Abstract
The endoplasmic reticulum (ER) is a massive cytoplasmic membrane network that functions primarily to ensure proper folding and post-translational modification of newly synthesized secretory and transmembrane proteins. Abnormal accumulation of unfolded proteins in this organelle causes a state of “ER stress”, which is a hallmark feature of various diseases including cancer, neurodegeneration and metabolic dysfunction. Cancer cells exploit the IRE1α-XBP1 arm of the ER stress response to efficiently adjust their protein-folding capacity and ensure survival under hostile tumor microenvironmental conditions. However, we recently found that dendritic cells (DCs) residing in the ovarian cancer microenvironment also experience sustained ER stress and demonstrate persistent activation of the IRE1α-XBP1 pathway. This previously unrecognized process disrupts metabolic homeostasis and antigen-presenting capacity in DCs, thereby crippling their natural ability to support the protective function of infiltrating anti-tumor T cells. In this review, we briefly discuss some of the mechanisms that fuel ER stress in tumor-associated DCs, the biological processes altered by aberrant IRE1α-XBP1 signaling in these innate immune cells, and the unique immunotherapeutic potential of targeting this pathway in cancer hosts.
BACKGROUND
Triggering IRE1α-XBP1 activation through the ER stress response
The endoplasmic reticulum (ER) is the primary organelle responsible for regulating intracellular calcium, lipid biosynthesis, and the proper glycosylation and folding of nascent transmembrane and secreted proteins. Numerous physiological stimuli often found within tumor microenvironments such as nutrient deprivation, calcium store depletion, oxidative stress, hypoxia, and inflammation can disrupt the protein folding capacity of the ER. When this intrinsic protein folding capacity is overwhelmed, the cell is considered to be in a state of “ER stress” and will initiate an unfolded protein response (UPR) via the ER transmembrane proteins IRE1α (encoded by Ern1), PERK (encoded by Eif2ak3), and ATF6 (encoded by Atf6) in an attempt to restore homeostasis (1). If the combined action of these three proteins is insufficient to ameliorate ER toxicity, the affected cell will undergo apoptosis.
The serine/threonine-protein kinase/endoribonuclease IRE1α represents the most ancient branch of this signaling pathway, and is highly conserved from yeast to humans. At steady state, the chaperone protein BiP holds IRE1α in its monomeric form, thereby precluding activation. However, upon the induction of ER stress, the accumulating misfolded proteins titrate BiP away from IRE1α, triggering IRE1α dimerization, autophosphorylation, and a conformational shift that licenses its C-terminal endoribonuclease domain to cytoplasmically cleave 26 nucleotides from the Xbp1 mRNA. This spliced transcript is subsequently re-ligated by the tRNA ligase RtcB (2), resulting in a critical reading frame shift that enables translation of the functionally active X-box binding protein 1 (XBP1). This multi-tasking transcription factor alleviates ER stress by upregulating a variety of chaperones, redox-dependent foldases, and glycosyltransferases. Beyond these canonical functions, several groups have demonstrated that XBP1 also modulates ER stress-independent, context-specific signaling events such as the hypoxia response (by dimerizing with HIF1α) (3), lipid metabolism (4), estrogen receptor activity (5) and the transcription of pro-inflammatory cytokines (6).
Biological functions for IRE1α-XBP1 signaling
Multiple groups have identified key roles for IRE1α-XBP1 signaling in a number of organs and cell types through the use of conditional mouse models. Germline Xbp1 deletion is embryonic lethal due to fetal liver failure (7). If this is rescued with a liver-specific Xbp1 transgene, the mice die shortly after birth due to insufficient exocrine pancreas function (8). However, selective deletion of Xbp1 or Ern1 in the liver of adult mice results in marked reduction in serum triglyceride and cholesterol levels (4, 9). Selective deletion of Xbp1 in pancreatic β cells results in mild hyperglycemia and glucose intolerance (10). In the hematopoietic system, XBP1 is a key, cell-intrinsic requirement for plasma cell (11) and eosinophil differentiation (12), and mice with dendritic cell-specific Xbp1 deletion exhibit reductions in splenic CD8α dendritic cells (13). Furthermore, XBP1 optimizes TLR-driven pro-inflammatory cytokine production in macrophages (6). Conditional deletion of Xbp1 in the intestinal epithelium triggers Paneth cell death and colitic lesions resembling inflammatory bowel disease (14). However, this pathology is significantly attenuated in conditional Ern1 knockout animals, suggesting that IRE1α hyperactivation leading to RIDD, which can occur after selective deletion of Xbp1, may be involved in exacerbating this inflammatory phenotype (15). Conditional deletion of Xbp1 in the brain is neuroprotective in mouse models of Huntington’s disease (16) and ALS (17), while XBP1-mediated control of hexosamine biosynthesis in cardiomyocytes is cardioprotective in models of ischemia-reperfusion (18). Finally, animals lacking Ern1 in all tissues except the placenta were viable and generally healthy, but displayed modest hyperglycemia and a reduction in serum antibody levels as predicted (19). The IRE1α-XBP1 signaling pathway therefore has a number of important physiological functions spanning multiple organ systems.
Cancer cell-intrinsic roles of IRE1α-XBP1 signaling
Malignant cells manage to survive under hostile conditions such as hypoxia and nutrient starvation via sustained activation of the IRE1α-XBP1 branch of the ER stress response (3, 20). Indeed, XBP1 expression is increased in breast cancer cells resistant to anti-estrogen therapy (21) and high levels of Xbp1s transcripts are significantly associated with poor outcomes in endocrine-treated breast tumors (22). In addition, it was recently demonstrated in vivo that XBP1 drives triple negative breast cancer (TNBC) progression by cooperating with HIF1α to support tumor-initiating cell function and metastatic capacity of cancer cells under harsh environmental conditions (3). Therapeutic silencing of XBP1 in TNBC cells led to suppression of tumor initiation, progression, metastasis and recurrence, and high expression of XBP1-dependent gene signatures was found to be associated with worse prognosis in TNBC patients (3). XBP1 has also been demonstrated to drive the pathogenesis of multiple myeloma (23), and has been implicated in cancer cell de-differentiation, susceptibility to oncovirus infection and the epithelial-to-mesenchymal transition (24). Seminal work by the group of C.C. Andrew Hu also demonstrated constitutive IRE1α-XBP1 activation in murine chronic lymphocytic leukemia (CLL) cells, which promoted their pathogenesis in vivo. Accordingly, targeting IRE1α signaling in vivo with the selective small molecule endoribonuclease inhibitor B-I09 showed significant therapeutic effects, especially when used in combination with targeted anti-leukemic agents such as ibrutinib (25). In a xenograft model of human glioma, inhibiting IRE1α function by overexpressing a dominant negative variant significantly increased overall survival by decreasing tumor growth rate and angiogenesis (26). Furthermore, recent in vivo studies have also indicated that IRE1α-XBP1 signaling supports the aggressiveness of pancreatic cancer cells, and abrogating IRE1α activity using a small molecule inhibitor induced apoptosis and consequently delayed pancreatic tumor growth in xenograft models (27). Increasing evidence hence demonstrates that sustained IRE1α-XBP1 activation operates directly in cancer cells to promote tumor growth and metastasis in vivo in a variety of aggressive cancer types, many of which currently lack targeted therapies.
Immune cell dysfunction driven by abnormal IRE1α-XBP1 signaling
While IRE1α-XBP1 signaling has been shown to positively influence the growth and survival of malignant cells, the role of this cellular pathway in shaping the cancer immunoenvironment and the anti-tumor immune response had not been explored. Aggressive cancers recruit a broad collection of immune cells and effectively manipulate their intrinsic protective activity as a fundamental pro-tumoral mechanism. This process is epitomized by ovarian carcinoma, a highly immunosuppressive and lethal cancer that exquisitely controls normal dendritic cell (DC) functions in order to abrogate the generation of protective T cell-based responses (28). We hypothesized that common adverse conditions in the ovarian cancer microenvironment that induce protein misfolding (e.g. hypoxia, nutrient deprivation and/or oxidative stress) could trigger ER stress and robust activation of the IRE1α-XBP1 pathway in tumor-associated DCs (tDCs), a process that might influence their normal activity. Unlike DCs in non-tumor sites, DCs residing in human and mouse ovarian cancers exhibited robust and sustained IRE1α-XBP1 activation and concomitant overexpression of XBP1-dependent genes involved in the ER stress response (13). Mechanistically, high levels of reactive oxygen species (ROS) in tDCs promoted intracellular lipid peroxidation and subsequent generation of reactive byproducts such as 4-hydroxynonenal (4-HNE), which induced ER stress by directly modifying critical ER-resident proteins and chaperones (13) (Figure 1). Treatment with antioxidants or pharmacological agents that efficiently sequester lipid peroxidation byproducts therefore prevented the induction of ER stress and IRE1α-XBP1 activation in DCs exposed to tumor-derived factors like those commonly present in malignant ovarian cancer ascites (13). We are currently defining the molecular mechanisms by which the tumor microenvironment fuels ROS accumulation and lipid peroxidation in tDC. Interestingly, lipid peroxidation byproducts have also been shown to promote vascular inflammation and atherogenesis by triggering ER stress in endothelial cells (29). Most importantly, ovarian cancer-bearing mice selectively lacking XBP1 in DCs demonstrated delayed progression of primary and metastatic ovarian tumors in three distinct preclinical models of disease (13). These effects correlated with enhanced intra-tumoral infiltration of activated, antigen-experienced T cells producing IFN-γ in situ (13), suggesting that tDC devoid of XBP1 were immunocompetent, rather than immunosuppressive. Global transcriptional profiling of tDCs revealed that constitutively active XBP1 not only promoted the expression of canonical XBP1-target genes involved in the ER stress response, but also induced a robust triglyceride biosynthetic program leading to abnormal lipid accumulation (Figure 1) (13). Interestingly, XBP1 had previously been demonstrated to drive hepatic lipogenesis by inducing the expression of key lipid biosynthetic genes (4). Seminal studies by the group of D. Gabrilovich had also uncovered that a major mechanism contributing to DC malfunction in cancer is indeed abnormal intracellular lipid accumulation. This dyslipidemia was shown to inhibit the efficient loading of antigenic peptides onto MHC-I molecules, thereby impairing optimal antigen cross-presentation to T cells by DCs (30). Consistent with this concept, XBP1-deficient tDC unable to accumulate intracellular lipid droplets demonstrated enhanced capacity to support T cell function both in vitro and in vivo, and memory (tumor-reactive) T cells generated in ovarian cancer-bearing mice selectively lacking XBP1 in DC demonstrated enhanced anti-tumor capacity when adoptively transferred into wild-type ovarian cancer hosts (13). We are currently exploring additional (lipid metabolism-independent) mechanisms by which sustained IRE1α-XBP1 activation promotes DC dysfunction in the tumor microenvironment.
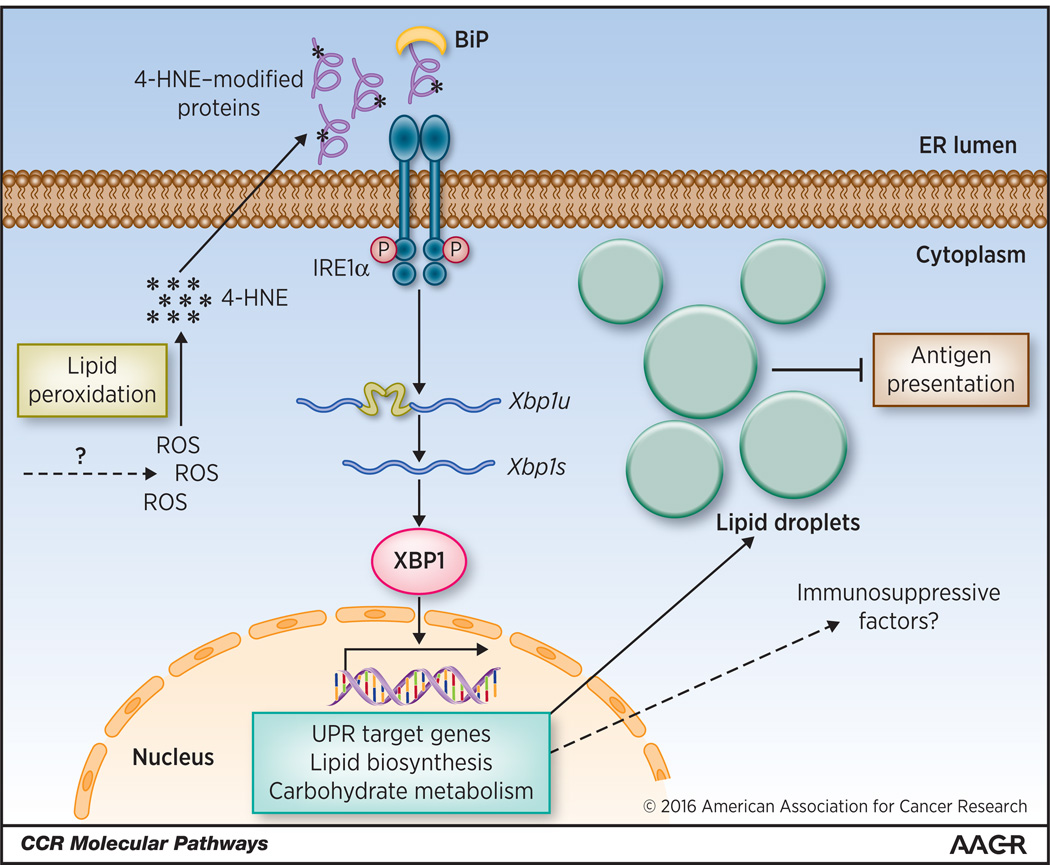
Figure 1. Activation and function of IRE1α-XBP1 in ovarian cancer-resident DCs.
High levels of ROS in tDCs promote intracellular lipid peroxidation and generation of reactive byproducts like 4-HNE. This diffusible aldehyde modifies ER resident proteins and triggers the ER stress response, thus inducing activation of IRE1α and subsequent cleavage of the Xbp1u mRNA to generate the spliced Xbp1s form. Constitutively active XBP1 promotes expression of genes involved in the UPR, carbohydrate metabolism and lipid biosynthesis. Aberrant lipid accumulation inhibits the capacity of tDCs to present local tumor antigens to infiltrating T cells.
Depleting or “licensing” tumor-associated myeloid cells in vivo has been widely used to restrain the optimal progression of several cancer types, but the precise microenvironmental conditions and molecular pathways that tumors exploit in these immune cells to co-opt their otherwise protective activity remain poorly understood. Our study provided the first evidence of a lethal cancer capable of co-opting IRE1α-XBP1 function in DCs of the tumor microenvironment as a strategy to evade immune control. This process may also orchestrate tolerance and immunosuppression in other lethal solid tumors that commonly rely on infiltrating innate immune cells to promote malignant progression. Future studies therefore aim at defining whether other cell types in the ovarian cancer immunoenvironment exhibit detrimental ER stress responses, and whether additional tumor types also rely on IRE1α-XBP1 signaling as a major immunosuppressive mechanism.
CLINICAL-TRANSLATIONAL ADVANCES
Small molecule inhibitors
Given that IRE1α-XBP1 signaling sustains both cancer cell-extrinsic immunosuppression and cancer cell-intrinsic growth and metastasis, there is significant interest in developing targeted therapies against this UPR pathway. While technical limitations preclude the development of direct small molecule XBP1 inhibitors, the formation of the active, spliced Xbp1 variant can be readily targeted via its dependency on IRE1α. The dual enzyme IRE1α is amenable to small molecule targeting, and multiple inhibitor classes have been identified from various independent small molecule screens. Several crystal structures of IRE1α in complex with either kinase inhibitors or hydroxyl-aryl-aldehyde endoribonuclease inhibitors have been published (31, 32), enabling rational development of novel IRE1α inhibitors.
Small molecule IRE1α inhibitors can be grouped into three main categories based on their structures and mode of action. The first group consists of inhibitors with indirect or unknown mechanisms of action, and include irestatin, trierixin (33) and quinotrierixin (34). These compounds were each identified by screening small molecule libraries against human cell lines expressing IRE1α endoribonuclease-driven luciferase reporter plasmids in the presence of chemical ER stressors such as thapsigargin or tunicamycin. In these reporter systems, the firefly luciferase cDNA is fused out of frame to a fragment of human XBP1 cDNA bearing IRE1α splicing recognition sites, and is only translated in-frame upon IRE1α-mediated RNA splicing. The IRE1α inhibitory capacity for each inhibitor was subsequently confirmed with luciferase-independent, PCR-based methods in human cell lines. However, the mechanisms underlying inhibitor activity remain poorly defined, and it is unclear whether these compounds specifically target IRE1α or whether they interfere with UPR activation more generally, as has been suggested for quinotrierixin (34).
The second and largest group of inhibitors is comprised of direct IRE1α endoribonuclease inhibitors. Some of these compounds were identified in high-throughput screens against the endoribonuclease activity of the purified IRE1α cytoplasmic domain, while others were developed during optimization efforts on pre-existing leads. Most of these compounds, including 3-ethoxy-5,6-dibromosalicylaldehyde (35), 4μ8C (36), MKC-3946 (37), and B-I09 (25), are salicylaldehyde and coumarin derivatives which generally share a core hydroxyl-aryl-aldehyde (HAA) structure. Crystallographic analyses have demonstrated that these HAA inhibitors bind covalently to lysine K907, which resides in a shallow, solvent-exposed pocket on the IRE1α endoribonuclease domain (31). However, this HAA motif is not an absolute structural requirement, as both STF-083010 (38) the nucleoside-type antibiotic analogue toyocamycin can also directly block the IRE1α endoribonuclease (39). All direct IRE1α endonuclease inhibitors dose-dependently reduced Xbp1 splicing in vitro in human cell lines without affecting IRE1 phosphorylation or signaling from the PERK and ATF6. Importantly, STF-083010 (38), MKC-3946 (37) and toyocamycin (39) demonstrated efficacy against multiple myeloma both in vitro and in xenograft survival studies, and B-I09 reduced tumor burden in a genetic mouse model of CLL driven by the Eμ-TCL1 transgene (25). Furthermore, daily intraperitoneal administration of 4μ8C significantly reduced pathological joint swelling in the KBxN serum transfer murine model of rheumatoid arthritis (40). Cumulatively, these reports validate IRE1α as an attractive clinical target and indicate that the endoribonuclease domain is chemically tractable.
The final group of small molecule IRE1α inhibitors is kinase inhibitors, which act allosterically to disrupt endoribonuclease function. Compared with the extensive and rapidly expanding collection of endoribonuclease inhibitors, IRE1α kinase inhibitors are considerably less well developed despite their significant therapeutic potential. This disparity may be due in part to nuances in how the IRE1α endoribonuclease domain responds to different classes of kinase inhibitors. When the IRE1α kinase DFG loop shifts into a “DFG-in” conformation, a structure stabilized by certain type I kinase inhibitors like sunitinib and a novel compound known as “Compound 3”, the endoribonuclease domain cleaves the Xbp1 mRNA in the absence of IRE1α autophosphorylation (41). However, upon adopting a “DFG-out” conformation, which can be enforced with certain type II kinase inhibitors such as KIRA6 and AD60 (42), both the kinase domain and the endoribonuclease domain are rendered inert (43). Interestingly, in male Ins2+/Akita mice, which express a mutated pro-insulin that causes chronic ER stress in pancreatic β-cells, twice daily administration of KIRA6 reduced plasma glucose levels and improved glucose tolerance test outcomes (43). Furthermore, intravitreal KIRA6 injection in the P23H transgenic rat model of retinitis pigmentosa preserved photoreceptor viability and function (43). These in vivo data are consistent with accumulating evidence suggesting that protein misfolding and ER stress may be linked to both metabolic dysfunction and retinal degeneration. Though kinase domains are highly structurally conserved, extremely selective IRE1α kinase inhibitors can be generated, as illustrated by the recently reported “Compound 18” and GSK2850163 (32, 44). However, the in vivo effects of these selective compounds were not reported. Type II kinase inhibition, but not type I kinase inhibition, therefore represents a second pharmacologically tractable strategy for globally blocking IRE1α endoribonuclease activity in the tumor microenvironment.
Immune cell-specific approaches
Due to unique properties of the immune system, other small molecule-independent strategies could also be utilized to disable IRE1α-XBP1 signaling selectively in DCs of cancer hosts. First, DCs within malignant ovarian cancer ascites have exceptional phagocytic capacity, rendering them excellent targets for nanoparticle-mediated RNAi therapeutics (45). As ovarian cancer metastasis is generally confined within the peritoneal cavity, intraperitoneal administration of siRNA-loaded nanoparticles targeting Ern1 or Xbp1 represents a novel and feasible immuno-oncology strategy. In animal models of established metastatic ovarian cancer, silencing Xbp1 expression using this approach rendered tDCs highly immunostimulatory and significantly extended host survival by stimulating T cell-mediated anti-tumor immunity (13).
As a second strategy, the genes encoding IRE1α or XBP1 could be ablated to enhance the efficacy of autologous DC adoptive transfer strategies. Despite the modest successes of adoptive DC therapy, ovarian cancer patients were refractory to similar tumor antigen-pulsed adoptive DC treatments (46). Genome editing technologies such as CRISPR/CAS9, zinc finger nucleases, or TALENs (47) should enable precise and efficient mutation of XBP1 or ERN1 in DCs prior to adoptive transfer, thereby protecting these transplanted DCs from the suppressive effects of aberrant ER stress responses induced by the tumor microenvironment. In proof-of-concept experiments, we demonstrated that transferring Xbp1-deficient BMDCs into mice with established primary ovarian cancer significantly delayed tumor progression compared with infusion of wild type BMDCs (13). Strikingly, transplanted Xbp1-deficient DCs were dominantly immunostimulatory over the endogenous (wild type) regulatory DCs residing in the tumor microenvironment. Hence, cutting-edge genetic methods for targeting IRE1α-XBP1 signaling would likely enhance the efficacy of current adoptive DC therapies in ovarian cancer.
To conclude, the IRE1α-XBP1 branch of the ER stress response is a novel and well-characterized pathway with significant therapeutic relevance in a variety of human cancers. This molecular pathway controls unique biological processes in the cancer cell and in tumor-infiltrating immune cells to ultimately promote tumor progression. While IRE1α-XBP1 signaling can be targeted through a variety of classical and non-classical methods (Figure 2), potent small molecule inhibitors represent an attractive strategy to simultaneously disable this pro-tumoral pathway in the cancer cell and in the innate immune system.
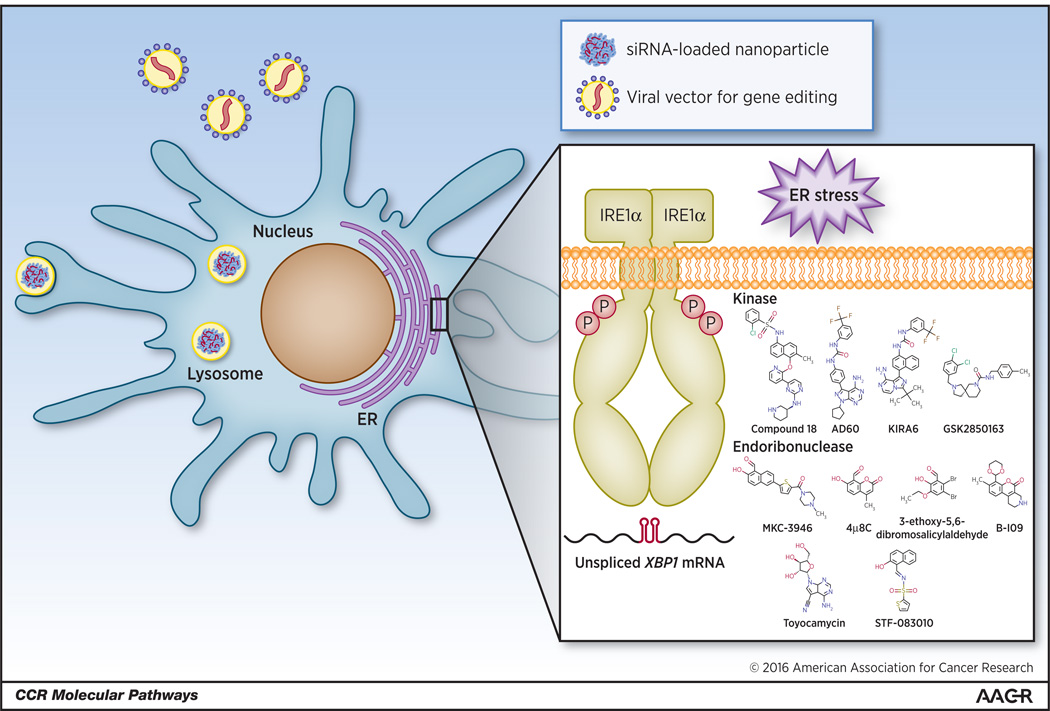
Figure 2. Potential therapeutic strategies for targeting IRE1α-XBP1 signaling in cancer-associated DCs.
The IRE1α kinase and endoribonuclease domains can be individually blocked with small molecules. Additionally, the constitutive activation of either XBP1 or IRE1α can be reduced using nanoparticles encapsulating siRNAs. In terms of DC-based vaccines or autologous DC transfer, IRE1α or XBP1 could be selectively ablated ex vivo through the use of virally-delivered DNA editing technologies such as zinc finger nucleases, TALENs, and CRISPR/CAS9. Genome edited DCs could then be transplanted back into patients to enhance standard therapeutic regimes.
Acknowledgments
GRANT SUPPORT
J.R. Cubillos-Ruiz was supported by The Irvington Institute Fellowship Program of the Cancer Research Institute. L.H. Glimcher was supported by the NIH under award numbers R01DK82448 and CA112663.
L.H. Glimcher is on the board of directors for and has ownership interest in Bristol-Myers Squibb.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed by the other authors.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES
- 1.Bettigole SE, Glimcher LH. Endoplasmic reticulum stress in immunity. Annual review of immunology. 2015;33:107–138. doi: 10.1146/annurev-immunol-032414-112116. [DOI] [PubMed] [Google Scholar]
- 2.Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Molecular cell. 2014;55:758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding L, Yan J, Zhu J, Zhong H, Lu Q, Wang Z, et al. Ligand-independent activation of estrogen receptor alpha by XBP-1. Nucleic acids research. 2003;31:5266–5274. doi: 10.1093/nar/gkg731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, et al. An essential role in liver development for transcription factor XBP-1. Genes & development. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. The EMBO journal. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.So JS, Hur KY, Tarrio M, Ruda V, Frank-Kamenetsky M, Fitzgerald K, et al. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8885–8890. doi: 10.1073/pnas.1105564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 12.Bettigole SE, Lis R, Adoro S, Lee AH, Spencer LA, Weller PF, et al. The transcription factor XBP1 is selectively required for eosinophil differentiation. Nature immunology. 2015;16:829–837. doi: 10.1038/ni.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, et al. ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell. 2015;161:1527–1538. doi: 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C, et al. Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Human molecular genetics. 2012;21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes & development. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR, et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156:1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwawaki T, Akai R, Kohno K. IRE1alpha disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PloS one. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nature reviews Drug discovery. 2013;12:703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 21.Gomez BP, Riggins RB, Shajahan AN, Klimach U, Wang A, Crawford AC, et al. Human X-box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. FASEB J. 2007;21:4013–4027. doi: 10.1096/fj.06-7990com. [DOI] [PubMed] [Google Scholar]
- 22.Davies MP, Barraclough DL, Stewart C, Joyce KA, Eccles RM, Barraclough R, et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. International journal of cancer Journal international du cancer. 2008;123:85–88. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]
- 23.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shajahan AN, Riggins RB, Clarke R. The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 2009;22:241–246. doi: 10.1358/dnp.2009.22.5.1378631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang CH, Ranatunga S, Kriss CL, Cubitt CL, Tao J, Pinilla-Ibarz JA, et al. Inhibition of ER stress-associated IRE-1/XBP-1 pathway reduces leukemic cell survival. J Clin Invest. 2014;124:2585–2598. doi: 10.1172/JCI73448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auf G, Jabouille A, Guerit S, Pineau R, Delugin M, Bouchecareilh M, et al. Inositol-requiring enzyme 1alpha is a key regulator of angiogenesis and invasion in malignant glioma. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15553–15558. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chien W, Ding LW, Sun QY, Torres-Fernandez LA, Tan SZ, Xiao J, et al. Selective inhibition of unfolded protein response induces apoptosis in pancreatic cancer cells. Oncotarget. 2014;5:4881–4894. doi: 10.18632/oncotarget.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scarlett UK, Rutkowski MR, Rauwerdink AM, Fields J, Escovar-Fadul X, Baird J, et al. Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J Exp Med. 2012 doi: 10.1084/jem.20111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vladykovskaya E, Sithu SD, Haberzettl P, Wickramasinghe NS, Merchant ML, Hill BG, et al. Lipid peroxidation product 4-hydroxy-trans-2-nonenal causes endothelial activation by inducing endoplasmic reticulum stress. J Biol Chem. 2012;287:11398–11409. doi: 10.1074/jbc.M111.320416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herber DL, Cao W, Nefedova Y, Novitskiy SV, Nagaraj S, Tyurin VA, et al. Lipid accumulation and dendritic cell dysfunction in cancer. Nat Med. 2010;16:880–886. doi: 10.1038/nm.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanches M, Duffy NM, Talukdar M, Thevakumaran N, Chiovitti D, Canny MD, et al. Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors. Nature communications. 2014;5:4202. doi: 10.1038/ncomms5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrington PE, Biswas K, Malwitz D, Tasker AS, Mohr C, Andrews KL, et al. Unfolded Protein Response in Cancer: IRE1alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability. ACS medicinal chemistry letters. 2015;6:68–72. doi: 10.1021/ml500315b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tashiro E, Hironiwa N, Kitagawa M, Futamura Y, Suzuki S, Nishio M, et al. Trierixin, a novel Inhibitor of ER stress-induced XBP1 activation from Streptomyces sp. 1. Taxonomy, fermentation, isolation and biological activities. The Journal of antibiotics. 2007;60:547–553. doi: 10.1038/ja.2007.69. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto K, Tashiro E, Imoto M. Quinotrierixin inhibited ER stress-induced XBP1 mRNA splicing through inhibition of protein synthesis. Bioscience, biotechnology, and biochemistry. 2011;75:284–288. doi: 10.1271/bbb.100622. [DOI] [PubMed] [Google Scholar]
- 35.Volkmann K, Lucas JL, Vuga D, Wang X, Brumm D, Stiles C, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286:12743–12755. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A. 2012;109:E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bricker NK, Raskin RE, Densmore CL. Cytochemical and immunocytochemical characterization of blood cells and immunohistochemical analysis of spleen cells from 2 species of frog, Rana (Aquarana) catesbeiana and Xenopus laevis. Veterinary clinical pathology / American Society for Veterinary Clinical Pathology. 2012;41:353–361. doi: 10.1111/j.1939-165X.2012.00452.x. [DOI] [PubMed] [Google Scholar]
- 40.Qiu Q, Zheng Z, Chang L, Zhao YS, Tan C, Dandekar A, et al. Toll-like receptor-mediated IRE1alpha activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32:2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi A, Newbatt Y, McAndrew PC, Stubbs M, Burke R, Richards MW, et al. Molecular mechanisms of human IRE1 activation through dimerization and ligand binding. Oncotarget. 2015;6:13019–13035. doi: 10.18632/oncotarget.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendez AS, Alfaro J, Morales-Soto MA, Dar AC, McCullagh E, Gotthardt K, et al. Endoplasmic reticulum stress-independent activation of unfolded protein response kinases by a small molecule ATP-mimic. eLife. 2015;4 doi: 10.7554/eLife.05434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Concha NO, Smallwood A, Bonnette W, Totoritis R, Zhang G, Federowicz K, et al. Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1alpha Endoribonuclease Activity. Molecular pharmacology. 2015;88:1011–1023. doi: 10.1124/mol.115.100917. [DOI] [PubMed] [Google Scholar]
- 45.Cubillos-Ruiz JR, Fiering S, Conejo-Garcia JR. Nanomolecular targeting of dendritic cells for ovarian cancer therapy. Future Oncol. 2009;5:1189–1192. doi: 10.2217/fon.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kandalaft LE, Powell DJ, Jr, Chiang CL, Tanyi J, Kim S, Bosch M, et al. Autologous lysate-pulsed dendritic cell vaccination followed by adoptive transfer of vaccine-primed ex vivo co-stimulated T cells in recurrent ovarian cancer. Oncoimmunology. 2013;2:e22664. doi: 10.4161/onci.22664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]