As an alternative to mitophagy, neutrophils spontaneously extrude mitochondrial (mt) DNA devoid of oxidized residues (Ox). Activated lupus neutrophils or healthy neutrophils treated with IFN/αRNP release ox-mtDNA bound to TFAM, which induces high levels of IFN-α in pDCs.
Abstract
Autoantibodies against nucleic acids and excessive type I interferon (IFN) are hallmarks of human systemic lupus erythematosus (SLE). We previously reported that SLE neutrophils exposed to TLR7 agonist autoantibodies release interferogenic DNA, which we now demonstrate to be of mitochondrial origin. We further show that healthy human neutrophils do not complete mitophagy upon induction of mitochondrial damage. Rather, they extrude mitochondrial components, including DNA (mtDNA), devoid of oxidized (Ox) residues. When mtDNA undergoes oxidation, it is directly routed to lysosomes for degradation. This rerouting requires dissociation from the transcription factor A mitochondria (TFAM), a dual high-mobility group (HMG) protein involved in maintenance and compaction of the mitochondrial genome into nucleoids. Exposure of SLE neutrophils, or healthy IFN-primed neutrophils, to antiribonucleotide protein autoantibodies blocks TFAM phosphorylation, a necessary step for nucleoid dissociation. Consequently, Ox nucleoids accumulate within mitochondria and are eventually extruded as potent interferogenic complexes. In support of the in vivo relevance of this phenomenon, mitochondrial retention of Ox nucleoids is a feature of SLE blood neutrophils, and autoantibodies against Ox mtDNA are present in a fraction of patients. This pathway represents a novel therapeutic target in human SLE.
Over the last 15 yr, genomic studies have highlighted the role of innate immunity in human SLE pathogenesis (Pascual et al., 2006). This was further supported by the demonstration that systemic lupus erythematosus (SLE) serum induces DC maturation in an IFN-dependent way (Blanco et al., 2001). Expression of IFN- and neutrophil-related transcripts correlates with disease activity, and common allelic gene variants within these pathways confer disease susceptibility (Bennett et al., 2003; Moser et al., 2009; Bentham et al., 2015). More recently, the identification of monogenic SLE caused by mutations in genes involved in intracellular and extracellular DNA degradation (Al-Mayouf et al., 2011; Crow, 2011) supports the hypothesis that improper disposal of nucleic acids might be an upstream event in human SLE.
SLE patients develop autoantibodies against double-stranded DNA (dsDNA) and RNA–protein complexes. Internalization of SLE immune complexes (ICs) containing nucleic acids by plasmacytoid DCs (pDCs) induces endosomal TLR activation and type I IFN production (Means et al., 2005). This takes place upon convergence of the phagocytic and autophagic pathways in a process called LC3-associated phagocytosis (Henault et al., 2012). SLE neutrophils also contribute to IFN production. Thus, their activation with anti-RPN/Sm (Garcia-Romo et al., 2011) or antimicrobial peptide (Lande et al., 2011) autoantibodies leads to extrusion of interferogenic DNA.
The nature of the DNA that initiates and perpetuates an immune response in SLE remains unknown, though genomic DNA (gDNA) released from dead cells is the most accepted candidate. Mitochondrial DNA (mtDNA), unlike gDNA, contains hypomethylated CpG motifs similar to bacterial DNA and might be highly inflammatory in vivo (Collins et al., 2004). mtDNA activates neutrophils through TLR9 engagement (Zhang et al., 2010) and, upon cytoplasmic leakage, leads to cell autonomous NLRP3 and TLR9 activation (Nakahira et al., 2011; Oka et al., 2012).
We have now characterized the interferogenic DNA released by SLE neutrophils in response to TLR7-agonistic autoantibodies. We show that this form of neutrophil activation interferes with the disassembly of mtDNA–transcription factor A mitochondria (TFAM) complexes, which is required to export oxidized (Ox) mtDNA into lysosomes for degradation, resulting in their retention within mitochondria and eventual extrusion. Ox mtDNA displays an exceptional capacity to activate pDCs. Furthermore, autoantibodies against this form of modified DNA are detected in SLE sera. Thus, lack of proper intra- or extracellular degradation of neutrophil Ox mtDNA might be central to SLE pathogenesis.
RESULTS
Live neutrophils spontaneously extrude mtDNA–protein complexes
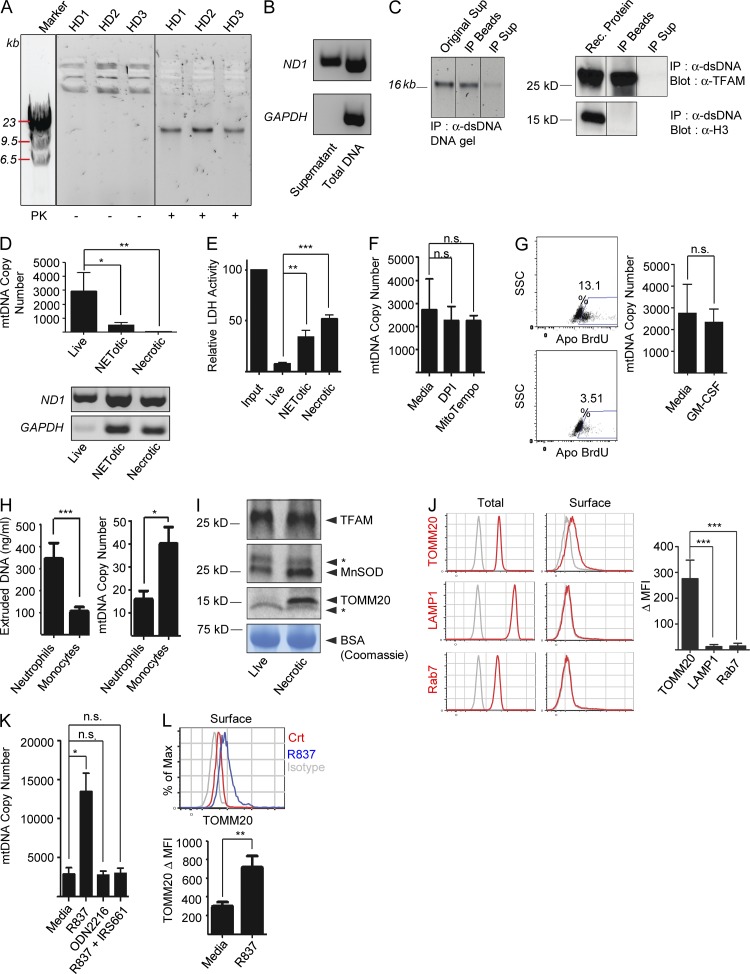
Short-term cell-free supernatants from healthy neutrophil cultures contain DNA–protein complexes in the absence of activation. Upon digestion with proteinase K, these complexes yield a 16-kb DNA band, the size of mtDNA (Fig. 1 A). This origin was confirmed by selective amplification of the mitochondrial-encoded gene ND1 (Fig. 1 B) and coimmunoprecipitation of DNA and the mitochondrial transcription factor TFAM, but not the chromatin protein Histone 3 (H3; Fig. 1 C).
Figure 1.
Live neutrophils extrude mtDNA–protein complexes. (A) Neutrophil supernatants from three healthy donors (HD) were run on agarose gels. High molecular weight complexes (mtC) yield a single DNA band of ∼16 Kb upon digestion with proteinase K (PK). (B) Amplification of the mitochondrial gene ND1, but not the nuclear gene GAPDH, from DNA isolated from live neutrophil supernatants. Total neutrophil DNA was used as control. (C) Neutrophil supernatant (Orig Sup) was immunoprecipitated with anti-dsDNA antibody. The beads (IP Beads) and the resulting supernatants (IP Sup) were analyzed by agarose gel (left) or by Western blot with anti-TFAM or anti-H3 antibodies (right). (D) Abundance of mitochondrial and genomic DNA was assessed on 5 ng of isolated DNA from live, NETotic, or necrotic neutrophil supernatants by Real-Time PCR (mtDNA Copy Number; top) or by conventional PCR (bottom). (E) LDH activity was measured in cell-free supernatants from live, NETotic, or necrotic neutrophil cultures. (F) Quantification of extruded mtDNA by Real-Time PCR in the presence of DPI (NADPH Inhibitor) or MT (mtROS scavenger). (G, left) Apoptosis progression in untreated or GM-CSF–treated neutrophils was assessed by TUNEL assay. The percentage of TUNEL+ cells is shown. (right) The amount of extruded mtDNA was assessed by Real-Time PCR (mtDNA Copy Number). (H, left) Extruded mtDNA was quantified by Picogreen. (right) mtDNA Copy Number in total cell DNA was quantified by Real-Time PCR. (I) The presence of TFAM, MnSOD, and TOMM20 was assessed in concentrated cell-free supernatants from live or necrotic neutrophils by Western blot. Coomassie staining of the BSA band was used as loading control. * indicates nonspecific band. (J) Surface expression of TOMM20, LAMP1, or Rab7 was assessed by flow cytometry on permeabilized (Total) or nonpermeabilized (Surface) neutrophils. Open gray histograms represent the isotype control. Δ MFI (Mean Fluorescence Intensity) = MFI antibody – MFI isotype control. (K–L) Extruded mtDNA (K) and TOMM20 plasma membrane expression (L) were measured in neutrophils treated with R837 (TLR7 agonist) in the presence or absence of IRS661 (TLR7 inhibitor) or with ODN2216 (TLR9 agonist). Data in B, C, and I are representative of three independent experiments with n = 3. Bars represent mean ± SD from at least three independent experiments, with n = 3–8. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Absence of gDNA (Fig. 1 D) and lactate dehydrogenase (LDH) activity (Fig. 1 E) in neutrophil supernatants supports the observation that mtDNA extrusion is not linked to the cell membrane disruption that characterizes necrosis or NETosis. Furthermore, in contrast to chromatin extrusion during NETosis (Brinkmann et al., 2004), steady-state extrusion of mtDNA is ROS independent, as neither DPI nor MitoTempo (MT) block it (Fig. 1 F). Addition of GM-CSF, a neutrophil prosurvival factor (Klein et al., 2000), does not affect the amount of extruded mtDNA (Fig. 1 G), supporting the belief that constitutive apoptosis does not drive this process. As opposed to neutrophils, monocytes extrude negligible amounts of mtDNA even though these cells have higher mtDNA content per cell (Fig. 1 H).
Western blot analysis of cell-free supernatants from healthy neutrophils reveals the presence of mitochondrial matrix proteins (TFAM and manganese superoxide dismutase [MnSOD]) but not of outer mitochondrial membrane proteins such as TOMM20 (Fig. 1 I), suggesting that neutrophils do not extrude full mitochondria. Although the mechanism underlying this process remains to be elucidated, we observed translocation of TOMM20, but not of different organelle markers, such as LAMP1 and Rab7, on the plasma membrane (Fig. 1 L). MtDNA extrusion is amplified by addition of TLR7 but not TLR9 (Fig. 1 M) or TLR4 (not depicted) agonists. Accordingly, activation with the TLR7 agonist R837 increases TOMM20 translocation to the cell membrane (Fig. 1 N).
Thus, neutrophils spontaneously release mtDNA–protein complexes (mtCs) in the absence of overt cell death and membrane disruption, and this process is enhanced by TLR7 engagement.
Extrusion of mtDNA–protein complexes as a neutrophil alternative to mitophagy
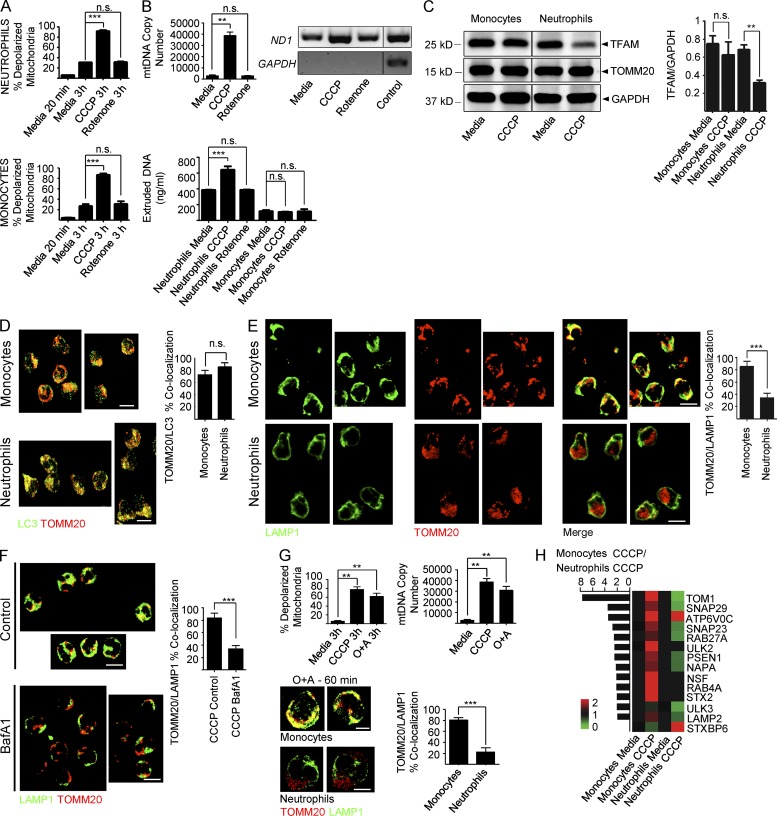
Neutrophil mtC extrusion could be the result of improper disposal of damaged mitochondria. To address this, we amplified mitochondrial damage in both neutrophils and monocytes by increasing either baseline depolarization with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or mtROS with Rotenone (Fig. 2 A). CCCP, but not Rotenone, increased mtDNA extrusion in neutrophils (Fig. 2 B). As expected, we observed the concomitant reduction of TFAM intracellular levels in these cells (Fig. 2 C).
Figure 2.
Neutrophils fail to complete mitophagy in response to mitochondrial depolarization. (A) Mitochondrial depolarization was measured in neutrophils (top) and monocytes (bottom) in the presence of the protonophore CCCP or the complex I inhibitor Rotenone. (B, top) Neutrophils were treated with CCCP or Rotenone and extruded mtDNA was quantified by Real-Time PCR (mtDNA copy number) or conventional PCR. (bottom) Neutrophils or monocytes were treated with CCCP or Rotenone and extruded mtDNA was quantified by Picogreen. (C) Neutrophils or monocytes were treated with media or CCCP in the presence of 50 µM of the protein synthesis inhibitor cycloheximide. Total cell lysates were then analyzed by Western blot. TFAM/GAPDH ratio is shown on the right. (D) Neutrophils and monocytes were treated with CCCP for 60 min and immunostained with anti-TOMM20 and anti-LC3B antibodies or (E) with anti-LAMP1 and anti-TOMM20 antibodies. Bars, 10 µm. (F) Monocytes were treated with CCCP for 60 min in the presence of the lysosomal inhibitor Bafilomycin A1 (BafA1) and immunostained with anti-TOMM20 and anti-LAMP1 antibodies. Bars, 10 µm. (G) Mitochondria depolarization and extruded mtDNA levels (top) and quantification of autophagosome/lysosome fusion (bottom) in neutrophils treated with the combination Oligomycin (10 µM) and Antimycin (1 µM; O+A). Bars, 5 µm. (H) Fold up- (red) or down-regulation (green) of mitophagy-related transcripts in monocytes and neutrophils upon CCCP treatment. Transcript expression ratio in monocytes over neutrophils is shown on the left. Data were normalized to media-treated cells. Bars represent mean ± SD from at least three independent experiments with n = 3–6. **, P < 0.01; ***, P < 0.001.
In most cells, damaged mitochondria are removed by mitophagy. This process requires autophagy activation, sequestration of damaged mitochondria into autophagosomes, and fusion of autophagosomes with lysosomes (Ashrafi and Schwarz, 2013). Upon exposure to CCCP, autophagy activation (detected by increased LC3B punctae staining) and cargo sequestration (detected by TOMM20-LC3B co-localization) were similar in neutrophils and monocytes (not depicted and Fig. 2 D). However, autophagosome–lysosome fusion (assessed by TOMM20-LAMP1 co-localization) was considerably impaired in neutrophils (Fig. 2 E). Autophagy arrest with Bafilomycin A1 (BafA1) reproduced this phenotype in monocytes (Fig. 2 F). Amplification of mitochondrial depolarization with oligomycin/antimycin (O+A) yielded results similar to those obtained with CCCP (Fig. 2 G). Notably, the mitochondrial protein content did not decrease in CCCP-treated monocytes (Fig. 2 C), which undergo complete mitophagy, because CCCP blocks lysosomal function (Padman et al., 2013).
Gene expression profiling revealed that in response to CCCP monocytes, but not neutrophils, up-regulated transcripts related to autophagy activation (ULK2), autophagosome trafficking (Rab27a and Rab4a), and fusion (NSF, NAPA, SNAP23, SNAP29, STX2, and LAMP2), as well as lysosome activation (Presenilin-1). TOM1, which participates in autophagosome maturation (Tumbarello et al., 2012), was significantly down-regulated in neutrophils (Fig. 2 H).
Thus, extrusion of mtCs by neutrophils might result from a constitutive inability to complete mitophagy.
Extruded neutrophil mtCs activate pDCs only when enriched in Ox mtDNA
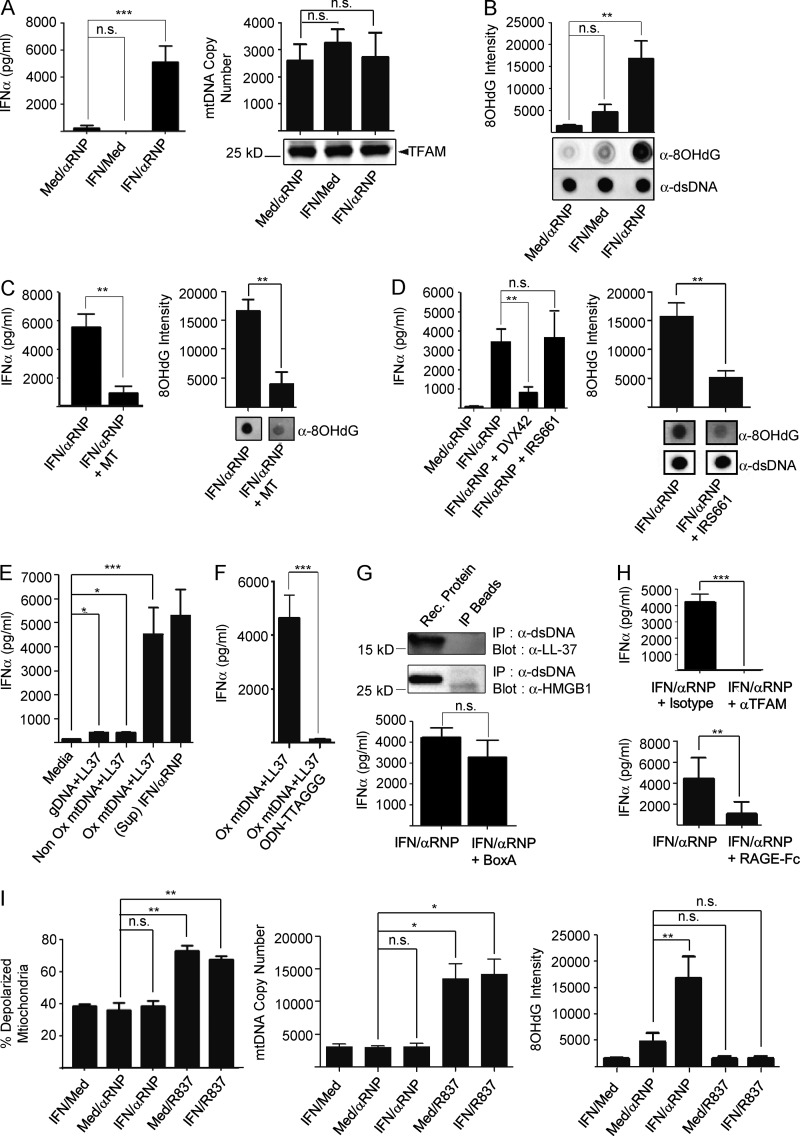
SLE neutrophils activated with anti-Sm/ribonucleotide protein (RNP) antibodies extrude interferogenic DNA (Garcia-Romo et al., 2011), whereas healthy neutrophils require IFN-α priming to recapitulate this effect (Fig. 3 A, top). Yet, we now show that all neutrophils extrude mtCs in the steady state (Fig. 1 A). Exposure to IFN-α and anti-Sm/RNP antibodies might alter the quality of mtC, as similar amounts of mtDNA and TFAM are found in neutrophil supernatants regardless of activation (Fig. 3 A, bottom). mtDNA is highly susceptible to oxidation (Yakes and Van Houten, 1997), a proinflammatory modification (Shimada et al., 2012). Indeed, high levels of 8-hydroxydeoxyguanosine (8OHdG), a marker of DNA oxidation (Kasai and Nishimura, 1984), were detected only in mtDNA extruded by IFN/αRNP-treated neutrophils (Fig. 3 B). The role of oxidation is further supported by the loss of interferogenicity of extruded DNA upon reduction of 8OHdG levels with the mitochondrial-targeted antioxidant MT (Fig. 3 C). Neutrophils require TLR7 engagement to release interferogenic mtDNA (Garcia-Romo et al., 2011). Consequently, the TLR7-specific antagonist IRS661, but not the TLR8 antagonists DVX42, reduced both the oxidation status and the interferogenicity of extruded mtDNA in response to IFN/αRNP antibodies (not depicted and Fig. 3 D).
Figure 3.
MtDNA oxidation is required for pDC activation. (A, top) pDCs were incubated with supernatants from neutrophils treated with or without IFN ± αRNP. IFN-α levels were quantified after 18 h culture. (bottom) Extruded mtDNA or TFAM quantification (mtDNA copy number) in the supernatants from neutrophils treated with or without IFN ± αRNP. (B) Dot blot analysis, using anti-8OHdG or anti-dsDNA (loading control) antibodies, of the DNA isolated from the supernatants of neutrophils treated with or without IFN ± αRNP. Bars represent the quantification of 8OHdG intensity. (C) Neutrophils were stimulated with IFN/αRNP in the absence or in the presence of the mtROS scavenger MitoTempo (MT). Extruded mtDNA was assessed for its interferogenic effect on pDCs (top) and for its oxidation status (bottom). (D, top) Neutrophils were treated with IFN/αRNP in the presence of IRS661 (TLR7 inhibitor) or DVX42 (TLR8 inhibitor) and the corresponding supernatants were assessed for their interferogenic effect on pDCs. (bottom) The oxidation status of extruded mtDNA was assessed by dot blot. (E) IFN-α production by pDCs stimulated with LL-37 added to genomic DNA, Non-Ox, or Ox mtDNA, or with supernatants from IFN/αRNP-treated neutrophils. (F) IFN-α production by pDCs stimulated with Ox mtDNA with our without the TLR9 inhibitor ODN-TTAGGG. (G, top) DNA from IFN/αRNP activated neutrophil supernatants were immunoprecipitated with anti-dsDNA antibody. The IP was analyzed by Western blot with anti-LL37 or anti-HMGB1 antibodies. (bottom) IFN-α production by pDCs stimulated with interferogenic neutrophil supernatants in the presence or absence of the HMGB1 inhibitor BoxA. (H) IFN-α production was assessed after pDC incubation with interferogenic neutrophil supernatants in the presence or absence of anti-TFAM antibodies or recombinant RAGE-Fc chimera. (I) Neutrophils were treated as shown and mitochondrial depolarization was assessed with MitoTracker DeepRed; the amount of extruded mtDNA was assessed by Real-Time PCR and its oxidation status by 8OHdG dot blot. Bars represent mean ± SD from at least three independent experiments with n = 3–8. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further establish that Ox mtDNA is a powerful pDC activator, Ox and Non Ox mtDNA fragments were generated using rt-PCR. Addition of the cationic peptide LL-37 to facilitate their uptake (Lande et al., 2007) resulted in a 10-fold increase in IFN-α levels in the presence of Ox mtDNA (Fig. 3 E). As we previously reported (Garcia-Romo et al., 2011), IFN production by pDCs under these activation conditions is TLR9 dependent (Fig. 3 F).
Although activated neutrophils secrete LL37 and HMGB1 (Garcia-Romo et al., 2011), neither protein is an integral part of the interferogenic mtC, as they do not co-immunoprecipitate with mtDNA. Furthermore, the HMGB1 antagonist BoxA does not block pDC activation (Fig. 3 G). In contrast, TFAM neutralization with either anti-TFAM antibodies or soluble RAGE-Fc chimera abrogates the interferogenicity of extruded mtC (Fig. 3 H), confirming that TFAM is the carrier responsible for neutrophil-derived mtDNA internalization (Julian et al., 2012).
As opposed to IFN/αRNP antibodies, exposure of neutrophils to synthetic TLR7 ligands amplifies mitochondrial depolarization and extrusion of mtDNA that is NonOx (Fig. 3 I). This different outcome might be caused by differential compartmentalization of TLR7 ligands in neutrophils, as previously reported for CpGA- versus anti-dsDNA IC-activation of TLR9 in pDCs (Henault et al., 2012).
Collectively, these data indicate that oxidation is essential for self-mtDNA to become a potent pDC activator.
Ox mtDNA is exported in steady-state neutrophils from mitochondria to lysosomes within cytosolic vesicles
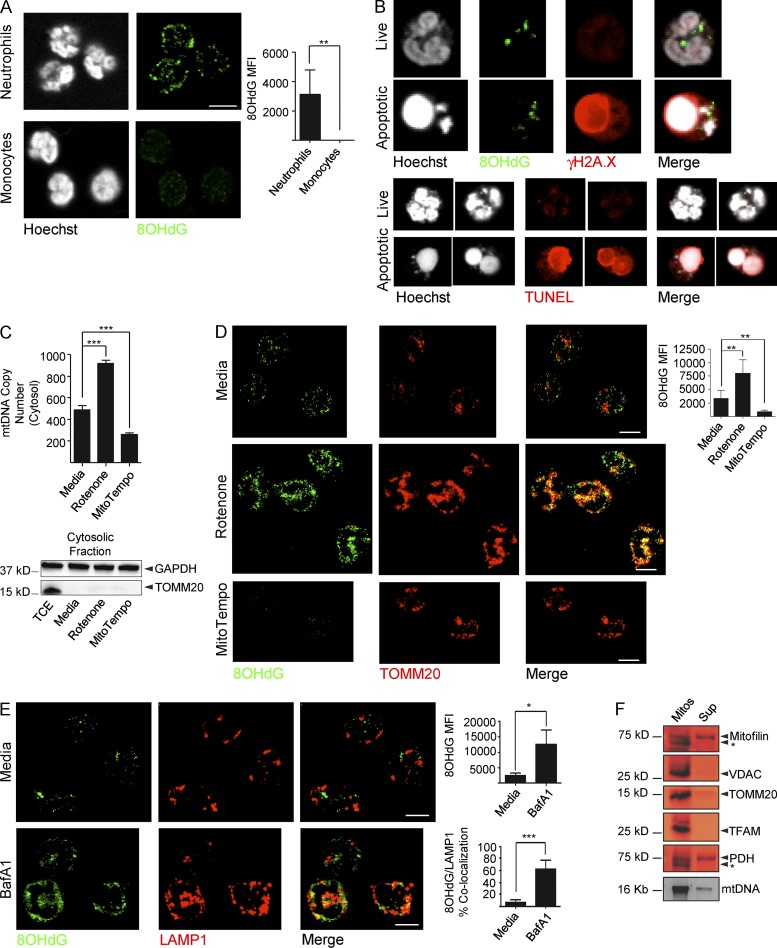
To address why neutrophils only extrude Ox mtDNA upon exposure to IFN/αRNP, we stained healthy neutrophils and monocytes with anti-8OHdG antibodies in the steady state. Cytosolic 8OHdG(+) DNA was detected only in neutrophils (Fig. 4 A).
Figure 4.
Ox mtDNA is exported from mitochondria to lysosomes under steady-state conditions. (A) Cytoplasmic 8OHdG(+) staining in neutrophils and monocytes. Bar, 10 µm. (B) Live or apoptotic neutrophils were stained with anti-8OHdG antibody and the damaged nuclear DNA marker γH2A.X (top) or with TUNEL assay (bottom). Bar, 5 µm. (C and D) Cytosolic mtDNA copy number (C) and 8OHdG staining and quantification (D) in neutrophils treated with media, Rotenone or MitoTempo. The purity of the cytosolic fraction is also shown in (C). TCE = total cell extract. TOMM20 was used as mitochondrial marker. Bars, 10 µm. (E) Neutrophils were treated with the lysosomal inhibitor Bafilomycin A1 (BafA1) and stained with anti-8OHdG and anti-LAMP1 antibodies. Bars, 10 µm. (F) Supernatant (Sup) from the mitochondrial budding assay was subjected to Western blot analysis (top) or analyzed for mtDNA content (bottom) by agarose gel. The mitochondrial fraction (Mitos) was loaded as control. * indicates nonspecific band. Data in B–F are representative of three independent experiments, with n = 3. Bars represent mean ± SD from at least three independent experiments, with n = 3–5. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Two recent studies showed that damaged gDNA fragments are constitutively exported to lysosomes for degradation (Ivanov et al., 2013; Lan et al., 2014). Lack of co-localization of cytosolic 8OHdG(+) DNA and damaged (γ.H2A.X) or fragmented (TUNEL) gDNA markers rules out its nuclear origin (Fig. 4 B). In support of the mitochondrial origin of cytosolic 8OHdG(+) DNA, addition of Rotenone increases and MT decreases both cytosolic mtDNA copy number (Fig. 4 C) and the amount of 8OHdG cytosolic staining per cell (Fig. 4 D).
Sorting of Ox cargo to lysosomes through mitochondria-derived vesicles (MDVs) has been described in HeLa cells as a mechanism whereby partially damaged mitochondria remove Ox components without engaging mitophagy (Soubannier et al., 2012a). Accordingly, cytosolic 8OHdG(+) DNA accumulates and merges with the LAMP1(+) compartment in the presence of BafA1 (Fig. 4 E).
To analyze the composition of 8OHdG(+) vesicles, we performed a mitochondrial cell–free budding assay (Soubannier et al., 2012b). Similar to lysosome-targeting MDVs (Soubannier et al., 2012a;b), we found that 8OHdG(+) vesicles selectively incorporate IMM proteins, the matrix enzyme pyruvate dehydrogenase (PDH), and mtDNA (Fig. 4 F). TFAM, however, is excluded (Fig. 4 F), suggesting that Ox mtDNA dissociates from this transcription factor before being exported into lysosomes.
Collectively, these data reveal that Ox mtDNA is steadily removed from neutrophil mitochondria and routed to lysosomes upon dissociating from TFAM.
IFN/αRNP block the routing of neutrophil Ox mtDNA to lysosomes
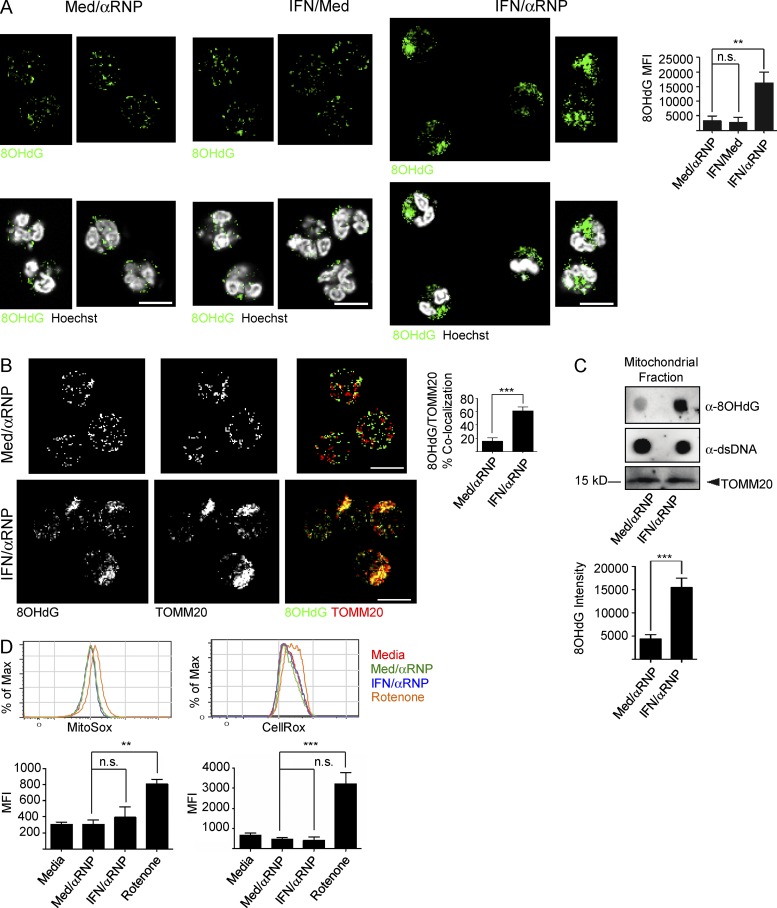
Our studies show that the IFN/αRNP combination induces neutrophils to extrude Ox mtDNA, which in the steady state is diverted into lysosomes. This form of activation also increases the total amount of intracellular Ox mtDNA (Fig. 5 A) and leads to its accumulation inside mitochondria (Fig. 5, B and C). Although a similar phenotype is observed upon exposure to Rotenone (Fig. 4 D), IFN/αRNP does not increase neutrophil ROS production (Fig. 5 D).
Figure 5.
IFN/αRNP activation of neutrophils induces intramitochondrial retention of Ox mtDNA. (A) Cytoplasmic 8OHdG(+) staining in neutrophils treated with or without IFN ± αRNP. 8OHdG quantification is also shown. Bars, 10 µm. (B) Neutrophils were treated with or without IFN ± αRNP and co-stained with anti-8OHdG and anti-TOMM20 antibodies. Bars, 10 µm. (C) Alternatively, mtDNA was extracted and analyzed by dot blot with anti-8OHdG and anti-dsDNA antibodies. TOMM20 was used as loading control. (D) Neutrophils were treated with or without IFN ± αRNP, and the production of mtROS or total cellular ROS was quantified by MitoSox or CellRox, respectively. Bars represent mean ± SD from at least three independent experiments, with n = 3–6. **, P < 0.01; ***, P < 0.001.
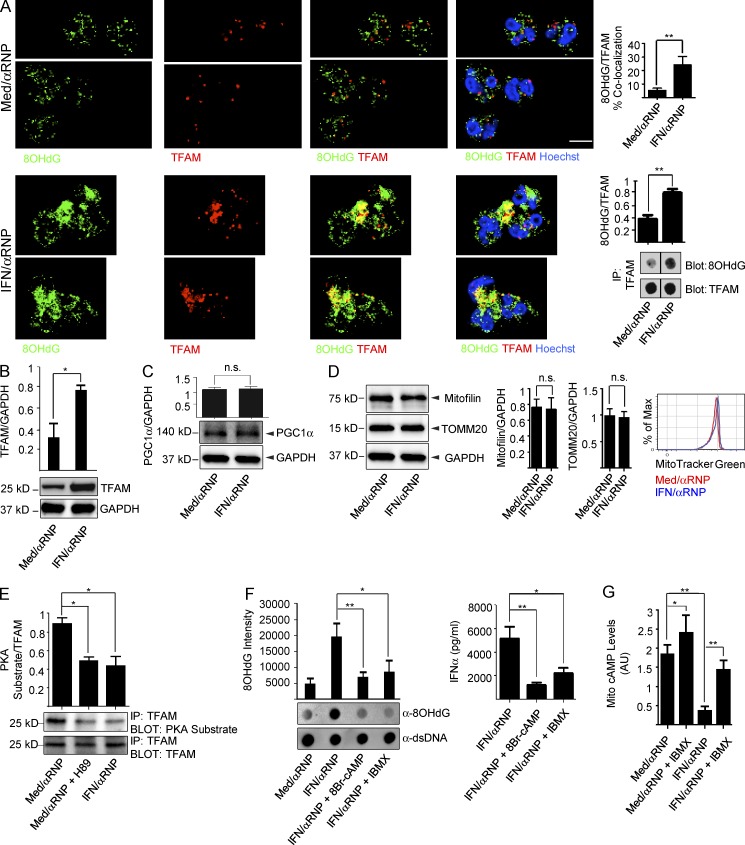
Ox mtDNA is never associated with TFAM in the cytoplasm of unstimulated neutrophils. Intracellular Ox mtDNA–TFAM complexes can be easily detected, however, upon activation with IFN/αRNP (Fig. 6 A). This activation also increases intracellular TFAM levels (Fig. 6 B), which reflects decreased TFAM degradation rather than increased biosynthesis, as supported by lack of up-regulation of the TFAM master regulator PGC1α (Hock and Kralli, 2009) and by increased mitogenesis (Fig. 6, C and D).
Figure 6.
IFN/αRNP inhibit the dissociation of Ox mtDNA and TFAM. (A) Neutrophils were treated as shown and co-stained with anti-8OHdG and anti-TFAM antibodies. Bars, 10 µm. Alternatively, cell lysates were subjected to IP with anti-TFAM antibody and the immunoprecipitates were analyzed by dot blot. (B–D) Neutrophils were treated as shown and cell lysates were analyzed by Western blot with antibodies against TFAM (B), PGC1α (C), Mitofilin or TOMM20 (D), and GAPDH as control. Total mitochondrial mass was assessed by MitoTracker Green (D). (E) Neutrophils were treated as shown and cell lysates were subjected to IP with anti-TFAM antibody. The immunoprecipitates were then blotted with antibodies against PKA-phosphorylated peptides (α-PKA sub) or TFAM (loading control). The PKA inhibitor H89 was used as a control. (F) Neutrophils were treated with αRNP ± IFN in the presence or absence of the cAMP analogue 8Br-cAMP or the PDE inhibition IBMX. Extruded mtDNA was analyzed by dot blot (left) and supernatants were assessed for their interferogenic capacity on pDCs (right). (G) Neutrophils were treated as shown and cAMP levels were measured in the enriched mitochondrial fraction. cAMP levels were normalized to the total protein content and expressed in arbitrary units (AU). Bars represent mean ± SD from at least three independent experiments, with n = 3–5. *, P < 0.05; **, P < 0.01.
We confirmed that, as previously reported (Lu et al., 2013), TFAM turnover requires dissociation from mtDNA and degradation by the Lon Protease in the steady state (unpublished data). The dissociation step requires TFAM phosphorylation by the matrix resident protein kinase A (PKA) (Lu et al., 2013). IFN/αRNP significantly reduces TFAM phosphorylation (Fig. 6 E) without directly inhibiting PKA, as its agonist 8Br-cyclic-AMP (cAMP) decreases the oxidation status as well as the interferogenic capacity of extruded mtDNA (Fig. 6 F). PKA is activated by cAMP, levels of which are modulated by adenylyl cyclase and phosphodiesterases (PDEs). Both enzymes are present in the mitochondrial matrix (Acin-Perez et al., 2009). Indeed, IFN/αRNP exposure reduces mitochondrial cAMP levels (Fig. 6 G). Consequently, the pan-PDE inhibitor IBMX decreases the oxidation status and the interferogenicity of extruded mtDNA (Fig. 6 F). Whether this is a result of adenylyl cyclase inhibition, PDE activation, or mitochondrial ATP level reduction remains to be addressed.
Thus, our results link neutrophil activation with IFN/αRNP to decreased mitochondrial cAMP levels, impaired PKA activation, and lack of mtDNA–TFAM disassembly. This reduces the mitochondrial removal of Ox mtDNA and promotes its extrusion.
SLE patients retain Ox mtDNA within their neutrophil mitochondria and develop anti–Ox mtDNA autoantibodies
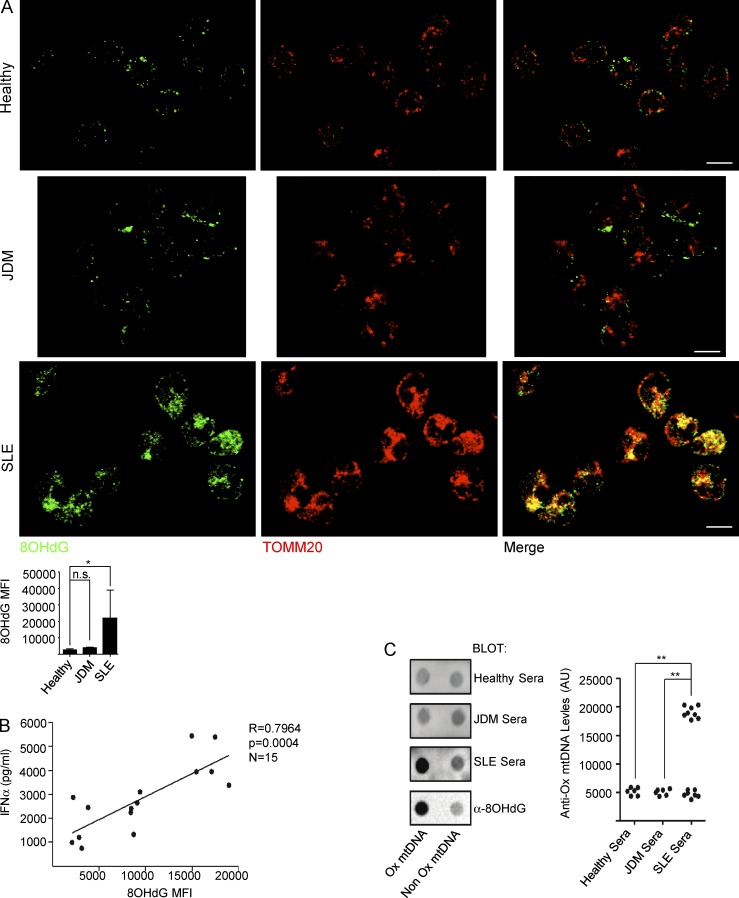
Development of TLR7 agonist autoantibodies and overexpression of IFN-inducible transcripts are hallmarks of human SLE. We therefore analyzed whether patient blood neutrophils would recapitulate the mitochondrial alterations induced by IFN/αRNP in vitro. IF analysis reveals significant intramitochondrial retention of Ox mtDNA in blood neutrophils from 50% SLE patients, but not in neutrophils from healthy controls or juvenile dermatomyositis (JDM) patients (Fig. 7 A). Furthermore, the presence of 8OHdG(+) aggregates within ex vivo SLE neutrophils correlates with the ability of the supernatants, recovered from the corresponding patient cells in culture, to activate pDCs (Fig. 7 B). Finally, anti–Ox mtDNA autoantibodies are detected in a fraction of SLE sera (Fig. 7 C).
Figure 7.
Intramitochondrial Ox mtDNA is a feature of SLE blood neutrophils. (A) Freshly isolated neutrophils from, healthy, JDM, or SLE blood were co-stained with anti-8OHdG and anti-TOMM20 antibodies. Bars, 10 µm. Quantification of 8OHdG staining in neutrophils from healthy donors (n = 6), JDM (n = 6), and SLE (n = 14) patients is also shown. (B) Correlation between Ox mtDNA levels in SLE neutrophils and IFN-α production by pDCs upon activation with corresponding SLE neutrophil supernatants. (n = 15). (C) Healthy (n = 6), JDM (n = 6), or SLE (n = 15) sera were assessed for the presence of anti-Ox mtDNA autoantibodies. Anti-8OHdG antibody was used as a control. *, P < 0.05; **, P < 0.01.
Thus, in addition to inducing pDC activation and IFN production, Ox mtDNA is an SLE autoantigen.
DISCUSSION
SLE neutrophils extrude interferogenic DNA when activated with anti-Sm/RNP antibodies in an FcγR, TLR7, and ROS-dependent manner (Garcia-Romo et al., 2011). This effect is recapitulated by healthy neutrophils upon priming with IFN-α. Although we previously reported that release of interferogenic DNA from lupus neutrophils correlated with NETosis (Garcia-Romo et al., 2011), subsequent analyses have led us to uncover that this process does not require neutrophil death. Furthermore, interferogenic DNA has a mitochondrial origin and requires oxidation to become a potent pDC activator.
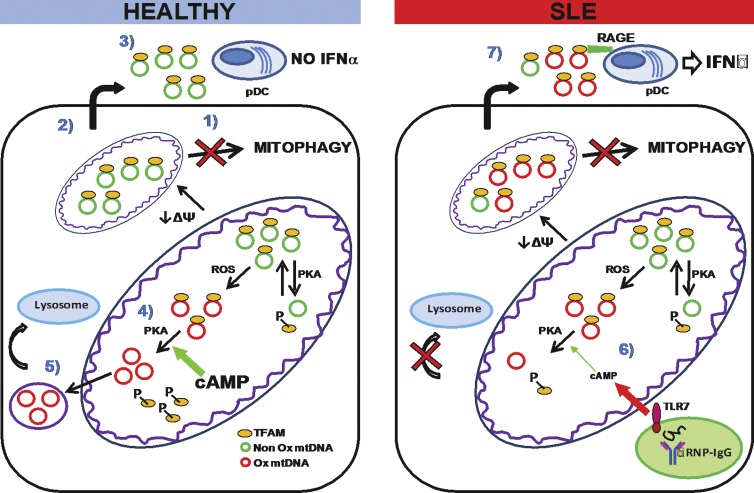
Our efforts to identify the nature of neutrophil-derived interferogenic DNA have led us to discover that human neutrophils display a constitutive defect in mitophagy that is compensated by two complementary pathways: extrusion of inner mitochondrial components, including two constituents of mitochondrial nucleoids (TFAM and mtDNA), devoid of Ox residues; and exportation of Ox mtDNA, upon dissociation from TFAM, into lysosomes for degradation. Exposure to IFN/αRNP disrupts the intracellular disposal of Ox mitochondrial products and leads to intramitochondrial accumulation and extracellular release of highly interferogenic Ox mtDNA bound to TFAM (Fig. 8).
Figure 8.
Proposed effect of IFN/αRNP on neutrophil mitochondria. (left) MtDNA is normally found in complex with TFAM in the mitochondrial matrix of healthy neutrophils. In the steady state, mitochondria can undergo oxidative damage and/or depolarization (↓ΔΨ). While most cells remove damaged mitochondria through mitophagy, this process does not take place in neutrophils (1). Instead, healthy neutrophils extrude matrix components into the extracellular space (2). These include mtDNA–TFAM complexes devoid of Ox DNA. These complexes do not activate pDCs (3). MtDNA undergoing oxidation is removed through an alternative route. Thus, TFAM dissociates from it upon phosphorylation by the matrix-resident PKA (4). TFAM is then degraded and Ox mtDNA is sorted into vesicles directed to lysosomes (5). (right) In SLE, neutrophil activation with TLR7-agonist autoantibodies leads to decreased mitochondrial cAMP levels (6) and reduced matrix PKA activity. As a result, TFAM is not efficiently disassembled from Ox mtDNA. This leads to intramitochondrial retention and extrusion of Ox nucleoids. Extracellular Ox nucleoids activate pDCs in a TFAM/RAGE-dependent manner (7).
Extrusion of mtDNA by granulocytes has been reported (Yousefi et al., 2008, 2009), but the mechanism remains elusive. Absence of extrusion of mitochondrial membrane-associated proteins suggests that it differs from exophagy, whereby the autophagosome content is released into the extracellular space (Duran et al., 2010; Manjithaya et al., 2010). Our preliminary observations show association between the lipidated form of LC3 (LC3-II) and neutrophil mitochondrial membranes, as well as co-localization of LC3 and TOMM20 on the plasma membrane (not depicted). This is reminiscent of the direct conjugation between LC3 and the phagosome membrane during LC3-associated phagocytosis (Sanjuan et al., 2007; Florey et al., 2011). Because of its highly fusogenic properties (Yang et al., 2013), LC3 might favor the direct fusion of mitochondrial membranes with the plasmalemma. Further studies are required, however, to confirm this hypothesis.
Extrusion of nucleoid components could be linked to the inability of neutrophils to complete mitophagy. In particular, we report that autophagosome–lysosome fusion is constitutively impaired in these cells. Autophagosome maturation is regulated by different proteins, including Rab7 (Jäger et al., 2004), presenilin-1 (Neely and Green, 2011), and UVRAG (Liang et al., 2008). In addition, TOM1, a constituent of the alternative endosomal sorting complex required for transport (ESCRT)-class 0, has been implicated in autophagosome–lysosome fusion (Tumbarello et al., 2012). Accordingly, we observe that neutrophils fail to up-regulate the transcription of several of the molecules involved in this process, including TOM1, upon chemical induction of mitochondrial damage. Although genetic manipulation of these pathways would strengthen our observations, blood neutrophils are terminally differentiated cells and therefore not amenable to these approaches.
Endosomal TLRs play a fundamental role in the IFN production and downstream immune alterations that characterize SLE (Guiducci et al., 2010; Rowland et al., 2014; Sisirak et al., 2014). The origin and nature of the NA ligands that trigger TLR sensors in this disease remain unclear. Here, we show that Ox mtDNA has a remarkable capacity to activate pDCs in a TLR9-dependent manner. The mechanism responsible for its increased interferogenicity compared with gDNA or NonOx mtDNA remains an open question. A recent study demonstrated that cytosolic Ox gDNA is less susceptible to TREX1-mediated degradation and contributes to STING activation in a cell-intrinsic manner (Gehrke et al., 2013). In our hands, Ox and Non Ox mtDNA display similar susceptibility to degradation by extracellular deoxyribonucleases (unpublished data). Likewise, we did not observe differences in uptake, intracellular compartmentalization, or stability of these two forms of mtDNA within pDCs (unpublished data). Unique TLR9-binding affinities and recruitment of specific adaptors might explain these findings.
Mitochondria use MDVs to deliver Ox cargo to lysosomes for degradation. MDVs contain damaged respiratory chain subunits, but were never found to incorporate mtDNA (Soubannier et al., 2012a,b). Our data reveal, for the first time, that neutrophil mitochondria remove Ox mtDNA in the steady state into lysosomes through a pathway resembling MDVs. This process seems to require nucleoid disassembly, as TFAM is never found in complex with Ox mtDNA either inside mitocondria or in the cytoplasm of healthy neutrophils.
The cationic protein TFAM binds RAGE on pDCs and facilitates mtC internalization. Steady-state dissociation from TFAM and exportation of Ox mtDNA into lysosomes might be in place to avoid that the highly interferogenic TFAM–Ox mtDNA complexes reach the extracellular space under conditions that lead to plasma membrane rupture, such as necrosis or NETosis. Nucleoid disassembly relies on TFAM phosphorylation by protein kinase A (PKA; Lu et al., 2013). We show that IFN/αRNP interfere with this step by decreasing the availability of cAMP, which regulates PKA activity in the mitochondrial matrix (Acin-Perez et al., 2009). This leads to intramitochondrial localization of TFAM–Ox mtDNA complexes and, eventually, to their extrusion.
To address the in vivo relevance of our findings, we analyzed neutrophils from SLE patient blood. In a large fraction of patients, these cells recapitulate the mitochondrial alterations observed in vitro, including aggregation and retention of Ox mtDNA. These alterations are not found in neutrophils from healthy children or JDM patients, who also overexpress IFN-inducible transcripts but do not carry the same TLR7 agonist autospecificities (Robinson and Reed, 2011). Furthermore, Ox mtDNA elicits an immune response in SLE, as autoantibodies against this form of modified DNA are detected in patient sera. Future studies will address whether neutrophil mitochondrial changes and/or the presence of anti–Ox mtDNA autoantibodies can be used as biomarkers to stratify SLE patients with unique clinical phenotypes and/or disease course.
Our data support that therapeutic efforts to increase extracellular and/or intracellular pathways involved in Ox mtDNA degradation should be explored in human SLE, a disease for which only one new drug has been approved in the past 50 yr.
MATERIALS AND METHODS
Patient information
This study was approved by the Institutional Review Boards of the University of Texas Southwestern Medical Center, Texas Scottish Rite Hospital for Children, and Baylor Scott & White Health Care Systems (Dallas, Texas). Informed consent was obtained from all patients or their parent/guardians. Blood samples were obtained from patients fulfilling the diagnosis of SLE according to the criteria established by the American College of Rheumatology and of JDM patients with biopsy-proven disease. Healthy pediatric controls were children visiting the clinic either for reasons not related to autoimmunity or for surgery not associated with any inflammatory diseases.
Antibodies, recombinant proteins, and chemicals
MitoTracker Green, MitoTracker DeepRed, MitoSox, and CellRox were purchased from Molecular Probes. Proteinase K was purchased from Roche. Recombinant human GM-CSF was purchased from BD. Recombinant human LL-37, R837, ODN-TTAGGG, and ODN-2216 were purchased from InvivoGen. FcR Blocking Reagent was purchased from Miltenyi Biotec. Antibody against 8OHdG (J-1; rabbit polyclonal IgG2b) and all chemicals were purchased from Santa Cruz Biotechnology, Inc. Antibodies against LL-37, TFAM (Clone 18G102B2E11), Mitofilin (Clone 2E4AD5), γH2A.X, TOMM20 (Clone 4F3), Pyruvate Dehydrogenase E2/E3bp (PDH), VDAC/Porin, Glyceraldehyde 3-phosphate dehydrogenase (GADPH), dsDNA, LAMP1 (Clone H4A3), and Rab7 were purchased from Abcam. Antibodies against MnSOD were purchased from Millipore. Antibodies against HMGB1, PGC1α, and Phospho-(Ser/Thr) PKA Substrate were purchased from Cell Signaling Technologies. Genomic DNA was obtained from BioChain. IRS661 and DVX41 were a gift from Dynavax Technologies Corporation. Specified neutrophils were cultured in the presence of αRNP-IgG (50 µg/ml), recombinant human GM-CSF (50 ng/ml), diphenylene iodonium (DPI; 10 µM), MT (10 µM), CCCP (25 µM), Rotenone (5 µM), Oligomycin (10 µM) plus Antimycin (1 µM; O+A), IRS661 (TLR7 antagonist; 1 µM), DVX42 (TLR8 antagonist; 1 µM), Bafilomycin A1 (BafA1; 1 µM), 8Br-cAMP (100 µM), IBMX (50 µM), or H89 (1 µM).
Anti-RNP/Sm autoantibodies (αRNP) isolation
Serum samples from SLE patients were filtered through a 0.45-µm polyvinylidene fluoride syringe. Anti-RNP/Sm and anti-dsDNA titers were measured using commercially available ELISA kits (GenWay Biotech). Samples positive for anti-RNP/Sm and negative for anti-dsDNA were selected and the total IgG fraction was purified using HiTrap Protein G HP column (GE Healthcare). Once purified, the total IgG fraction (αRNP) was desalted, dialyzed against PBS (Phosphate-Buffered Saline, pH 7.4), and quantified.
Cell isolation and culture
Blood from healthy donors was collected in ACD tubes (Vacutainer; BD). Neutrophils were then immediately isolated as previously described (Garcia-Romo et al., 2011). All neutrophil cultures were performed in complete RPMI supplemented with 2% FBS at cell density 1.25 million cells/500 µl. For IFN priming, neutrophils were preincubated with IFNα2β (2,000 U/ml; Schering Corp.) for 90 min at 37°C before stimulation. To study the effect of TLR ligands on mtDNA extrusion, neutrophils were cultured with R837 (TLR7 agonist; 1 µg/ml) alone or in combination with IRS661 (TLR7 antagonist; 1 µM) or with ODN2216 (TLR9 agonist; 1 µg/ml). Neutrophils were made necrotic by culturing the cells for 48 h. Cells were made apoptotic by UV irradiation or were made NETotic by PMA treatment (25 nM). Monocytes were obtained from healthy PBMCs after magnetic enrichment using negative selection (Stem Cell Technology). For pDC isolation, the total DC fraction was obtained from healthy buffy coats by magnetic cell sorting with the pan-DC Enrichment kit (Stem Cell Technology). Highly pure (>99%) plasmacytoid DCs (Lin− HLADR+ CD11c− CD123+) were then isolated from this fraction by FACS sorting as previously described (Garcia-Romo et al., 2011). PDCs were cultured (3 × 105 cells/100 µl) with 40% neutrophil supernatants. Alternatively, 400 ng/ml of synthetic Ox or nonOx mtDNA or genomic DNA, all preincubated with LL-37 (50 µg/ml), were used to stimulate pDCs. After 18 h, IFN-α levels in the corresponding supernatants were measured with the Human IFN-α Flex Set kit (BD). For blocking experiments, pDCs were preincubated, for 30 min at 37°C, with anti-TFAM (7 µg/ml; Cell Signaling Technology) or the corresponding isotype control, Recombinant Human RAGE-Fc Chimera (10 µg/ml; R&D Systems), BoxA (10 µg/ml; HMGBiotech), or the TLR9 inhibitor ODN-TTAGGG (5 µM).
Supernatant analysis
Cell supernatants were collected after 3 h of culture, centrifuged for 10 min at 1,400 g to remove cellular debris, and stored at −80C. For agarose gel electrophoresis, crude supernatants where treated with proteinase K (PK; 1 mg/ml) for 60 min at 60°C, and then 20 µl of the digested supernatants was loaded on 1% agarose gel and DNA was visualized with GelRed Nucleic Acid Stain (Phenix Research Products). For Western blot analysis, crude supernatants were concentrated three times with Concentrators PES Spin Columns (MWCO 3K; Thermo Fisher Scientific), and then boiled in 5× Lane Marker Reducing Sample Buffer (Thermo Fisher Scientific) for 5 min at 100°C before SDS-PAGE/Western blot analysis. Coomassie staining of the BSA band was used to assure that equal amounts of supernatants were loaded in each well.
Immunofluorescence microscopy (IF)
Neutrophils or monocytes were settled on poly-l-lysine–coated glass coverslips and cultured for 3 h, or for 60 min for CCCP and O+A experiments. Cells where then rinsed with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were permeabilized with 0.05% Triton X-100 in PBS for 5 min at room temperature, and then treated with Blocking Buffer (1% goat serum, 1% FcR Blocking Reagent and 1% BSA in PBS) for 30 min at room temperature. Primary and secondary antibody stainings were performed in Staining Buffer (1% BSA in PBS). Isotype specific anti–mouse or anti–rabbit Alexa Fluor 488 or Alexa Fluor 568 were used as secondary antibodies. Counterstaining of cell nuclei was performed with the Hoechst stain (Molecular Probes). Samples were embedded in ProLong Gold Antifade Reagent (Molecular Probes) and examined with a TCS SP5 confocal laser-scanning microscope equipped with a 63×/1.4 oil objective (Leica). ImageJ software (Version 1.47t; National Institutes of Health) was used for analysis. For the TUNEL assay, the Click-iT TUNEL Imaging Assay kit (Molecular Probes) was used according to the manufacturer’s instructions. The percentage of co-localization was calculated from the Manders' Overlap Coefficient using the Co-localization analysis plugin (ImageJ).
Flow cytometry
For mitochondrial mass assessment, cells were labeled with MitoTracker Green (25 nM) for 30 min at 37°C, and then analyzed immediately by flow cytometry. Apoptosis progression was assessed with the APO-BrdU TUNEL assay kit (Molecular Probes) following the manufacturer’s instructions. For TOMM20, LAMP1, or Rab7 surface expression, cells were cultured for 3 h, washed with FACS buffer (PBS + 1%FBS), and then stained with the corresponding antibodies or isotype controls. For total TOMM20, LAMP1, or Rab7 expression, cells were permeabilized with Cytofix/Cytoperm (BD) before proceeding with antibody staining. For mitochondria depolarization, cells were incubated for 3 h at 37°C. MitoTracker DeepRed (25 nM) was added the last 30 min of culture. Cells were then washed in PBS and analyzed immediately by flow cytometry. For ROS quantification, cells were loaded for 30 min at 37°C with MitoSox (2.5 µM) or CellRox (2.5 µM). Cells were then washed, cultured for 3 h as described, and then subjected to flow cytometry analysis.
Assessment of cell lysis
The degree of cell lysis was assessed by quantifying the activity of the cytosolic enzyme LDH using the Lactate Dehydrogenase Activity Assay kit (Sigma-Aldrich) according to the manufacturer’s instructions. Results were normalized to the enzyme activity in the total cell lysate (Input).
SDS-PAGE and Western blot
Cultured cells were washed in PBS, and then lysed in RIPA buffer in the presence of Halt Protease and Phosphates Inhibitor Cocktail (Thermo Fisher Scientific). Samples were incubated on ice for 30 min and then centrifuged (13,000 g for 10 min at 4°C). The supernatants containing the protein fraction were collected and stored at −80°C until further analysis. Protein concentration was estimated using the BCA kit (Thermo Fisher Scientific) following the manufacturer’s instructions. 40 µg of proteins were resuspended in 5× Lane Marker Reducing Sample Buffer (Thermo Fisher Scientific), boiled for 5 min at 100°C, and then subjected to electrophoresis with Mini-PROTEAN TGX Precast Gel (Bio-Rad Laboratories). The proteins were then transferred to nitrocellulose membranes, blocked for 1 h in 5% nonfat dry milk in TBST (Tris Buffered Saline containing 0.1% Tween-20), and incubated overnight at 4°C with the primary antibodies. After washing in TBST, the membranes were incubated for 1 h at room temperature with Poly HRP-conjugated anti–rabbit or anti–mouse IgG (Thermo Fisher Scientific). ECL Plus reagents (GE Healthcare) were used for detection. Digital images were acquired with ChemiDoc MP System (Bio-Rad Laboratories) and analyzed with Image Lab Software (Bio-Rad Laboratories).
Immunoprecipitation (IP) and co-IP
For co-IP of mtDNA–protein complexes, 1 ml crude neutrophil supernatants was precleared with 20 µl protein A/G plus agarose (Santa Cruz Biotechnology, Inc.) for 1 h at 4°C. IP was performed overnight at 4°C with anti-dsDNA antibody (10 µg/ml), followed by addition of 20 µl of protein A/G plus agarose for another 4 h. Immunoprecipitates (IP beads) were collected and washed five times with PBS, resuspended in 5× Lane Marker Reducing Sample Buffer, boiled for 5 min at 100°C, and then subjected to SDS-PAGE/Western blot analysis. Alternatively, washed IP beads were treated with PK (1 mg/ml) for 60 min at 60°C. The digested material was then loaded on 1% agarose gel and DNA was visualized with GelRed Nucleic Acid Stain. For intracellular for TFAM–8OHdG complexes or phospho-TFAM detection cells were gently lysed in ice-cold IP Lysis/Wash Buffer (Thermo Fisher Scientific) supplemented with Halt Protease and Phosphatase Inhibitor Cocktails (Thermo Fisher Scientific). Cell lysate (75 µg proteins) was incubated overnight with anti-TFAM antibody (10 µg/ml). Subsequently, 20 µl of protein A/G plus agarose were added for additional 6 h. The beads were then washed five times with 10 mM Tris HCl/20 mM NaCl (for TFAM–8OHdG complexes) or with PBS (for phospho-TFAM detection) and the associated complexes/proteins were released from the immunocomplexes by incubation for 5 min at 100°C with 2% SDS (for TFAM–8OHdG complexes) or with 5× Lane Marker Reducing Sample Buffer (for phospho-TFAM detection). The dissociated complexes/proteins were then collected by centrifugation and dot blotted to a nitrocellulose membrane (for TFAM–8OHdG complexes) or subjected to SDS-PAGE for Western blot analysis (for phospho-TFAM detection). To avoid interference of heavy and light antibody chains, HRP-conjugated Clean-Blot IP Detection Reagent (Thermo Fisher Scientific) was used as a detection reagent.
Isolation of DNA from supernatants and analysis
Supernatants (500 µl) were digested with PK (1 mg/ml) for 60 min at 60°C and then mixed with 1 vol UltraPure Phenol/Chloroform/Isoamyl Alcohol (25:24:1 vol/vol/vol; Invitrogen). DNA was precipitated from the aqueous phase with Ammonium Acetate and 100% Ethanol. Glycogen (20 µg/ml) was added as a carrier. The DNA pellet was washed twice with 75% Ethanol, vacuum-dried, and then resuspended in 40 µl TE Buffer, pH 8. DNA concentration was assessed with Quant-iT Picogreen dsDNA Assay kit (Invitrogen). PCR was performed with 5 ng of isolated DNA, AmpliTaq Gold 360 (Invitrogen) and 0.5 µM of the following primers: mtDNA encoded NADH dehydrogenase subunit 1 (ND1; 5′-GCATTCCTAATGCTTACCGAAC-3′ and 5′-AAGGGTGGAGAGGTTAAAGGAG-3′); genomic DNA encoded Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 5′-AGGCAACTAGGATGGTGTGG-3′ and 5′-TTGATTTTGGAGGGATCTCG-3′).
The PCR conditions were as follows: 95°C for 10 min; 30 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 60 s with a final extension of 72°C for 7 min. PCR products were resolved on a 3% agarose gel. For dot blot assay, 5 ng of DNA was blotted on a positively charged nylon membrane using the Bio-Dot Microfiltration System (Bio-Rad Laboratories), and then cross-linked by UV irradiation for 3 min. The membranes were blocked with 5% nonfat dry milk in TBST for 2 h at room temperature before overnight incubation, at 4°C, with the primary antibodies (diluted 1:200 in TBST) or with patients’ sera (diluted 1:200 in 1% nonfat dry milk in TBST). After washing in TBST, the membranes were incubated for 1 h at room temperature with Poly HRP-conjugated anti–rabbit, anti–mouse, or anti–human IgG. ECL Plus Western Blotting Detection Reagent (GE Healthcare) was used for detection.
Quantification of extruded mtDNA
3 ng of DNA isolated from cell-free supernatants were subjected to Real-Time PCR with Power SYBR Green PCR Master Mix (Invitrogen) and 0.5 µM of the primers described above. Results were expressed as mtDNA copy number (Venegas and Halberg, 2012). When the DNA concentration was too low to perform ethanol precipitation/Real-Time PCR, the amount of extruded mtDNA was measured in the PK-digested supernatants with Quant-iT Picogreen dsDNA Assay kit (Invitrogen) as previously described (Yousefi et al., 2009).
Quantification of mtDNA copy number in total cell extract
Total DNA was extracted from neutrophils or monocytes with the DNAzol Reagent (Invitrogen). The abundance of genomic and mtDNA were assessed by Real-Time PCR and the results were expressed as mtDNA copy number as described in the previous section.
In vitro mtDNA generation and labeling
mtDNA was amplified using two overlapping fragments, each ∼8.5 kb, with previously reported primers (Fendt et al., 2009). Amplification reaction was performed against human genomic DNA using elongase enzyme mix (Invitrogen). Ox mtDNA was generated by performing PCR reaction in presence of 200 µM 8-Oxo-2'-dGTP (TriLink). Amplicons were purified from residual primers and dNTPs by MSB Spin PCRapace (B-Bridge International).
Microarray analysis
Cells were cultured with medium or CCCP (25 µM) for 60 min, and then lysed with RLT Lysis Buffer (QIAGEN). Total RNA was isolated using the RNeasy kit (QIAGEN), amplified, and then labeled with Illumina TotalPrep RNA amplification kit (Invitrogen). Agilent 2100 Analyzer (Agilent Technologies) was used to assess RNA integrity. Biotinylated complementary RNA (cRNA) was hybridized to Human-6 Beadchip Array version 2 and scanned on Beadstation 500 (both from Illumina). Fluorescent hybridization signals were assessed with Beadstudio software (Illumina), and statistical analysis and hierarchical clustering were performed with GeneSpring 7.3.1 software (Agilent Technologies). Function and interactions of differentially expressed genes were inferred using Ingenuity Pathways Analysis (IPA 8.5). Raw microarray data were submitted to Gene Expression Omnibus under accession no. GSE78243.
Mitochondria isolation and Western/dot blot assay
Mitochondria were isolated from 107 neutrophils using the Mitochondria Isolation kit (Thermo Fisher Scientific) following the manufacturer’s instructions. For Western blot analysis, the mitochondrial pellet was resuspended in 5× Lane Marker Reducing Sample Buffer (Thermo Fisher Scientific), boiled for 10 min at 100°C, and then subjected to SDS-PAGE/Western blotting as described in the SDS-Page and Western blot section. For Ox DNA dot blot assay, mitochondrial pellet was digested with PK (1 mg/ml) and 0.5% SDS for 60 min at 60°C. MtDNA was then precipitated, quantified, and subjected to dot blot assay as described before.
Reconstitution of MDV formation in vitro
Mitochondrial budding assay was done as previously described (Soubannier et al., 2012b). In brief, purified mitochondria from 200 million neutrophils were incubated in 100 µl of an osmotically controlled, buffered environment, including an energy regenerating system, where the final concentrations of the reagents were: 50 µM antimycin, 220 mM mannitol, 68 mM sucrose, 80 mM KCl, 0.5 mM EGTA, 2 mM magnesium acetate, 20 mM Hepes, pH 7.4, 1 mM ATP, 5 mM succinate, 80 µM ADP, and 2 mM K2HPO4, pH 7.4. After 60 min at 37°C, the intact mitochondria were removed from the mixture by two sequential centrifugations at 7,400 g at 4°C. For Western blot, the supernatants containing the MDVs fraction were treated with 0.5 mg/ml trypsin for 10 min at 4°C. After trypsin treatment, loading buffer was added and the samples were separated by SDS-PAGE, transferred to nitrocellulose membranes, and blotted. For mtDNA content, the supernatants containing the MDVs fraction were incubated 20 min at 4°C in the presence of 25 U/ml of DNase I (Roche) to degrade unprotected mtDNA. Thereafter, the supernatants were digested with 1 mg/ml PK and 0.5% SDS for 60 min at 60°C. MtDNA was then precipitated and subjected to agarose gel electrophoresis, as described in the Supermatant Analysis section.
Mitochondrial cAMP assay
Neutrophil-enriched mitochondrial fraction was obtained as previously described (Maianski et al., 2004). cAMP levels were measured with the Cyclic AMP XP Assay kit (Cell Signaling Technologies) after the manufacturer’s instructions and results were then normalized to the protein content.
Measurement of mtDNA in the cytosolic extracts
Cytosolic extracts were generated largely as described previously (Holden and Horton, 2009). In brief, 2 × 107 neutrophils were resuspended in 700 µl buffer containing 150 mM NaCl, 50 mM Hepes, pH 7.4, and 25 µg/ml digitonin. The homogenates were incubated end over end for 10 min, to allow selective plasma membrane permeabilization, and then centrifuged at 1,400 g for 4 min two times to pellet intact cells. The cytosolic supernatants were transferred to fresh tubes and spun at 13,000 g for 8 min to pellet any remaining cellular debris and organelles. Protein concentration and volume of the cytosol extracts were normalized and DNA was isolated from 500 µl of the extracts as described above in the Isolation of DNA from supernatants and analysis section. mtDNA copy number was measured by RT-PCR with the same volume of the DNA solution.
Statistics
Results are shown as the mean ± SD. Statistical significance of differences was determined by Student’s t test or analysis of variance (ANOVA; Tukey-Kramer’s post-hoc test). P < 0.05 was considered significant. Correlation between variables was determined by using the Spearman test.
ACKNOWLEDGMENTS
The authors wish to thank Drs. Yong-Jun Liu, Nicole Baldwin, and Patrick Blanco for helpful discussions, as well as our patients, healthy donors, and their parents.
This work was supported by National Insitutes of Health grants P50 AR054083-01 and U19 AI082715 (to V. Pascual) and by the Baylor-Scott & White Health Care Research Foundation.
C. Guiducci and M. Gong are full-time employee of Dynavax. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- 8OHdG
- 8-hydroxydeoxyguanosine
- cAMP
- cyclic-AMP
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- dsDNA
- double-stranded DNA
- IC
- immune complex
- JDM
- juvenile dermatomyositis
- LDH
- lactate dehydrogenase
- MDV
- mitochondria-derived vesicle
- MnSOD
- manganese superoxide dismutase
- MT
- MitoTempo
- mtC
- mitochondrial complex
- mtDNA
- mitochondrial DNA
- Ox mtDNA
- oxidized mtDNA
- pDC
- plasmacytoid DC
- PDE
- phosphodiesterase
- PKA
- protein kinase A
- RNP
- ribonucleotide protein
- SLE
- systemic lupus erythematosus
- TFAM
- transcription factor A mitochondria
References
- Acin-Perez R., Salazar E., Kamenetsky M., Buck J., Levin L.R., and Manfredi G.. 2009. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 9:265–276. 10.1016/j.cmet.2009.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mayouf S.M., Sunker A., Abdwani R., Abrawi S.A., Almurshedi F., Alhashmi N., Al Sonbul A., Sewairi W., Qari A., Abdallah E., et al. 2011. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet. 43:1186–1188. 10.1038/ng.975 [DOI] [PubMed] [Google Scholar]
- Ashrafi G., and Schwarz T.L.. 2013. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20:31–42. 10.1038/cdd.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L., Palucka A.K., Arce E., Cantrell V., Borvak J., Banchereau J., and Pascual V.. 2003. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197:711–723. 10.1084/jem.20021553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham J., Morris D.L., Cunninghame Graham D.S., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L., et al. 2015. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47:1457–1464. 10.1038/ng.3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco P., Palucka A.K., Gill M., Pascual V., and Banchereau J.. 2001. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 294:1540–1543. 10.1126/science.1064890 [DOI] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., and Zychlinsky A.. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Collins L.V., Hajizadeh S., Holme E., Jonsson I.M., and Tarkowski A.. 2004. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 75:995–1000. 10.1189/jlb.0703328 [DOI] [PubMed] [Google Scholar]
- Crow Y.J. 2011. Type I interferonopathies: a novel set of inborn errors of immunity. Ann. N. Y. Acad. Sci. 1238:91–98. 10.1111/j.1749-6632.2011.06220.x [DOI] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., and Malhotra V.. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188:527–536. 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt L., Zimmermann B., Daniaux M., and Parson W.. 2009. Sequencing strategy for the whole mitochondrial genome resulting in high quality sequences. BMC Genomics. 10:139 10.1186/1471-2164-10-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O., Kim S.E., Sandoval C.P., Haynes C.M., and Overholtzer M.. 2011. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13:1335–1343. 10.1038/ncb2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Romo G.S., Caielli S., Vega B., Connolly J., Allantaz F., Xu Z., Punaro M., Baisch J., Guiducci C., Coffman R.L., et al. 2011. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci. Transl. Med. 3:73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke N., Mertens C., Zillinger T., Wenzel J., Bald T., Zahn S., Tüting T., Hartmann G., and Barchet W.. 2013. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 39:482–495. 10.1016/j.immuni.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Guiducci C., Gong M., Xu Z., Gill M., Chaussabel D., Meeker T., Chan J.H., Wright T., Punaro M., Bolland S., et al. 2010. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 465:937–941. 10.1038/nature09102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J., Martinez J., Riggs J.M., Tian J., Mehta P., Clarke L., Sasai M., Latz E., Brinkmann M.M., Iwasaki A., et al. 2012. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 37:986–997. 10.1016/j.immuni.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock M.B., and Kralli A.. 2009. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 71:177–203. 10.1146/annurev.physiol.010908.163119 [DOI] [PubMed] [Google Scholar]
- Holden P., and Horton W.A.. 2009. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes. 2:243 10.1186/1756-0500-2-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A., Pawlikowski J., Manoharan I., van Tuyn J., Nelson D.M., Rai T.S., Shah P.P., Hewitt G., Korolchuk V.I., Passos J.F., et al. 2013. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 202:129–143. 10.1083/jcb.201212110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., and Eskelinen E.L.. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848. 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- Julian M.W., Shao G., Bao S., Knoell D.L., Papenfuss T.L., VanGundy Z.C., and Crouser E.D.. 2012. Mitochondrial transcription factor A serves as a danger signal by augmenting plasmacytoid dendritic cell responses to DNA. J. Immunol. 189:433–443. 10.4049/jimmunol.1101375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H., and Nishimura S.. 1984. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 12:2137–2145. 10.1093/nar/12.4.2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J.B., Rane M.J., Scherzer J.A., Coxon P.Y., Kettritz R., Mathiesen J.M., Buridi A., and McLeish K.R.. 2000. Granulocyte-macrophage colony-stimulating factor delays neutrophil constitutive apoptosis through phosphoinositide 3-kinase and extracellular signal-regulated kinase pathways. J. Immunol. 164:4286–4291. 10.4049/jimmunol.164.8.4286 [DOI] [PubMed] [Google Scholar]
- Lan Y.Y., Londoño D., Bouley R., Rooney M.S., and Hacohen N.. 2014. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Reports. 9:180–192. 10.1016/j.celrep.2014.08.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B., Cao W., Wang Y.H., Su B., Nestle F.O., et al. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 449:564–569. 10.1038/nature06116 [DOI] [PubMed] [Google Scholar]
- Lande R., Ganguly D., Facchinetti V., Frasca L., Conrad C., Gregorio J., Meller S., Chamilos G., Sebasigari R., Riccieri V., et al. 2011. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci. Transl. Med. 3:73ra19 10.1126/scitranslmed.3001180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., and Jung J.U.. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10:776–787. 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Lee J., Nie X., Li M., Morozov Y.I., Venkatesh S., Bogenhagen D.F., Temiakov D., and Suzuki C.K.. 2013. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 49:121–132. 10.1016/j.molcel.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maianski N.A., Geissler J., Srinivasula S.M., Alnemri E.S., Roos D., and Kuijpers T.W.. 2004. Functional characterization of mitochondria in neutrophils: a role restricted to apoptosis. Cell Death Differ. 11:143–153. 10.1038/sj.cdd.4401320 [DOI] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., and Subramani S.. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188:537–546. 10.1083/jcb.200911149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means T.K., Latz E., Hayashi F., Murali M.R., Golenbock D.T., and Luster A.D.. 2005. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J. Clin. Invest. 115:407–417. 10.1172/JCI23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser K.L., Kelly J.A., Lessard C.J., and Harley J.B.. 2009. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 10:373–379. 10.1038/gene.2009.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., et al. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12:222–230. 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely K.M., and Green K.N.. 2011. Presenilins mediate efficient proteolysis via the autophagosome-lysosome system. Autophagy. 7:664–665. 10.4161/auto.7.6.15448 [DOI] [PubMed] [Google Scholar]
- Oka T., Hikoso S., Yamaguchi O., Taneike M., Takeda T., Tamai T., Oyabu J., Murakawa T., Nakayama H., Nishida K., et al. 2012. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 485:251–255. 10.1038/nature10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padman B.S., Bach M., Lucarelli G., Prescott M., and Ramm G.. 2013. The protonophore CCCP interferes with lysosomal degradation of autophagic cargo in yeast and mammalian cells. Autophagy. 9:1862–1875. 10.4161/auto.26557 [DOI] [PubMed] [Google Scholar]
- Pascual V., Farkas L., and Banchereau J.. 2006. Systemic lupus erythematosus: all roads lead to type I interferons. Curr. Opin. Immunol. 18:676–682. 10.1016/j.coi.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Robinson A.B., and Reed A.M.. 2011. Clinical features, pathogenesis and treatment of juvenile and adult dermatomyositis. Nat. Rev. Rheumatol. 7:664–675. 10.1038/nrrheum.2011.139 [DOI] [PubMed] [Google Scholar]
- Rowland S.L., Riggs J.M., Gilfillan S., Bugatti M., Vermi W., Kolbeck R., Unanue E.R., Sanjuan M.A., and Colonna M.. 2014. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med. 211:1977–1991. 10.1084/jem.20132620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., and Green D.R.. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 450:1253–1257. 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Shimada K., Crother T.R., Karlin J., Dagvadorj J., Chiba N., Chen S., Ramanujan V.K., Wolf A.J., Vergnes L., Ojcius D.M., et al. 2012. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 36:401–414. 10.1016/j.immuni.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisirak V., Ganguly D., Lewis K.L., Couillault C., Tanaka L., Bolland S., D’Agati V., Elkon K.B., and Reizis B.. 2014. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 211:1969–1976. 10.1084/jem.20132522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V., McLelland G.L., Zunino R., Braschi E., Rippstein P., Fon E.A., and McBride H.M.. 2012a A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22:135–141. 10.1016/j.cub.2011.11.057 [DOI] [PubMed] [Google Scholar]
- Soubannier V., Rippstein P., Kaufman B.A., Shoubridge E.A., and McBride H.M.. 2012b Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 7:e52830 10.1371/journal.pone.0052830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello D.A., Waxse B.J., Arden S.D., Bright N.A., Kendrick-Jones J., and Buss F.. 2012. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat. Cell Biol. 14:1024–1035. 10.1038/ncb2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas V., and Halberg M.C.. 2012. Measurement of mitochondrial DNA copy number. Methods Mol. Biol. 837:327–335. 10.1007/978-1-61779-504-6_22 [DOI] [PubMed] [Google Scholar]
- Yakes F.M., and Van Houten B.. 1997. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA. 94:514–519. 10.1073/pnas.94.2.514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Li Y., Pantoom S., Triola G., and Wu Y.W.. 2013. Semisynthetic lipidated LC3 protein mediates membrane fusion. ChemBioChem. 14:1296–1300. 10.1002/cbic.201300344 [DOI] [PubMed] [Google Scholar]
- Yousefi S., Gold J.A., Andina N., Lee J.J., Kelly A.M., Kozlowski E., Schmid I., Straumann A., Reichenbach J., Gleich G.J., and Simon H.U.. 2008. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 14:949–953. 10.1038/nm.1855 [DOI] [PubMed] [Google Scholar]
- Yousefi S., Mihalache C., Kozlowski E., Schmid I., and Simon H.U.. 2009. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 16:1438–1444. 10.1038/cdd.2009.96 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., and Hauser C.J.. 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 464:104–107. 10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]