Abstract
Tumors are dynamic organs that evolve during disease progression with genetic, epigenetic, and environmental differences among tumor cells serving as the foundation for selection and evolution in tumors. Tumor-initiating cells (TICs) that are responsible for tumorigenesis are a source of functional cellular heterogeneity while chromosomal instability (CIN) is a source of karyotypic genetic diversity. However, the extent that CIN contributes to TIC genetic diversity and its relationship to TIC function remains unclear. Here we demonstrate that glioblastoma TICs display chromosomal instability with lagging chromosomes at anaphase and extensive non-clonal chromosome copy number variations. Elevating the basal chromosome mis-segregation rate in TICs both decreases proliferation and the stem-like phenotype of TICs in vitro. Consequently tumor formation is abolished in an orthotopic mouse model. These results demonstrate that TICs generate genetic heterogeneity within tumors but that TIC function is impaired if the rate of genetic change is elevated above a tolerable threshold.
Keywords: chromosomal instability, CIN, glioblastoma, tumor-initiating cells, and intra-tumor heterogeneity
Introduction
Co-existing within a tumor are diverse populations of cells with extensive genetic and functional heterogeneity. The cancer stem cell hypothesis posits that in tumors there is a functional cellular hierarchy with tumor-initiating cells (TICs) (also referred to as cancer stem cells) at the apex. In this model, TICs are the population of cells responsible for tumor formation and for sustaining tumorigenesis. In accordance, TICs are functionally distinct from non-TICs and share several common characteristics with stem cells including the ability to generate non-TICs analogous to stem cell differentiation and the ability to self-renew leading to serial tumor formation (1). Importantly, TICs are also thought to contribute to therapeutic resistance and tumor relapse because TICs are resistant to ionizing radiation and chemotherapeutics (2,3).
The functional diversity arising between TICs and non-TICs can derive from genetic, epigenetic, and environmental differences among tumor cells. In particular, tumor cells are genetically heterogeneous with respect to chromosome complement. Over 90% of solid tumors are reported to be aneuploid (4). Whole chromosome aneuploidy is a state defined as an abnormal chromosome complement that deviates from a multiple of the haploid number. Moreover, many aneuploid tumor cells also exhibit chromosomal instability (CIN). CIN is a persistent rate of chromosome mis-segregation that leads to random chromosome losses and/or gains generating genetic diversity among cancer cells. The most common cause of CIN is lagging chromosomes at anaphase that result from the persistence of erroneous merotelic chromosome microtubule attachments (a chromatid attached to microtubules from both spindle poles) during mitosis (5). These errors in mitosis contribute to both whole chromosome aneuploidy and structural chromosome alterations due to DNA damage incurred on the lagging chromosome during cytokinesis (6).
Importantly, similar to TICs, CIN correlates with therapy resistance and poor patient prognosis (7,8). CIN generates continual karyotype heterogeneity in tumor cells providing substrates for selection and evolution in tumors and may confer resistance due to the selection of advantageous chromosome complements (8,9). Ultimately, both CIN, as a source of genetic intra-tumor heterogeneity, and TICs, as a source of functional intra-tumor heterogeneity, provide tumors with adaptability and pose significant challenges for effective treatment. Despite these commonalities, the relationship between CIN and TICs is unclear and unexplored.
Here we investigate the extent of karyotype heterogeneity and CIN within Glioblastoma (GBM) TICs and whether CIN influences TIC function. Glioblastoma is one of the most lethal cancers with only a 2% five-year survival rate for patients treated with radiation (10). Importantly, extensive experimental evidence has demonstrated that glioblastoma follows the cancer stem cell model with a population of TICs driving tumorigenesis and therapy resistance (2,11–14). Previous work has established that primary glioblastoma tumor cells are genetically heterogeneous with extensive whole chromosome and gene copy number variations (15–18). However, these studies relied on methods that analyzed bulk tumor samples and did not isolate TICs. Thus, we specifically analyzed glioblastoma TICs for mitotic defects and chromosome copy number variations to determine the chromosomal stability of these cells. Further, using an orthotopic mouse model, we investigated whether altering the rate of glioblastoma TIC chromosome mis-segregation impacted tumor development.
Results
Glioblastoma TICs Display CIN
To determine if TICs from glioblastoma tumors are chromosomally unstable, we measured the prevalence of chromosome segregation defects during mitosis (Figure 1A) (5). Initially, we examined a series of TICs derived from patients with glioblastoma that were previously established and named glioblastoma neural stem (GNS) cells (19). These cells were cultured in serum-free conditions to enrich for TICs and to ensure that cells maintain genetic and phenotypic similarity to primary tumor cells (20). We verified that a majority of TICs grown in these conditions express the stem cell markers NESTIN (Figures S1A–B) and SOX2 (Figures S2A–B). GNS cells grown under these conditions were also multi-potent in vitro and differentiated along astrocyte (Figures S3A–B) and neuron lineages (Figures S3C–D). These properties were stable through long-term culturing as the fraction of cells expressing the stem cell markers NESTIN and SOX2 and differentiating in vitro did not change after twenty or more passages in culture relative to the early passage cells (Figures S1–S3).
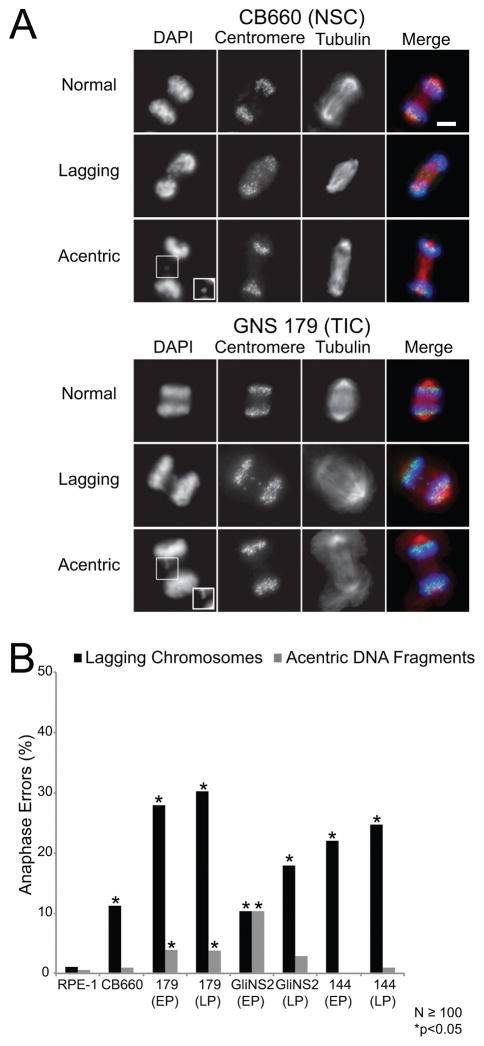
Figure 1. GNS cells have anaphase defects.
(A) Representative images of anaphases that were categorized as normal, those with lagging chromosomes, or those with acentric DNA fragments for control CB660 neural stem cells (NSCs) and GNS 179 TICs. Lagging chromosomes have centromeres localized between the two separating daughter nuclei while acentric DNA fragments do not have detectable centromere signal. Shown in the images are DNA (blue), centromeres (green), and microtubules (red). The insets are contrasted images to enhance the acentric DNA fragments. Scale bar, 5μm. (B) Percentage of anaphase defects in control RPE-1 and CB660 cells and the TICs GNS 179, GNS 144, and GliNS2. EP indicates early passage and LP late passage that is 20 or more passages in culture relative to the early passage cells. For the CB660 and GNS cells, 8 or more replicate coverslips were analyzed for N≥100 anaphases scored and *p<0.05, Fisher’s exact test compared with control RPE-1 cells.
We measured the frequency of mitotic defects in GNS cells (Figure 1A) relative to both retinal epithelial cells (RPE-1) grown in the presence of serum, which are diploid, chromosomally stable, and non-transformed and human neural stem cells (CB660), which are diploid, chromosomally stable, and cultured under the same serum-free conditions as the GNS cells (21). The GNS cells had an increased frequency of lagging chromosomes compared to RPE-1 cells characteristic of CIN (Figure 1B) and consistent with a previous study demonstrating that GNS 179 cells had lagging chromosomes when defects in chromosome microtubule attachments were experimentally induced (22). The lagging chromosomes at anaphase are unlikely to be caused by a defective spindle assembly checkpoint because the mitotic index of GNS cells increased when treated with the microtubule poison nocodazole (Figures S4A–B); however this assay does not rule out that the cells have weakened checkpoint activity. Surprisingly, CB660 cells also had an increased frequency of lagging chromosomes in anaphase compared to RPE-1 cells suggesting that CB660 neural stem cells also potentially have an elevated rate of chromosome mis-segregation (Figure 1B).
Karyotypic diversity in a population can only arise from CIN if the high rate of chromosome mis-segregation is coupled to the continued propagation of cells following the completion of a defective mitosis. To assess the karyotypic diversity in populations of CB660 and GNS cells, we used fluorescent in-situ hybridization (FISH) with centromeric DNA probes. Chromosome mis-segregation is random with respect to which chromosome(s) is mis-segregated, so we chose probes to chromosomes 2, 3, 7, and 10 that encompass a range of chromosomes sizes to determine the modal chromosome copy number and the percentage of the population that deviates from that mode for each chromosome (Figures 2A and S4C). For all four chromosomes analyzed, CB660 cells had a modal chromosome copy number of two and a low percentage of mode deviation as expected for diploid cells (Figures 2B–C). In contrast, all the GNS cells were aneuploid for most chromosomes (Figures 2B–C). For example, GNS 179 cells had a modal chromosome copy number of four, three, five, and two for chromosomes 2, 3, 7, and 10, respectively (Figures 2B–C). Moreover, the GNS cells showed significantly higher percentages of deviation from the mode compared to CB660 cells (Figure 2C). This indicates that GNS cells have a high rate of chromosome mis-segregation and that these cells continue to propagate after mis-segregating chromosomes during mitosis.
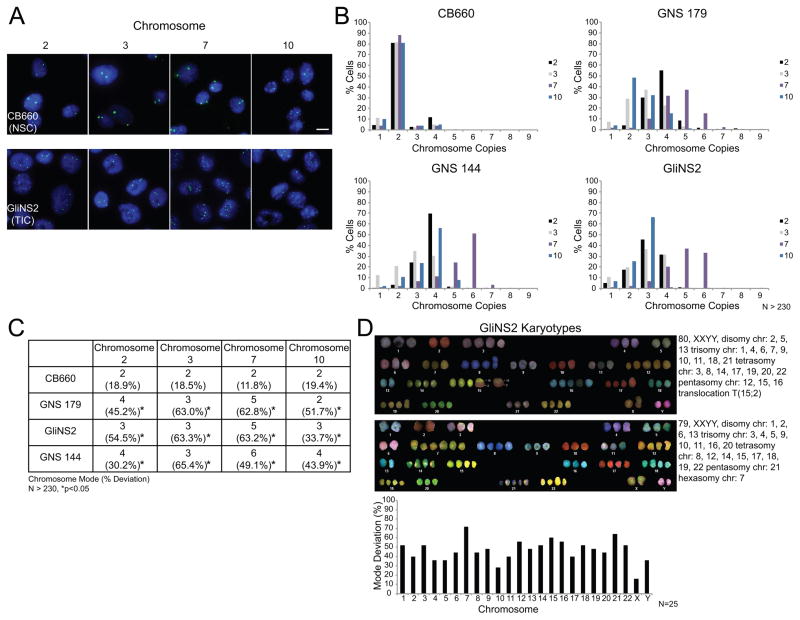
Figure 2. GNS cells are aneuploid and have a CIN phenotype.
(A) Representative images of FISH analysis for control CB660 NSCs and GliNS2 TICs. Chromosome copy number variations were determined using centromeric DNA probes for chromosomes 2, 3, 7, and 10. Two independent experiments were performed for each cell line. Scale bar, 10μm. (B) Histograms showing the distribution of chromosome copies with data pooled from two independent experiments. N>230 total nuclei scored per probe. (C) Summary table derived from the data in panel B displaying the modal chromosome copy numbers and the percentages of cells that deviate from the modal chromosome number. The asterisks indicate p<0.05, Fisher’s exact test compared with control CB660 cells. (D) Two representative karyotypes from SKY analysis of GliNS2 cells and a graph displaying the mode deviation for all chromosomes. N=25 cells.
FISH analysis is limited to determining the copy number of single chromosomes and fails to identify structural chromosome abnormalities. Thus, we performed spectral karyotyping (SKY) on GliNS2 cells to identify both whole chromosome and structural aneuploidy (Figures 2D and S5). In GliNS2 cells, we analyzed 25 metaphase chromosome spreads and found the karyotype varied from 48 to 87 chromosomes. Importantly, no two metaphase spreads had the same karyotype and all cells had multiple deviations in whole chromosome copy numbers. Further, all chromosomes showed copy number variability. Although these cells also displayed acentric DNA fragments during mitosis (Figure 1B) that are proposed to generate structural aneuploidy (23), only 40% of the metaphase spreads showed evidence of structural chromosome aneuploidy and 80% of these were an apparently clonal translocation between chromosomes 15 and 2. Combined, these FISH and SKY analyses demonstrate that GNS cells are chromosomally unstable and that deviations in whole chromosome copy numbers are the dominant karyotypic feature.
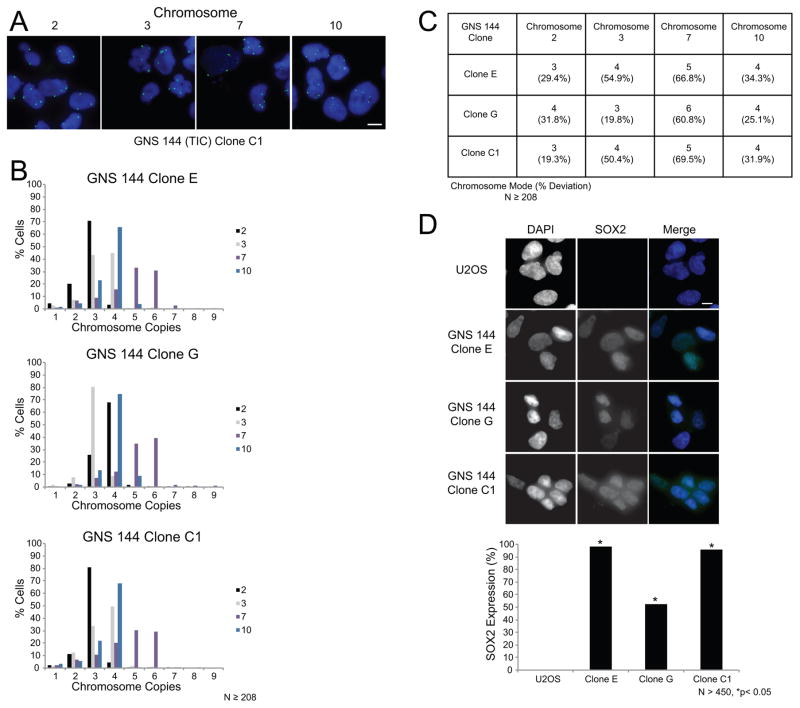
In addition to determining the modal chromosome copy numbers and percentage mode deviations in populations of GNS cells, we also analyzed cells derived from single cell clones. Three independent GNS 144 single cell clones were generated and analyzed by FISH for the modal chromosome copy numbers and percentage deviations (Figures 3A and S4C). Each clone was aneuploid as expected, but there were differences in modal chromosome copy numbers between the clones (e.g. chromosome 2 is trisomic in clones E and C1 but tetrasomic in clone G). Moreover, the clones displayed a high percentage of deviation from the mode for most chromosomes indicative of CIN (Figures 3B–C). Importantly, these single cell clones retain expression of the stem cell marker SOX2 indicating that they maintain characteristics of TICs (Figure 3D).
Figure 3. GNS 144 single cell clones are aneuploid and have a CIN phenotype.
(A) Representative images of FISH analysis for the TIC GNS 144 single cell clone C1. Chromosome copy number variations were determined using centromeric DNA probes for chromosomes 2, 3, 7, and 10. Two independent experiments were performed for each clone. Scale bar, 10μm. (B) Histograms showing the distribution of chromosome copies with data pooled from two independent experiments. N 208 total nuclei scored per probe. (C) Summary table of the modal chromosome copy numbers and mode deviations derived from the data in panel B. (D) Representative images of SOX2 expression in negative control U2OS cells that are non-tumor initiating osteosarcoma cells, and GNS 144 single cell clones. Shown in the images are DNA (blue) and SOX2 (green). Scale bar, 10μm. Bar graph shows the percent of cells expressing SOX2 with data pooled from duplicate coverslips. N>450 cells scored and *p<0.05, Fisher’s exact test compared with negative control U2OS cells.
Lastly, we analyzed the p53 pathway in CB660 and GNS cells because loss of p53 function is an important determinant that allows cells to propagate after chromosome mis-segregation (24). In response to DNA damage induced by doxorubicin treatment, control RPE-1 and CB660 cells up-regulated p53 and p21CIP1/WAF1 levels to initiate growth arrest as expected (Figure S6A). In contrast, GNS cells had an abundance of p53 in the absence or presence of DNA damage and p21 levels failed to increase, demonstrating that the p53 pathway is aberrant in GNS cells (Figure S6A). Genomic DNA sequencing revealed mutations in the DNA binding domain sequence of p53 in the GNS 179 and GliNS2 cells that are commonly found in tumors and in agreement with previous DNA sequencing of p53 from glioblastoma tumors (Figures S6B–C) (25,26). Although we did not identify a commonly found homozygous point mutation in the p53 gene in GNS 144 cells, the lack of a p53 response to DNA damage suggests that the pathway is mutated. Taken together, these data show that the GNS cells have an aberrant p53 response that accompanies their persistent rate of chromosome mis-segregation during mitosis fulfilling the requirements of a CIN phenotype. In contrast, CB660 neural stem cells possess a functional p53 pathway that would act to limit the propagation of cells that mis-segregate chromosomes and prevent the generation of an aneuploid population. Taken all together the data demonstrates that GNS cells have a CIN phenotype that generates extensive genetic heterogeneity within the TIC population.
Glioblastoma Patient-Derived Xenograft TICs Display CIN
Much ambiguity and controversy surround the identification and methods used to culture TICs in vitro (14,27–29). We specifically selected the GNS TICs to study because these cells were isolated without selection of an expression marker. To validate our initial findings in GNS cells, we expanded our analyses to include glioblastoma patient-derived xenograft (PDX) cells that were cultured as xenograft tumors in mice to maintain characteristics of the parental tumors from which the cells were derived. TICs were prospectively isolated using the CD133/PROM-1 glycosylated surface epitope and minimally propagated in culture while the cells lacking CD133 expression were isolated as the non-TIC population.
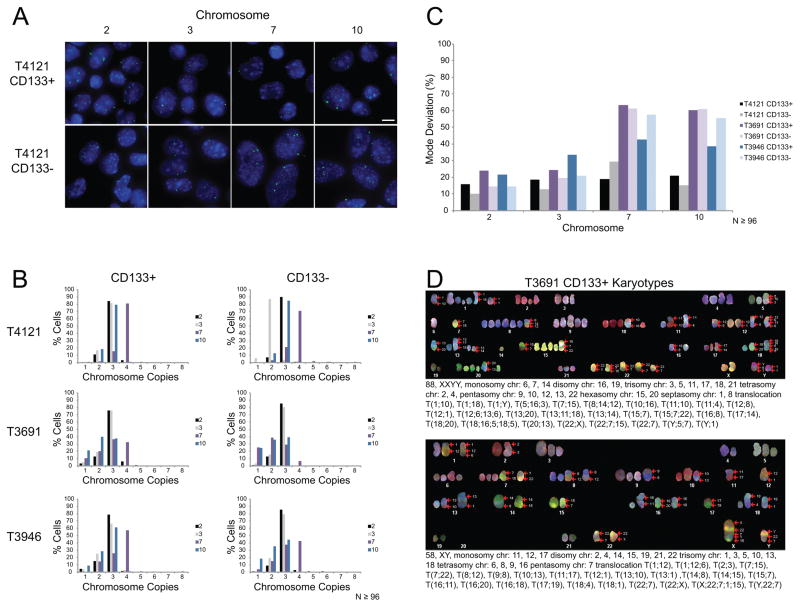
In total, we analyzed three previously characterized glioblastoma PDX lines designated T4121, T3691, and T3946 for the modal chromosome copy number and percentage of mode deviation to assess their chromosomal stability (30). These cells were harvested from xenograft tumors and sorted into populations enriched and functionally validated as TICs (CD133+) and populations of non-TICs (CD133−). The TICs were minimally expanded (five passages or less) as neurospheres while the non-TICs were minimally expanded as adherent monolayers in serum to obtain an adequate number of cells for subsequent analysis. The modal chromosome copy number and percentage mode deviation were determined for both populations by FISH using centromeric DNA probes for chromosomes 2, 3, 7, and 10 (Figures 4A–C and S7A–B). For all three PDX lines both the TICs and non-TICs were aneuploid for most chromosomes (Figure 4B). TICs and non-TICs from T3691 and T3946 also had comparable percentages of mode deviation to GNS cells indicative of CIN (Figures 4B–C and S7B). The percentage of mode deviation in T4121 TICs and non-TICs was comparable to the CB660 cells despite their aneuploid karyotype (Figures 4B–C and S7B). Furthermore, the percentage mode deviation between the TICs and non-TICs was similar in each PDX line suggesting that both populations are chromosomally unstable (Figure 4C).
Figure 4. PDX cells are aneuploid and have a CIN phenotype.
(A) Representative images of FISH analysis for the glioblastoma PDX line T4121. PDX cells were sorted into CD133+ TICs and CD133- non-TICs. Chromosome copy number variations were determined using centromeric DNA probes for chromosomes 2, 3, 7, and 10. Scale bar, 10μm. (B) Histograms showing the distribution of chromosome copies. N≥96 total nuclei scored per probe. (C) Bar graph showing the percentage of cells that deviate from the modal chromosome copy number derived from data in panel B. (D) Two representative karyotypes from SKY analysis of T3691 CD133+ TICs.
To further validate the CIN phenotype, we performed SKY on T3691 TICs and non-TICs to obtain complete karyotypes of individual cells. In total, we examined metaphase spreads from 5 TIC and 5 non-TIC cells (Figures 4D and S7C). Both the TICs and non-TICs from this patient had whole chromosome copy number variations and structural chromosome alterations. Similar to the GliNS2 cells, no two karyotypes were the same demonstrating extensive genetic heterogeneity within these cells. It is improbable that this level of karyotypic heterogeneity arose during the brief expansion of cells in culture prior to analysis. Overall, the modal chromosome copy numbers, the percentage of chromosomes deviating from the mode, and the karyotypes of individual cells demonstrate that both GNS and PDX TICs are heterogeneous with respect to chromosome complement and that TICs display CIN.
Manipulating the Chromosome Mis-Segregation Rate in GNS Cells
To explore the relationship between CIN and TICs, we altered the rate of chromosome mis-segregation in TICs. Previous studies have shown that the root cause of CIN is lagging chromosomes in anaphase that result from the persistence of incorrect merotelic chromosome microtubule attachments (5,31). Further, it was shown that expression of wild-type or mutant versions of MCAK (Kif2C), a member of the kinesin-13 family of microtubule depolymerases, decrease or increase the rate of lagging chromosomes in anaphase, respectively (5,31–33). Overexpression of the microtubule depolymerase MCAK decreases the stability of chromosome microtubule attachments and improves the correction of erroneous microtubule attachments (31,32). Conversely, overexpression of MCAK mutants that lack depolymerase activity displace endogenous MCAK from centromeres thereby increasing the stability of chromosome microtubule attachments and eroding the efficiency of error correction (33). Thus, to explore the relationship between TIC function and the rate of chromosome mis-segregation, we constructed GNS 179 stable cell lines expressing GFP-MCAK or a dominant negative GFP-MCAK mutant that lacks depolymerase activity (designated as MCAK Hypir) (33). As controls, we constructed GNS 179 stable cell lines expressing GFP or GFP-Kif2A, a related kinesin-13 that does not affect chromosome microtubule attachments or the rate of chromosome mis-segregation (32).
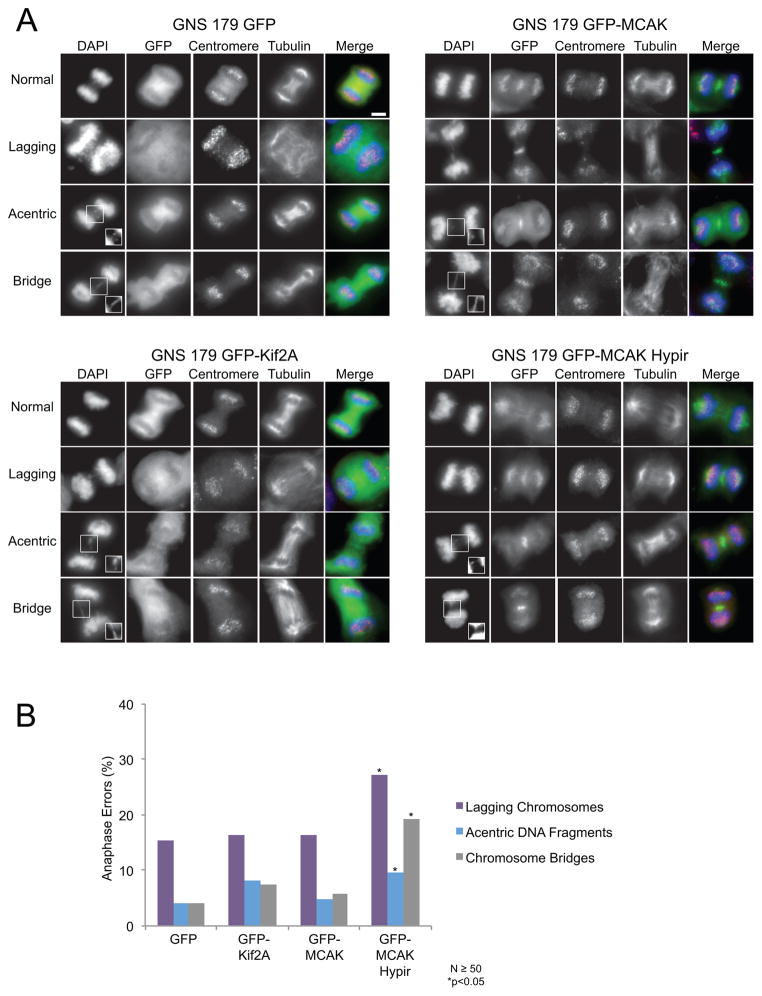
First, we measured the frequency of chromosome segregation defects including lagging chromosomes, acentric DNA fragments, and chromosome bridges in these cells (Figures 5A–B). Expression of GFP or GFP-Kif2a had little effect on the rates of chromosome mis-segregation as expected. Surprisingly, the expression of GFP-MCAK did not significantly reduce chromosome segregation defects relative to control cells as previously shown in other cell lines (32). The reason that TICs are refractory to MCAK over-expression compared to other cancer cells is unknown, although it may be related to the fact that MCAK expression is up-regulated in glioblastoma tumors compared to normal brain tissue (34). In contrast, expression of GFP-MCAK Hypir significantly increased all types of chromosome segregation defects including nearly doubling the rate of lagging chromosomes in anaphase (Figure 5B). Thus, these cells continue to propagate after their rate of chromosome mis-segregation in mitosis is experimentally increased. This indicates that TICs can tolerate a stable increase in the chromosome mis-segregation rate.
Figure 5. Expression of the MCAK-Hypir mutant elevates the rate of CIN.
(A) Representative images of anaphases that were categorized as normal, those with lagging chromosomes, those with acentric DNA fragments, or those with chromosome bridges for GNS 179 cells expressing GFP, GFP-Kif2A, GFP-MCAK, or the dominant negative GFP-MCAK Hypir mutant. Lagging chromosomes have centromeres localized between the two separating daughter nuclei while acentric DNA fragments do not have detectable centromere signal. Chromosome bridges are stretches of DNA connecting the two separating daughter nuclei. Shown in the images are DNA (blue), the GFP expression construct (green), centromeres (red), and microtubules (grayscale). The insets are contrasted images to enhance the acentric DNA fragments and bridges. Scale bar, 5μm. (B) Percentage of anaphase defects in GNS 179 cells. N≥50 anaphases scored from 11 or more replicate coverslips and *p<0.05, Fisher’s exact test compared with control GFP expressing cells.
Elevating the Rate of Chromosomal Instability Impairs GNS Cell
Proliferation and Stem-Like Properties
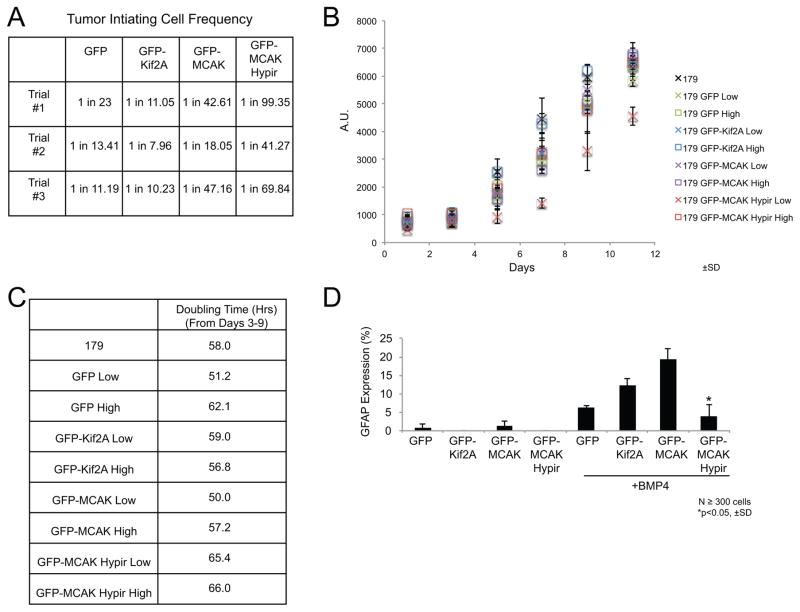
We next tested if altering the rate of chromosome mis-segregation influenced the functional properties of TICs in vitro. First, we analyzed the cell lines expressing the GFP-tagged fusion proteins for expression of the stem cell markers NESTIN and SOX2. Cells expressing GFP-MCAK Hypir did not show a significant difference in the percent of cells that expressed NESTIN or SOX2 or in the total levels of NESTIN or SOX2 (Figures S8A-D) compared to the other cell lines demonstrating that TICs expressing GFP-MCAK Hypir retain stem cell marker expression. However, TIC function is explicitly defined by the ability of TICs to self-renew and to differentiate analogous to normal stem cells. To test if the rate of CIN influences self-renewal, we performed in vitro limiting dilution assays. Cells expressing GFP-MCAK Hypir had a decreased frequency of in vitro self-renewal (Figures 6A and S9) compared to the cells expressing the other GFP-tagged fusion proteins.
Figure 6. Increasing CIN leads to a loss of TIC function.
(A) Estimated frequency of tumor-initiating cells determined from in vitro limiting dilution assays for GNS 179 cells expressing the GFP-tagged fusion proteins. N=3 trials. (B) Cell proliferation assay comparing the growth of parental GNS 179 cells, GNS 179 GFP, GFP-Kif2A, GFP-MCAK, and GFP-MCAK Hypir expressing cells. The cell lines expressing the GFP-tagged fusion proteins were sorted into populations of low and high GFP expressing cells. Growth was then monitored every other day for 11 days total with an alamarBlue® assay. Three independent replicates were performed and error bars represent ±SD. (C) A table of the population doubling time during the exponential growth phase (days 3–9) of the various cell lines. (D) The astrocyte differentiation frequency of GNS 179 GFP, GFP-Kif2A, GFP-MCAK, and GFP-MCAK Hypir expressing cells as measured by the mean percentage of cells expressing the astrocyte marker GFAP after growth in the presence of 10ng/ml BMP-4 for 8 days. N≥300 total GFP+ cells scored from three independent experiments. Errors bars represent ±SD and *p<0.05, Student’s T-test compared with control GFP-Kif2A expressing cells.
Limiting dilution assays are a measure of both self-renewal and the proliferation rate of TICs. To determine if the rate of CIN influenced one or both properties, we measured the proliferation of the cell lines in a long-term growth assay. First, the cell lines were sorted into populations of low and high GFP expressing cells (Figure S10). The growth of the GFP expressing cells and the parental GNS 179 cells was then measured every other day for 11 days (Figure 6B). From the growth curves, population doubling time was calculated during the exponential growth phase from days 3–9 (Figure 6C). All the cells had a doubling time over 50 hours with GFP-MCAK Hypir expressing cells displaying a slightly increased doubling time compared to the other cell lines (Figure 6C). Overall, the in vitro limiting dilution and growth rate data suggests that increasing the chromosome mis-segregation rate impairs both TIC self-renewal and proliferation.
In addition to self-renewal, TICs are also defined by their ability to differentiate. To assess whether CIN influences TIC differentiation, we measured the frequency of astrocyte differentiation in the cell lines expressing the GFP-tagged fusion proteins. Cells expressing GFP-MCAK Hypir had the lowest frequency of astrocyte differentiation (Figures 6D and S11) compared to the other cell lines. Thus, elevating chromosome mis-segregation impairs TIC proliferation, self-renewal, and differentiation suggesting that increasing chromosome mis-segregation may lead to a loss of tumor initiating function in these cells.
Elevating the Rate of Chromosomal Instability Inhibits Tumor Formation
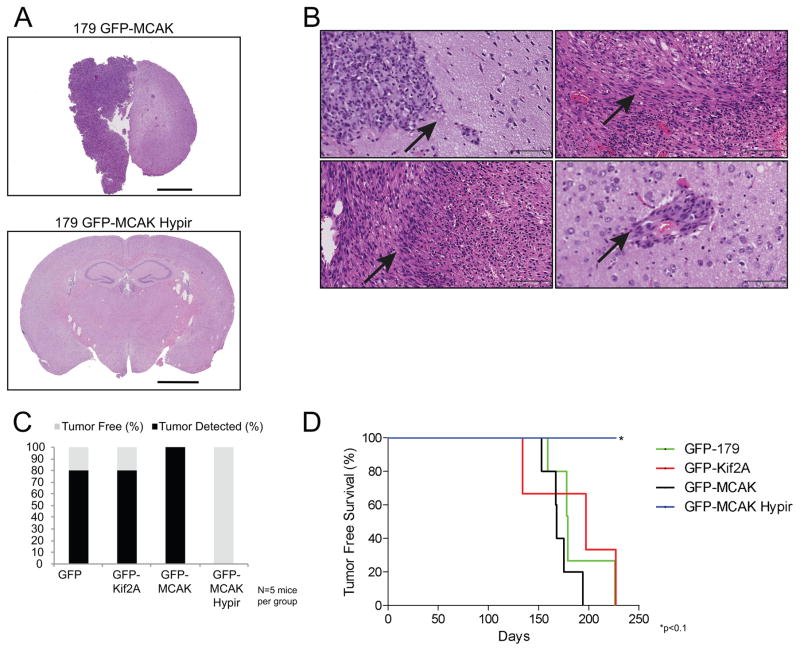
To test whether altering the rate of chromosome mis-segregation impacts the tumorigenicity of TICs, we intracranially injected the stable GNS 179 cell lines into immune-compromised mice. The cells were sorted for GFP expression immediately prior to injection to ensure equal numbers of GFP+ cells were injected for each group. Five mice were injected with 100,000 total cells per model, and mice were sacrificed at the first signs of tumor burden. Ultimately, any surviving mice were sacrificed at 312 days post-injection and were examined for evidence of a brain tumor.
Multiple animals injected with the GNS 179 GFP, GFP-Kif2A, and GFP-MCAK expressing cells developed brain tumors, and in agreement with previous results, the tumors that developed recapitulated common features of human glioblastoma tumors including desmoplasia, infiltration, and pseudopalisading (Figures 7A–B) (19). In total, 80% of the mice injected with cells expressing GFP or GFP-Kif2A and 100% of the mice injected with cells expressing GFP-MCAK developed tumors by the 312 day endpoint of the experiment (Figure 7C). In striking contrast, none of the mice injected with cells expressing GFP-MCAK Hypir showed detectable evidence of brain tumors upon histological examination at any time point (Figures 7A and 7C). For animals that were sacrificed prior to the 312 day endpoint, there was no significant difference between the tumor free survival curves of the mice injected with the control GFP expressing cells and the GFP-Kif2A or GFP-MCAK expressing cells (p>0.2, log-rank test) (Figure 7D). There was a significance difference with a threshold of p<0.1 (p=0.07, log-rank test) in the tumor free survival curves for the mice injected with the control GFP expressing cells compared to mice injected with the GFP-MCAK Hypir expressing cells (Figure 7D). Taken together, these data demonstrate that TICs derived from glioblastoma tumors display CIN, and both contribute to and propagate genetic diversity within a tumor. Further, increasing the rate of chromosome mis-segregation reduces the ability of TICs to initiate tumor formation demonstrating that TICs have an upper tolerable limit for the rate of genetic change.
Figure 7. Increasing CIN inhibits tumor formation.
(A) Representative hematoxylin and eosin stained brain sections of mice injected with GNS 179 GFP-MCAK (top panel) or GFP-MCAK Hypir (bottom panel) cells. A tumor is clearly evident in the brain of the animal injected with GFP-MCAK expressing cells while there is no indication of a tumor in the brain of the animal injected with GFP-MCAK Hypir expressing cells. In total, N=5 mice were injected with 100,000 GNS 179 GFP, GFP-Kif2A, GFP-MCAK, or GFP-MCAK Hypir expressing cells. Scale bar, 2000μm. (B) Representative hematoxylin and eosin stained sections highlighting the pathological features of the intracranial tumors that developed in mice injected with GNS 179 GFP, GFP-Kif2A, and GFP-MCAK cells. The upper left panel shows tumor tissue adjacent to normal brain tissue (arrow). The upper right panel is an example of desmoplasia that developed in tumors (arrow). The bottom left panel is an example of pseudopalisading in tumors (arrow). The bottom right panel is an example of tumor cells infiltrating into normal brain tissue (arrow). Scale bar, 25x-100μm. (C) Percentage of brain tumors detected in mice injected with GNS 179 GFP, GFP-Kif2A, GFP-MCAK, or GFP-MCAK Hypir expressing cells by the 312 day endpoint of the experiment. (D) Tumor free survival curves of mice that were sacrificed prior to the 312 day endpoint of the experiment. *p<0.1, Log-rank (Mantel-Cox) test compared to mice injected with GNS 179 GFP expressing cells.
Discussion
CIN is a Source of Genetic Heterogeneity in TICs
Tumor progression is commonly based upon principles of evolutionary theory. From a vast heterogeneous genetic landscape within populations of tumor cells, local environmental conditions promote the selection of individual cells that have acquired unique capacities to outcompete their neighbors fueling tumor progression and generating intra-tumor heterogeneity. This process plays out iteratively so that tumors and their metastatic progeny can be represented as trees with genetically definable trunks and branches with differing levels of genetic disparity. A central premise of this model is that tumor cells possess a capacity to tolerate genetic change (35). Here we demonstrate that glioblastoma TICs have a CIN phenotype resulting in extensive karyotype diversity within the population and providing an ongoing source for genetic changes. Indeed, our SKY data show that no two cells in the TIC populations that we assayed have the same chromosomal karyotype. These data support a model of genetic intra-tumor heterogeneity arising from both TICs and non-TICs and provides a direct link between a mechanism that drives genetic change (i.e. CIN) to the genetic diversity within the TIC population. However since TICs are responsible for tumor recurrence after treatment, these cells can propagate genetic changes while the non-TICs cannot. To our knowledge this is the first demonstration of CIN in TICs.
Previous studies have demonstrated both whole chromosome and gene copy number intra-tumor heterogeneity in primary glioblastoma tumor tissue but did not distinguish TICs from non-TICs (16–18). Further studies have analyzed different types of genetic diversity within TICs and showed that glioblastoma TICs and non-TICs had abnormal karyotypes but reported that these karyotypes were clonal and that single cell clones were genetically homogeneous (12,14,36–38). In contrast, we demonstrate that glioblastoma TICs have extensive sub-clonal karyotype heterogeneity and that single cell clones do not maintain a homogenous karyotype. In agreement with our results, other studies that analyzed gene mutations and gene copy number variations arrived at the same conclusion showing extensive sub-clonal genetic heterogeneity in TICs, but these studies did not provide a mechanism for the generation of such sub-clonal heterogeneity (39,40). From these sub-clonal patterns of genetic heterogeneity it was suggested that TICs follow a “back to Darwin” evolutionary model with TICs having variegated genotypes that selection can act upon (39). However, unlike individual gene mutations or copy number variations, the potential of whole chromosome mis-segregation and CIN to be agents for selection and to create phenotypic diversity may be larger because the copy number of hundreds to thousands of genes is simultaneously altered upon whole chromosome mis-segregation (41,42) and CIN is a source of continual genetic re-shuffling. An important extension of this work would be to use single cell methods to simultaneously test the tumor initiating capacity of a cell as well as its specific karyotype and rate of chromosome mis-segregation. Unfortunately, karyotypic analysis is lethal to cells so there are no methods currently available to combine these strategies for single cells.
TICs Have an Upper Tolerable Limit for CIN
The prevalence and complexity associated with intra-tumor heterogeneity is a major obstacle for current therapeutics and poses challenges for new drug development (43). In agreement, CIN is correlated with therapy resistance and poor patient prognosis (7,8). According to evolutionary theory, CIN may contribute to resistance by creating genetic diversity to provide substrates for the subsequent selection of cells with advantageous karyotypes. Alternatively, resistance may be an intrinsic property associated with a CIN phenotype (8,9). Similarly, TICs are resistant to a variety of treatments including ionizing radiation and chemotherapeutics and therefore are thought to be the population of cells responsible for tumor relapse (2,3). This combined tolerance to both genetic and environmental stress is an attribute of TICs that is likely to contribute to tumor recurrence.
Yet, our results demonstrate that there is an upper limit to the rate of chromosome mis-segregation that TICs can tolerate and still maintain their TIC function as defined by their ability to initiate tumors. If we assume that chromosome mis-segregation scales linearly with the frequency of mitotic errors that we detected following expression of a dominant negative MCAK mutant, then TICs lose their functional properties following an approximately 2-fold increase in the chromosome mis-segregation rate. Surprisingly, unlike previous studies demonstrating that elevating chromosome mis-segregation leads to cell death (44–46), this change in the chromosome mis-segregation rate is not uniformly lethal and does not alter stem cell marker expression (NESTIN and SOX2). This reveals that the functional capacity of TICs to drive tumor formation is separable from expression of those stem cell markers. Also, this raises the possibility that TICs give rise to non-TICs within the tumor population through mitotic events that confer karyotypes onto daughter cells that are incompatible with a tumor initiating phenotype rather than solely occurring through processes akin to stem cell differentiation. The loss of TIC function may result from transcriptional changes and/or imbalances. Evidence from aneuploid cells shows that a gain in a whole chromosome leads to an increase in mRNA expression levels for the majority genes on the gained chromosome demonstrating that transcription scales with chromosome copy number (41,42). Therefore, whole chromosome changes created by CIN may substantially alter transcriptional programs leading to a loss of TIC function.
Increasing CIN in TICs as a Therapeutic Target
These data reveal a new potential therapeutic strategy to eliminate TIC function. Previous studies have suggested inhibiting the mitotic checkpoint to cause massive chromosome mis-segregation and lethality in tumor cells (22,44,45). Our data indicates that a significant therapeutic response could be achieved by elevating chromosome mis-segregation rates to levels that simultaneously decrease proliferation and erode TIC function, however, this is below a level needed to cause lethality in all tumor cells.
This strategy would hinge on driving TICs through the cell cycle and into an error-prone mitosis. We envision a two-step strategy to eliminate TIC function and tumor development. Current treatments kill the bulk of tumor cells, however after treatment it has been shown that quiescent TICs re-enter the cell cycle to re-populate the tumor (11). It is at this time that TICs would be vulnerable to treatments that would modestly increase chromosome mis-segregation rates. Proteins that ensure faithful chromosome segregation would be candidate drug targets to achieve this outcome. MCAK is one such candidate and is a particularly attractive target for glioblastoma as mRNA expression analysis of glioblastoma tumor samples shows that it is up-regulated compared to normal brain tissue (34). Another candidate is the protein kinase MPS1 that is also required for faithful chromosome segregation. The MPS1 inhibitor, MPS1-IN-3, has been shown to pro-long survival in mouse models of glioblastoma when combined with the anti-mitotic vincristine as a first-line treatment (47). Importantly, our data suggests treatments elevating chromosome mis-segregation will be most effective as a secondary treatment after the bulk of tumor cells have been eliminated and quiescent TICs re-enter mitosis re-initiating tumor growth.
In conclusion, we show that intra-tumor heterogeneity stems from genetic heterogeneity within the population of TICs due to CIN. Although CIN generates continual heterogeneity that serves as a foundation for selection, adaptation, and evolution in a dynamic tumor environment, there is an upper tolerable limit that can be exploited as a therapeutic strategy.
Material and Methods
Cell Lines
The CB660, GliNS2, and GNS 144 cell lines were all obtained directly from the lab of S. Pollard in 2010 where these cell lines were previously characterized (19). The GNS 179 cell line was also obtained directly from S. Pollard in 2011 where this cell line was previously characterized (19). For determining the frequency of anaphase errors and chromosome copy numbers by FISH, GNS 179 cells ranged in passage number from 26–31 (early) and 47–51 (late). GliNS2 cells ranged in passage number from 25–35 (early) and 50 (late). GNS 144 cells ranged in passage number from 39–42 (early) and 61–76 (late). CB660 cells ranged in passage number from 39–44 in these experiments. The PDX cells were obtained directly from the lab of J. Rich in 2014 where these cells were previously validated and characterized (2,30,48,49). The GNS and PDX cell lines were previously characterized and no further authentication was performed for this study. RPE-1 (CRL-4000) and U2OS (HTB-96) cells were obtained from ATCC. For further details on the isolation of single cell GNS 144 clones, GNS cell astrocyte differentiation, and the construction of the stable GNS 179 cell lines, please see supplemental methods.
Cell Culture
Cells were maintained at 37°C in a humidified 5% CO2 atmosphere. GNS 179, GNS 144, GliNS2, and CB660 cells were a kind gift from S. Pollard and were maintained as previously described (19). Briefly, GNS and CB660 cells were cultured in DMEM/F12 (Life Technologies) supplemented with 1x non-essential amino acids (NEAA; Life Technologies), 0.5x N2 (Life Technologies), 45mM Hepes (Sigma), 0.5x B27 (Life Technologies), 29mM glucose (Sigma), 0.12mg/ml BSA (Sigma), 55μM 2-mercaptoethanol (Life Technologies), 20ng/ml FGF (Peprotech), 20ng/ml EGF (Peprotech), 1μg/ml laminin (Sigma), 100units/ml penicillin (Life Technologies), and 100μg/ml streptomycin (Life Technologies). For plasmid selection, GNS 179 cells were maintained with 250μg/ml of G418 (Invivogen). RPE-1 and U2OS cells were maintained in Dulbecco’s modified medium (DMEM; Mediatech) containing 10% fetal calf serum (Hyclone), 50units/ml penicillin (Lonza), 50units/ml streptomycin (Lonza), and 0.25μg/ml fungizone (Hyclone). T4121, T3691, and T3946 CD133+ TICs were maintained as previously described (30). T4121, T3691, and T3946 CD133- non-TICs were maintained in DMEM (Mediatech or Cleveland Clinic Media Productions Core) containing 10% fetal calf serum (Hyclone or Life Technologies), 50units/ml penicillin (Lonza), and 50units/ml streptomycin (Lonza) and validated to be of tumor origin (49). GNS, CB660, and PDX cells were dissociated using Accutase (Life Technologies) and RPE-1 and U2OS cells were dissociated using 0.05% Trypsin (Mediatech).
Immunofluorescence
GNS and CB660 cells were plated on 12mm poly-D-lysine/laminin coated coverslips (Corning) prior to fixation. RPE-1 and U2OS cells were plated on standard 18mm coverslips prior to fixation. For measuring the frequency of chromosome segregation errors, cells were fixed with ice-cold methanol for 3 mins or 3.5% paraformaldehyde, pH 6.8 for 5 mins (experiments with GNS 179 GFP cell lines), washed 2 x 5 mins TBS-0.1% triton, and blocked with “Abdil” (TBS-0.1% triton and 2% BSA) for 30 mins. Primary antibodies were diluted in Abdil at 1:4000 mouse DM1α (Sigma) and 2μg/ml rabbit CENP-A (kind gift A. Straight) and coverslips were incubated for 1 hour. Subsequently, coverslips were washed 4 x 5mins Abdil and incubated with secondary antibodies diluted in Abdil at 1:1000 Alexa Fluor anti-mouse 594 and Alexa Fluor anti-rabbit 488 or Alexa Fluor anti-mouse 647 and Alexa Fluor anti-rabbit 594 (experiments with GNS 179 GFP cell lines) for 1 hour (Alexa Fluor reagents Life Technologies). Coverslips were washed 4 x 5 mins Abdil, DNA stained with DAPI for 15 mins, washed 3 x 5 mins Abdil, washed 3 x 5 mins TBS-0.1% triton, and mounted with Pro-Long Gold Anti-Fade (Life Technologies).
For SOX2 immunofluorescence, cells were fixed in 3.5% paraformaldehyde, pH 6.8 for 5 mins, washed 2 x 5 mins TBS-0.1% triton, and blocked with Abdil + 10% donkey serum. The primary antibody was used at 10 μg/ml mouse SOX2 (R&D Systems).
For astrocyte differentiation experiments with GNS 179 GFP cell lines, cells were fixed with 3.5% paraformaldehyde, pH 6.8 for 5 mins and the following primary antibodies were used: mouse GFAP at 5μg/ml (Sigma), 1:1000 rabbit GFP (kind gift of W. Wickner), and 1:2000 human ACA (Geisel School of Medicine).
Images were acquired with a cooled CCD camera (Andor Technology) mounted on a microscope (Eclipse Ti; Nikon). 0.2μm or 0.5μm optical sections in the z axis were collected with a planApo 60× 1.4 NA oil immersion objective at room temperature. For experiments comparing the frequency of expression of proteins of interest, images for each cell line were acquired with the same acquisition parameters and exposure times. For additional details on immunofluorescence protocols, please see supplemental methods.
FISH
Cells were washed with PBS and resuspended in 5mls 75mM KCL for 10–15mins at 37°C. Cells were then fixed and washed twice in 3:1 methanol:acetic acid. FISH was performed using centromeric DNA probes to chromosomes 2, 3, 7, and 10 (Cytocell).
SKY Analysis
GliNS2 cells were treated with 100ng/ml nocodazole (Acros) for 16 hours to enrich for mitotic cells. Cells were washed and fixed as described for FISH analysis except cells were incubated in 10mls 75mM KCL for 20 mins at 37°C and fixed twice with 3:2 methanol:acetic acid. T3691 CD133+ TICs and CD133- non-TICs were treated with 100ng/ml nocodazole for 12 hours to enrich for mitotic cells. Cells were washed and fixed as described for FISH analysis except cells were incubated in 5mls 75mM KCL for 15 mins at 37°C. SKY analysis was performed by the Jackson Laboratory Imaging Services (Bar Harbor, ME).
Cell Proliferation Assay
GNS 179 cells, GNS 179 GFP, GFP-Kif2A, GFP-MCAK, and GFP-MCAK Hypir cells were sterile sorted to plate cells into 24 well Primaria tissue culture treated plates at 12,500 cells per well. Additionally, the GFP expressing cell lines were sorted into plates of low and high GFP expression. Cell sorting was performed by the Immunoassay and Flow Cytometry Shared Resource at the Geisel School of Medicine (Hanover, NH).
Cell growth was measured using alamarBlue® according to the manufacturer instructions (Thermo Fisher Scientific). Briefly, wells were incubated with alamarBlue® reagent for 6 hrs at 37°C. Cell growth was then measured using a fluorescent plate reader with an excitation wavelength of 530nm and an emission wavelength of 580nm. Cell growth measurements were repeated every other day for a total of 11 days.
From the growth curves, population doubling time was calculated during the exponential phase of growth from days 3–9. Population doubling time was calculated using the following equation: DT=Tln2/ln(Xe/Xb) were T=incubation time hours, Xe=ending fluorescent intensity, and Xb=beginning fluorescent intensity.
Animals and Orthotopic Injections
All animal studies described were approved by the Cleveland Clinic Foundation Institutional Animal Care and Use Committee and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Human tissues were acquired from primary human brain tumor patient specimens in accordance with appropriate approved Institutional Review Board protocols. Tumor grade and available cytogenetic information for each specimen has been previously described (2,48,49). PDX tumor specimens were maintained through subcutaneous xenografts in the flanks of NSG mice. Subcutaneous tumors were initiated via injection of 100,000 bulk tumor cells and harvested when the tumor reached ~1.0 cm in diameter.
For intracranial implantation studies to evaluate tumor initiation by TICs, 100,000 viable TICs (GFP, GFP-Kif2a, GFP-MCAK or GFP-MCAK Hypir) were implanted into the right frontal lobes of female NSG mice (n = 5 mice per group). Mice were monitored daily for neurological impairment at which time they were sacrificed and brains removed to evaluate for tumor development. After harvesting and fixation, the brains were sectioned and hematoxylin and eosin stained by the Pathology Translational Research Shared Resources at the Geisel School of Medicine. The H&E sections were than blindly scored for the presence of brain tumor by a neuropathologist.
In Vitro Limiting Dilution Assay
Genetically modified TICs (GFP, GFP-Kif2a, GFP-MCAK, and GFP-MCAK Hypir) were cell sorted based on viability (Live/Dead dye blue, Life Technologies) and GFP positivity using a FACS Aria II Cell Sorter (BD Biosciences) to plate the cells into 96 well plates at a final cell number per well of 1 (38 wells/plate), 5 (24 wells/plate), 10, 20, or 50 (all at 12 wells/plate). Colony formation was evaluated 14 days after sorting and wells were scored positive or negative for the presence of at least one colony. The estimated stem cell frequency was calculated using extreme limiting dilution analysis (50).
TIC cell surface sorting via CD133
To isolate PDX TICs, subcutaneous flank tumors were dissociated using a papain dissociation system (Worthington Biochemical). TICs were isolated based on surface expression of glycosylated CD133 enriched either by FACS (anti-CD133/2 (293C3)-APC, human) or magnetic activated cell sorting (anti-CD133/1 (AC133), human) as per manufacturers recommendations (MACS; Miltenyi Biotec) and grown as tumorspheres or adherently plated on GelTrex (Life Technologies). After isolation, all cells were utilized in experiments in fewer than 5 passages.
Statistical Analysis
A Fisher’s exact two-tailed test was performed for anaphase error rates, chromosome mode deviations, and SOX2 expression. A student’s two-tailed t-test was performed for the frequency of astrocyte differentiation. Kaplan-Meier tumor free survival curves were constructed using Prism 5 software with subjects that did not show evidence of a brain tumor upon histological examination censored. A Log-rank (Mantel-Cox) test was used to compare survival curves.
Supplementary Material
Significance.
Genetic heterogeneity among tumor-initiating cells (TICs) may produce advantageous karyotypes that lead to therapy resistance and relapse, however; we found that TICs have an upper tolerable limit for chromosomal instability. Thus, increasing the chromosome mis-segregation rate offers a new therapeutic strategy to eliminate TICs from tumors.
Acknowledgments
Financial Support: This work was supported by US National Institutes of Health (R37GM051542 to D.A.C., 5 P30 CA023108-36 and 8 P30 GM103415-14 to DartLab, CA154130, CA169117, CA171652, NS087913, NS089272 to J.N.R), the Research Programs Committees of Cleveland Clinic (J.N.R), the James S. McDonnell Foundation (J.N.R), and the American Cancer Society (PF-12-103-01-CCG to K.M.G.).
We thank members of the Compton lab for their thoughtful comments. Also, we kindly thank Dr. Stephen Pollard (University of Edinburgh) for providing the GNS cell lines, Dr. A. Straight (Stanford University) for providing the CENP-A antibody and modified pEGFP vector, Dr. L. Wordeman (University of Washington) for the MCAK Hypir plasmid, Dr. W. Wickner (Geisel School of Medicine at Dartmouth) for the GFP antibody, the Pathology Translational Research Shared Resources at the Geisel School of Medicine, and the DartLab: Immunoassay and Flow Cytometry Shared Resource at the Geisel School of Medicine at Dartmouth.
Footnotes
Disclosure of Potential Conflicts of Interest: These authors declare no competing financial interests.
Author Contributions:
K.M.G. designed and performed experiments and wrote the manuscript. D.A.C. designed experiments and wrote the manuscript. M.V. and J.N.R. designed experiments. M.V. and Q.W. performed the orthotopic injections. M.V. performed the LDA assays, monitored the mice daily for neurological signs, and harvested the brains. K.D.M. assisted with the SKY. W.F.H. scored mice for brain tumors. M.V., J.N.R., and K.D.M. provided insightful comments on the manuscript.
References
- 1.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 2.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 3.Oravecz-Wilson KI, Philips ST, Yilmaz OH, Ames HM, Li L, Crawford BD, et al. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell. 2009;16:137–48. doi: 10.1016/j.ccr.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–67. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 7.Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:7704–11. doi: 10.1158/1078-0432.CCR-11-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Molecular oncology. 2014;8:1095–111. doi: 10.1016/j.molonc.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 13.Lathia JD, Gallagher J, Myers JT, Li M, Vasanji A, McLendon RE, et al. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS ONE. 2011;6:e24807. doi: 10.1371/journal.pone.0024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 15.Hecht BK, Turc-Carel C, Chatel M, Grellier P, Gioanni J, Attias R, et al. Cytogenetics of malignant gliomas: I. The autosomes with reference to rearrangements. Cancer Genet Cytogenet. 1995;84:1–8. doi: 10.1016/0165-4608(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 16.Loeper S, Romeike BF, Heckmann N, Jung V, Henn W, Feiden W, et al. Frequent mitotic errors in tumor cells of genetically micro-heterogeneous glioblastomas. Cytogenet Cell Genet. 2001;94:1–8. doi: 10.1159/000048773. [DOI] [PubMed] [Google Scholar]
- 17.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–80. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Pollard S, Conti L, Toselli M, Biella G, Parkin G, et al. Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Mol Cell Neurosci. 2008;38:245–58. doi: 10.1016/j.mcn.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Ding Y, Hubert CG, Herman J, Corrin P, Toledo CM, Skutt-Kakaria K, et al. Cancer-Specific requirement for BUB1B/BUBR1 in human brain tumor isolates and genetically transformed cells. Cancer Discov. 2013;3:198–211. doi: 10.1158/2159-8290.CD-12-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–55. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 26.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen A-J, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–33. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son MJ, Woolard K, Nam D-H, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–52. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–75. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 30.Venere M, Hamerlik P, Wu Q, Rasmussen RD, Song LA, Vasanji A, et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Nature Publishing Group. 2013:1–12. doi: 10.1038/cdd.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–42. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wordeman L, Wagenbach M, Dassow von G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–79. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Science Translational Medicine. 2015;7:304ra143. doi: 10.1126/scitranslmed.aac6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 36.Baronchelli S, Bentivegna A, Redaelli S, Riva G, Butta V, Paoletta L, et al. Delineating the cytogenomic and epigenomic landscapes of glioma stem cell lines. PLoS ONE. 2013;8:e57462. doi: 10.1371/journal.pone.0057462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci USA. 2015:201320611. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stieber D, Golebiewska A, Evers L, Lenkiewicz E, Brons NHC, Nicot N, et al. Glioblastomas are composed of genetically divergent clones with distinct tumourigenic potential and variable stem cell-associated phenotypes. Acta Neuropathol. 2014;127:203–19. doi: 10.1007/s00401-013-1196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–61. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 40.Piccirillo SGM, Colman S, Potter NE, van Delft FW, Lillis S, Carnicer M-J, et al. Genetic and functional diversity of propagating cells in glioblastoma. Stem Cell Reports. 2015;4:7–15. doi: 10.1016/j.stemcr.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 43.Gerlinger M, Swanton C. British Journal of Cancer. Vol. 103. Nature Publishing Group; 2010. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine; pp. 1139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kops GJPL, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101:8699–704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janssen A, Kops GJPL, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–13. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci USA. 2013;110:E4134–41. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tannous BA, Kerami M, Van der Stoop PM, Kwiatkowski N, Wang J, Zhou W, et al. Effects of the selective MPS1 inhibitor MPS1-IN-3 on glioblastoma sensitivity to antimitotic drugs. J Natl Cancer Inst. 2013;105:1322–31. doi: 10.1093/jnci/djt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eyler CE, Wu Q, Yan K, MacSwords JM, Chandler-Militello D, Misuraca KL, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.