Abstract
Purpose
The expression of programmed cell death ligand 1 (PD-L1) provides limited predictive value in identifying patients most likely to respond to immunotherapy. Since the heterogeneity of PD-L1 expression may lead to sampling error and the misclassification of PD-L1 status, we assessed the distribution of PD-L1 expression in paired, resected multifocal lung cancers.
Experimental Design
PD-L1 was assessed by immunohistochemistry. Paired lesions were defined as independent primaries or related lesions using mate-pair next-generation sequencing. Agreement statistics were used for analysis.
Results
Sixty-seven multifocal lung cancers from 32 patients were sequenced and stained for PD-L1. There was agreement of PD-L1 expression by the tumor cells in paired lesions of 20 patients, and disagreement of PD-L1 expression by the tumor cells in paired lesions of 12 patients (kappa=0.01). Sequencing identified that 23 patients had independent primary lung cancers and that nine patients had related cancers. In paired lesions of patients with independent cancers, there was agreement of PD-L1 expression by the tumor cells in 12 patients, and disagreement in 11 patients (kappa=0.31). In paired lesions of patients with related lung cancers, there was agreement of PD-L1 expression by the tumor cells in 8 patients, and disagreement in 1 patient (kappa=0.73).
Conclusions
The expression of PD-L1 is heterogeneous amongst paired independent lung cancers, but there are high levels of agreement in intrapulmonary metastasis.
Keywords: Lung Cancer, Tumor Immunology, PD-L1, Heterogeneity, Metastasis
Introduction
Immunotherapy is rapidly being adopted for the treatment of multiple cancers. Recently, inhibitors of negative co-stimulatory pathways (immune checkpoints) such as programmed cell death 1 (PD-1) or its ligand (PD-L1) have been shown to improve outcomes for patients with lung cancer (1–3). Our current understanding of which patients may benefit from immune checkpoint inhibitors is limited. While there are multiple assays to detect the presence of PD-L1, expression levels do not correlate well between these assays. It also remains uncertain if expression of PD-L1 by tumor cells, tumor-associated immune cells, a combination thereof, or another marker, is the best predictor of response to these therapies. Furthermore, it is uncertain if PD-L1 expression at one tumor site is representative of expression elsewhere. Thus sampling error may lead to misclassification of PD-L1 expression status and may partially explain why some patients without detected PD-L1 expression have responded to PD-1 or PD-L1 inhibitors.
Multifocal lung cancer is an increasingly common and challenging clinical scenario (4, 5). The adoption of lung cancer screening has led to the detection of multiple synchronous or metachronous tumors in up to 20% of patients with screen detected lung cancers (4, 6–11). These lesions may represent independent primary tumors or intrapulmonary metastases. Currently there is no gold standard for clinicians to distinguish between these scenarios. In the absence of metastatic disease to lymph nodes or elsewhere, local measures such as surgery or stereotactic body radiotherapy may be recommended. Alternatively, systemic, palliative chemotherapy may be recommended when metastatic disease is suspected. Given the difficulties of determining which patients with multifocal lung cancer have independent primaries or metastatic disease, our clinical group systematically evaluates these patients for local therapies (NCT01946100). When a surgical approach is feasible and no distant disease or contraindications to surgery are identified, surgical resection may be offered. A subset of these resected tumors have frozen tissue available for research. For this study we identified patients with multifocal lung cancer and available tissue who underwent surgical resection and characterized their tumors using next generation sequencing with a mate-pair library approach (12). This technique allowed us to assess the lineage relationships of these tumors and to categorize them as independent primaries or related lesions (intrapulmonary metastases). We then determined PD-L1 expression amongst these lesions to assess heterogeneity among these robustly defined populations.
Materials and Methods
Patients
The Tissue Registry and Lung Specimen Registry at Mayo Clinic were searched to identify samples of multifocal lung cancers available for use in this study. Specimens were rapidly frozen upon collection. Two pulmonary pathologists performed independent reviews, blinded to clinical and genomic data. Based on morphology, using criteria as previously suggested (13), a case was predicted as independent or favor independent primaries, as related or favor intrapulmonary metastasis, or indeterminate if the pathologists did not agree. Mate-pair next-generation sequencing was used to make the final determination of tumor relatedness as per below (12). Mayo Clinic’s Institutional Review Board approved this study.
Laser capture microdissection of frozen tissue specimens
Histological review of H&E stained fresh frozen sections was performed for quality control. Laser capture microdissection (LCM) was performed on 10-micron frozen sections and pure populations of tumor cells were isolated using the Arcturus PixCell II microscope and CapSure Macro LCM caps (Arcturus Carlsbad, CA; LCM 0211). DNA was extracted directly from LCM captured cells using a previously described single-step whole genome amplification (WGA) procedure (12, 14). Four individual 50μl WGA reactions were pooled for each sample. DNA was quantified by Quant-iT-PicoGreen analysis (Invitrogen, Eugene, OR; P7581).
Next Generation Sequencing
Mate Pair (MP) sequencing tiles the genome with larger spanning (~3kb) fragments than conventional paired end next generation sequencing to increase the probability of spanning a genomic breakpoint. MP libraries were assembled from WGA DNA, according to a previously published protocol (12, 14) using the Illumina mate pair kit. Two multiplexed libraries were loaded per lane of an Illumina flow cell and sequenced to 101x2 paired-end reads on an Illumina HiSeq2000. Base calling was performed using Illumina Pipeline v1.5.
Data analysis
Bioinformatics protocols to rapidly and efficiently process NGS MP data using a 32-bit binary indexing of the Hg19 reference genome have been previously published from our laboratory (15, 16). The algorithm maps both MP reads successively to the whole genome, selecting reads < 15Kb apart allowing up to 10 mismatches, with the lowest cumulative mismatch count sent to the output. Discordant MPs mapping >30kb apart or in different chromosomes were selected for further analysis. Algorithmic filters to determine lineage relationships were set to minimize the effects of both false positives (FP) and false negatives (FN). Namely, the lowest limit of MP associates to call an event was set at 7, where the FP rate was practically zero, and a mask of breakpoints was used to eliminate common variants and discordant mate-pairs from experimental or algorithmic errors. The combined nucleotide distance to cluster associates to an event was set to 3,000, thereby eliminating closely related but not identical breakpoints from being called as shared. Breakpoints near gaps of reference genome sequence were also eliminated. The FN rate was estimated to be less than 15%, dictated by the incompleteness of the reference genome and by regions that are difficult to map. Using a probability statistic, we estimated that the probability of relatedness between two samples is less than 0.15n when the expected number of shared breakpoints is n, and no shared events are found.
Immunohistochemistry (IHC)
Tissue sectioning and IHC staining was performed at the Pathology Research Core (Mayo Clinic, Rochester, MN) using the Leica Bond RX stainer (Leica, Buffalo, IL). Tissues were sectioned at 5 microns. Normal tonsil was used as positive control and normal tonsil without primary antibody was used as a negative control. These tonsil specimens were de-identified samples from Mayo Clinic’s frozen laboratory, reviewed and confirmed by a pathologist, and processed with the tumor specimens. IHC staining was performed on-line; tissue slides were dewaxed using Bond Dewax (Leica, Buffalo, IL). Slides for PD-L1 stain were retrieved for 20 minutes using Epitope Retrieval 2 (EDTA; Leica, Buffalo, IL). PD-L1, Rabbit Monoclonal (Clone E1L3N; Cell Signaling #13684) was diluted in Background Reducing Diluent (Dako, Carpinteria, CA) to 1:600 and incubated for 15 minutes. The detection system used was a Polymer Refine Detection System (Leica, Buffalo, IL). This system includes the hydrogen peroxidase block, post primary and polymer reagent, DAB, and Hematoxylin. Immunostaining visualization was achieved by incubating slides 10 minutes in DAB and DAB buffer (1:19 mixture) from the Bond Polymer Refine Detection System. To this point, slides were rinsed between steps with 1X Bond Wash Buffer (Leica, Buffalo, IL). Slides were counterstained for five minutes using Schmidt hematoxylin and molecular biology grade water (1:1 mixture), followed by several rinses in 1X Bond wash buffer and distilled water, this is not the hematoxylin provided with the Refine kit. Once the process was completed, slides were removed from the stainer and rinsed in tap water for five minutes. Slides were dehydrated in increasing concentrations of ethyl alcohol and cleared in 3 changes of xylene prior to permanent coverslipping in xylene-based medium.
PD-L1 Expression Scoring
PD-L1 was considered as expressed in tumor cells only if membranous or membranous and cytoplasmic staining was present. The scoring of PD-L1 in tumor cells was expressed as a percentage of stained cells in the overall section of tumor and estimated in increments of 5%. Immune cells, both intra-tumoral and peri-tumoral at the interface between tumor and lung, positive for PD-L1 were also scored. A low-power magnification area with greatest intensity of staining was identified. The percent of positive immune cells was estimated over the surface area in increments of 5%. Patients with at least moderate 5% or greater PD-L1 staining of tumor cells or immune cells were considered positive.
Statistics
Descriptive statistics were used to summarize patient demographics and results. Agreement statistics were used to assess heterogeneity of expression between paired lesions. There were three patients with three lesions whereas all of the other patients had two. These three cases were considered discrepant if all specimens did not share staining characteristics for purposes of the agreement analysis. The mate-pair classification was used for the comparisons between independent primaries and related, intrapulmonary metastases. JMP 10.0.0 (SAS Institute Cary, NC) was used for all analyses. The limits of agreement were displayed with Prism (GraphPad Software, Inc. La Jolla, CA). Circos diagrams were created using an online interface (17) and modified for clarity with Adobe Photoshop CC 2014.
Results
Patient demographics
There were 67 multifocal lung cancers obtained from 32 patients available for our study (Table 1; Supplemental Table S1). These included 57 adenocarcinomas, 8 squamous cell carcinomas, 1 large cell neuroendocrine carcinoma and 1 sarcomatoid carcinoma. Mate-pair sequencing identified that 23 patients had independent primary lung cancers and that nine patients had related cancers, or intrapulmonary metastases.
Table 1.
Patient Characteristics
| n (%) or median (IQR*) | |
|---|---|
| Gender | |
| Female | 14 (44%) |
| Male | 18 (56%) |
| Age at diagnosis (years) | 66 (56–77) |
| Tobacco use at diagnosis | |
| Current | 11 (34%) |
| Former | 16 (50%) |
| Never | 5 (16%) |
| Histologies | |
| Adenocarcinoma | 57 (85%) |
| Squamous Cell | 8 (12%) |
| Large Cell Carcinoma | 1 (1.5%) |
| Sarcomatoid Carcinoma | 1 (1.5%) |
interquartile range
The patient characteristics of those included in this study are reported above.
PD-L1 expression
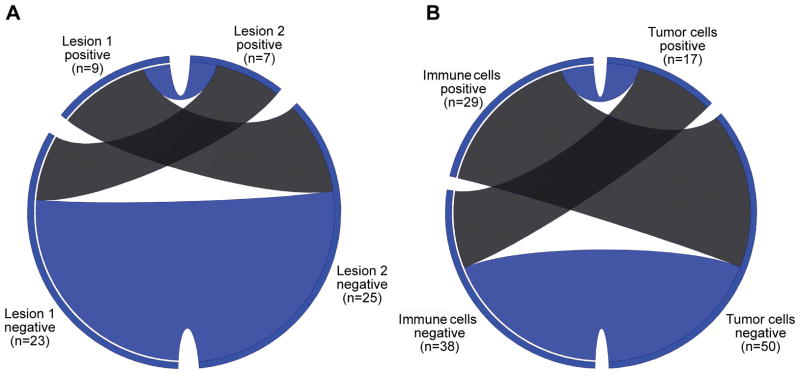
Tumor cells with 5% or greater expression of PD-L1 were observed in 17 (25% of total) of the lung cancers, while the remaining 50 lung cancers were negative for PD-L1 expression (75% of total). Amongst all 32 patients there was agreement of PD-L1 expression by the tumor cells in paired lesions of 20 patients, and disagreement in 12 patients. Two of the 20 concordant tumors were both positive, while 18 were both negative for PD-L1 expression (kappa=0.01, standard error 0.18, 95% confidence interval −0.34 to 0.35; Figure 1A; Supplemental Table S2). In comparison, there were 29 cases with immune cells that were positive for PD-L1 (43% of total) and 38 cases without expression by immune cells (57% of total). There was agreement of PD-L1 expression in the immune cells in paired lesions of 17 patients, and disagreement in 15 patients (kappa=0.11, standard error=0.18, 95% confidence interval −0.23 to 0.46). Agreement of PD-L1 expression between the tumor cells and immune cells was observed in 35 of the 67 tumors. PD-L1 expression was detected in the tumor cells and immune cells of seven patients, the other 28 cases in agreement had no PD-L1 expression. Disagreement of PD-L1 expression between the tumor cells and immune cells was observed in 32 cases where PD-L1 expression was detected in the tumor cells of 10 patients but not in the immune cells or vice versa in 22 cases (kappa=−0.02, standard error 0.11, 95% confidence interval −0.24 to 0.20; Figure 1B; Supplemental Table S3).
Figure 1. Circos diagrams of agreement in PD-L1 expression.
These circos diagrams display the agreement of PD-L1 expression by tumor cells between all paired lesions (A), and between tumor cells and immune cells for each specimen (B). There were three subjects who had a third resected lesion and these lesions were excluded from Figure 1A for simplicity. For Figure 1A, the left side of the circos diagram represents the first lesion, and the right side represents the second lesion. Similarly, for Figure 1B the left side of the circos diagram represents the immune cell expression of PD-L1 for each specimen, and the right side represents the tumor cell expression of PD-L1 for each specimen. For both circos diagrams, the positive and negative specimens are demonstrated by the labeled segments. Ribbons within the circos diagram connect paired specimens. Blue ribbons represent agreement in PD-L1 status, and the gray ribbons represent heterogeneous expression.
Heterogeneity amongst independent primary lung cancers
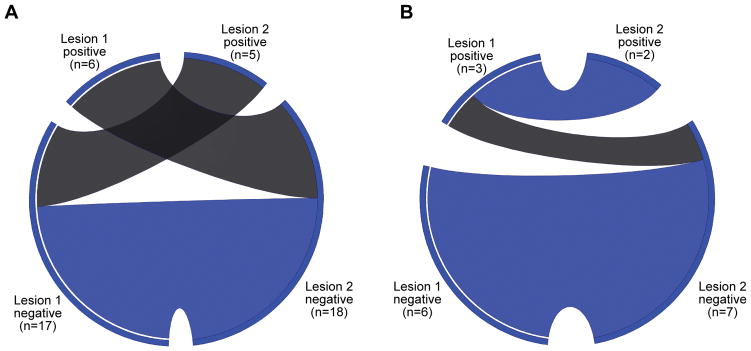
Amongst the 23 patients with independent cancers, there was agreement of PD-L1 expression by the tumor cells in paired lesions of 12 patients, and disagreement in 11 patients. None of the independent paired lesions were both positive for PD-L1 expression, whereas 12 paired lesions were both negative (kappa=−0.31, standard error 0.09, 95% confidence interval −0.49 to −0.13; Figure 2; Figure 3; Supplemental Table S4). Amongst these patients, there was agreement of PD-L1 expression in the immune cells in paired lesions of 12 patients, and disagreement in 11 patients. Four paired lesions were both positive and eight were both negative, the remainder were discrepant (kappa=0.02, standard error 0.21, 95% confidence interval −0.39 to 0.42; Supplemental Table S5).
Figure 2. Circos diagram of agreement in by type of lesion.
These circos diagrams display the agreement of PD-L1 expression by tumor cells between paired independent lesions (A), and between paired related lesions (B). There were three subjects who had a third resected lesion and these lesions were excluded from the figure. The left sides of the circos diagrams represent the first lesions, and the right sides represent the paired second lesions. The positive and negative specimens are demonstrated by the labeled segments. Ribbons within the circos diagram connect paired specimens. Blue ribbons represent agreement in PD-L1 status, and the gray ribbons represent heterogeneous expression.
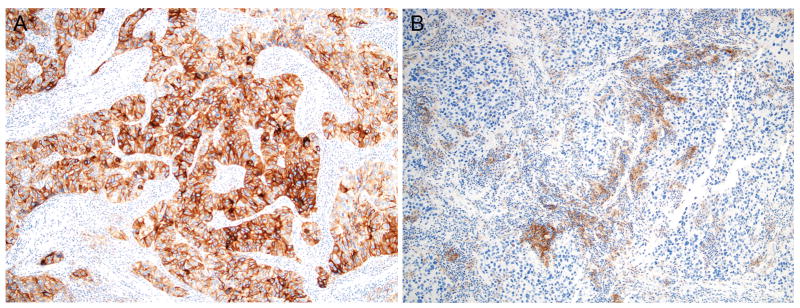
Figure 3. PD-L1 expression in independent primary lung cancers.
This case represents paired independent lung cancers from the same patient with the tumor from the right middle lobe (A) showing tumor cell expression of PD-L1 while the tumor from the left upper lobe (B) showed expression in the immune cells (Immunohistochemistry with PD-L1 antibody, 100X).
Heterogeneity amongst related, intrapulmonary metastases
Amongst the nine patients with related lung cancers, there was agreement of PD-L1 expression by the tumor cells in paired lesions of 8 patients, and disagreement in 1 patient. Two paired lesions were both positive and six were both negative (kappa=0.73, standard error 0.25, 95% confidence interval 0.24 to 1.0; Figure 2; Figure 4; Supplemental Table S6). Amongst these patients, there was agreement of PD-L1 expression by the immune cells in paired lesions of 6 patients, and disagreement in 3 patients. Three paired lesions were both positive and three were both negative (kappa=0.34, standard error 0.30, 95% confidence interval −0.25 to 0.94; Supplemental Table S7).
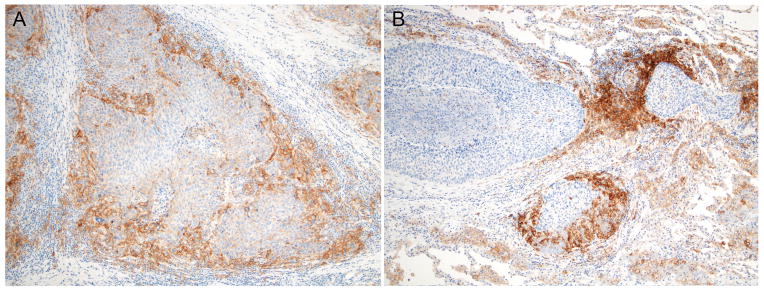
Figure 4. PD-L1 expression in related lung cancers.
This case represents paired lesions from a patient with intrapulmonary metastasis. Both tumors (A. right middle and lower lobes and B. left lower lobe) show concordant PD-L1 expression in the immune cells (Immunohistochemistry with PD-L1 antibody, 100X).
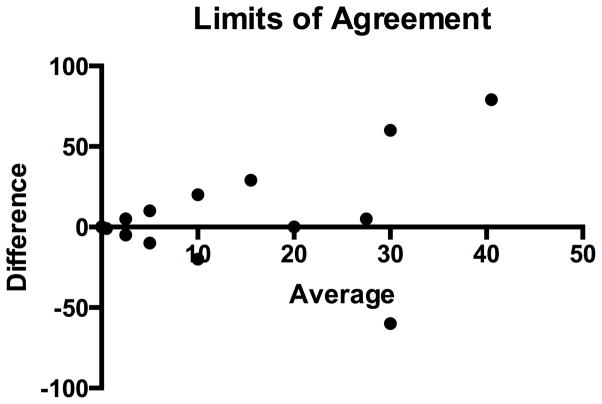
Limits of agreement of PD-L1 expression
The percent of tumor cells that expressed PD-L1 was compared between paired lesions using limits of agreement analysis (also known as a Bland-Altman analysis) (Figure 5). The degree of positivity of PD-L1 expression by tumors, based on the percent of expression, can vary widely. In other words, the degree of positivity of one lesion does not predict the degree of positivity in a paired lesion.
Figure 5. Limits of Agreement.
The differences in PD-L1 expression by tumor cells were plotted against the mean of PD-L1 expression between paired lesions. Amongst all paired lesions there was a mean difference of −3.5 (95% limits of agreement −40 to 47 as displayed with the dotted horizontal lines).
Discussion
In this study we assessed the agreement of PD-L1 expression by the tumor cells and immune cells of paired lesions from patients with surgically resected multifocal lung cancers. In order to contrast the level of PD-L1 expression between the synchronous lesions, it was important to define the lineage relationship of the associated tumors. A mate-pair next generation sequencing approach was therefore used to robustly define the lineage relationships of paired lesions from these patients as either independent primary lesions or related, metastatic lesions. The agreement of PD-L1 expression was then characterized between these groups. There was poor agreement of PD-L1 expression (based on expression by tumor cells or tumor associated immune cells) between paired independent lesions. In contrast, we observed strong agreement in PD-L1 expression by tumor cells amongst related, metastatic multifocal lung cancers. Furthermore, there was poor agreement of PD-L1 expression between tumor cells and their adjacent immune cells. This heterogeneity of PD-L1 expression by tumor cells or immune cells in either group of patients may impact diagnostic strategies that utilize tissue biopsies to determine eligibility for treatment with an immune checkpoint inhibitor. In other words, although PD-L1 expression agreement was strong between related metastatic lesions, tissue sampling of a single lesion may not reflect the PD-L1 expression of the remaining lesions. This finding may account for the suboptimal correlation between PD-L1 expression and disease response to PD-1 or PD-L1 blockade and some of the mixed responses that have been observed clinically. As it is not practical or feasible to biopsy multiple lesions to determine PD-L1 expression, our results emphasize the need for novel strategies to detect PD-L1 expression such as peripheral blood biomarkers and imaging (18).
Even though there are preclinical data on the dynamics PD-L1 expression (19, 20) and limited clinical data on the heterogeneity of PD-L1 expression (21), very few have evaluated the agreement of PD-L1 expression between independent primary and metastatic lesions. Another study looked at agreement of PD-L1 expression between paired primary lesions and lymph nodes in patients with squamous cell carcinoma of the lung (22). They identified agreement in PD-L1 expression in 70% (52/74) of these paired lesions (60% of paired lesions were both negative, and 10% of paired lesions were both positive). Similarly, amongst all of our patients, we identified agreement of PD-L1 expression between paired specimens in 63% (20/32) of our patients; however, we observed even greater agreement in related, metastatic lesions (89%, 8/9). It remains uncertain if the tumor microenvironment (i.e. lung compared to lymph node) and the local expression of interferon-γ or other factors plays a role in PD-L1 expression discrepancies, but our two studies suggest that a portion of paired lesions from the same patient will have discrepant PD-L1 expression. Similarly, one group identified that half (23 of 46 patients) of the patients with longitudinally obtained primary and metastatic melanoma specimens had discordant PD-L1 expression patterns (23). PD-L1 expression and the tumor microenvironment were also reported to be heterogeneous between cerebral and extra-cerebral metastases in patients with melanoma (24). Since our specimens were resected from the lung, we are not able to comment on the heterogeneity of expression of PD-L1 in lung cancer between tissue sources. Overall, the heterogeneity of PD-L1 expression between paired lesions suggests that patients with “negative” biopsies may have separate lesions that express PD-L1 and this may explain why some tumor responses to PD-1 or PD-L1 inhibition have been observed in this patient population.
Our study is limited by a few factors. First, we have a relatively small sample size; however, these specimens were robustly characterized genetically for lineage relationships and represent one of the largest and unique datasets of multiple, paired, fully resected and frozen lesions from patients with multifocal lung cancer. Second, we used the Cell Signalling antibody for detection of PD-L1. Detection of PD-L1 is not consistent amongst all of the available antibodies (25, 26), so our results must be interpreted within the context of the detection of PD-L1 with this antibody. Regardless, our rate of detection of PD-L1 expression by tumor cells is similar to that of another series (26). Third, we used a threshold of 5% for the determination of positivity. We are concerned that a lower threshold may not distinguish true staining from background staining; however, others have used lower thresholds. Fourth, these patients did not receive immunotherapy (namely PD-1 or PD-L1 inhibitors), as these agents were not available or were not part of the standard of care for their stages disease when their specimens were obtained. Fifth, we were not able to robustly identify the subtypes of tumor-associated immune cells that expressed PD-L1; novel techniques such as multiplex immunofluorescence may help overcome the limitations of serial sectioning and determination of co-expression. Regardless, multifocal lung cancer provides a model for the study of heterogeneity of expression that has important diagnostic implications.
In conclusion, we have observed that the expression of PD-L1 is heterogeneous amongst paired multifocal lung cancers, but there are high levels of agreement amongst related, metastatic lung cancers. Analysis of a single lesion may not appropriately identify PD-L1 expression.
Supplementary Material
Statement of Translational Relevance.
The expression of programmed cell death-ligand 1 (PD-L1) is being used to select which patients receive inhibitors of programmed cell death 1 (PD-1) or PD-L1; however, there are reports of patients without PD-L1 expression who have responded to PD-1 inhibitors. While preclinical work has demonstrated that PD-L1 expression is dynamic, very little is known about the PD-L1 expression between different lung lesions in multifocal lung cancer. We compared the expression of PD-L1 between paired fully resected lung cancer lesions from patients with multifocal lung cancer. Overall, the expression of PD-L1 was heterogeneous amongst paired independent primary lung cancers, but there were high levels of agreement amongst related, intra-pulmonary metastases. Our data highlight that a single biopsy in patients with multifocal lung cancer may not accurately capture PD-L1 expression status and emphasize the need for novel methods of patient selection for immunotherapy.
Acknowledgments
This work was supported in part by Oncospire, Mayo Clinic’s Center for Individualized Medicine, the National Cancer Institutes of Health [K12CA090628 to A.S.M. and K23CA159391 to TP]. We thank the Mayo Clinic Biobank for providing normal genomic samples used in establishing our germline rearrangement mask; the efforts of Kathy Laivell and Eric Edell, MD, in selection and acquisition of clinical samples from the Mayo Clinic Lung Specimen Registry; and Robert Sikkink and Bruce Eckloff of the Mayo Medical Genome Facility for next-generation sequencing.
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. The New England journal of medicine. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. The New England journal of medicine. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai J, Tsuchiya T, Oikawa M, et al. Clinical and molecular analysis of synchronous double lung cancers. Lung cancer. 2012;77:281–287. doi: 10.1016/j.lungcan.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Gazdar AF, Minna JD. Multifocal lung cancers--clonality vs field cancerization and does it matter? Journal of the National Cancer Institute. 2009;101:541–543. doi: 10.1093/jnci/djp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang YL, Wu CT, Lee YC. Surgical treatment of synchronous multiple primary lung cancers: experience of 92 patients. The Journal of thoracic and cardiovascular surgery. 2007;134:630–637. doi: 10.1016/j.jtcvs.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Deschamps C, Pairolero PC, Trastek VF, Payne WS. Multiple primary lung cancers. Results of surgical treatment The Journal of thoracic and cardiovascular surgery. 1990;99:769–777. discussion 777–768. [PubMed] [Google Scholar]
- 8.Kim TJ, Goo JM, Lee KW, et al. Clinical, pathological and thin-section CT features of persistent multiple ground-glass opacity nodules: comparison with solitary ground-glass opacity nodule. Lung cancer. 2009;64:171–178. doi: 10.1016/j.lungcan.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Nakata M, Sawada S, Yamashita M, et al. Surgical treatments for multiple primary adenocarcinoma of the lung. The Annals of thoracic surgery. 2004;78:1194–1199. doi: 10.1016/j.athoracsur.2004.03.102. [DOI] [PubMed] [Google Scholar]
- 10.Pommier RF, Vetto JT, Lee JT, Johnston KM. Synchronous non-small cell lung cancers. American journal of surgery. 1996;171:521–524. doi: 10.1016/S0002-9610(96)00019-0. [DOI] [PubMed] [Google Scholar]
- 11.Verhagen AF, Tavilla G, van de Wal HJ, et al. Multiple primary lung cancers. The Thoracic and cardiovascular surgeon. 1994;42:40–44. doi: 10.1055/s-2007-1016453. [DOI] [PubMed] [Google Scholar]
- 12.Murphy SJ, Aubry MC, Harris FR, et al. Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:4050–4058. doi: 10.1200/JCO.2014.56.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard N, Deshpande C, Lau C, et al. Comprehensive histologic assessment helps to differentiate multiple lung primary nonsmall cell carcinomas from metastases. The American journal of surgical pathology. 2009;33:1752–1764. doi: 10.1097/PAS.0b013e3181b8cf03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy SJ, Cheville JC, Zarei S, et al. Mate pair sequencing of whole-genome-amplified DNA following laser capture microdissection of prostate cancer. DNA research : an international journal for rapid publication of reports on genes and genomes. 2012;19:395–406. doi: 10.1093/dnares/dss021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drucker TM, Johnson SH, Murphy SJ, et al. BIMA V3: an aligner customized for mate pair library sequencing. Bioinformatics. 2014;30:1627–1629. doi: 10.1093/bioinformatics/btu078. [DOI] [PubMed] [Google Scholar]
- 16.Vasmatzis G, Johnson SH, Knudson RA, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. 2012;120:2280–2289. doi: 10.1182/blood-2012-03-419937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heskamp S, Hobo W, Molkenboer-Kuenen JD, et al. Noninvasive Imaging of Tumor PD-L1 Expression Using Radiolabeled Anti-PD-L1 Antibodies. Cancer research. 2015;75:2928–2936. doi: 10.1158/0008-5472.CAN-14-3477. [DOI] [PubMed] [Google Scholar]
- 19.Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nature medicine. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 20.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nature medicine. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 21.Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim MY, Koh J, Kim S, et al. Clinicopathological analysis of PD-L1 and PD-L2 expression in pulmonary squamous cell carcinoma: Comparison with tumor-infiltrating T cells and the status of oncogenic drivers. Lung cancer. 2015;88:24–33. doi: 10.1016/j.lungcan.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Madore J, Vilain RE, Menzies AM, et al. PD-L1 expression in melanoma shows marked heterogeneity within and between patients: implications for anti-PD-1/PD-L1 clinical trials. Pigment cell & melanoma research. 2015;28:245–253. doi: 10.1111/pcmr.12340. [DOI] [PubMed] [Google Scholar]
- 24.Kluger HM, Zito CR, Barr ML, et al. Characterization of PD-L1 Expression and Associated T-cell Infiltrates in Metastatic Melanoma Samples from Variable Anatomic Sites. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:3052–3060. doi: 10.1158/1078-0432.CCR-14-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leventakos K, Mansfield AS. Reflections on immune checkpoint inhibition in non-small cell lung cancer. Translational lung cancer research. 2014;3:411–413. doi: 10.3978/j.issn.2218-6751.2014.09.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velcheti V, Schalper KA, Carvajal DE, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Laboratory investigation; a journal of technical methods and pathology. 2014;94:107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.