the purified VHHASC to cyanogen bromide-activated Sepharose beads. Incubation of this VHHASC matrix with lysates of THP-1 cells that overexpress EGFP allowed recovery of a single predominant polypeptide with an apparent molecular weight of 20 kD (Fig. 1 A). The identity of this polypeptide as ASC was confirmed by mass spectrometry (80% sequence coverage; unpublished data). A similarly immobilized GFP-specific VHH, VHHEnhancer (Kirchhofer et al., 2010), recovered EGFP but not ASC from the same THP-1 lysates. VHHASC thus interacts specifically with ASC in THP-1 cell lysates.
Figure 1.
VHHASC specifically binds human ASC CARD. (A) Resins with covalently coupled VHHASC and VHHEnhancer were used to immunoprecipitate the respective VHH targets from THP-1 cells expressing EGFP. Proteins bound to the resin were eluted with acidic buffers, separated by SDS-PAGE, and stained with colloidal Coomassie. The highlighted protein bands were cut out and subjected identification by mass spectrometry. (B) Differentiated THP-1 cells were left untreated or treated with LPS and nigericin to trigger NLRP3 inflammasome activation. Cells were subjected to immunofluorescence staining with rabbit anti-ASC and Alexa Fluor 647–coupled VHHASC or VHH NP1. Maximum intensity projections of representative images of at least three experiments are presented. Bars, 10 µm. (C) A549 cells transiently expressing HA-tagged VHHs and the designated bait proteins fused to Renilla luciferase were lysed to immunoprecipitate VHHs with immobilized anti-HA. Renilla luciferase activity of the coimmunoprecipitated proteins was measured and normalized by the activity in the input. Mean values ± SEM from three independent experiments are displayed. (D) MBP-ASCCARD was incubated with an excess of VHHASC and complexes separated from monomeric VHHASC by size-exclusion chromatography. MBP was cleaved off purified complexes with TEV protease, and the yielded protein samples were subjected to size-exclusion chromatography as well. Samples of the fractions were analyzed by SDS-PAGE and Coomassie staining. (E) A549 cells were transiently transfected with expression vectors for EGFP, EGFP-ASC, EGFP-ASCPYD, or EGFP-ASCCARD. 24 h after transfection, cells were lysed and subjected to SDS-PAGE and immunoblot with anti-EGFP, anti-ASC (raised against the PYD of ASC), and VHHASC combined with the respective HRP-coupled secondary antibodies. Representative immunoblots of at least three experiments are shown.
The VHH expression vectors encode VHHs equipped with a sortase motif to facilitate site-specific labeling with organic fluorophores. Fluorescently labeled VHHASC specifically stained ASC foci in THP-1 cells evoked by treatment with the NLRP3 activators LPS and nigericin, but did not stain any distinct structures in untreated cells (Fig. 1 B). A fluorescently labeled control VHH specific for the influenza A virus nucleoprotein, VHH NP1 (Ashour et al., 2015), did not stain ASC foci.
We applied the LUMIER assay (Barrios-Rodiles et al., 2005) to test interactions of HA-tagged VHHASC with Renilla luciferase fused to human and mouse full-length ASC, as well as fusions of Renilla with the individual domains of human ASC, ASCPYD, and ASCCARD. Full-length human ASC and ASCCARD interacted with VHHASC, whereas murine ASC and the PYD of human ASC did not (Fig. 1 C). We concluded that VHHASC recognizes the CARD of human ASC. VHHASC also recognized ASC CARD in immunoblots (Fig. 1 E), indicating that the epitope recognized by VHHASC is retained in SDS-treated ASCCARD. Moreover, purified VHHASC, when mixed with MBP-ASCCARD, engaged in complex formation and co-eluted with MBP-ASCCARD upon size-exclusion chromatography (Fig. 1 D). Size-exclusion chromatography showed co-elution of ASCCARD and VHHASC at an estimated 1:1 stoichiometry. We concluded that VHHASC specifically binds to the CARD of human ASC, but not mouse ASC.
Structures of VHHASC alone and the ASCCARD–VHHASC complex
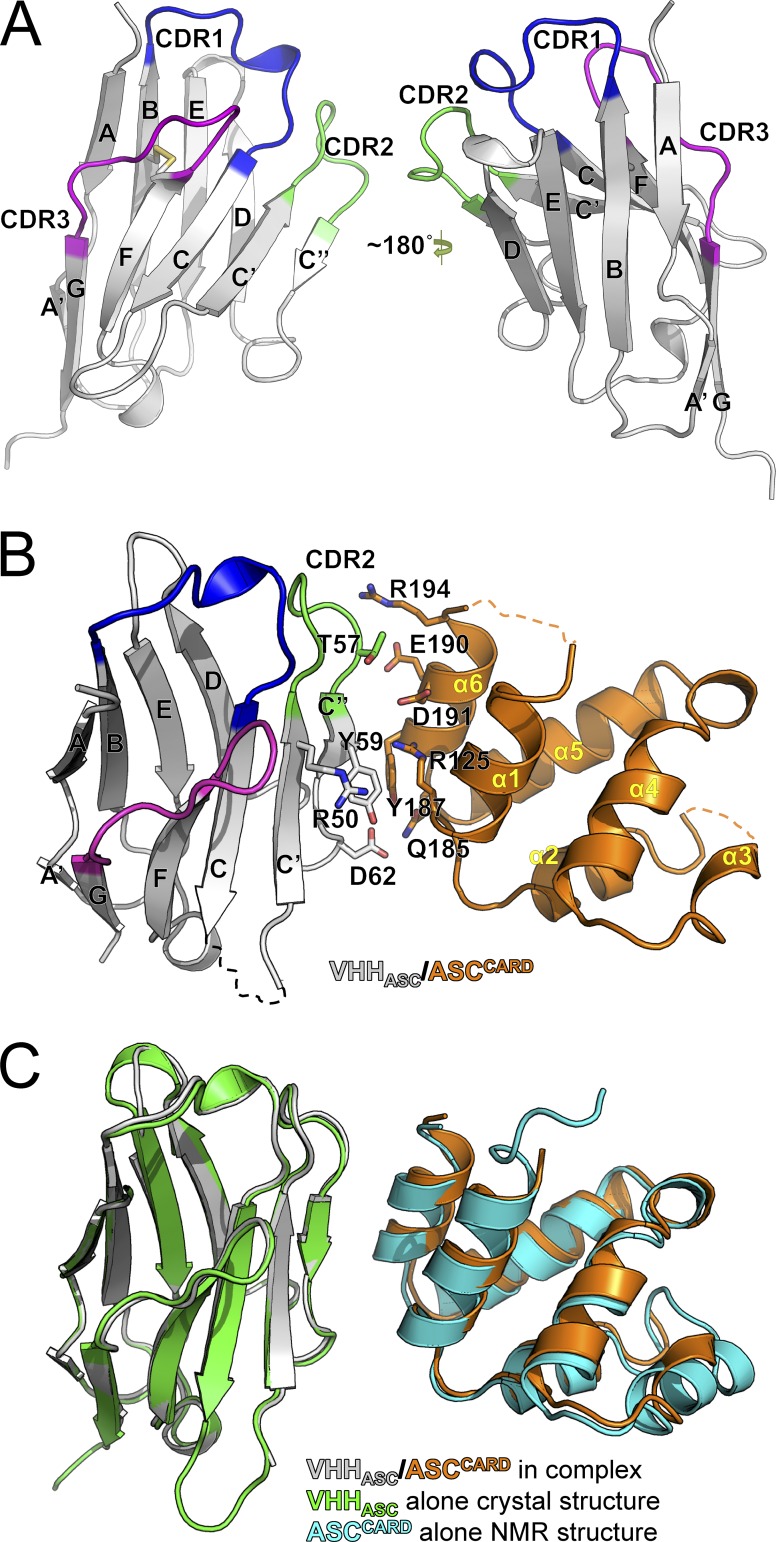
To map the molecular interactions between VHHASC and ASC, we determined the crystal structures of VHHASC alone and in complex with ASCCARD at 1.9 and 4.2 Å resolution, respectively (Table S1 and Table S2). The VHHASC structure shows the typical features of a V domain fold, comprised of nine β-strands stacked into a four-stranded β-sheet and a five-stranded β-sheet (Fig. 2 A). The four-stranded β-sheet is formed by β-strands A/A′, B, E, and D, and the five-stranded β-sheet is formed by β-strands C, C′, C′′, F, and G, which are connected by intervening loops. The loop regions connecting β-strands B and C, strands C′ and C′′, and strands F and G correspond to the complementarity-determining regions CDR1, CDR2, and CDR3, respectively. Compared with other VHHs, VHHASC exhibits an unusually short CDR3. We inferred that the very short CDR3 in VHHASC is unlikely to make a major contribution to binding its target.
Figure 2.
VHHASC binds to an interface of ASCCARD that is required for CARD–CARD interactions. (A) Structure of VHHASC at 1.9 Å resolution. (B) Structure of VHHASC in complex with the N128A/E130R monomeric mutant of ASCCARD at 4.2 Å resolution. Residues at the interaction interface are indicated. Because the resolution of the complex was not sufficient to resolve the displayed amino acid side chains, we used the side chain conformations from the VHH crystal structure at 1.9 Å resolution and the full-length ASC NMR structure. (C) Comparisons of the structures of the complex of VHHASC/ASCCARD N128A/E130R with VHHASC and ASCCARD alone.
To determine the binding interface with ASC, we determined the crystal structure of VHHASC in complex with ASCCARD. We were not successful in crystallizing the wild-type ASCCARD–VHHASC complex, and instead turned to a monomeric mutant (N128A/E130R) of ASCCARD. These mutations were predicted to impair CARD–CARD interactions via the type IIb interface based on the rather conserved interaction surfaces in the death domain family (Lin et al., 2010; Ferrao and Wu, 2012). The mutant ASCCARD remained monomeric, could still form a complex with VHHASC, and crystallized into needle-shaped crystals, which allowed us to solve the structure (Fig. 2 B).
Compared with the crystal structure of VHHASC alone and the NMR structure of ASC (Research Collaboratory for Structural Bioinformatics Protein Data Base code [RCSB PDB]: 2KN6; de Alba, 2009), neither VHHASC nor ASCCARD in the VHHASC–ASCCARD complex shows significant conformational changes (Fig. 2 C). Even though the structure of the complex was determined only to near-atomic resolution, it nonetheless allowed identification of residues in contact. Surprisingly, among the CDRs, only the CDR2 loop participates in ASC recognition (Fig. 2 B). In addition, the C′ strand and the C′′ strand of VHHASC, which correspond to frameworks 2 and 3 of the domain, respectively, constitute the majority of the interface with ASC (Fig. 2 B). On the side of ASCCARD, the last helix α6 and, less extensively, α1, directly contact VHHASC. Residues Thr57, Tyr59, Arg50, and Asp62 of VHHASC interact with residues Arg194, Glu190, Asp191, Arg125, Tyr187, and Gln185 of ASCCARD to form the binding interface. Importantly, the exposed Tyr187 on ASCCARD is completely masked by VHHASC, and this hydrophobic interaction with a large buried surface area presumably contributes significantly to the binding affinity (Figs. 2 B and 3 A). Collectively, our structural analysis shows that VHHASC mostly binds to the C-terminal region of human ASC, using an unusual combination of CDR2 and framework residues.
Figure 3.
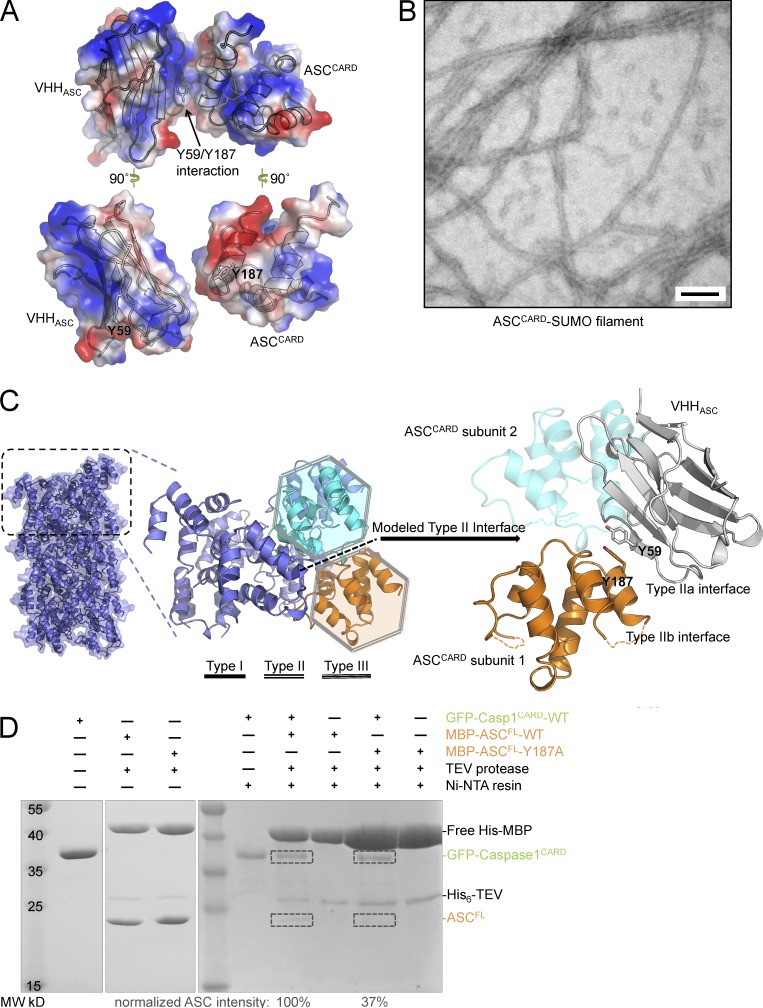
Binding of VHHASC to ASCCARD sterically occludes ASCCARD type II interface. (A) Electrostatic surface representation of the VHHASC–ASCCARD complex, highlighting tyrosine 59 in VHHASC and tyrosine 187 in ASCCARD. (B) The MBP-ASCCARD-SUMO fusion protein was incubated with TEV protease to cleave off the MBP protein, which led to polymerization of ASCCARD-SUMO. A representative electron micrograph of the resulting fibers subjected to negative staining is shown. Bar, 100 nm. (C) Representation of the ASCCARD filament modeled using the known MAVSCARD filament structure (left), a zoomed view of the type II interface of two highlighted ASCCARD subunits (middle), and illustration of the complex of VHHASC with ASCCARD subunit 1 (right). The VHH sterically clashes with ASCCARD subunit 2 bound via the type II interface (shown in transparent cyan). (D) Interaction of ASC WT and type II mutant Y187A with His-tagged caspase-1. The indicated combinations of His6-GFP-caspase-1CARD and WT or mutant His6-MBP-ASC were incubated. Where indicated, His6-MBP was cleaved off ASC using TEV protease, and His-tagged proteins were purified using Ni-NTA resin. Samples of the protein mixtures were analyzed by SDS-PAGE and Coomassie staining. Positions of the indicated proteins are marked on the right. The intensity of the ASCFL bands was determined from the scanned gel and normalized by the band intensity of GFP-caspase-1CARD.
Binding of VHHASC to ASCCARD sterically occludes ASCCARD type II interface
To understand the molecular interaction between ASCCARD and VHHASC, as well as the implications for the effect of VHHASC in inhibiting ASCCARD-mediated interactions, we attempted to identify the surfaces of ASCCARD that mediate interactions with itself and with caspase-1CARD. We first investigated the quaternary structure assumed by oligomers of ASCCARD. We constructed a sandwich-tagged MBP-ASCCARD-SUMO fusion protein. Upon removal of MBP, ASCCARD-SUMO oligomerized into extended filaments (Fig. 3 B). The SUMO fusion prevents aggregation of filaments and increased overall solubility after oligomerization. Irrespective of whether endogenous ASCCARD forms extended filaments or short filamentous assemblies under physiological conditions in cells, the ASCCARD filaments formed in vitro serve as a proxy that helps reveal structural details of CARD–CARD interactions. No structural information on the filamentous structure of ASCCARD exists, but CARD domains exhibit a sufficient degree of conservation to allow meaningful structural predictions based on other CARD structures. We therefore modeled the structure of an ASCCARD filament based on the 3.6 Å resolution cryo-EM structure of the MAVSCARD filament (Wu et al., 2014). We aligned copies of ASCCARD to each unit of the MAVSCARD filament, effectively imposing the one-stranded helical parameters (101.1° twist angle and 5.13 Å axial rise per unit) onto ASCCARD. We then superimposed the VHHASC–ASCCARD complex structure on the predicted ASCCARD filament structure (Fig. 3 C). Thus, the surface of ASCCARD used in the VHHASC interaction overlaps with its type IIb surface; the bound VHHASC sterically clashes with the adjacent subunit in the ASCCARD filament along the type II direction, suggesting that binding of VHHASC to ASCCARD interferes with CARD–CARD interactions in ASCCARD self-oligomerization via the type II interface.
ASCCARD and Caspase-1CARD also interact through type II interfaces
To probe the importance of the type II interface of ASCCARD, we analyzed the effect of mutations at this interface on ASCCARD interaction with pro–caspase-1CARD. The VHHASC–ASCCARD complex structure shows a hydrophobic surface patch near α6, primarily contributed by the exposed Tyr187 (Fig. 3 A). We generated the Y187A mutant in full-length ASC fused to the C terminus of solubilizing MBP, and cleaved off MBP in the presence of His6-GFP-caspase-1CARD. When we purified His6-GFP-caspase-1CARD with Ni-NTA resin, the amount of the ASC mutant co-purified was reduced by 63% compared with wild-type ASC (Fig. 3 D). We concluded that type II interactions are essential in mediating the ASCCARD–caspase-1CARD association. Blocking this interface by VHHASC was thus expected to abolish ASCCARD self-oligomerization, as well as its ability to recruit pro–caspase-1.
VHHASC directly inhibits ASCCARD-nucleated Caspase-1CARD and ASCCARD polymerization
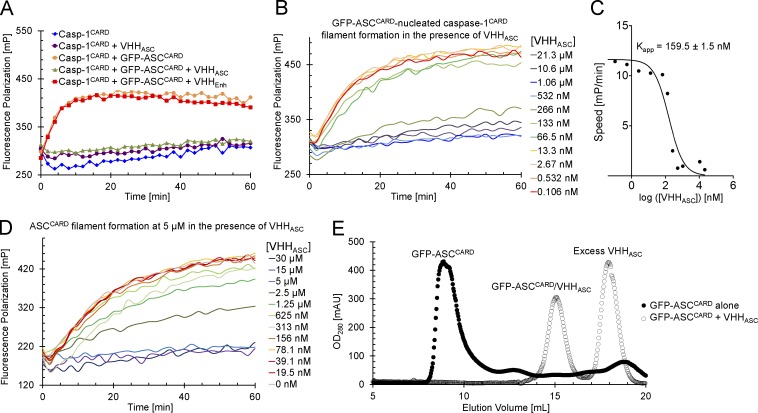
We can reconstitute the polymerization of inflammasome components in vitro using purified proteins, including the ASCCARD-nucleated polymerization of caspase-1CARD (Lu et al., 2014). We used a fluorescence polarization assay to monitor nucleation of caspase-1CARD filaments by oligomeric ASCCARD in the presence and absence of VHHASC (Fig. 4 A). This assay exploits the fact that fluorescence anisotropy increases when a fluorescent monomeric species polymerizes. In short, we labeled monomeric caspase-1CARD (N-terminal MBP, C-terminal SUMO) by means of a sortase reaction (Guimaraes et al., 2013), followed by purification of the labeled monomers (Lu et al., 2014). We added substoichiometric amounts of oligomeric GFP-ASCCARD to seed the growth of caspase-1CARD filaments upon tobacco etch virus (TEV) protease cleavage of the MBP fusion partner. We preincubated the GFP-ASCCARD seeds with either VHHASC or the anti-GFP VHHEnhancer. Incubation of the GFP-ASCCARD seeds with equimolar amounts of VHHASC abolished their ability to nucleate caspase-1CARD filament formation (Fig. 4 A). Incubation of GFP-ASCCARD with VHHEnhancer did not affect caspase-1CARD polymerization. Binding of ASCCARD by VHHASC thus directly and specifically interferes with the ASCCARD–capase-1CARD interaction.
Figure 4.
VHHASC prevents ASCCARD-nucleated polymerization of caspase-1CARD and ASCCARD. (A) ASC-nucleated caspase-1CARD polymerization in the presence of VHHASC and VHHEnhancer. Fluorescently labeled monomeric MBP-caspase-1CARD-SUMO (4 µM) was mixed with oligomeric GFP-ASCCARD (1.1 µM) preincubated with VHHASC or VHHEnhancer (1.1 µM) where indicated. MBP was cleaved from fluorescent caspase-1CARD-SUMO to allow ASCCARD-nucleated polymerization of caspase-1CARD. Polymerization was followed by measuring fluorescence polarization over time. (B) ASC-nucleated caspase-1CARD polymerization in presence of different VHHASC concentration. Polymerization of caspase-1CARD-SUMO by oligomeric GFP-ASCCARD was measured by fluorescence polymerization assays as described in A, except that GFP-ASCCARD was preincubated with the indicated concentrations of VHHASC. (C) Apparent inhibition constant of VHHASC. The initial slope of the fluorescence polarization processes in B was plotted against the logarithm of the VHHASC concentration. A single-site antagonist model was fitted to this plot to extract the apparent inhibition constant Kapp. (D) ASCCARD polymerization. Fluorescently labeled monomeric MBP-ASCCARD-SUMO (5 µM) was preincubated with VHHASC at the indicated concentrations. MBP was cleaved off fluorescent ASCCARD-SUMO to self-nucleated polymerization of ASCCARD. Polymerization was followed by measuring fluorescence polarization over time. (E) Stability of ASCCARD oligomers in presence of VHHASC. Oligomeric GFP-ASCCARD was incubated with excess VHHASC for 30 min at room temperature or left untreated. Protein samples were subsequently subjected to size-exclusion chromatography. OD280 profiles are displayed.
To determine the stoichiometry of inhibition by VHHASC, we preincubated a fixed, substoichiometric amount of ASCCARD with various amounts of VHHASC, and subsequently monitored caspase-1CARD filament formation (Fig. 4 B). We fitted a single-site antagonist model to the obtained initial slopes of the fluorescence polarization traces and determined the apparent binding constant (Kapp) between VHHASC and GFP-ASCCARD to be 159.5 ± 1.5 nM (Fig. 4 C). Kapp is of the same order of magnitude as the concentration of GFP-ASCCARD, suggesting that 1:1 binding of VHHASC inhibits ASCCARD–caspase-1CARD interaction.
In addition, we assayed the effect of VHHASC directly on ASCCARD oligomerization. We used fluorescently labeled monomeric MBP-ASCCARD-SUMO in an experiment similar to the caspase-1CARD polymerization assay, but exploited the fact that, at sufficient concentrations, ASCCARD-SUMO polymerizes in the absence of any nucleators. Addition of an approximately equimolar amount of VHHASC to the polymerizing ASCCARD was required to block polymerization (Fig. 4 D), suggesting that 1:1 binding of VHHASC to ASCCARD also inhibits its self-oligomerization. To compare the affinity of VHHASC for ASCCARD with the affinities of ASC CARD–CARD interactions, we incubated oligomeric GFP-ASCCARD with excess VHHASC. GFP-ASCCARD on its own behaves as an oligomer as assayed by size-exclusion chromatography. After incubation with excess VHHASC, however, only smaller dimeric complexes of ASCCARD–VHHASC as well as monomeric VHHASC, were observed (Fig. 4 E), suggesting that VHHASC may even disassemble already activated inflammasome complexes by competing with ASC CARD–CARD interactions. Collectively, the data demonstrate that VHHASC—by occluding the type IIb interface of ASCCARD—perturbs CARD–CARD interactions of ASCCARD with both ASCCARD and caspase-1CARD.
VHHASC blocks NLRP3- and AIM2-dependent inflammasome activation
In contrast to conventional antibodies, VHHs can retain their binding properties in the reducing milieu of the cytosol. VHHs can therefore be expressed cytosolically to perturb protein function under physiological conditions and at endogenous concentrations of the wild-type form of a VHH target (Ashour et al., 2015; Helma et al., 2015). In this sense, VHH-mediated perturbations are fundamentally distinct from analyses of mutant forms of the target protein, or from siRNA or Cas9/CRISPR-based approaches where the goal is a reduction in expression levels of the target protein.
To study the role of ASC CARD during inflammasome assembly in human cells, we generated derivatives of the human monocyte-like cell line THP-1 that inducibly express HA-tagged VHHASC or the control VHH NP1 (Ashour et al., 2015) upon addition of doxycycline. Both VHHs were induced to similar levels of expression (Fig. 5 A), did not alter levels of ASC (Fig. 5 B), and did not affect LPS-induced TNF secretion in phorbol-12-myristate-13-acetate (PMA)–differentiated cells (Fig. 5 C). To test the effect of VHHASC on NLRP3 inflammasome activation, we differentiated THP-1 cells with PMA over night, and then induced VHH expression for 24 h. For activation, doxycycline-induced cells were treated with LPS for 3 h and with the potassium ionophore nigericin for 45 min, both in the absence of doxycycline.
Figure 5.
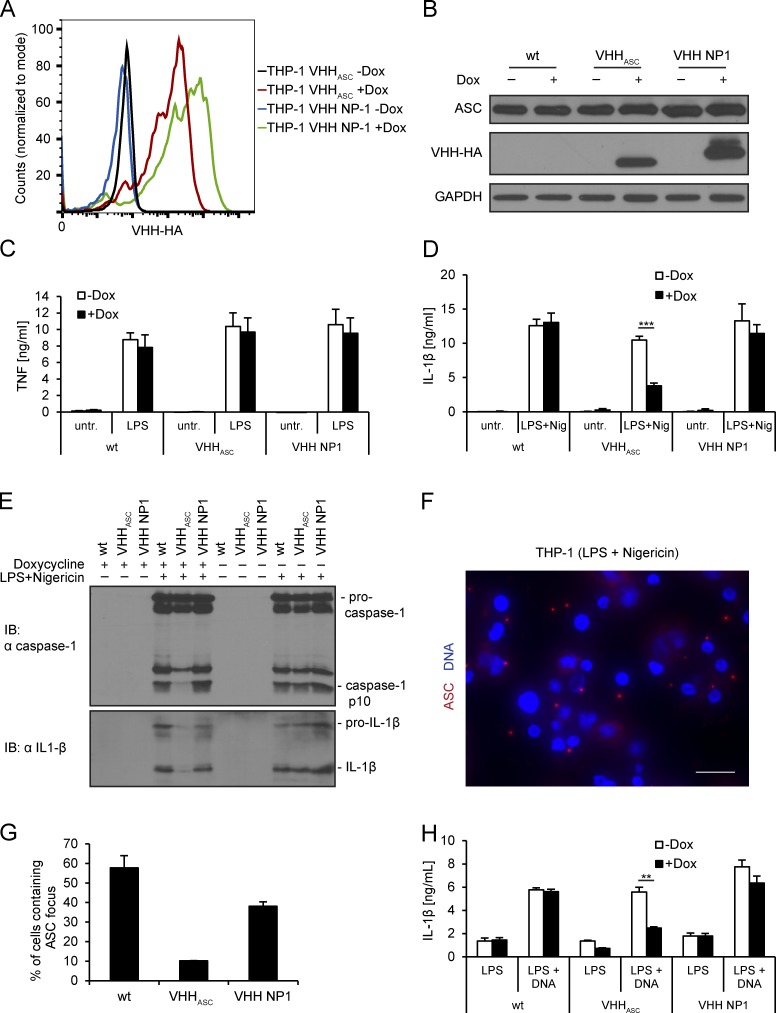
VHHASC prevents activation of NLRP3 and AIM2 inflammasomes in THP-1 cells. (A) THP-1 cell lines inducibly expressing VHHASC-HA or VHH NP1-HA were cultivated for 24 h in the absence or presence of doxycycline, fixed, stained for HA, and analyzed by flow cytometry. (B–E) Wild-type THP-1 cells and the cell lines described in A were differentiated and VHH expression induced for 24 h. (B) Cells were treated with 200 ng/ml LPS for 3 h, harvested, and cell lysates analyzed by immunoblot with anti-ASC, anti-HA, and anti-GAPDH. (C) Cells were treated with 200 ng/ml LPS for 4 h or left untreated. Supernatants were analyzed by TNF ELISA. (D and E) Cells were treated with LPS for 3 h and nigericin for 45 min, or left untreated. Supernatants were analyzed by IL-1β ELISA (D) and by immunoblots with anti–IL-1β and anti-caspase-1 p10 (E). Immunoblots representative of three experiments are shown. The asterisk indicates a statistically significant difference (Student‘s t test; P < 0.001). (F and G) Cells treated as in D were fixed and stained for DNA and ASC; images were recorded by wide field fluorescence microscopy (F) and the fraction of cells containing ASC foci was quantified (G). Data from three independent experiments with at least 500 cells per condition ± SEM is shown. Bars, 20 µm. (H) THP-1 cell lines cultivated as described in B were treated with LPS for 3 h and transfected with poly (dA:dT) or transfection agent only. Supernatants were harvested after 4 h and IL-1β levels were quantified by ELISA. The asterisk indicates a statistically significant difference (Student‘s t test; P = 0.002). Data from three independent experiments was quantified for all ELISA results and mean values ± SEM are shown.
Potassium efflux activates the NLRP3 inflammasome. This involves the recruitment and polymerization of ASC and pro–caspase-1, autocatalytic activation of caspase-1, and subsequent maturation of IL-1β and -18. As assessed by immunoblot and ELISA, parental THP-1 cells secreted processed caspase-1 and IL-1β in response to NLRP3 triggers (Fig. 5, D and E). Doxycycline treatment did not affect caspase-1 activation or IL-1β conversion. Moreover, on average, 58% of the cells contained an ASC focus as detected by immunofluorescence microscopy using anti-ASC antibodies (Fig. 5, F and G). Although VHHASC cell lines behaved like unmodified THP-1 cells in the absence of doxycycline, caspase-1 cleavage and IL-1β secretion were substantially reduced in cells induced to express VHHASC. Only 10% of the treated cells contained an ASC focus when expression of VHHASC was induced. Induction of the control VHH NP1 neither changed secretion of caspase-1 and IL-1β, nor substantially impaired ASC focus formation. VHHASC thus specifically reduces the assembly of ASC foci and perturbs the recruitment and autocatalytic cleavage of pro–caspase-1 and, consequently, IL-1β secretion.
When activated by DNA, AIM2 employs ASC to recruit and activate pro–caspase-1 similar to NLRP3. When THP-1 cell lines were transfected with poly(dA:dT), IL-1β release was substantially reduced by expression of VHHASC, but not by the control VHH NP1 (Fig. 5 H). Consistent with our interpretation of the in vitro data, VHHASC masks the CARD of soluble ASC molecules in living cells and thus prevents CARD–CARD interactions between ASC molecules and recruitment of pro–caspase-1.
VHHASC blocks NAIP/NLRC4-dependent inflammasome activation
Whereas NLRP3 and AIM2 receptors depend on ASC to recruit pro–caspase-1 to nascent inflammasomes, NAIP–NLRC4 inflammasome complexes can directly engage pro–caspase-1 via homotypic CARD–CARD interactions. In mouse cells, NLRC4-dependent secretion of IL-1β is more efficient in the presence of ASC (Broz et al., 2010a). To investigate the role of ASC in NAIP/NLRC4 inflammasomes in human cells, we differentiated THP-1 cell lines and activated NAIP/NLRC4 inflammasomes with the Shigella flexneri needle protein MxiH, which directly binds to NAIP and thus activates NAIP/NLRC4 inflammasomes (Yang et al., 2013). MxiH was delivered into the cytosol using the anthrax toxin delivery system (Arora and Leppla, 1993; Young and Collier, 2007). In brief, cells were treated with the anthrax toxin protective antigen (PA) and a fusion protein of MxiH and the N terminus of the anthrax lethal factor, LFn. PA binds to the anthrax receptor at the cell surface, is proteolytically activated to form the prepore, and then recruits LFn-MxiH. The complex is endocytosed and LFn-MxiH is translocated into the cytosol through the PA pore upon acidification of endosomes.
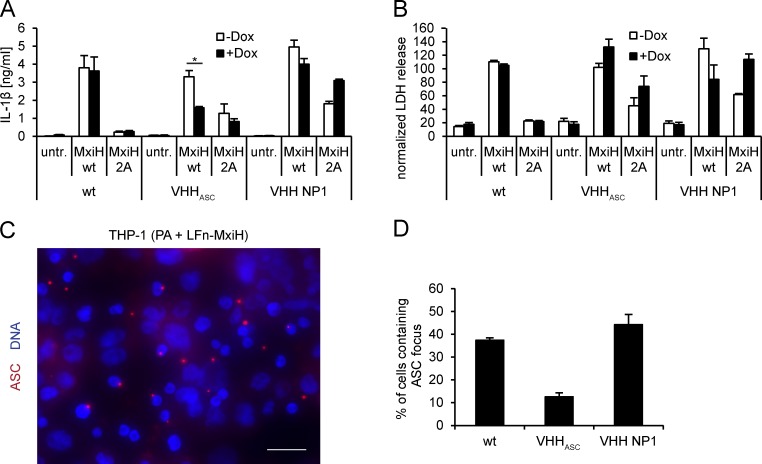
In parental THP-1 cells, translocation of MxiH to the cytosol induced robust IL-1β secretion and pyroptotic cell death, as judged by release of cytosolic LDH (Fig. 6, A and B). To show that the observed IL-1β secretion and pyroptotic cell death were indeed dependent on MxiH, and not caused by bacterial contaminants contained in our protein preparations, we used a mutant version of MxiH. The mutant MxiH 2A, in which two conserved hydrophobic residues in the needle protein helical hairpin region were changed to alanine (Yang et al., 2013), was indeed substantially less potent than MxiH in activating IL-1β secretion and cell death. In cells expressing VHHASC, cytokine secretion was clearly diminished in response to MxiH delivery, whereas control cells that expressed VHH NP1 behaved like the parental THP-1 line. Human NAIP/NLRC4 inflammasome activation thus requires ASC for efficient cytokine maturation. IL-1β secretion was diminished slightly in THP-1 VHHASC cells in which VHH expression was not induced. This is most likely caused by leakiness of the inducible expression system.
Figure 6.
VHHASC prevents IL-1β secretion, but not cell death, by NAIP/NLRC4 inflammasomes in THP-1 cells. (A and B) WT THP-1 cells and cell lines inducibly expressing VHHASC-HA or VHH NP1-HA were differentiated and VHH expression induced for 24 h. Cells were treated with PA and LFn-MxiH WT or 2A for 3 h or left untreated. IL-1β in the supernatants was quantified by ELISA (A), LDH activity was quantified and normalized to cells lysed in 1% Triton X-100 (B). Data from three independent experiments was quantified and mean values ± SEM are shown. The asterisk indicates a statistical significant difference (Student‘s t test; P = 0.039). (C and D) THP-1 cell lines were differentiated and induced as described in A, and subsequently treated with PA and MxiH WT or 2A for 2 h in the presence of 50 µM z-YVAD-cmk. Cells were fixed and stained for DNA and ASC; images were recorded by wide field fluorescence microscopy (C) and the fraction of cells containing ASC foci was quantified (D). Bars, 20 µm. Data from three independent experiments with at least 500 cells per condition ± SEM is shown.
In parental THP-1 cells and THP-1 cell lines expressing VHH NP1, we observed ASC foci in 37 and 44% of the treated cells (Fig. 6, C and D). In THP-1 cells that express VHHASC, the fraction of cells containing ASC foci was reduced to 13%. As is the case for NLRP3 inflammasomes, VHHASC acted both at the level of ASC focus formation and in the recruitment of pro–caspase-1 to perturb activation of NAIP/NLRC4 inflammasomes.
In contrast to cytokine secretion, cell death by pyroptosis occurred at comparable levels in THP-1, THP VHHASC, and THP VHH NP1, irrespective of whether VHH expression was induced or not. This suggests that human ASC is dispensable for pyroptosis. Direct pro–caspase-1 recruitment to NLRC4 may be sufficient for cell death, as has been proposed for mouse macrophages infected with Salmonella (Broz et al., 2010b). In summary, our data suggest that NLRC4-dependent IL-1β maturation and secretion requires ASC. Caspase-1 activation very likely takes place at the macromolecular assemblies visualized as ASC foci. In contrast, cell death by pyroptosis does not require ASC or ASC foci, and is presumably initiated in smaller assemblies.
VHHASC-EGFP assembles along ASC filaments in response to NLRP3 and AIM2 activators
In the presence of VHHASC-HA, we observed reduced numbers of ASC foci in response to NLRP3 triggers. Given that VHHASC binds to the CARD of ASC, and that NLRP3 activation induces the formation of ASCPYD filaments in vitro, we were surprised that we were unable to stain any macromolecular structures containing ASC or VHHASC-HA in cells that lacked ASC foci. Because we used conventional indirect immunofluorescence microscopy with primary antibodies against the PYD of ASC or against HA in combination with fluorescently-labeled secondary antibodies, we reasoned that impaired accessibility of antigens for staining with conventional antibodies, or the small number of molecules involved might be the explanation.
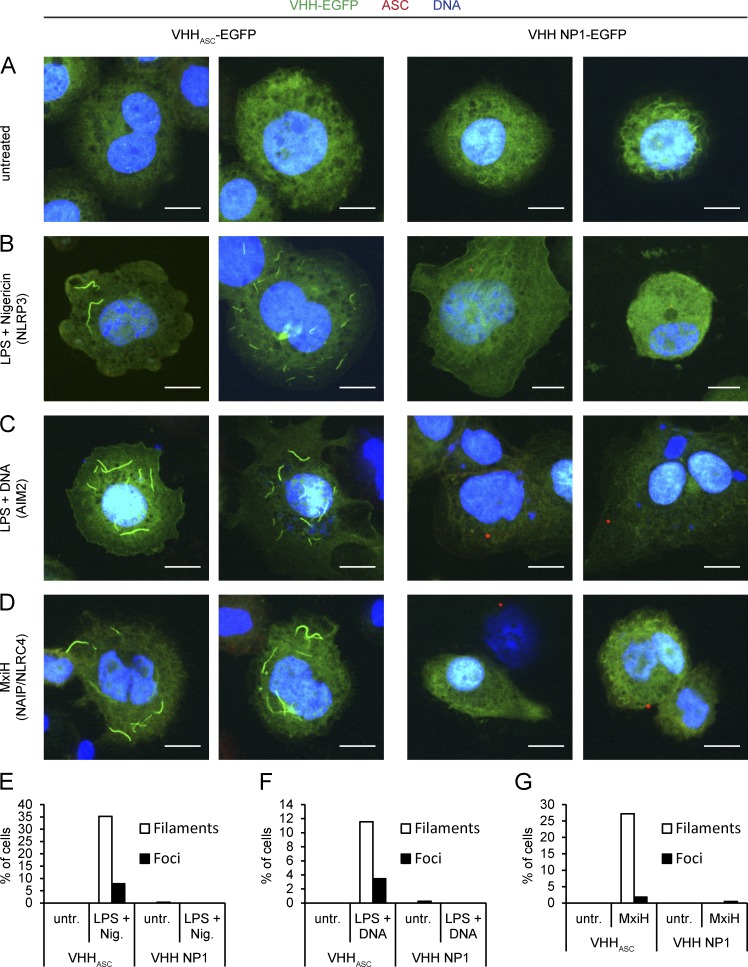
To address these issues, we generated THP-1 cell lines that inducibly expressed a VHHASC-EGFP fusion protein or the control VHH NP1-EGFP. When THP-1 VHHASC-EGFP cells were treated with LPS and nigericin to activate NLRP3 inflammasomes, we observed two different outcomes (Fig. 7, B and E): few cells contained ASC foci that could be stained with conventional anti-ASC antibodies and that were EGFP positive—they presumably represent the fraction of cells in which VHH levels were too low to impair ASC focus formation, but in which VHHASC may still impair pro–caspase-1 recruitment. We also observed many cells that exhibited one, a few, or multiple filamentous EGFP structures of variable length that could not be stained with ASC PYD antibodies. In the absence of inflammasome triggers, cells that expressed VHHASC-EGFP lacked such structures (Fig. 7, A and E). Filamentous structures were absent also from cells that expressed the control VHH NP1-EGFP, even when treated with NLRP3 inflammasome triggers. Filamentous EGFP was thus observed only upon meeting two conditions: (1) when ASCPYD could build filaments in response to inflammasome triggers and (2) when VHHASC-EGFP was expressed. We conclude that these EGFP-positive structures were formed along endogenous ASC filaments, assembled upon NLRP3-nucleated polymerization of ASCPYD. We hypothesize that an excess of VHHASC-EGFP thus masked ASCCARD, but still permitted the formation of ASCPYD filaments. This is the first time that ASC filaments could be visualized at physiological ASC levels and therefore cannot be attributed to overexpression artifacts. These filaments must represent an intermediate phenotype, as ASCCARD was masked by VHHASC and could not complete its physiological function. That ASC formed long, unbranched filaments, as opposed to densely packed ASC foci, suggests that the organization of multiple ASC filaments into a single focus requires accessible ASCCARD. The ASCCARD, beyond its role in the recruitment of pro–caspase-1, thus serves a function in the organization of multiple ASC filaments into a single dense ASC focus, possibly by cross-linking ASC filaments assembled through the ASCPYD.
Figure 7.
VHHASC-EGFP assembles along ASC filaments in response to inflammasome triggers. (A–D) THP-1 cell lines inducibly expressing VHHASC-EGFP or VHH NP1-EGFP were differentiated and VHH expression induced for 24 h. Cells were left untreated (A), treated with LPS for 3 h and nigericin for 45 min to trigger NLRP3 inflammasomes (B), treated with LPS for 3 h and transfected with DNA for 4 h to trigger AIM2 inflammasomes (C), or treated with PA and LFn-MxiH WT in the presence of 30 µM z-YVAD-cmk for 2 h to trigger NAIP/NLRC4 inflammasomes (D). Cells were fixed and stained for DNA and ASC; images of single planes were recorded by spinning disc confocal microscopy. Bars, 10 µm. Representative images from at least three experiments are shown. (E–G) THP-1 cell lines were differentiated, VHH expression induced, and NLRP3, AIM2, and NLRC4 inflammasomes activated or cells left untreated as in A–D, but in the presence of 50 µM z-YVAD-cmk. Cells were fixed and mounted with mounting medium containing DAPI; Z stacks were recorded by spinning disc confocal microscopy. At least 300 cells per condition were analyzed and cells containing green-fluorescent filaments or foci were manually counted. One typical experiment representative of at least three independent experiments was quantified.
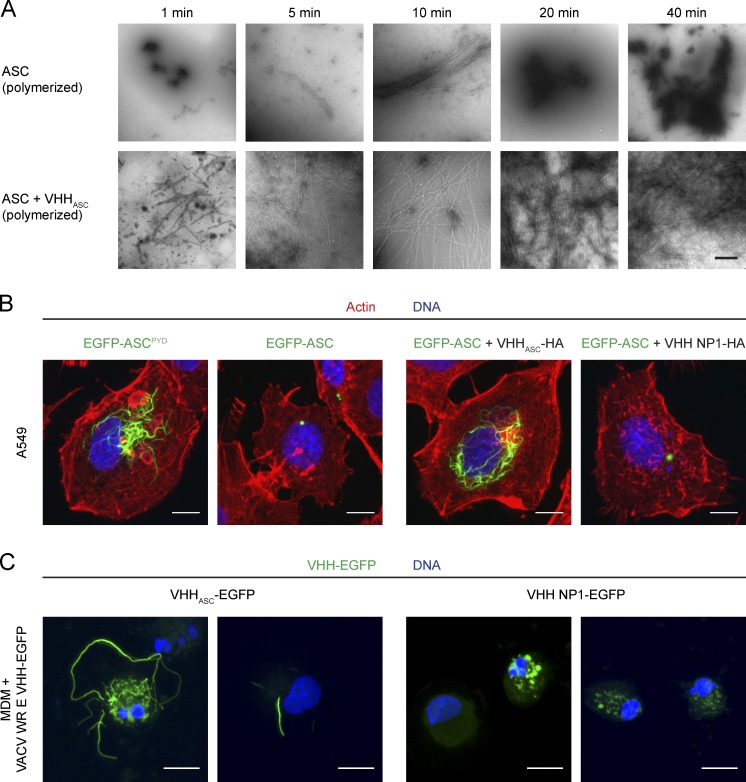
In vitro, full-length ASC assembles into highly cross-linked filaments, while ASCPYD assembled into long, linear filaments (Cai et al., 2014; Lu et al., 2014). If VHHASC-EGFP indeed prevented cross-linking of full-length ASC filaments (vide supra), VHHASC should also prevent cross-linking of ASC filaments in vitro. To test the effect of VHHASC on the quaternary structure of full-length ASC, we formed a complex of VHHASC and recombinantly expressed full-length ASC fused to MBP, in which MBP prevents ASC oligomerization. When MBP was removed from the complex by TEV protease cleavage, ASC/VHHASC polymerized into long, linear filaments as observed by negative-stain EM after only 5 min (Fig. 8 A). These filaments have a diameter of >20 Å, corresponding to a core PYD filament (≈9 Å; Lu et al., 2014) decorated by ASCCARD–VHHASC monomers. Importantly, these filaments are not cross-linked, and thus resemble ASCPYD filaments. When MBP was cleaved from MBP-ASC in the absence of VHHASC, ASC polymerized as well, but formed highly cross-linked bundled structures which precipitated over time. After 20 min, essentially no more filamentous structures were recognized and ASC formed an amorphous, dense mass. By masking the ASCCARD, VHHASC therefore may prevent cross-linking of full-length ASC filaments in vitro, in support of the model deduced from our microscopy studies.
Figure 8.
VHHASC stabilizes ASC filaments formed through ASCPYD polymerization in vitro and in cells. (A) MBP-ASC or MBP-ASC preincubated with VHHASC was incubated with TEV protease to remove the MBP fusion and allow polymerization. At the indicated times, the reaction was stopped and the resulting macromolecular structures were visualized by negative stain electron microscopy. Bars, 500 nm. (B) A549 cells were transiently transfected with expression vectors for EGFP-ASCPYD or EGFP-ASC in the absence or presence of expression vectors for VHHASC or VHH NP1. 24 h after transfection, cells were fixed and stained for DNA and filamentous actin. Z-stacks of representative cells were recorded by spinning disc confocal microscopy and maximum intensity projections are displayed. Bars, 10 µm. (C) Human monocyte-derived macrophages were differentiated in the presence of 10% FBS and 100 ng/ml M-CSF for 5 d and subsequently infected with mature virions of vaccinia virus WR E VHHASC-EGFP L mCherry or WR E VHH NP1-EGFP L mCherry at an MOI of 20. Cells were fixed 18 h after infection and stained for DNA. Z stacks of representative cells were recorded by spinning disc confocal microscopy and images of maximum intensity projections are shown. Bars, 10 µm.
To more directly visualize ASC in the observed filamentous assemblies, we took advantage of the propensity of overexpressed EGFP-ASC to assemble into ASC foci when overexpressed in epithelial cells (Masumoto et al., 2001a). Although nucleation of EGFP-ASC foci themselves is artificial and does not depend on any upstream nucleators, formation of the macromolecular structures seems to be governed by the PYD/PYD and CARD–CARD interactions that are assumed to underlie the assembly of endogenous ASC foci (Lu et al., 2014; Sahillioglu et al., 2014). This includes the finding that EGFP-ASCPYD assembles into filaments, as it does in vitro. When EGFP-ASC or EGFP-ASCPYD was overexpressed in epithelial A549 cells, we observed foci and filaments, respectively (Fig. 8 B). When full-length EGFP-ASC was expressed in the presence of VHHASC, however, we did observe EGFP-ASC filaments but no ASC foci. These filaments resembled the EGFP-ASCPYD filaments, indicating that in masking ASCCARD, VHHASC blocked further compaction of ASC filaments formed by PYD polymerization.
Activation of AIM2 by DNA-binding nucleates the formation of ASC PYD filaments in vitro, too (Lu et al., 2014). To validate our model, we tested whether DNA—as an alternative activator of ASC PYD filaments—would also lead to the assembly of VHHASC-EGFP along endogenous ASC structures in a cellular context (Fig. 7, C and F). Indeed, we observed filamentous EGFP in poly(dA:dT)-transfected THP-1 cells expressing VHHASC-EGFP, but not in cells expressing the control VHH NP1-EGFP. The occurrence of filamentous VHHASC-EGFP structures in response to two different activators of ASC PYD polymerization thus confirmed that filamentous VHHASC-EGFP assemblies are readouts for ASC PYD filament formation.
VHHASC-EGFP assembles along ASC filaments in response to NAIP/NLRC4 triggers
Efficient cytokine maturation in response to the NAIP/NLRC4 trigger MxiH involves multimolecular ASC complexes because ASC foci accrued upon stimulation and VHHASC reduced IL-1β secretion. Unlike NLRP3 and AIM2, NLRC4 does not contain a PYD and therefore cannot directly nucleate the oligomerization of ASC filaments by PYD–PYD interactions. Two scenarios that allow NLRC4 to induce the formation of ASC filaments are conceivable: NLRC4 may directly nucleate the formation of ASC filaments via the ASCCARD. Indeed, both overexpressed ASCCARD in cells (Masumoto et al., 2001b), as well as purified ASCCARD in vitro (Fig. 3 B), can oligomerize into filament-like structures. Alternatively, NLRC4 may bind one or several ASC molecules through its CARD, which in turn changes the conformation of the bound ASC molecules, so that ASC itself can nucleate the formation of ASC filaments using its PYD. This scenario would resemble the situation observed for NLRP3 and AIM2. To test which of the two possibilities applies, we triggered THP-1 VHHASC-EGFP and THP-1 VHH NP1-EGFP with cytosolically delivered MxiH in the presence of the caspase-1 inhibitor z-YVAD-cmk to reduce pyroptotic cell death. When cells expressing VHHASC-EGFP were treated with MxiH, we observed filamentous EGFP structures in many cells (Fig. 7, D and G). The EGFP structures were similar to those found when NLRP3 was activated by K+ efflux (nigericin), or when AIM2 was activated by DNA. Filamentous assemblies did not occur in the absence of inflammasome triggers and were never observed in cells expressing VHH NP1-EGFP. Based on the specificity of VHHASC for ASCCARD, the EGFP structures seen upon NLRP3/AIM2 activation must be comprised of VHHASC-EGFP molecules aligned along ASCPYD filaments, by binding to the free ASCCARD. We therefore reasoned that NAIP/NLRC4 activation also involves the formation of ASC filaments polymerized via ASCPYD. In that case, one or several initiator ASC molecules are presumably activated by NLRC4 and turned into nucleators of ASCPYD polymerization.
In summary, our data on VHHASC-EGFP filament formation demonstrate that the activation of NAIP/NLRC4 inflammasomes involves the formation of ASC filaments that assemble by polymerization of ASCPYD. As in the case of NLRP3, the ASCCARD is likely to also have a dual role in inflammasome activation: it mediates cross-linking of linear ASC fibers, and recruits the effector pro–caspase-1 to the macromolecular complex.
VHHASC-EGFP assembles along ASC filaments in vaccinia virus–infected human macrophages
To rule out that the observed VHHASC-EGFP assemblies along ASCPYD filaments constitute an artifact of the monocytic leukemia cell line THP-1, we analyzed inflammasome activation in differentiated human macrophages derived from peripheral blood monocytes. To express VHHASC-EGFP in macrophages and at the same time activate inflammasomes, we generated a recombinant vaccinia virus (VACV) strain, which expresses VHHASC-EGFP or VHH NP1-EGFP under the control of an early viral promoter and mCherry under the control of a late viral promoter. Infection of mouse macrophages and THP-1 cells with this DNA virus induces inflammasome activation in an AIM2 or NLRP3-dependent manner (Delaloye et al., 2009; Hornung et al., 2009). Consistently, THP-1 cells infected with mature virions of VACV E VHHASC-EGFP exhibited filamentous EGFP-positive assemblies (unpublished data). Many monocyte-derived macrophages infected with VACV E VHHASC-EGFP contained filamentous assemblies of EGFP, whereas control cells infected with VACV E VHH NP1-EGFP did not (Fig. 8 C). We did not observe mCherry expression in the vast majority of cells, suggesting that virus entry or early gene expression is sufficient for inflammasome activation. Of note, most VACV-infected human monocyte–-derived macrophages exhibited signs of pyroptotic or apoptotic cell death, including nuclear fragmentation. In conclusion, we found that VHHASC-EGFP assemblies along ASCPYD filaments also occur in primary human macrophages and therefore visualize a genuine intermediate in inflammasome activation.
DISCUSSION
Inflammasomes are critical information hubs of the innate immune system. A lack of appropriate tools to perturb inflammasomes in physiological conditions has hampered a more complete molecular understanding of the underlying cell biology and of the molecular interactions involved. We generated and characterized VHHASC, an alpaca single domain antibody that recognizes the CARD of the human inflammasome adapter ASC. Its introduction into the cytoplasm of living cells allowed us to occlude a key interaction interface of ASC, while leaving the function of its ASCPYD unperturbed. This approach proved to be more informative than genetic ablation of ASC altogether. The application of VHHs to explore events in the cytoplasm complements more conventional genetic approaches: without having to resort to deletions or point mutations, protein–protein interactions can be perturbed conditionally. The ability of VHHs to serve as crystallization chaperones for proteins that have proven difficult to crystallize, too, allows identification of the exact molecular interfaces concerned. VHHs thus provide new mechanistic insights.
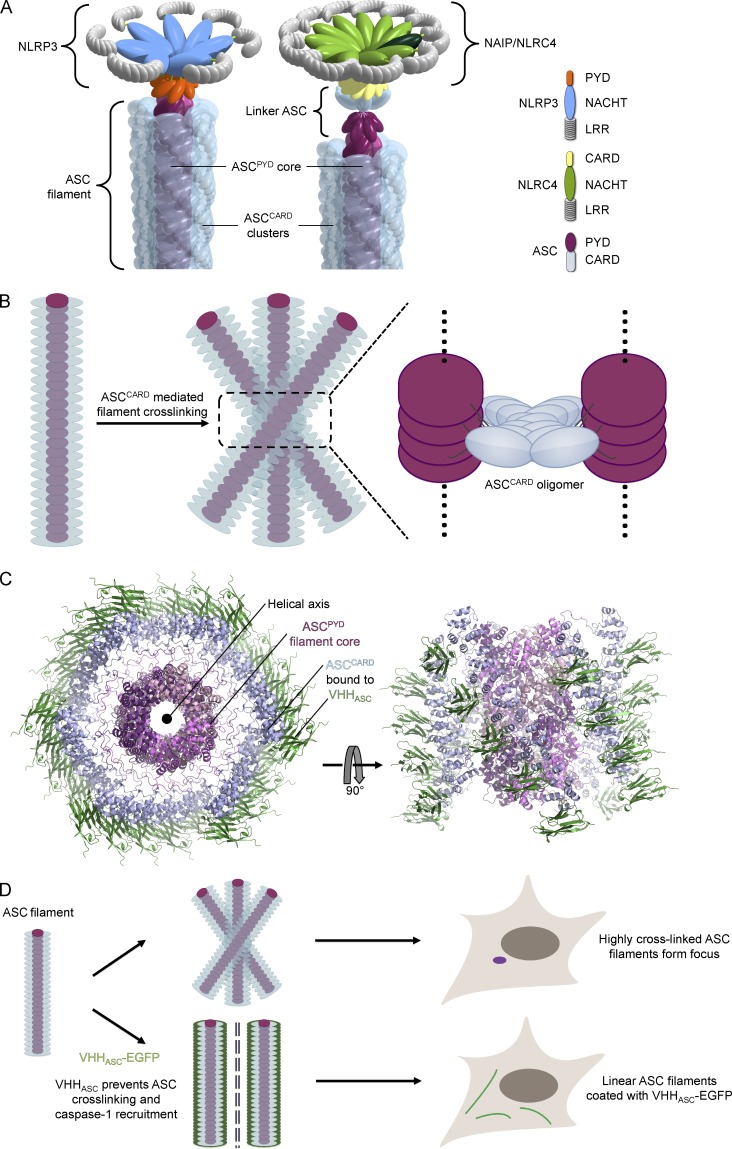
Using VHHASC, we demonstrated the role of ASC in NLRP3, AIM2, and NAIP/NLRC4-dependent IL-1β secretion. VHHASC also allowed us to tease apart the functions of the ASC domains involved: ASCPYD forms filaments in response to its direct nucleation by NLRP3PYD and AIM2PYD, or in response to the engagement of linker ASC molecules by NLRC4CARD (Fig. 9 A). ASCCARD appears critical in cross-linking the multiple ASCPYD filaments formed (Fig. 9 B), which consolidates all ASC molecules into a single supramolecular organizing center (Kagan et al., 2014; Hauenstein et al., 2015), also known as ASC speck or focus. Moreover, we confirmed the role of ASCCARD in the recruitment and subsequent activation of pro–caspase-1.
Figure 9.
Molecular model of inflammasome activation. (A) Mechanism of ASC filament formation nucleated by NLRP3 (left) and NAIP/NLRC4 (right). (B) Model of ASC filament cross-linking mediated by ASCCARD. (C) Modeled structure of ASC filament covered with VHHASC. Relative arrangement of ASCPYD, ASCCARD, and VHHASC is shown. ASCCARD–VHHASC unlikely follows the exact helical symmetry of the PYD filament core as a result of the flexible linker between PYD and CARD. (D) Schematic representation of ASC focus formation in unperturbed conditions, and formation of ASC filaments coated with VHHASC-EGFP.
Identification and characterization of VHHASC included solving the structure of the VHHASC alone and in complex with ASCCARD. We thus established that VHHASC occludes the type II interface of the ASCCARD. Immunoprecipitation experiments demonstrate that VHHASC readily interacts with ASC from unstimulated cells, suggesting that coexpressed VHHASC likely forms stable complexes with ASC in cells. We further showed that the type II interface of ASC is required for homotypic CARD–CARD interactions with other ASC molecules and pro–caspase-1, and that both interactions were perturbed by VHHASC in well-defined in vitro polymerization assays.
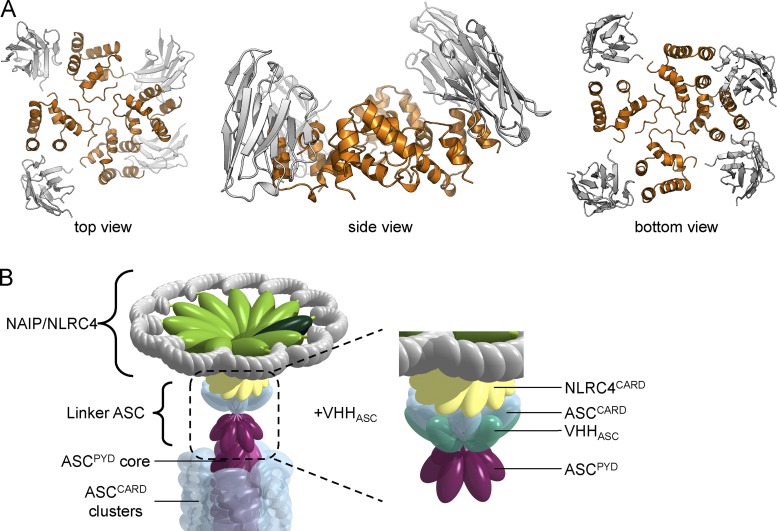
Our findings—in vitro, in cell lines, and in primary cells—expand a model of inflammasome activation that involves the local polymerization of inflammasome components, recently put forward based on in vitro studies and yeast prion assays (Cai et al., 2014; Lu et al., 2014). We demonstrate that endogenous ASC can form ASCPYD filaments at endogenous ASC levels in the relevant cell type, ruling out any artifacts that may be caused by overexpression or by a lack of regulatory factors. Once NLRP3 is activated, NLRP3PYD directly nucleates ASCPYD filaments by inducing an ASC conformation that drives polymerization. Assembly of NAIP/NLRC4 inflammasomes is a more intriguing case: activation of NAIP/NLRC4 by bacterial needle proteins yields ASCPYD filaments, as well. We therefore reasoned that NLRC4CARD must use linker molecules of ASC via its CARD. This is in line with yeast prion assays, in which NLRC4CARD-triggered formation of ASC–Sup35C prions was dependent on both oligomerization-competent PYD and CARD (Cai et al., 2014). In a mechanism that requires further structural characterization, NLRC4CARD induces an active conformation of ASC by binding its ASCCARD, which then nucleates PYD-mediated ASC filaments (Fig. 9 A). Mere engagement of the ASC CARD was not sufficient for activation, as binding of ASCCARD by VHHASC neither induced the formation of filaments nor led to IL-1β secretion. Similarly, in yeast experiments, caspase-1CARD was insufficient to induce polymerization of ASC–Sup35C prions (Cai et al., 2014). In light of our structural understandings, the VHHASC–ASC complex would presumably retain its ability to interact with NLRC4. Because complex formation occludes only the type IIb interface, the intact type I, III, and IIa interfaces would allow the VHHASC-bound ASC to interact with activated NLRC4 by forming a small layer of ASC to nucleate ASCPYD filament formation (Fig. 10).
Figure 10.
Model of small oligomer of ASCCARD complexed by VHHASC. (A) An ASCCARD oligomer (orange) with bound VHHASC (gray) was modeled based on the MAVSCARD filament symmetry and the ASCCARD/VHHASC structure. VHHASC occludes type IIb interface of ASCCARD, but does not interfere with small oligomer assembly. (B) A few ASC molecules bound to activated NLRC4 are sufficient to nucleate the formation of ASCPYD filaments even in the presence of VHHASC.
ASC is dispensable for NAIP/NLRC4-dependent pyroptosis in human cells, because VHHASC does not impair LDH release in response to MxiH treatment. Similarly, Salmonella-induced pyroptosis of mouse macrophages does not require ASC, whereas it is necessary for efficient cytokine release (Broz et al., 2010b). Direct recruitment of pro–caspase-1 by NLRC4 may be sufficient to trigger caspase-1–dependent cell death. However, neutrophils are insensitive to pyroptosis when NAIP/NLRC4-dependent cytokine secretion was induced by Salmonella infection (Chen et al., 2014). The fact that pyroptosis did not occur in neutrophils from ASC−/− mice, despite the presence of activated NLRC4 and pro–caspase-1, shows that regulation of pyroptosis is more complex than currently understood.
By masking the type II interface of ASCCARD, but not ASCPYD, we stabilized an intermediate state in inflammasome activation (Fig. 9 C). We often observed multiple ASCPYD filaments at the same time, indicating that nucleation can occur at multiple sites. Typically, only a single ASC focus is observed per cell, which suggests that several ASC filaments coalesce into a single microscopic structure during inflammasome assembly. This requires ASCCARD, which is in line with our observation that in vitro inclusion of VHHASC prevents ASC CARD–CARD interactions and blocks the cross-linking of full-length ASC filaments (Fig. 9 D). We cannot rule out that ASCCARD might also facilitate a transport mechanism that brings multiple ASC filaments together. A coalescence step involving the cross-linking of multiple ASC filaments would elegantly explain the formation of inflammasomes involving different NLRs, such as the mixed NLRP3/NLRC4 inflammasomes observed in Salmonella-infected mouse macrophages (Man et al., 2014). Activation of different NLRs may trigger polymerization of individual ASC filaments that later join to form a single ASC focus. Lastly, masking ASCCARD with VHHASC also prevents the recruitment of caspase-1 and therefore prevents the ultimate outcome of inflammasome activation, the autocatalytic activation of caspase-1. This work has implications for the development of antiinflammatory agents that could work by targeting the type II interaction face on ASCCARD and may pave the way for further perturbation studies using single domain antibodies as research tools.
MATERIALS AND METHODS
Primary cells
Human monocytes were isolated from human whole blood buffy coat obtained from VWR International. Cells were purified using a Ficoll-Paque Plus (GE Healthcare) cushion, and monocytes adhered to coverslips in RPMI for 2 h. Cells were differentiated in RPMI supplemented with 10% FBS and 100 ng/ml recombinant human M-CSF (BioLegend) for 5 d.
Cell lines
Human epithelial A549 cells were grown in DMEM supplemented with 10% FBS; human monocyte-like THP-1 cells were cultivated in RPMI medium supplemented with 10% FBS and 50 µM 2-mercaptoethanol. THP-1 cell lines inducibly expressing VHHASC-HA, VHH NP-1-HA, VHHASC-EGFP, VHH NP-1-EGFP, and EGFP were generated using lentiviruses produced with derivatives of pInducer20 (Meerbrey et al., 2011) and selected in THP-1 medium with 500 µg/ml geneticin.
Virus strains
Vaccinia virus strains Western Reserve (WR) E VHHASC-EGFP L mCherry and WR E VHH NP1-EGFP L mCherry express VHH-EGFP fusions under the control of the J2R early promoter and mCherry under the control of the F17R late promoter. The virus strains were derived from WR WT using plasmids based on pJS4 (Chakrabarti et al., 1997) to insert transgenes into the tk locus. Recombinant viruses were identified based on mCherry expression and purified through three rounds of plaque purification. Mature virions were produced in BSC-40 cells and purified from cytoplasmic extracts through a 16% sucrose cushion in 20 mM Tris, pH 9.0.
Reagents
Doxycycline hyclate was purchased from Sigma-Aldrich; E. coli K12 LPS, nigericin, and poly(dA:dT) were obtained from InvivoGen. PMA was purchased from Santa Cruz Biotechnology, Inc.; TAK-242 was obtained from EMD Millipore; and z-YVAD-cmk was purchased from Bio-Techne. Mouse anti-HA.11 (clone 16B12) was acquired from BioLegend, mouse anti-EGFP (clone JL-8) was purchased from Takara Bio Inc., mouse anti–IL-1β (clone 3A6) was obtained from Cell Signaling Technology, rabbit anti–caspase-1 p10 (C-20) was purchased from Santa Cruz Biotechnology, Inc., and rabbit anti-ASC (AL177) was obtained from Adipogen (all antibodies against the human homologues). HRP-coupled sheep anti–mouse IgG was purchased from GE Healthcare, HRP-coupled goat anti–rabbit IgG was obtained from SouthernBiotech, HRP-coupled goat anti–llama IgG was obtained from Bethyl Laboratories, peroxidase-coupled rat anti-HA (clone 3F10) was obtained from Sigma Aldrich, and HRP-coupled mouse anti-GAPDH (clone 6C5) was purchased from Abcam. Fluorescent secondary antibodies were acquired from Life Technologies.
VHH library generation
To raise heavy chain-only antibodies against human ASC, one alpaca was immunized five times with a mixture of proteins, including MBP-ASC. A VHH plasmid library in the M13 phagemid vector pJSC was generated as previously described (Maass et al., 2007; Sosa et al., 2014). In brief, RNA from peripheral blood lymphocytes was extracted and used as a template to generate cDNA using three sets of primers (random hexamers, oligo[dT], and primers specific for the constant region of the alpaca heavy chain gene). VHH coding sequences were amplified by PCR using VHH-specific primers, cut with AscI and NotI, and ligated into pJSC linearized with the same restriction enzymes. E. coli TG1 (Agilent) cells were electroporated with the ligation reaction and the obtained ampicillin-resistant colonies (∼3 × 106) harvested, pooled, and stored as glycerol stocks.
VHH phage display and panning
ASC-specific VHHs were obtained by phage display and panning with a protocol modified from a previous study (Sosa et al., 2014). E. coli TG1 cells containing the VHH library were infected with helper phage VCSM13 to produce phages displaying the encoded VHHs as pIII fusion proteins. Phages in the supernatant were purified and concentrated by precipitation; phages displaying MBP-specific VHHs were removed in a depletion step with MBP immobilized to the bottom of tissue culture flasks. Phages presenting ASC-specific VHHs were enriched using biotinylated MBP-ASC immobilized to MyOne Streptavidin T1 Dynabeads (Life Technologies) in the presence of excess soluble MBP and 500 mM NaCl. The obtained phages were used to infect E. coli ER2738 and subjected to a second round of panning. 96 E. coli ER2837 colonies yielded in the second panning were grown in 96-well plates and VHH expression was induced with IPTG. VHHs leaked into the supernatant were tested for specificity using ELISA plates coated with MBP or MBP-ASC; bound VHHs were detected with HRP-coupled rabbit anti–E-Tag antibodies (Bethyl Laboratories) and the chromogenic substrate TMB. The amino acid sequence of VHHASC is: QLQLVESGGG LVQPGGSLKL SCAASGFTFS RYAMSWYRQA PGKERESVAR ISSGGGTIYY ADSVKGRFTI SREDAKNTVY LQMNSLKPED TAVYYCYVGG FWGQGTQVTV SS.
Protein expression and purification
For periplasmic bacterial expression, VHH coding sequences were cloned into a derivative of pHEN6 (Conrath et al., 2001) encoding a C-terminal sortase recognition site (LPETG), followed by a His6-tag. VHHASC-LPETG-His6 and VHHEnhancer-LPETG-His6 were expressed in E. coli WK6 cells and purified from periplasmic extracts using Ni-NTA affinity purification and size-exclusion chromatography with a HiLoad 16/600 Superdex 75 pg column. To fluorescently label VHHs using sortase, proteins were incubated with sortase and GGG-Alexa Fluor 647, as previously described (Guimaraes et al., 2013), followed by removal of His-tagged sortase and size-exclusion chromatography.
For cytoplasmic expression, VHHASC was cloned into pETDuet-1, expressed in E. coli BL21(DE3), and purified by Ni-NTA purification and size-exclusion chromatography with a Superdex 200 10/300 GL column. ASCCARD (N128A/E130R) was expressed using a pET28a-based vector in E. coli BL21(DE3) and purified as described for the cytoplasmic expression of VHHASC.
For fluorescence polarization assay, MBP-ASCCARD-SUMO and MBP-ASC-caspase-1CARD-SUMO constructs were expressed and purified as described in our previous study (Lu et al., 2014). In short, ASCCARD (107–195) or caspase-1 (1–95) was cloned into modified pDB-His-MBP vector containing the sequence of SUMO C-terminal to the inserted gene. For GFP-ASCCARD expression, pET28a was modified to obtain an N-terminal GFP (monomeric) fusion protein, followed by insertion of ASCCARD in frame to GFP. The MBP-ASCFL was described previously (Lu et al., 2014). The Y187A mutation on MBP-ASC was introduced by QuikChange site-directed mutagenesis (Agilent Technologies). All proteins were expressed in BL21(DE3) cells and purified by Ni-NTA affinity chromatography, followed by size-exclusion chromatography.
For expression of Bacillus anthracis PA, E. coli BL21(DE3) cells were transformed with pGEX-6P-1 PA. GST-PA was expressed and bound to glutathione-S agarose, followed by release of PA with PreScission protease (GE Healthcare), as previously described (Wu et al., 2010). PA was further purified by anion exchange chromatography with a Mono Q column. His6-LFn-MxiH WT and His6-LFn-MxiH 2A (containing point mutations V58A and I78A) were expressed in E. coli BL21(DE3) using vectors derived from pET-15b LFN-DTA, a gift from J. Collier (Harvard Medical School, Boston, MA)(Addgene plasmid #11075; Milne et al., 1995), and purified by Ni-NTA affinity purification and size-exclusion chromatography with a HiLoad 16/600 Superdex 75 pg column. Endotoxins were removed from PA and LFn-MxiH preparations using two extractions with Triton X-114, followed by removal of remaining detergent with Bio-Beads SM-2 beads (Bio-Rad Laboratories; Aida and Pabst, 1990).
X-ray crystallography
VHHASC crystals grew in a condition containing 0.1 M citric acid, pH 3.5, and 35% PEG3350. 5 µl of sample at ∼15 mg/ml was mixed with 5 µl of the reservoir condition in a 24-well sitting drop tray (Hampton Research). Crystals appeared during overnight incubation. Crystals were recovered and washed in reservoir condition supplemented with 25% glycerol. The dataset was collected at the APS 24-ID-C beamline at 0.979 Å.
For structure determination, molecular replacement was used for phase determination with a predicted model from SWISS-MODEL (based on a VHH structure deposited in RCSB PDB under code: 3UX9). Manual structure refinement was done using the COOT program (Emsley and Cowtan, 2004), and iterative automatic refinement was performed in PHENIX (Adams et al., 2010). Ramachandran plots indicate 96.4% (3.6%) in the favored (allowed) region.
VHHASC–ASCCARD (N128A/E130R) complex crystals were grown in a condition containing 0.1 M citric acid at pH 3.5 and 3 M sodium chloride. Ni-NTA–purified ASCCARD (N128A/E130R) was mixed and concentrated with over-stoichiometric amounts of Ni-NTA-purified VHHASC. Excess VHH was removed from the complex by size-exclusion chromatography on a Superdex 75 column. The complex sample was concentrated to >30 mg/ml and mixed 1 µl:1 µl with reservoir condition in a hanging-drop vapor diffusion experiment. Needle clusters appeared after 4 d of incubation. Crystals were separated into individual needles and frozen in reservoir condition supplemented with 25% glycerol. The dataset was collected at the APS 24-ID-C beamline at 0.979 Å. Phase determination was performed in Phaser using nanobody crystal structure and the NMR structure of ASCCARD (RCSB PDB code: 2KN6; de Alba, 2009) as search models. Both structures were first trimmed to poly-alanine sequence using the Chainsaw in CCP4. Subsequent refinements were performed in PHENIX using the ASC and VHHASC structures as reference models to provide restrains. Manual model adjustment, including replacement of residues at interface, was done in COOT. Ramachandran plots indicate 96% (4%) in the favored (allowed) region.
The structures of VHHASC and the complex VHHASC–ASCCARD (N128A/E130R) have been deposited in the RCSB Protein Data Bank (PDB) with the accession codes 5H8D and 5H8O.
Immunoprecipitation
For the immunoprecipitation of endogenous ASC, 4 mg VHHASC and VHHEnhancer were covalently coupled to 0.25 g CNBr-activated Sepharose 6B (Sigma-Aldrich). 8 × 107 THP-1 EGFP cells (grown in the presence of 1 µg/ml doxycycline for 24 h to induce EGFP expression) were lysed in a high-salt lysis buffer (50 mM Tris, pH 7.4, 500 mM NaCl, and 1% NP-40, and protease inhibitor cocktail [Roche]), and lysates were bound to the VHH beads in the cold. After extensive washes, bound proteins were eluted with 0.2 M glycine pH 2.0 and analyzed by SDS-PAGE. The major bands were cut out of the gel and the contained proteins identified by mass spectrometry.
LUMIER assay
Protein interactions in transfected A549 cells were quantified using the LUMIER assay according to a protocol modified from Taipale et al. (2012). A549 cells in 24-well trays were transfected with 0.5 µg bait expression vectors (pCAGGS VHHASC-HA or pCAGGS VHH NP1-HA), and 0.5 µg prey expression vectors (empty vector, pEXPR hASC-Renilla, pEXPR hASCCARD-Renilla, pEXPR hASCPYD-Renilla, or pEXPR mASC-Renilla, all derived from pcDNA3-ccdB-Renilla; a gift from M. Taipale and S. Lindquist, Whitehead Institute, Cambridge, MA) using Lipofectamine LTX (Life Technologies). 2 d after transfection, cells were lysed in 120 µl LUMIER IP buffer (50 mM Hepes, pH 7.9, 150 mM NaCl, 2 mM EDTA, pH 8.0, 0.5% Triton X-100, 5% glycerol, and protease inhibitor cocktail [Roche]). 90 µl of the lysates were transferred to blocked LUMITRAC 600 plates (Greiner) coated with mouse anti-HA.11 and incubated at 4°C for 3 h. After extensive washing steps with IP buffer, incubated wells (or 10 µl lysate) were incubated with Coelenterazine-containing Renilla luciferase substrate mix (BioLux Gaussia Luciferase Assay kit; New England BioLabs) and light emission was quantified using a SpectraMax M3 microplate reader (Molecular Devices). Renilla luciferase activity in the immunoprecipitation samples was normalized by Renilla luciferase activity in the lysates.
Fluorescence polarization assays
The detailed protocol for fluorescence polarization assays is described in our previous study (Lu et al., 2014). In short, MBP-ASCCARD-SUMO and MBP-caspase-1CARD-SUMO containing a C-terminal sortase motif were fluorescently labeled as previously described (Guimaraes et al., 2013). Monomeric proteins were incubated with TEV protease to remove the MBP fusion. The rate of polymerization was enhanced by addition of the oligomeric GFP-ASCCARD. To assay for inhibitory effect, VHHASC was added to the mixture before MBP removal.
NLRP3 activation assays
To assay NLRP3 inflammasome activation, THP-1 cell lines were differentiated in the presence of 50 ng/ml PMA for 16 h, followed by incubation in THP-1 medium with or without 1 µg/ml doxycycline for 24 h. Cells were then incubated with 200 ng/ml LPS in THP-1 medium for 3 h, followed by treatment with 5 µM nigericin in OptiMEM for 45 min, both in the absence of doxycycline. Supernatants were cleared by centrifugation (1,000 g, 4°C, 10 min) and analyzed by ELISA or immunoblot.
AIM2 activation assays
To assay AIM2 inflammasome activation, THP-1 cells were differentiated and gene expression was induced as described for NLRP3 activation assays. Cells were incubated with 200 ng/ml LPS in THP-1 medium for 3 h, followed by transfection of 1 µg/ml poly (dA:dT) with 1.75 µl/ml Lipofectamine 2000 (final concentrations on transfected cells indicated). Supernatants were harvested and cleared after 4 h as described for NLRP3 activation assays.
NAIP/NLRC4 activation assays
To assay NAIP/NLRC4 inflammasome activation, THP-1 cell lines were differentiated and gene expression induced as described for NLRP3 activation assays. Cells were incubated with 1 µg/ml PA and 0.1 µg/ml LFn-MxiH WT or 2A in THP-1 medium supplemented with 20 µM of the TLR4 inhibitor TAK-242. After 3 h, supernatants were harvested and cleared as described for NLRP3 activation assays.
Vaccinia virus infection assays
To infect cells with vaccinia virus, mature virions were diluted in DMEM and used to infect cells at a multiplicity of infection (MOI) of 20. 30 min after infection, the inoculum was removed and cells were cultivated in full medium for 17.5 h.
IL-1β and TNF ELISAs
Concentrations of mature IL-1β and TNF in cell culture supernatants were quantified using the OptEIA Human IL-1β ELISA Set II or the OptEIA Human TNF ELISA Set (both BD) according to the manufacturer’s instructions. Typically, 2 × 105 cells were covered with 0.5 ml medium and 1:100 dilutions of the cleared supernatants were used for the ELISA. Statistical significance of differences was evaluated using the Student’s t test.
LDH activity assay
Lactate dehydrogenase activity in cell culture supernatants was quantified using the LDH Cytotoxicity Detection kit (Takara Bio Inc.). Typically, 1:2 dilutions of cleared supernatants (see IL-1β ELISA) were used for the assay according to the manufacturer’s instructions. LDH activity was corrected by the background LDH activity in the medium and normalized to the LDF activity of a well of cells lysed with 1% Triton X-100.
Immunoblots
To analyze secretion and maturation of caspase-1 and IL-1β by immunoblot, cleared supernatants from 2 × 106 THP-1 cells (covered with 1 ml Opti-MEM) were concentrated by methanol/chloroform precipitation, as described by Jakobs et al. (2013). Proteins were separated by SDS-PAGE and transferred to PVDF membranes. IL-1β was detected with mouse anti–IL-1β (clone 3A6, 1:1,000 in 5% BSA/TBS-T) and HRP-coupled sheep anti–mouse IgG; caspase-1 was detected with rabbit anti-caspase-1 p10 (1:200 in 5% milk/TBS-T) and HRP-coupled goat anti–rabbit IgG.
Immunofluorescence microscopy
To stain proteins for immunofluorescence microscopy, cells were fixed with 4% formaldehyde and permeabilized in permeabilization buffer (PS; 0.05% saponin, 1% BSA, and 0.05% NaN3 in PBS) for 20 min. Samples were incubated with rabbit anti-ASC (1:200 in PS) for 2 h, washed with PBS, and subsequently incubated with Alexa Fluor 594–coupled goat anti–rabbit IgG (1:1,000), Hoechst 33342 (1:5,000), and, where indicated, with Alexa Fluor 647–coupled VHHASC or VHH NP1 (1 µg/ml) for 1 h. Samples were washed with PBS and H2O, and mounted with Fluoromount-G (Southern Biotech) or Duolink In Situ Mounting Medium with DAPI (Sigma-Aldrich). Images were acquired using a PerkinElmer Ultraview Spinning Disk Confocal microscope or a Nikon Eclipse Ti wide field microscope. ASC foci and nuclei in wide field microscopy images were quantified using the spot detection function of the Imaris software package (Bitplane).
Online supplemental material
Tables S1 and S2 show crystallographic information. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151790/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Raimond Stroe for help with protein purifications, Tian-min Fu and Li Wang for crystal data collection, Eric L. Spooner of the Whitehead Institute proteomics core facility for mass spectrometric analysis of samples, and Mikko Taipale and Georgios Karras for help with LUMIER assays. Confocal microscopy experiments were performed in the W.M. Keck Facility for Biological Imaging at Whitehead Institute.
This work is supported by a National Institutes of Health Pioneer award to H.L. Ploegh (as well as additional funding from Fujifilm/MediVector); F.I. Schmidt was supported by an Advanced Postdoc Mobility Fellowship from the Swiss National Science Foundation.
H.L. Ploegh is a paid consultant to MediVector, which provided financial support for this work. Neither Fujifilm nor MediVector derive any rights from the work reported here. The authors declare no additional competing financial interests.
Author contributions: F.I. Schmidt, A. Lu, J.W. Chen, and C. Tang performed experiments and analyzed the data; J. Ruan provided the monomeric ASCCARD mutant; and F.I. Schmidt, A. Lu, H. Wu, and H.L. Ploegh conceived the study and wrote the manuscript.
Footnotes
Abbreviations used:
- CARD
- caspase recruitment domain
- MBP
- maltose-binding protein
- PA
- protective antigen
- PMA
- phorbol-12-myristate-13-acetate
- PYD
- Pyrin domain
- WR
- Western Reserve
- TEV
- tobacco etch virus
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida Y., and Pabst M.J.. 1990. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods. 132:191–195. 10.1016/0022-1759(90)90029-U [DOI] [PubMed] [Google Scholar]
- Antonin W., Holroyd C., Fasshauer D., Pabst S., Von Mollard G.F., and Jahn R.. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453–6464. 10.1093/emboj/19.23.6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora N., and Leppla S.H.. 1993. Residues 1-254 of anthrax toxin lethal factor are sufficient to cause cellular uptake of fused polypeptides. J. Biol. Chem. 268:3334–3341. [PubMed] [Google Scholar]
- Ashour J., Schmidt F.I., Hanke L., Cragnolini J., Cavallari M., Altenburg A., Brewer R., Ingram J., Shoemaker C., and Ploegh H.L.. 2015. Intracellular expression of camelid single-domain antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. J. Virol. 89:2792–2800. 10.1128/JVI.02693-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios-Rodiles M., Brown K.R., Ozdamar B., Bose R., Liu Z., Donovan R.S., Shinjo F., Liu Y., Dembowy J., Taylor I.W., et al. 2005. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 307:1621–1625. 10.1126/science.1105776 [DOI] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., and Monack D.M.. 2010a Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755. 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010b Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg J.S., Ingram J.R., Venkatakrishnan A.J., Jude K.M., Dukkipati A., Feinberg E.N., Angelini A., Waghray D., Dror R.O., Ploegh H.L., and Garcia K.C.. 2015. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 347:1113–1117. 10.1126/science.aaa5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Chen J., Xu H., Liu S., Jiang Q.X., Halfmann R., and Chen Z.J.. 2014. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 156:1207–1222. 10.1016/j.cell.2014.01.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Sisler J.R., and Moss B.. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques. 23:1094–1097. [DOI] [PubMed] [Google Scholar]
- Chen K.W., Groß C.J., Sotomayor F.V., Stacey K.J., Tschopp J., Sweet M.J., and Schroder K.. 2014. The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Reports. 8:570–582. 10.1016/j.celrep.2014.06.028 [DOI] [PubMed] [Google Scholar]
- Conrath K.E., Lauwereys M., Galleni M., Matagne A., Frère J.M., Kinne J., Wyns L., and Muyldermans S.. 2001. β-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 45:2807–2812. 10.1128/AAC.45.10.2807-2812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alba E. 2009. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J. Biol. Chem. 284:32932–32941. 10.1074/jbc.M109.024273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloye J., Roger T., Steiner-Tardivel Q.G., Le Roy D., Knaup Reymond M., Akira S., Petrilli V., Gomez C.E., Perdiguero B., Tschopp J., et al. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480 10.1371/journal.ppat.1000480 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Doxsey S.J., Brodsky F.M., Blank G.S., and Helenius A.. 1987. Inhibition of endocytosis by anti-clathrin antibodies. Cell. 50:453–463. 10.1016/0092-8674(87)90499-5 [DOI] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Wu J., Yu J.W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., and Alnemri E.S.. 2007. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14:1590–1604. 10.1038/sj.cdd.4402194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R., and Wu H.. 2012. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 22:241–247. 10.1016/j.sbi.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes C.P., Witte M.D., Theile C.S., Bozkurt G., Kundrat L., Blom A.E., and Ploegh H.L.. 2013. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8:1787–1799. 10.1038/nprot.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., and Hamers R.. 1993. Naturally occurring antibodies devoid of light chains. Nature. 363:446–448. 10.1038/363446a0 [DOI] [PubMed] [Google Scholar]
- Hauenstein A.V., Zhang L., and Wu H.. 2015. The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr. Opin. Struct. Biol. 31:75–83. 10.1016/j.sbi.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helma J., Cardoso M.C., Muyldermans S., and Leonhardt H.. 2015. Nanobodies and recombinant binders in cell biology. J. Cell Biol. 209:633–644. 10.1083/jcb.201409074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., and Fitzgerald K.A.. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 458:514–518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.W., Sui S.F., and Chai J.. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 350:399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Irannejad R., Tomshine J.C., Tomshine J.R., Chevalier M., Mahoney J.P., Steyaert J., Rasmussen S.G., Sunahara R.K., El-Samad H., Huang B., and von Zastrow M.. 2013. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 495:534–538. 10.1038/nature12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs C., Bartok E., Kubarenko A., Bauernfeind F., and Hornung V.. 2013. Immunoblotting for active caspase-1. Methods Mol. Biol. 1040:103–115. 10.1007/978-1-62703-523-1_9 [DOI] [PubMed] [Google Scholar]
- Kagan J.C., Magupalli V.G., and Wu H.. 2014. SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 14:821–826. 10.1038/nri3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C.S., Cardoso M.C., et al. 2010. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17:133–138. 10.1038/nsmb.1727 [DOI] [PubMed] [Google Scholar]
- Kortmann J., Brubaker S.W., and Monack D.M.. 2015. Cutting edge: inflammasome activation in primary human macrophages Is dependent on flagellin. J. Immunol. 195:815–819. 10.4049/jimmunol.1403100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtenberg B.C., Mace P.D., and Riedl S.J.. 2014. Structural mechanisms in NLR inflammasome signaling. Curr. Opin. Struct. Biol. 29:17–25. 10.1016/j.sbi.2014.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Lo Y.C., and Wu H.. 2010. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 465:885–890. 10.1038/nature09121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R., Schröder G.F., Fitzgerald K.A., Wu H., and Egelman E.H.. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 156:1193–1206. 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D.R., Sepulveda J., Pernthaner A., and Shoemaker C.B.. 2007. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J. Immunol. Methods. 324:13–25. 10.1016/j.jim.2007.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Hopkins L.J., Nugent E., Cox S., Glück I.M., Tourlomousis P., Wright J.A., Cicuta P., Monie T.P., and Bryant C.E.. 2014. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA. 111:7403–7408. 10.1073/pnas.1402911111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto J., Taniguchi S., Nakayama J., Shiohara M., Hidaka E., Katsuyama T., Murase S., and Sagara J.. 2001a Expression of apoptosis-associated speck-like protein containing a caspase recruitment domain, a pyrin N-terminal homology domain-containing protein, in normal human tissues. J. Histochem. Cytochem. 49:1269–1275. 10.1177/002215540104901009 [DOI] [PubMed] [Google Scholar]
- Masumoto J., Taniguchi S., and Sagara J.. 2001b Pyrin N-terminal homology domain- and caspase recruitment domain-dependent oligomerization of ASC. Biochem. Biophys. Res. Commun. 280:652–655. 10.1006/bbrc.2000.4190 [DOI] [PubMed] [Google Scholar]
- Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., et al. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 108:3665–3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne J.C., Blanke S.R., Hanna P.C., and Collier R.J.. 1995. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol. 15:661–666. 10.1111/j.1365-2958.1995.tb02375.x [DOI] [PubMed] [Google Scholar]
- Muyldermans S. 2013. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82:775–797. 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- Paalanen M.M., Ekokoski E., El Khattabi M., Tuominen R.K., Verrips C.T., Boonstra J., and Blanchetot C.. 2011. The development of activating and inhibiting camelid VHH domains against human protein kinase C epsilon. Eur. J. Pharm. Sci. 42:332–339. 10.1016/j.ejps.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Pardon E., Laeremans T., Triest S., Rasmussen S.G., Wohlkönig A., Ruf A., Muyldermans S., Hol W.G., Kobilka B.K., and Steyaert J.. 2014. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 9:674–693. 10.1038/nprot.2014.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahillioglu A.C., Sumbul F., Ozoren N., and Haliloglu T.. 2014. Structural and dynamics aspects of ASC speck assembly. Structure. 22:1722–1734. 10.1016/j.str.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Sborgi L., Ravotti F., Dandey V.P., Dick M.S., Mazur A., Reckel S., Chami M., Scherer S., Huber M., Böckmann A., et al. 2015. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 112:13237–13242. 10.1073/pnas.1507579112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A.R., Wellington D.A., Kumar P., Kassa H., Booth C.J., Cresswell P., and MacMicking J.D.. 2012. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 336:481–485. 10.1126/science.1217141 [DOI] [PubMed] [Google Scholar]
- Sosa B.A., Demircioglu F.E., Chen J.Z., Ingram J., Ploegh H.L., and Schwartz T.U.. 2014. How lamina-associated polypeptide 1 (LAP1) activates Torsin. eLife. 3:e03239 10.7554/eLife.03239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K.D., Karras G.I., and Lindquist S.. 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 150:987–1001. 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja S.K., Rathinam V.A., and Fitzgerald K.A.. 2015. Mechanisms of inflammasome activation: recent advances and novel insights. Trends Cell Biol. 25:308–315. 10.1016/j.tcb.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R.E. 2015. The NAIP/NLRC4 inflammasomes. Curr. Opin. Immunol. 32:84–89. 10.1016/j.coi.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Peisley A., Tetrault D., Li Z., Egelman E.H., Magor K.E., Walz T., Penczek P.A., and Hur S.. 2014. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol. Cell. 55:511–523. 10.1016/j.molcel.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Feng C., Hong Y., Guo A., Cao S., Dong J., Lin L., and Liu Z.. 2010. Soluble expression and purification of the anthrax protective antigen in E. coli and identification of a novel dominant-negative mutant N435C. Appl. Microbiol. Biotechnol. 87:609–616. 10.1007/s00253-010-2495-5 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.A., and Collier R.J.. 2007. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 76:243–265. 10.1146/annurev.biochem.75.103004.142728 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 350:404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.