Naip knockout mice provide genetic evidence for the specificity of NAIP1, 2, and 5 in recognizing bacterial T3SS needle protein, rod protein, and flagellin, respectively. Naip1−/−, Naip2−/−, and Naip5−/− mice underscore the physiological contribution of the NAIP proteins in innate defense against cytosolic bacteria.
Abstract
Biochemical studies suggest that the NAIP family of NLR proteins are cytosolic innate receptors that directly recognize bacterial ligands and trigger NLRC4 inflammasome activation. In this study, we generated Naip5−/−, Naip1−/−, and Naip2−/− mice and showed that bone marrow macrophages derived from these knockout mice are specifically deficient in detecting bacterial flagellin, the type III secretion system needle, and the rod protein, respectively. Naip1−/−, Naip2−/−, and Naip5−/− mice also resist lethal inflammasome activation by the corresponding ligand. Furthermore, infections performed in the Naip-deficient macrophages have helped to define the major signal in Legionella pneumophila, Salmonella Typhimurium and Shigella flexneri that is detected by the NAIP/NLRC4 inflammasome. Using an engineered S. Typhimurium infection model, we demonstrate the critical role of NAIPs in clearing bacterial infection and protecting mice from bacterial virulence–induced lethality. These results provide definitive genetic evidence for the important physiological function of NAIPs in antibacterial defense and inflammatory damage–induced lethality in mice.
Canonical inflammasomes activate caspase-1 through diverse biochemical mechanisms, serving as cytosolic immunity against infection or danger signals (Zhao and Shao, 2016). Activated caspase-1 processes IL-1β/18 for its maturation and secretion and also triggers pyroptotic cell death through cleavage of the gasdermin D protein (Kayagaki et al., 2015; Shi et al., 2015). Assembly of the inflammasome is mediated by a stimulus-responsive pattern recognition receptor (PRR) that specifies the nature and function of the inflammasome. The PRR then recruits an inflammasome adaptor ASC (apoptosis-associated speck-like protein containing a CARD) or NLRC4, forming a large platform for caspase-1 activation (Hauenstein et al., 2015). PRRs functioning as inflammasome sensors include NLRP3, AIM2, NAIPs, NLRP1, and Pyrin, all of which can detect or exclusively recognize bacterial signals (Hagar and Miao, 2014; Zhao and Shao, 2016).
The NAIP/NLRC4 inflammasome is best characterized with a 3D structure determined (Hu et al., 2015; Zhang et al., 2015; Zhao and Shao, 2015). NAIPs are the only NLRs proved to be inflammasome receptors (Kofoed and Vance, 2011; Zhao et al., 2011; Yang et al., 2013; Tenthorey et al., 2014). One activated NAIP molecule recruits and activates one NLRC4 molecule that then propagates its active conformation to 8–10 more NLRC4 molecules to form a wheel-shaped complex (Hu et al., 2015; Zhang et al., 2015). The NAIP inflammasome is critical for antibacterial immunity (Zhao and Shao, 2015). Human NAIP recognizes the needle component of the bacterial type III secretion system (T3SS; Zhao et al., 2011). The T3SS is evolutionarily related to the flagellar export system. Mice have seven Naips located in the same genomic locus, among which Naip1, 2, 5, and 6 are expressed in C57BL/6 mice. NAIP1 is the orthologue of human NAIP, and NAIP2 recognizes the rod component of the T3SS; NAIP5 and 6 are cytosolic receptors for bacterial flagellin. The NAIP/NLRC4 inflammasome responds to a broad spectrum of pathogens, including Salmonella, Shigella, Legionella, and Pseudomonas spp., in which NAIPs exhibit high ligand specificity (Kofoed and Vance, 2011; Zhao et al., 2011; Rayamajhi et al., 2013; Yang et al., 2013; Tenthorey et al., 2014). Studies using Nlrc4−/− mice indicate a critical role of the NAIP/NLRC4 inflammasome in protecting mice from lethal infection with environmental bacteria such as Chromobacterium violaceum and Burkholderia thailandensis or an engineered Salmonella Typhimurium ectopically expressing flagellin or its T3SS rod protein (Miao et al., 2010; Aachoui et al., 2015; Maltez et al., 2015). In this process, neutrophil-mediated killing coordinated with IL-18–driven natural killer cell responses is proposed as the effector mechanism. Other studies suggest an epithelium-intrinsic function of the NAIP/NLRC4 inflammasome that expels infected enterocytes, thereby inhibiting bacterial replication in the intestine and colitis progression (Nordlander et al., 2014; Sellin et al., 2014). NLRC4-driven IL-1β production from intestinal phagocytes can also discriminate between pathogenic and commensal bacteria in the gut (Franchi et al., 2012).
Hyperactivation of the NLRC4–caspase-1 axis is lethal to mice, and the lethality is caused by severe systemic inflammation, similar to excessive activation of the caspase-11–gasdermin D pathway in endotoxic shock (Shi et al., 2014, 2015; Kayagaki et al., 2015). One study has shown that extraintestinal outgrowth of a multidrug-resistant Escherichia coli strain in antibiotics plus dextran sulfate sodium–treated mice causes sepsis-like lethality in an NAIP5/NLRC4-dependent manner (Ayres et al., 2012). Lethal NLRC4 inflammasome activation is best modeled by peritoneal injection of LFn-flagellin that efficiently delivers the flagellin protein into the cytosol (Zhao et al., 2011; Yang et al., 2013). This defined assay has recorded a critical role of Naip5, Nlrc4, and caspase-1 in the lethal response (von Moltke et al., 2012).
Although the function of Nlrc4 is extensively studied, the in vivo role of Naips in antibacterial defense and inflammatory damage are not fully defined yet. In this study, using macrophages derived from Naip1−/−, Naip2−/−, and Naip5−/− mice, we confirmed that NAIP1, 2, and 5 specifically recognize bacterial T3SS needle protein, rod protein, and flagellin, respectively, and examined the differential contribution of each NAIP in sensing various bacterial infections. The Naips are also required for lethal inflammasome activation in a ligand-specific manner. Finally, using the S. Typhimurium infection model (Miao et al., 2010), we show that NAIP2/5 but not NAIP1 activation can clear the infection and promote the survival of infected mice.
RESULTS AND DISCUSSION
Generation of Naip1−/−, Naip2−/−, and Naip5−/− mice
To assess the genetic function of different NAIP family members, we generated Naip1−/−, Naip2−/−, and Naip5−/− mice. Given the high polymorphism of the Naip locus in different mouse inbred strains (Wright et al., 2003), the same genetic background (C57BL/6) was used to make the mice. The Naip5−/− allele was obtained in C57BL/6-derived embryonic stem (ES) cells through homologous recombination (Fig. S1 A; see Materials and methods). The largest exon 9–encoding residues, Asp388 to Glu1092 (covering the entire nucleotide-binding oligomerization domain and two leucine-rich repeat units), were replaced with a neomycin-resistant cassette. The targeting was validated by PCR and subsequent DNA sequencing (Fig. S1 B). Naip1−/− and Naip2−/− mice were generated using transcription activator–like effector nuclease (TALEN)–based genome editing similarly as described previously (Xu et al., 2014); the targeting sequences and validation of the KO alleles are detailed in Materials and methods. One homozygous Naip1 mutant allele (Fig. S1 C) and two homozygous Naip2 alleles (Fig. S1 D), all expected to produce a nonfunctional protein, were subjected to further functional analyses.
Macrophages from Naip−/− mice resist stimulation by the corresponding bacterial ligand
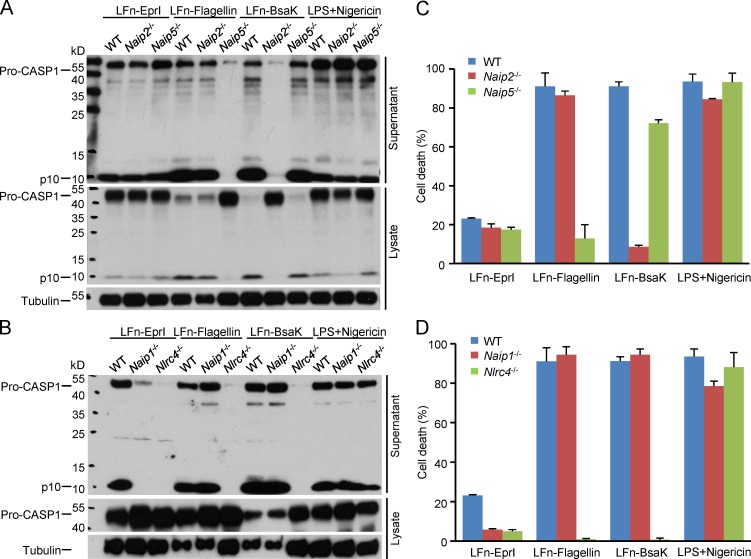
Primary BMDMs were derived from the Naip1−/−, Naip2−/−, and Naip5−/− mice as well as the WT and Nlrc4−/− C57BL/6 mice as the positive and negative control, respectively. The cells were stimulated with different NAIP ligands or LPS plus nigericin and subjected to inflammasome activation assays. LPS plus nigericin, an activator of the NLRP3 inflammasome, triggered similar extents of caspase-1 activation in WT and all four KO BMDMs (Fig. 1, A and B), confirming the intact inflammasome responses in these genetically modified cells. Flagellin (Pseudomonas aeruginosa), the T3SS rod protein (Bsak from B. thailandensis), and needle protein (EprI from enterohemorrhagic E. coli [EHEC]) were delivered into the BMDMs using the LFn fusion strategy. As expected from previous studies (Franchi et al., 2006; Miao et al., 2006; Lightfield et al., 2008; Kofoed and Vance, 2011; Zhao et al., 2011; Yang et al., 2013), all three bacterial ligands did not activate caspase-1 in Nlrc4−/− BMDMs (Fig. 1 B), and Naip5−/− cells showed no caspase-1 activation in response to LFn-flagellin among the three bacterial ligands (Fig. 1 A). In contrast, Naip5−/− BMDMs exhibited normal caspase-1 activation upon stimulation by LFn-BsaK and LFn-EprI. Similarly, Naip2−/− and Naip1−/− BMDMs only resisted LFn-BsaK and LFn-EprI stimulation, respectively (Fig. 1 A). When pyroptosis was examined in the aforementioned BMDMs, a perfect correlation between caspase-1 activation and pyroptosis was observed (Fig. 1, C and D). Collectively, these definitive genetic evidences support that NAIP1, NAIP2, and NAIP5 are specific intracellular receptors for bacterial T3SS needle protein, rod protein, and flagellin, respectively.
Figure 1.
Naips play nonredundant roles in specifically recognizing the corresponding bacterial ligands. Flagellin (type b from P. aeruginosa), the T3SS rod protein (Bsak from B. thailandensis), and the needle protein (EprI from EHEC) were delivered into primary BMDMs derived from WT or the indicated knockout KO mice (C57BL/6 background) by using the LFn fusion strategy. LPS plus nigericin, an activator of the NLRP3 inflammasome, served as a control. (A and B) Macrophage supernatants or lysates were collected for anti–caspase-1 and/or anti-tubulin immunoblotting. (C and D) Lactate dehydrogenase (LDH) release–based cell death assays (mean values ± SD) are from three technical replicates. All data shown are representative of three independent experiments. Pro-CASP1, caspase-1 precursor; p10, mature caspase-1.
Naips mediate lethal inflammasome activation in mice in a ligand-specific manner
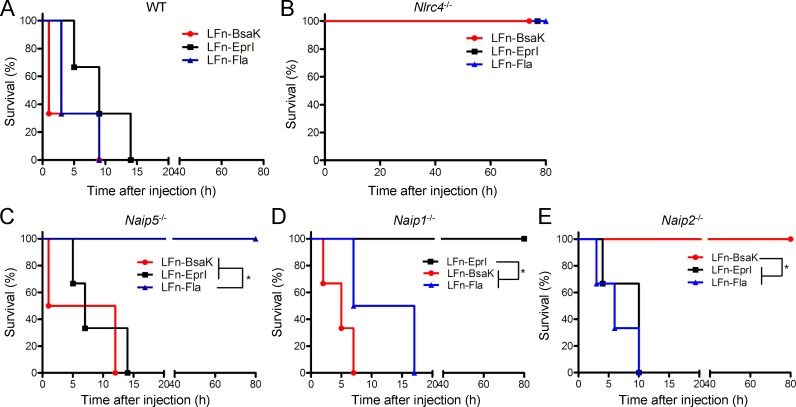
Intraperitoneal injections of LFn-flagellin could kill the mice within several hours as a result of systemic hyperactivation of the NAIP5/NLRC4 inflammasome (Fig. 2 A; von Moltke et al., 2012).The lethality was completely abrogated in Nlrc4−/− and Naip5−/− mice (Fig. 2, B and C). We further observed that injection of LFn-BsaK or LFn-EprI caused the same lethal effect, which was completely disrupted in the Nlrc4−/− mice (Fig. 2, A and B). The T3SS rod protein has the most potent biochemical activity in activating the NLRC4 inflammasome (Zhao et al., 2011; Yang et al., 2013). Consistently, mice injected with LFn-BsaK died around 30 min, whereas the lethality in LFn-flagellin– or LFn-EprI–injected mice took several hours to develop (Fig. 2 A). Notably, Naip5−/− gave no protection from LFn-BsaK– and LFn-EprI–induced lethality (Fig. 2 C), but the Naip1−/− and Naip2−/− mice showed complete resistance to LFn-EprI and LFn-BsaK injection, respectively (Fig. 2, D and E). In contrast, both Naip1−/− and Naip2−/− mice remained sensitive to the other two bacterial ligands (flagellin and Bsak for Naip1−/− and flagellin and EprI for Naip2−/−). Similar results were obtained with other T3SS rod and needle proteins, such as PrgJ from S. Typhimurium and MxiH from S. flexneri, respectively (not depicted). These data agree well with the specific recognition of the T3SS rod and needle protein by NAIP2/1 observed in the cell culture system. The results also highlight the nonredundant function of NAIPs in mediating lethal NLRC4 inflammasome activation in mice by the bacterial ligands.
Figure 2.
Naips mediate lethal inflammasome activation in the ligand-specific manner. (A–E) Lethal doses of LFn-tagged T3SS rod protein (Bsak from B. thailandensis), needle protein (EprI from EHEC), and P. aeruginosa flagellin (Fla) were peritoneally injected into WT, Naip1−/−, Naip2−/−, Naip5−/−, or Nlrc4−/− mice (C57BL/6 background) together with equal amounts of the protective antigen protein. The mouse survival curve analysis (six mice for each group) was performed in Prism 5. *, P ≤ 0.05. All data shown are representative of three independent experiments.
The role of Naips in macrophage responses to various bacterial infections
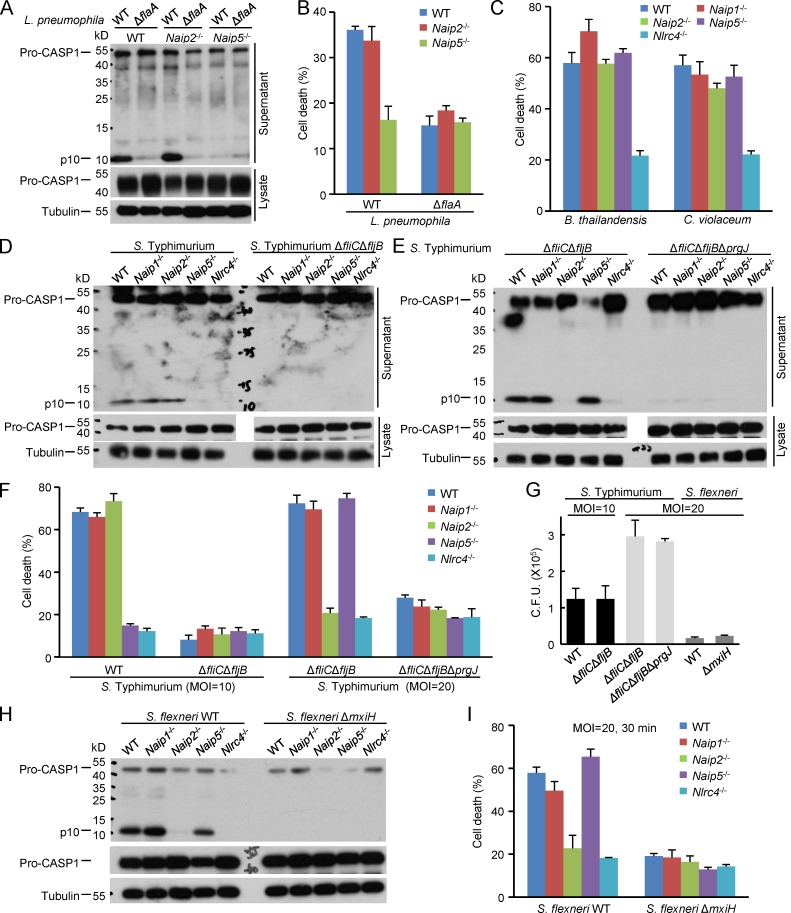
Having defined the ligand specificity of different NAIPs using the LFn-mediated delivery system, we then examined the role of each Naip in sensing various bacterial infections. Monomeric flagellin can be secreted out of the bacteria through the T3SS or type 4 secretion system. The T3SS rod and needle proteins, essential for assembly of a functional T3SS, can also be secreted into host cytosol during infection. Mutation of genes encoding flagellin and T3SS rod/needle proteins are expected to affect one another and cause other unintended interference on bacterial physiology (Yang et al., 2013). However, the individual Naip−/− cells allowed for dissecting the contribution of a particular bacterial ligand to infection-induced inflammasome activation. Five different pathogenic bacteria, including B. thailandensis, C. violaceum, Legionella pneumophila, S. Typhimurium, and S. flexneri, which can all activate caspase-1 in an NLRC4-dependent manner (Zhao and Shao, 2015), were used to infect the Naip−/− BMDMs. Consistent with a previous study (Lightfield et al., 2008), Naip5−/− abolished caspase-1 activation and pyroptosis in response to L. pneumophila infection (Fig. 3, A and B). Deletion of the flagellin-encoding gene flaA resulted in similar disruption of caspase-1 activation and pyroptosis in infected WT BMDMs. L. pneumophila harbors no T3SS and triggered the same level of inflammasome activation in Naip2−/− BMDMs as in WT cells (Fig. 3, A and B). Different from that in L. pneumophila infection, deletion of any single Naip caused little inhibition on B. thailandensis– and C. violaceum–induced pyroptosis despite the markedly reduced death in Nlrc4−/− BMDMs (Fig. 3 C). Given that NAIPs are the only known sensors upstream of NLRC4, this suggests that multiple NAIPs play a redundant role in recognizing the two bacteria. However, though less likely, it is possible that NLRC4 may have a NAIP-independent function in activating caspase-1.
Figure 3.
The role of Naips in macrophage responses to various bacterial infections. (A–C) Primary BMDMs derived from WT, Naip1−/−, Naip2−/−, or Naip5−/− mice (C57BL/6 background) were infected with L. pneumophila, B. thailandensis, or C. violaceum. (D–I) The BMDM cells were also infected with an indicated S. Typhimurium or S. flexneri strain at an MOI of 10 for 10 (D) or 30 min (F and G) or a higher MOI of 20 for 2 h (E–G) or 30 min (H and I). Macrophage supernatants or lysates were subjected to anti–caspase-1 and/or anti-tubulin immunoblotting (A, D, E, and H). Lactate dehydrogenase (LDH) release–based cell death assays (mean values ± SD) are from three technical replicates (B, C, F, and I). CFUs of S. Typhimurium and S. flexneri strains recovered from infected WT macrophages are in G (n = 3; mean ± SD). ΔflaA denotes the flagellin-deficient strain of L. pneumophila. ΔfliCΔfljB and ΔfliCΔfljBΔprgJ denote the flagellin-deficient and the flagellin/rod protein double-deficient S. Typhimurium strains, respectively. ΔmxiH is the needle protein–deficient S. flexneri strain. All data shown are representative of three independent experiments. Pro-CASP1, caspase-1 precursor; p10, mature caspase 1.
S. Typhimurium and S. flexneri are two extensively studied models for enteric bacteria. The virulence factors in S. Typhimurium are tightly regulated during infection; its Salmonella pathogenicity island (SPI)–1 T3SS is activated upon contact with the host cell whereas the SPI-2 T3SS is not turned on until successful invasion into the cytosol (Coburn et al., 2007). We observed that at the early time point after S. Typhimurium infection (multiplicity of infection [MOI] = 10), Naip5−/− BMDMs, compared with WT BMDMs, showed drastically reduced caspase-1 activation and pyroptosis, which also occurred in Nlrc4−/− but not Naip1−/− and Naip2−/− cells (Fig. 3, D and F). Under the same condition, deletion of flagellin-encoding genes (ΔfliCΔfljB) produced a similar effect in WT macrophages. Thus, NAIP5 recognition of flagellin dominates the early inflammasome response in S. Typhimurium–infected macrophages. When the infection was performed at a higher MOI of 20 for 120 min, evident caspase-1 activation and pyroptosis could still be detected in S. Typhimurium ΔfliCΔfljB-infected BMDMs (Fig. 3, E and F). This activation was diminished by additional deletion of prgJ encoding the SPI-I T3SS rod protein. As PrgJ is essential for SPI-1 T3SS assembly and the SPI-1 is critical for S. Typhimurium invasion and infection, results of the prgJ mutant could not tell the bacterial signal that was directly recognized by the inflammasome. Importantly, S. Typhimurium ΔfliCΔfljB-induced caspase-1 activation and pyroptosis were largely abolished in Naip2−/− cells but remained intact in Naip1−/− and Naip5−/− cells (Fig. 3, E and F). The absence of flagellin and PrgJ did not affect S. Typhimurium infection and intracellular growth within the time period of infection (Fig. 3 G), suggesting that the decreased pyroptosis observed in mutant bacteria is not a result of differences in the amount of intracellular bacteria. These data suggest that the SPI-I T3SS rod protein PrgJ but not the needle protein PrgI is a major target of the NAIP/NLRC4 inflammasome at the late stage of S. Typhimurium infection. This is likely because NAIP1 is expressed poorly in BMDMs (Yang et al., 2013).
S. flexneri is nonflagellar and contains one T3SS important for pathogenesis. siRNA knockdown experiments have shown that recognition of the T3SS needle protein MxiH by NAIP1 and human NAIP is an important defense response upon S. flexneri infection of mouse BM dendritic cells and human THP1 cells, respectively (Yang et al., 2013). In mouse BMDMs, deletion of either mxiH or mxiI (encoding the T3SS rod protein) from S. flexneri both disrupted caspase-1 activation and pyroptosis (Fig. 3, H and I; Suzuki et al., 2014). However, only Naip2−/− BMDMs and not Naip1−/− and Naip5−/− cells exhibited defective inflammasome activation in response to WT S. flexneri infection (Fig. 3, H and I). The extent of inhibition by Naip2−/− was comparable with that by Nlrc4−/−. Similar to that in S. Typhimurium, ΔmxiH did not affect S. flexneri intracellular growth (Fig. 3 G). Thus, cytosolic S. flexneri is mainly detected by NAIP2 in mouse BMDMs. Collectively, these results suggest that the role of a particular NAIP in cytosolic innate immunity is highly dependent on the bacterial species and the type of cells assayed as well as the infection condition.
The role of Naips in host defenses against systemic infection by engineered S. Typhimurium in mice
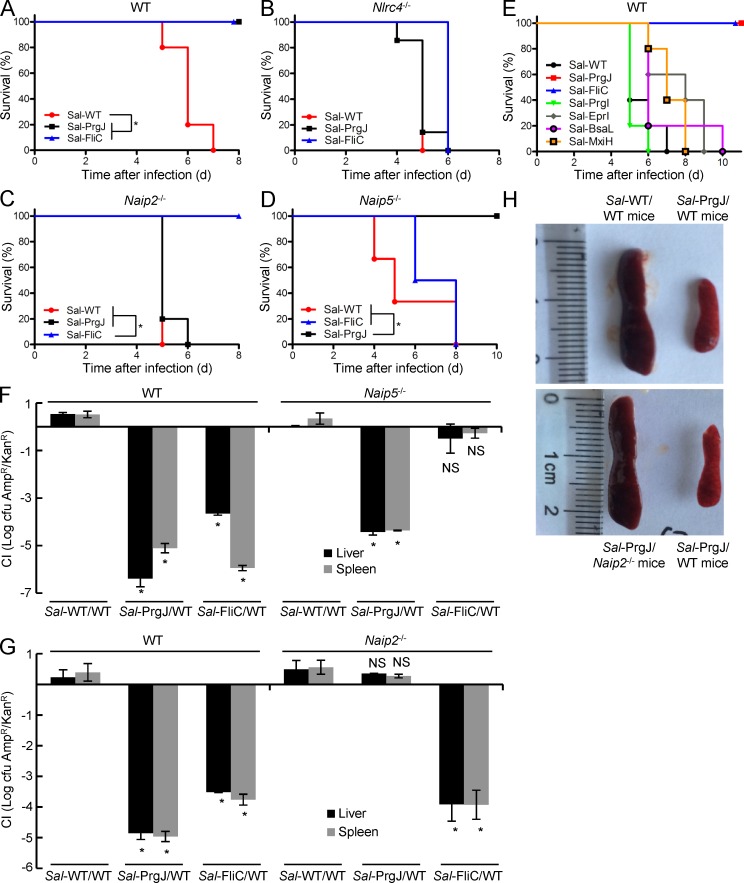
S. Typhimurium is widely used for examining NLRC4-mediated inflammasome defense against bacterial infection. Mice succumb to peritoneal S. Typhimurium infection within a week because of the high virulence of the genus. Consistent with a previous study (Miao et al., 2010), ectopic expression of flagellin or PrgJ under the SPI-II promoter in the bacteria protected the mice from S. Typhimurium–induced lethality (Fig. 4 A). This effect was diminished in Nlrc4−/− mice (Fig. 4 B), confirming the critical role of the NLRC4-mediated inflammasome responses in this process. Naip2−/− could also reverse the protection induced by enforced expression of PrgJ, but not that by flagellin (Fig. 4 C). Similarly, Naip5−/− mice remained resistant to PrgJ-expressing S. Typhimurium but succumbed to infection by the flagellin-expressing strain (Fig. 4 D). We also performed the same assay with S. Typhimurium expressing a T3SS needle protein such as PrgI, EprI, MxiH, or BsaL (from B. thailandensis), but in all cases, we observed no protection from infection-induced lethality (Fig. 4 E). This is consistent with the needle protein being generally less active than the rod protein and flagellin in activating the inflammasome (Yang et al., 2013), likely because of the poor expression of NAIP1 in mice.
Figure 4.
The role of Naips in mouse defenses against systemic infection by engineered S. Typhimurium. (A–E and H) WT, Naip2−/−, Naip5−/−, or Nlrc4−/− mice (C57BL/6 background) were subjected to peritoneal infection with engineered S. Typhimurium strains. The numbers of mice used were five (A, C, and E), six (D), and seven (B). Sal-WT denotes WT S. Typhimurium harboring an empty vector (ampicillin resistant); Sal-PrgJ, Sal-FliC, and Sal-PrgI/EprI/BsaL/MxiH denote S. Typhimurium–expressing PrgJ, FliC, or indicated T3SS needle proteins, respectively, under the promoter of an SPI-II T3SS effector gene, sseJ. The mouse survival curve analysis was performed in Prism 5. *, P ≤ 0.05. Representative photos of the spleens of the infected mice from A and C are in H. (F and G) Mice were infected similarly as those in A–D except that a WT S. Typhimurium strain harboring a kanamycin-resistant gene was included as an internal reference. The CI value of S. Typhimurium replication in the spleen and liver of mice (mean values ± SD) was determined 4–5 d after the infection. *, P < 0.05 (Student’s t test). All data shown are representative of three independent experiments.
We further investigated whether the protection observed with the engineered S. Typhimurium results from inhibition of bacterial replication. For this, we measured the competitive index (CI) in infected mice, similarly as previously performed in Nlrc4−/− mice (Miao et al., 2010). When the liver and the spleen from the infected mice were examined, bacterial loads in these organs from infection with the flagellin or PrgJ-expressing S. Typhimurium were found to be lower than that of the coinfected reference WT strain by more than three orders of magnitude (Fig. 4, F and G). Comparable levels of WT bacteria bearing different antibiotic resistance were recovered from the mice, suggesting that different antibiotic markers used in the engineered strain and the reference strain were not the cause for the observed lower bacterial loads. Remarkably, the lowered bacterial loads were specifically blocked in Naip5−/− and Naip2−/− mice for flagellin- and PrgJ-expressing S. Typhimurium, respectively (Fig. 4, F and G). Moreover, we consistently observed markedly enlarged spleens and abnormal livers in mice that failed to clear the bacteria (Fig. 4 H and not depicted). This is consistent with the notion that a higher load of bacteria could cause pathological effects and disrupt normal tissue homeostasis. These data suggest that Naips play critical roles in clearing bacterial infection by recognizing the cognate bacterial ligand and activating inflammasome-mediated defenses.
In summary, using Naip1−/−, Naip2−/−, and Naip5−/− mice, we provided definitive genetic evidences that highlight the in vivo function of the NAIPs in innate defense against cytosolic bacteria as well as in mediating lethal inflammasome activation by flagellin and the T3SS apparatus. Our Naip−/− mice also allowed for demonstrating the high specificity of NAIP1, 2 and, 5 in recognizing bacterial T3SS needle protein, rod protein, and flagellin, respectively. Recent studies have depicted a structural model for how ligand binding to a single NAIP receptor relays the activation signal to the NLRC4 adaptor and triggers the assembly of the NAIP–NLRC4 complex (Hu et al., 2015; Zhang et al., 2015). Because of the limited resolution of the cryo-electron microscopy model, further studies, ideally crystal structure of the ligand–NAIP complex, are needed to develop full mechanistic understanding of the ligand specificity of NAIP receptors. This is particularly interesting given that NAIPs do not use the presumed leucine-rich repeat domain to bind to the ligands (Tenthorey et al., 2014).
The Naip−/− mice generated in our study are also useful for dissecting the differential contributions of different bacterial signals to host defenses against a particular bacterial infection. It is worth mentioning that mice also express NAIP6. NAIP6 shares the highest sequence identity of 94.7% with NAIP5, and the two NAIPs are biochemically indistinguishable in binding to certain flagellin and activating NLRC4 in cell culture assays (Zhao et al., 2011). NAIP6 has a much lower expression than NAIP5 in BMDM cells. Our genetic experiments, as well as a previous study (Lightfield et al., 2008), appear to suggest that NAIP5 accounts for all observed inflammasome responses induced by the flagellin assayed. Thus, NAIP6 may have a specific cell type–dependent function in response to certain flagellin variants. Generating Naip6−/− mice and profiling a larger panel of flagellins from different bacteria shall help to address NAIP6 function in inflammasome-mediated antibacterial defenses.
MATERIALS AND METHODS
KO mice
C57BL/6 WT mice were purchased from Vital River Laboratory Animal Technology Co. Nlrc4−/− mice on the 129s background were provided by V. Dixit (Genentech) and backcrossed for >10 generations to the C57BL/6 background in our facility. All animal experiments were conducted according to the Ministry of Health national guidelines for housing and care of laboratory animals and performed in accordance with institutional regulations after review and approval by the Institutional Animal Care and Use Committee at National Institute of Biological Sciences, Beijing.
Naip5−/− mice were generated through conventional ES cell–based targeting. The targeting vector was constructed by using recombineering technology. In brief, a Naip5 genomic fragment containing both homologous arms and the targeted region was retrieved from a bacterial artificial chromosome plasmid (BAC RP23-434I9; obtained from the BACPAC Resources Center, Children’s Hospital Oakland, Oakland, CA) and cloned into a targeting vector by homologous recombination. A neomycin-coding cassette was then inserted into the targeting vector, also through homologous recombination, to replace the largest exon 9 (illustrated in Fig. S1 A). The targeting vector was linearized and electroporated into the C57BL/6-derived Bruce4 ES cell. Neomycin-resistant clones were picked and validated by PCR and DNA sequencing. Targeted ES cells were injected into recipient blastocysts, and germline transmission was screened by PCR and DNA sequencing. Naip1−/− and Naip2−/− mice were generated by TALEN-mediated genome editing in fertilized eggs (C57BL/6 background). The TALEN constructs were assembled according to a previously published protocol (Wang et al., 2012; the Naip1 TALEN construct was provided by J. Xi from Peking University, Beijing, China). The targeting sequences were 5′-TAAGAAATCTTCAGAGG-3′ (left) and 5′-TCCCTCATAGCCTTGGAT-3′ (right) for Naip1 and 5′-TAGAGACCAGGACCACAGT-3′ (left) and 5′-TAGGTCCCACTGGATG-3′ (right) for Naip2. Detailed protocols for generating the TALEN KO mice were the same as previously described for the Mefv−/− mice (Xu et al., 2014). All biallelic KO mice were obtained by intercrossing the offspring of the founders. The genotyping primers used were 5′-CAACACCCTCCCCAACAAAC-3′ (Naip1-F), 5′-CCCATGCTGTCTGCAGATGTT-3′ (Naip1-R), 5′-CTAGGAGGTTTGAATGCATTTCAA-3′ (Naip2-F), 5′-GGAGAAGACCTCAGGGATCGTC-3′ (Naip2-R), 5′-ACCCACAATCTCAGAACCAGAAAACC-3′ (Naip5-F), and 5′-TGTTGGTAGAGAGTCATTGTTGGCCT-3′ (Naip5-R).
Bacterial strains and infections
Primary BMDM cells used for bacterial infection were prepared from WT and the indicated KO mice (C57BL/6 background) according to a previously described protocol (Zhao et al., 2011). All bacterial strains used in this study have also been described in our previous publications (Zhao et al., 2011; Yang et al., 2013). S. Typhimurium and S. flexneri strains were cultured in the 2xYT medium, and L. pneumophila were cultured on buffered charcoal yeast extract agar supplemented with 0.1 mg/ml−1 thymidine (BCYET). For infection, L. pneumophila were scraped from the agar plate and diluted in sterile water. Overnight cultures of S. Typhimurium ATCC 14028s (WT or indicated mutant strains; provided by E. Miao, University of North Carolina at Chapel Hill, Chapel Hill, NC), S. flexneri 2457T, or other indicated bacteria were diluted by 1:100 and grown for another 3 h to induce T3SS expression. The infection was performed at the indicated MOI, facilitated by centrifugation of 1,000 g for 10 min at 30°C and stopped at indicated time points. Cells were washed and cultured in media containing 100 µg/ml−1 gentamicin to kill the extracellular bacteria. The CFU numbers measuring the amount of intracellular bacteria were determined by dilution plating. For systemic infection in mice, overnight culture of S. Typhimurium SL1344 (provided by B. Finlay, University of British Columbia, Vancouver, BC, Canada) was diluted in PBS, and a total of 1,000 bacteria were used for intraperitoneal infection. Survival of the mice was monitored every 12 h after the infection.
CI assay of bacterial replication in mice
The CI assay of S. Typhimurium replication was performed according to a previous publication (Miao et al., 2010). All mice were on the C57BL/6 background WT mice were from Vital River Laboratory Animal Technology Co. In brief, an ampicillin-resistant vector (pWSK29) expressing PrgJ or flagellin under the promoter of the SPI-II effector SseJ was transformed into S. Typhimurium. WT S. Typhimurium transformed with an empty vector harboring a kanamycin resistance gene was used as the internal reference. An equal number of experimental bacteria and reference bacteria (500 bacteria for each strain) were intraperitoneally injected into the mice. The spleens and livers of mice from the control group and experimental group, sacrificed at the same time point, were homogenized, and the CFUs were determined. CI results were presented as log (experimental/reference CFUs).
LFn fusion protein purification and injection into mice
LFn-tagged recombinant flagellin (type b from P. aeruginosa), T3SS rod proteins (BsaK from B. thailandensis and PrgJ from S. Typhimurium), T3SS needle proteins (EprI from EHEC, PrgI from S. Typhimurium, BsaL from B. thailandensis, and MxiH from S. flexneri), and protective antigen proteins were expressed and purified as described in our previous publications (Zhao et al., 2011; Yang et al., 2013). For injection into mice, proteins were diluted in 200 µl of PBS; the dosages used were 2 µg for the rod protein and 5 µg for flagellin and the needle protein per gram of mice. Cytosolic delivery was achieved by co-injecting equal amounts of the protective antigen. Injected mice were observed every few hours, and the survival rate was calculated. All mice were on the C57BL/6 background, and the WT mice were purchased from Vital River Laboratory Animal Technology Co.
Statistical analyses
Mouse survival data after bacterial infection or protein injection were analyzed in Prism 5 (GraphPad Software), in which the log-rank (Mantel-Cox) test was used for statistical analyses of the survival curve (p-values ≤0.05 were considered significant). The two-tailed unpaired Student’s t test was used for statistical analyses of the CI data, and p-values <0.05 were considered significant.
Online supplemental material
Fig. S1 shows the generation of Naip1, Naip2, and Naip5 KO mice. The strategy for removing the exon 9 of mouse Naip5 by homologous recombination is shown in A, and genotyping results of the Naip5−/− mice are in B. C and D show the sequence mutations of Naip1−/− and Naip2−/− mice generated by TALEN-mediated genome editing. Online supplemental materials are available at http://www.jem.org/cgi/content/full/jem.20160006/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Dixit for Nlrc4−/− mice, B. Finlay for S. Typhimurium SL1344 strain, J. Xi for the Naip1 TALEN construct, and E. Miao for plasmids and technical assistance. We thank members of the Shao laboratory for discussions and technical assistance.
This work was supported by the National Natural Science Foundation of China Program for Distinguished Young Scholars (31225002) and the Program for International Collaborations (31461143006), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08020202), and the National Basic Research Program of China 973 Program (2012CB518700 and 2014CB849602) to F. Shao. The research was also supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and by the Beijing Scholar Program to F. Shao.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CI
- competitive index
- ES
- embryonic stem
- MOI
- multiplicity of infection
- PRR
- pattern recognition receptor
- SPI
- Salmonella pathogenicity island
- T3SS
- type III secretion system
- TALEN
- transcription activator–like effector nuclease
References
- Aachoui Y., Kajiwara Y., Leaf I.A., Mao D., Ting J.P., Coers J., Aderem A., Buxbaum J.D., and Miao E.A.. 2015. Canonical inflammasomes drive IFN-γ to prime caspase-11 in defense against a cytosol-invasive bacterium. Cell Host Microbe. 18:320–332. 10.1016/j.chom.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S., Trinidad N.J., and Vance R.E.. 2012. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 18:799–806. 10.1038/nm.2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Sekirov I., and Finlay B.B.. 2007. Type III secretion systems and disease. Clin. Microbiol. Rev. 20:535–549. 10.1128/CMR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Franchi L., Kamada N., Nakamura Y., Burberry A., Kuffa P., Suzuki S., Shaw M.H., Kim Y.G., and Núñez G.. 2012. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 13:449–456. 10.1038/ni.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar J.A., and Miao E.A.. 2014. Detection of cytosolic bacteria by inflammatory caspases. Curr. Opin. Microbiol. 17:61–66. 10.1016/j.mib.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenstein A.V., Zhang L., and Wu H.. 2015. The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr. Opin. Struct. Biol. 31:75–83. 10.1016/j.sbi.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.W., Sui S.F., and Chai J.. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 350:399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfield K.L., Persson J., Brubaker S.W., Witte C.E., von Moltke J., Dunipace E.A., Henry T., Sun Y.H., Cado D., Dietrich W.F., et al. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171–1178. 10.1038/ni.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltez V.I., Tubbs A.L., Cook K.D., Aachoui Y., Falcone E.L., Holland S.M., Whitmire J.K., and Miao E.A.. 2015. Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity. 43:987–997. 10.1016/j.immuni.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Leaf I.A., Treuting P.M., Mao D.P., Dors M., Sarkar A., Warren S.E., Wewers M.D., and Aderem A.. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11:1136–1142. 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlander S., Pott J., and Maloy K.J.. 2014. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunol. 7:775–785. 10.1038/mi.2013.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D.E., Chavarria-Smith J., Vance R.E., and Miao E.A.. 2013. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., and Shao F.. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 514:187–192. 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F.. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526:660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Suzuki S., Franchi L., He Y., Muñoz-Planillo R., Mimuro H., Suzuki T., Sasakawa C., and Núñez G.. 2014. Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcδ. PLoS Pathog. 10:e1003926 10.1371/journal.ppat.1003926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenthorey J.L., Kofoed E.M., Daugherty M.D., Malik H.S., and Vance R.E.. 2014. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell. 54:17–29. 10.1016/j.molcel.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N.J., Moayeri M., Kintzer A.F., Wang S.B., van Rooijen N., Brown C.R., Krantz B.A., Leppla S.H., Gronert K., and Vance R.E.. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 490:107–111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li J., Huang H., Wang G., Jiang M., Yin S., Sun C., Zhang H., Zhuang F., and Xi J.J.. 2012. An integrated chip for the high-throughput synthesis of transcription activator-like effectors. Angew. Chem. Int. Ed. Engl. 51:8505–8508. 10.1002/anie.201203597 [DOI] [PubMed] [Google Scholar]
- Wright E.K., Goodart S.A., Growney J.D., Hadinoto V., Endrizzi M.G., Long E.M., Sadigh K., Abney A.L., Bernstein-Hanley I., and Dietrich W.F.. 2003. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr. Biol. 13:27–36. 10.1016/S0960-9822(02)01359-3 [DOI] [PubMed] [Google Scholar]
- Xu H., Yang J., Gao W., Li L., Li P., Zhang L., Gong Y.N., Peng X., Xi J.J., Chen S., et al. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 513:237–241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 350:404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., and Shao F.. 2015. The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol. Rev. 265:85–102. 10.1111/imr.12293 [DOI] [PubMed] [Google Scholar]
- Zhao Y., and Shao F.. 2016. Diverse mechanisms for inflammasome sensing of cytosolic bacteria and bacterial virulence. Curr. Opin. Microbiol. 29:37–42. 10.1016/j.mib.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.