We have recently shown that susceptibility to childhood B-cell acute lymphoblastic leukaemia (B-ALL) is influenced by the presence of HLA-C encoded ligands C1 and C2 for killer cell immunoglobulin-like receptors (KIR), (Babor et al, 2014). B-ALL patients exhibited an increased frequency of the C2 ligand and, moreover, C2 was associated with increased risk of late relapse. The study suggested that the expression of HLA class I-encoded KIR ligands on tumour cells influences disease susceptibility and Natural Killer (NK) cell-mediated control of B-ALL. In order to extend our knowledge about expression of the major inhibitory NK cell receptor ligands in childhood B-ALL, Bw4, HLA-C-encoded C1 and C2 epitopes, and HLA-E were assessed at the surface and mRNA level in leukaemic cells. To this end, we employed antibodies for specific detection of Bw4 (MUS4H4) and Bw6 (OUW4F11) as previously described (Verheyden et al, 2009), for HLA-C/E (DT9), (Braud et al, 1998, Thomas et al, 2009) and HLA-E (3D12, BioLegend, San Diego, CA, USA) in peripheral blood samples in 31 children (age 0.3–19.3 years, mean 7.2) with newly diagnosed B-ALL (common ALL: n=18; preB-ALL: n= 11; proB-ALL: n=2). In order to accommodate for possible differences in cell size between leukaemic blasts and unaffected B cells, mean fluorescence intensity was normalized to mean forward scatter levels of the respective cell population. All HLA class I-specific antibodies were unconjugated and subsequently stained with secondary fluorescein isothiocyanate-coupled anti-IgG antibodies to avoid a systematic bias of directly conjugated antibodies due to differences in coupling efficiencies and flow cytometric compensation.
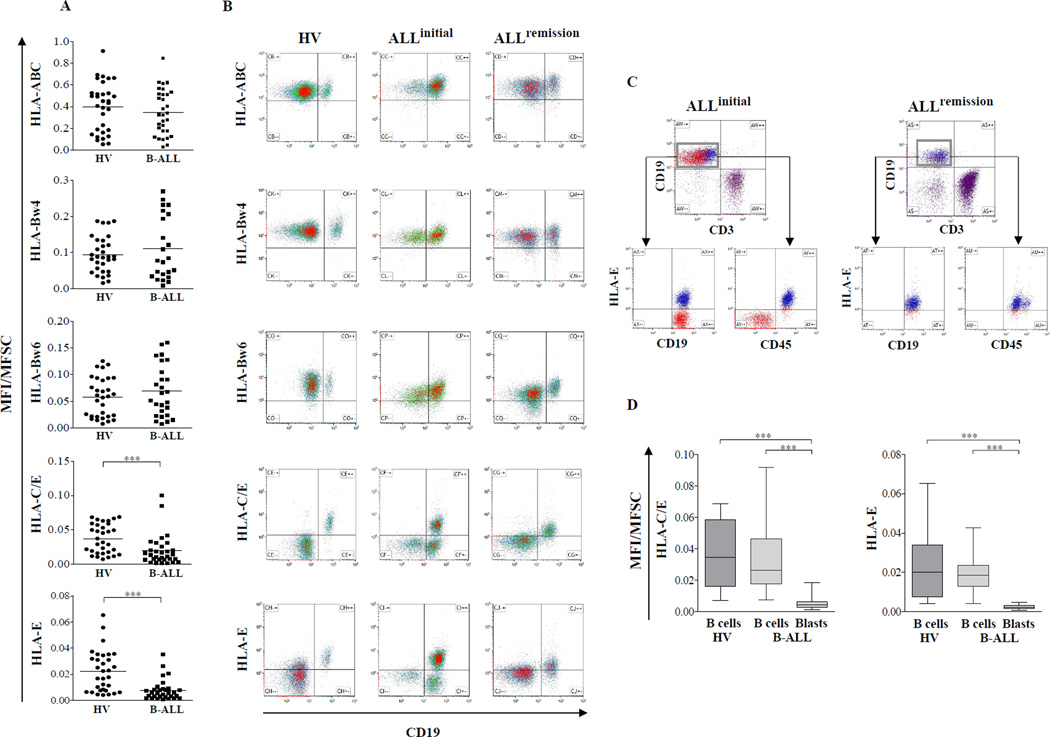
Overall levels of HLA class I surface expression, which were measured with a panHLA class I antibody (W6/32, BioLegend), were comparable and within the same range in CD19+ B cells of patients and healthy controls (Fig 1A). Similarly, we could not detect B-ALL-specific changes in overall expression levels of HLA-B allotypes carrying the Bw4 epitope, which constitutes the ligand for the inhibitory KIR3DL1 receptor, nor the Bw6 epitope, which is not recognized by inhibitory KIR. In all cases, the flow cytometry results were consistent with the HLA class I typing status, confirming the specificity of the Bw4 and Bw6 reagents (data not shown). Whereas panHLA class I and Bw4/Bw6 expression remained unchanged, strong and consistent downregulation of HLA-C and HLA-E surface expression was observed in patients (Fig 1A). At initial diagnosis, leukaemic patients but not healthy controls exhibited B cell populations with very low HLA-C and -E expression levels (Fig 1B) that could be consistently allocated to the CD19+CD45− leukaemic subset (Fig 1C). The differences in HLA-C and -E expression levels between non-leukaemic and leukaemic B cells were highly significant (Fig 1D). Upon complete remission, B cell populations with low HLA-C and HLA-E levels were no longer detectable, consistent with the disappearance of the leukaemic blast population (Fig 1B, C). In contrast, Bw4- and Bw6-bearing allotypes led to homogenous staining in leukaemic and control samples (Fig 1B).
Figure 1. Selective downregulation of HLA-C and HLA-E in paediatric B-ALL patients.
Peripheral blood mononuclear cells (PBMC) from 31 newly diagnosed B-ALL patients and 32 healthy volunteers (HV) were stained with HLA class I-specific human monoclonal antibodies (mAbs) for HLA-ABC, HLA-Bw4, HLA-Bw6, HLA-C/E and HLA-E. Measurements of all HLA class I-specific antibodies were made with the same instrument settings. The mean fluorescence intensity (MFI) was normalized to mean forward scatter (MFI/MFSC) to adjust for possible differences in cell size. (A) Plots representing the surface expression of the respective HLA class I molecules on B cells (CD19+ subset) of healthy volunteers and leukaemic patients. Each dot represents one individual and horizontal bars represent mean values. (B) Flow cytometric data are shown for a representative leukaemic patient (middle panel) at initial diagnosis and complete remission (right panel) following chemotherapy in comparison to a healthy volunteer (left panel). (C) Representative flow cytometric analysis illustrating the presence of two CD19+ subsets with either high (CD45+ non-leukaemic B cells) or low (CD45− leukaemic blasts) HLA-E expression at initial diagnosis (upper panel) and the subsequent disappearance of the leukaemic HLA-E-negative B cell subset following chemotherapy and complete remission (lower panel). (D) Box plots representing surface expression of HLA-C and HLA-E for non-leukaemic and leukaemic B cells in comparison to B cells from healthy volunteers. Horizontal bars represent median values. Statistical significance was determined by Mann-Whitney U test (***= p<0.001).
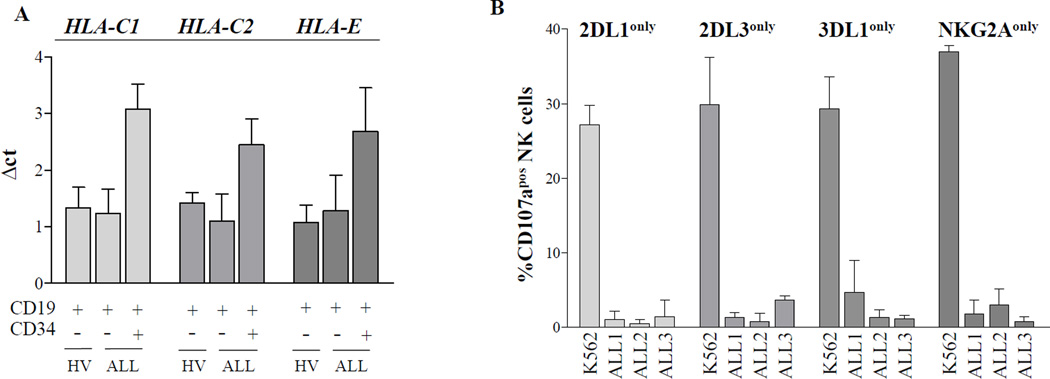
To determine whether downregulation of HLA-C and –E expression on the cell surface was due to repression of transcription, B cells were separated into leukaemic and non-leukaemic subsets by flow cytometric cell sorting and subsequently subjected to reverse transcription polymerase chain reaction analysis. Indeed, the mRNA levels for C1 and C2-bearing HLA-C alleles as well as for HLA-E were clearly lower in CD34+ leukaemic blasts than in the residual non-leukaemic B cells (Fig 2A). Nonetheless, except one case where no HLA-E message could be detected (data not shown), the reduction of mRNA levels was moderate (2- to 4-fold compared to normal B cells) pointing to additional post-transcriptional mechanisms that might explain the many cases exhibiting an almost complete lack of surface expression. Notably, the distribution of the well-characterized HLA-E dimorphism at position 107 (Geraghty et al, 1992) was not different from the control cohort (data not shown).
Figure 2. Reduced HLA-C and HLA-E mRNA expression and lack of NK cell activation by leukaemic cells.
(A) Semiquantitative reverse transcription (RT) PCR analysis of HLA-C1, HLA-C2 and HLA-E mRNA expression. Leukaemic blasts (CD34+CD19+) and B cells (CD34− CD19+) from acute lymphoblastic leukaemia (ALL) patients as well as B cells from healthy volunteers were separated by flow cytometric cell sorting. mRNA was isolated using RNeasy Mini Kit (Qiagen, Hilden, Germany) followed by cDNA synthesis (Moloney murine leukaemia virus, Promega, Madison, WI, USA). Transcriptional levels were analysed by SYBR green-based real-time PCR (StepOnePlus PCR-System, Applied Biosystems, Foster City, CA, USA) and normalized to expression levels of ACTB. Bars represent the mean expression level of three healthy (far left bar in each grouping) and three leukaemic individuals (middle and right bars in each grouping), each heterozygous for HLA-C1 and HLA–C2, respectively; error bars represent standard error of the mean (SEM). Primers for HLA-C-encoded ligands were from (Babor et al, 2014). Primers for HLA-E were as follows: forward: 5’-GCACATGGCACGTGTATCTC-3’, reverse: 5’-CCTTCTGGAGAGGAGCAGAG-3’. (B) CD107a mobilization assay of healthy NK cells stimulated overnight with interleukin 2 (1000 u/ml) against paediatric B-ALL blasts (frequency >80% of peripheral blood mononuclear cells) indicated as ALL1-3 or against K562 (erythroleukaemic HLA-class I deficient cell line) as a positive control (effector/target ratio of 10:1). CD107a monoclonal antibody (mAb) (H4A3, BioLegend) was added prior to incubation. Subsequent flow cytometric staining included mAbs for CD56 (HCD56, Biolegend), CD3 (UCHT1, BioLegend), CD159a (NKG2A, Z199, Beckman Coulter, Brea, CA, USA), CD158a/h (KIR2DL1/S1, 11PB6, Miltenyi Biotec, Bergisch Gladbach, Germany), CD158b1/b2/j (KIR2DL2/L3/S2, GL183, Beckman Coulter) and CD158e1 (KIR3DL1, DX9, BioLegend). CD107a expression was determined on CD56dimCD3− NK cells expressing the indicated inhibitory receptor. Bars represent the mean expression of three healthy individuals; error bars represent standard error of the mean (SEM).
This is the first comprehensive analysis of HLA-C and HLA–E-encoded NK cell ligands in childhood ALL. It is important to mention that the HLA-C-specific reagent (DT9) cross-reacts with HLA-E, which might partially account for the decrease in DT9 signals. Moreover, the HLA-E-specific antibody (3D12) weakly recognizes certain HLA-B and HLA-C alleles (Corrah et al, 2011). However, these cross-reactions would not explain our observation that both reagents gave vastly diminished signals on leukaemic blasts whereas the same leukaemic cells showed normal levels of HLA-A and -B expression. Our observations are in line with a previous study showing downregulation of HLA-C in a small ALL cohort, employing a panel of antibodies for single HLA-C allotypes (Verheyden et al, 2009). Furthermore, our data show that HLA-E is expressed at the mRNA but not cell surface level on leukaemic blasts, which is in contrast to a previous study, which reported complete absence of HLA-E mRNA level (Majumder et al, 2006).
The downregulation of HLA-C and HLA-E as reported here should theoretically lead to ‘missing-self’ recognition of leukaemic cells by NK cells expressing the respective cognate inhibitory receptor. However, CD107 mobilization of short-term stimulated NK cells from healthy donors remained low, even on NK cell subsets expressing a single inhibitory KIR for C2 (KIR2DL1), C1 (KIR2DL1), Bw4 (KIR3DL1) or HLA-E (NKG2A) (Fig 2B). The observation that B-ALL blasts are not efficiently recognized by NK cells despite their consistent downregulation of NK cell receptor ligands might be explained by the lack of appropriate ligands for stimulatory receptors, such as NCR, NKG2D, and DNAM1 (Pende et al, 2005). However, the driving force behind the downregulation of HLA-C and HLA-E is yet unknown. Possible explanations involve the escape from T cells that recognize peptides presented in the context of HLA-C or HLA-E. A thought-provoking but speculative explanation would be that the presence of HLA-C and HLA–E-negative leukaemic cells in haematopoietic stem cell niches, such as bone marrow, might drive NK cells into a hyporesponsive state. In this regard, it was shown that TAP-deficient patients that express almost no HLA class I (Sleiman et al, 2014) have NK cells that are not licensed and exhibit decreased cytotoxicity and cytokine production. Further experiments analysing the functional state of patient NK cells will hopefully help to clarify the underlying mechanism behind the specific downregulation of NK cell receptor ligands in childhood B-ALL.
Acknowledgments
This work was supported by the Deutsche Krebshilfe e.V. (M.U. and A.B., project 110351), the Forschungskommission of the Medical Faculty of the Heinrich-Heine-Universität Duesseldorf (F.B.). F.B. received a Habilitation stipend from the TRANSAID - Stiftung für krebskranke Kinder. This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the National Institutes of Health, Frederick National Laboratory, Center for Cancer Research.
Footnotes
Author contributions
F.B. and M.U. contributed equally to this work. S.B.R. and A.R.M. designed the project, performed the experiments and wrote the manuscript; J.E. and J.C.F performed experiments and reviewed the manuscript; A.M., F.H.C., and M.C. contributed essential reagents and reviewed the manuscript, A.B. contributed clinical samples and reviewed the manuscript; F.B. and M.U. designed the project and wrote the paper.
Conflict of interest
The authors state that they have no conflict of interest.
References
- Babor F, Manser AR, Fischer JC, Scherenschlich N, Enczmann J, Chazara O, Moffett A, Borkhardt A, Meisel R, Uhrberg M. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood. 2014;124:2248–2251. doi: 10.1182/blood-2014-05-572065. [DOI] [PubMed] [Google Scholar]
- Braud VM, Allan DS, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1998;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- Corrah TW, Goonetilleke N, Kopycinski J, Deeks SG, Cohen MS, Borrow P, McMichael A, Brackenridge S. Reappraisal of the relationship between the HIV-1-protective single-nucleotide polymorphism 35 kilobases upstream of the HLA-C gene and surface HLA-C expression. J Virol. 2011;85:3367–3374. doi: 10.1128/JVI.02276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty DE, Stockschleader M, Ishitani A, Hansen JA. Polymorphism at the HLA-E locus predates most HLA-A and -B polymorphism. Hum Immunol. 1992;33:174–184. doi: 10.1016/0198-8859(92)90069-y. [DOI] [PubMed] [Google Scholar]
- Majumder D, Bandyopadhyay D, Chandra S, Mukherjee N, Banerjee S. Lack of HLA-E surface expression is due to deficiency of HLA-E transcripts in the malignant hematopoietic cells of leukemic patients. Leuk Res. 2006;30:242–245. doi: 10.1016/j.leukres.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, Falco M, Lanino E, Pierri I, Zambello R, Bacigalupo A, Mingari MC, Moretta A, Moretta L. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112) Blood. 2005;105:2066–2073. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- Sleiman M, Brons NH, Kaoma T, Dogu F, Villa-Forte A, Lenoble P, Hentges F, Kotsch K, Gadola SD, Vilches C, Zimmer J. NK cell killer Ig-like receptor repertoire acquisition and maturation are strongly modulated by HLA class I molecules. J Immunol. 2014;192:2602–2610. doi: 10.4049/jimmunol.1302843. [DOI] [PubMed] [Google Scholar]
- Thomas R, Apps R, Qi Y, Gao X, Male V, O'HUigin C, O'Connor G, Ge D, Fellay J, Martin JN, Margolick J, Goedert JJ, Buchbinder S, Kirk GD, Martin MP, Telenti A, Deeks SG, Walker BD, Goldstein D, McVicar DW, Moffett A, Carrington M. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyden S, Ferrone S, Mulder A, Claas FH, Schots R, De Moerloose B, Benoit Y, Demanet C. Role of the inhibitory KIR ligand HLA-Bw4 and HLA-C expression levels in the recognition of leukemic cells by Natural Killer cells. Cancer Immunol Immunother. 2009;58:855–865. doi: 10.1007/s00262-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]