Abstract
We introduce a generative probabilistic model for segmentation of brain lesions in multi-dimensional images that generalizes the EM segmenter, a common approach for modelling brain images using Gaussian mixtures and a probabilistic tissue atlas that employs expectation-maximization (EM) to estimate the label map for a new image. Our model augments the probabilistic atlas of the healthy tissues with a latent atlas of the lesion. We derive an estimation algorithm with closed-form EM update equations. The method extracts a latent atlas prior distribution and the lesion posterior distributions jointly from the image data. It delineates lesion areas individually in each channel, allowing for differences in lesion appearance across modalities, an important feature of many brain tumor imaging sequences. We also propose discriminative model extensions to map the output of the generative model to arbitrary labels with semantic and biological meaning, such as “tumor core” or “fluid-filled structure”, but without a one-to-one correspondence to the hypo-or hyper-intense lesion areas identified by the generative model.
We test the approach in two image sets: the publicly available BRATS set of glioma patient scans, and multimodal brain images of patients with acute and subacute ischemic stroke. We find the generative model that has been designed for tumor lesions to generalize well to stroke images, and the generative-discriminative model to be one of the top ranking methods in the BRATS evaluation.
I. Introduction
Gliomas are the most frequent primary brain tumors. They originate from glial cells and grow by infiltrating the surrounding tissue. The more aggressive form of this disease is referred to as “high-grade” glioma. The tumor grows fast and patients often have survival times of two years or less, calling for immediate treatment after diagnosis. The slower growing “low-grade” disease comes with a life expectancy of five years or more, allowing the aggressive treatment to be delayed. Extensive neuroimaging protocols are used before and after treatment, mapping different tissue contrasts to evaluate the progression of the disease or the success of a chosen treatment strategy. As evaluations are often repeated every few months, large longitudinal datasets with multiple modalities are generated for these patients even in routine clinical practice. In spite of the need for accurate information to guide decision making process for an treatment, these image series are primarily evaluated using qualitative criteria – indicating, for example, the presence of characteristic hyperintense intensity changes in contrast-enhanced T1 MRI – or relying on quantitative measures that are as basic as calculating the largest tumor diameter that can be recorded in a set of axial images.
While an automated and reproducible quantification of tumor structures in multimodal 3D and 4D volumes is highly desirable, it remains difficult. Glioma is an infiltratively growing tumor with diffuse boundaries and lesion areas are only defined through intensity changes relative to surrounding normal tissues. As a consequence, the outlines of tumor structures cannot be easily delineated – even manual segmentations by expert raters show a significant variability [1] – and common MR intensity normalization strategies fail in the presence of extended lesions. Tumor structures show a significant amount of variation in size, shape, and localization, precluding the use of related mathematical priors. Moreover, the mass effect induced by the growing lesion may lead to displacements of the normal brain tissues, as well as a resection cavity that is present after treatment, limits the reliability of prior knowledge available for the healthy parts of the brain. Finally, a large variety of imaging modalities can be used for mapping tumor-related tissue changes, providing different types of biological information, such as differences in tissue water (T2-MRI, FLARI-MRI), enhancement of contrast agents (post-Gadolinium T1-MRI), water diffusion (DTI), blood perfusion (ASL-, DSC-, DCE-MRI), or relative concentrations of selected metabolites (MRSI). A segmentation algorithm must adjust to any of these, without having to collect large training sets, a common limitation for many data-driven learning methods.
Related Prior Work
Brain tumor segmentation has been the focus of recent research, most of which is dealing with glioma [2], [3]. Few methods have been developed for less frequent and less aggressive tumors [4], [5], [6], [7]. Tumor segmentation methods often borrow ideas from other brain tissue and other brain lesion segmentation methods that have achieved a considerable accuracy [8]. Brain lesions resulting from traumatic brain injuries [9], [10] and stroke [11], [12] are similar to glioma lesions in terms of size and multimodal intensity patterns, but have attracted little attention so far. Most automated algorithms for brain lesion segmentation rely on either generative or discriminative probabilistic models at the core of their processing pipeline. Many encode prior knowledge about spatial regularity and tumor structures, and some offer longitudinal extensions for 4D image volumes to exploit longitudinal image sets that are becoming increasingly available [13], [14].
Generative probabilistic models of spatial tissue distribution and appearance have enjoyed popularity for tissue classification as they exhibit good generalization performance [15], [16], [17]. Encoding spatial prior knowledge for a lesion, however, is challenging. Tumors may be modeled as outliers relative to the expected shape [18], [19] or to the image signal of healthy tissues [16], [20]. In [16], for example, a criterion for detecting outliers is used to generate a tumor prior in a subsequent EM segmentation that treats the tumor as an additional tissue class. Alternatively, the spatial prior for the tumor can be derived from the appearance of tumor-specific markers, such as Gadolinium enhancements [21], [22], or from using tumor growth models to infer the most likely localization of tumor structures for a given set of patient images [23]. All these segmentation methods rely on registration to align images and the spatial prior. As a result, joint registration and tumor segmentation [17], [24] and joint registration and estimations of tumor displacement [25] have been investigated, as well as the direct evaluation of the deformation field for the purpose of identifying the tumor region [7], [26].
Discriminative probabilistic models directly learn the differences between the appearance of the lesion and other tissues from the data. Although they require substantial amounts of training data to be robust to artifacts and variations in intensity and shape, they have been applied successfully to tumor segmentation tasks [27], [28], [29], [30], [31]. Discriminative approaches proposed for tumor segmentation typically employ dense, voxel-wise features from anatomical maps [32] or image intensities, such as local intensity differences [33], [34] or intensity profiles, that are used as input to inference algorithms such as support vector machines [35], decision trees ensembles [32], [36], [37], or deep learning approaches [38], [39]. All methods require the imaging protocol to be exactly the same in the training set and in the novel images to be segmented. Since local intensity variation that is characteristic of MRI is not estimated during the segmentation process, as in most generative mixture models, calibration of the image intensities becomes necessary which is already a difficult task in the absence of lesions [40], [41], [42].
Advantageous properties of generative and discriminative probabilistic models have been combined for a number of applications in medical imaging: generative approaches can be used for model-driven dimensional reduction to form a low-dimensional basis for a subsequent discriminative method, for example, in whole brain classification of Alzheimer’s patients [43]. Vice versa, a discriminative model may serve as a filter to constrain the search space for employing complex generative models in a subsequent step, for example, when fitting biophysical metabolic models to MRSI signals [44], or when fusing evidence across different anatomical regions in the analysis of contrast-enhancing structures [45]. Other approaches improve the output of a discriminative classification of brain scans by adding prior knowledge on the location of subcortical structures [46] or the skull shape [47] through generative models. The latter approach for skull stripping showed superior robustness in particular on images of glioma patients [48]. To the best of our knowledge no generative-discriminative model has been used for tumor analysis so far, although the advantages of employing a secondary discriminative classifier on the probabilistic output of a first level discriminative classifier, which considers prior knowledge on expected anatomical structures of the brain, has been demonstrated in [32].
Spatial regularity and spatial arrangement of the 3D tumor sub-structure is used in most generative and discriminative segmentation techniques, often in a postprocessing step and with extensions along the temporal dimension for longitudinal tasks: Local regularity of tissue labels can be encoded via boundary modeling within generative [16], [49] and discriminative methods [27], [28], [50], [49], or by using Markov random field priors [30], [31], [51]. Conditional random fields help to impose structures on the adjacency of specific labels and, hence, impose constraints on the wider spatial context of a pixel [29], [35]. 4D extensions enforce spatial contiguity along the time dimension either in an undirected fashion [52], or in a directed one when imposing monotonic growth constraints on the segmented tumor lesion acting as a non-parametric growth model [13], [53], [14]. While all these segmentation models act locally, more or less at the pixel level, other approaches consider prior knowledge about the global location of tumor structures. They learn, for example, the relative spatial arrangement of tumor structures such as tumor core, edema, or enhancing active components, through hierarchical models of super-voxel clusters [54], [34], or by relating image patterns with phenomenological tumor growth models that are adapted to the patient [25].
Contributions
In this paper we address three different aspects of multi-modal brain lesion segmentation, extending preliminary work we presented earlier in [55] [56], [57]:
We propose a new generative probabilistic model for channel-specific tumor segmentation in multi-dimensional images. The model shares information about the spatial location of the lesion among channels while making full use of the highly specific multimodal, i.e., multivariate, signal of the healthy tissue classes for segmenting normal tissues in the brain. In addition to the tissue type, the model includes a latent variable for each voxel encoding the probability of observing a tumor at that voxel, similar to [49], [50]. The probabilistic model formalizes qualitative biological knowledge about hyper- and hypo-intensities of lesion structures in different channels. Our approach extends the general EM segmentation algorithm [58], [59] using probabilistic tissue atlases [60], [15], [61] for situations when specific spatial structures cannot be described sufficiently through population priors.
We illustrate the excellent generalization performance of the generative segmentation algorithm by applying it to MR images of patient with ischemic stroke, which – to the best of our knowledge – is one of the first automated segmentation algorithms for this major neurological disease.
We extend the generative model to a joint generative-discriminative method that compensates for some of the shortcomings of both the generative and the discriminative modeling approach. This strategy enables us to predict clinically meaningful tumor tissue labels and not just the channel-specific hyper- and hypo-intensities returned by the generative model. The discriminative classifier uses the output of the generative model, which improves its robustness against intensity variation and changes in imaging sequences. This generative-discriminative model defines the state-of-the-art on the public BRATS benchmark data set [1].
In the following we introduce the probabilistic model (Section II), derive the segmentation algorithm and additional biological constraints, and we describe the discriminative model extensions (Section III). We evaluate the properties and performance of the generative and the generative-discriminative methods on a public glioma dataset (Sections IV and V, respectively), including an experiment on the transfer of the generative model to images from stroke patients. We conclude with a discussion of the results and of future research directions (Section VI).
II. A Generative Brain Lesion Segmentation Model
Generative models consider prior information about the structure of the observed data and exploit such information to estimate latent structure from new data. The EM segmenter, for example, models the image of a healthy brain through three tissue classes [60], [15], [61]. It encodes their approximate spatial distribution through a population atlas generated by aligning a larger set of reference scans, segmenting them manually, and averaging the frequency of each tissue class in a given voxel within the chosen reference frame. Moreover, it assumes that all voxels of a tissue class have about the same image intensity which is modeled through a Gaussian distribution. This segmentation method, whose parameters can be estimated very efficiently through the expectation maximization (EM) procedure, treats image intensities as nuisance parameters which makes it robust in the presence of the characteristic variability of the intensity distributions of MR images. Moreover, since the method formalizes the image content explicitly through the probabilistic model, it can be combined with other parametric transformations, for example, for registration [62] or bias field correction [15], and account for the related changes in the observed data. Generative models with tissue atlases used as spatial priors are at the heart of most advanced image segmentation models in neuroimaging [63], [64].
Population atlases cannot be generated for tumors as their location and extensions vary significantly across patients. Still, the tumor location is similar in different MR images of the same patient and a patient-specific atlas of the lesion class could be generated. Segmentation and atlas building can be performed simultaneously, in a joint estimation procedure [50]. Here, the key idea is to model the lesions through a separate latent atlas class. Combined with the standard population atlas of the normal tissues and the standard EM segmentation framework, this extends the EM segmenter to multimodal or longitudinal images of patients with a brain lesion. The generative model is illustrated in Fig. 1.
Fig. 1.
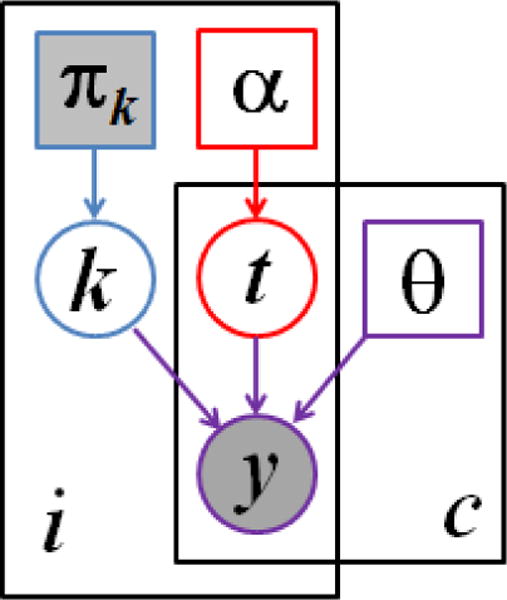
Graphical model for the proposed segmentation approach. Voxels are indexed with i, channels are indexed by c. The known prior πk determines the label k of the normal healthy tissue. The latent atlas α determines the channel-specific presence of tumor t. Normal tissue label k, tumor labels t, and intensity distribution parameters θ jointly determine the multimodal image observations y. Observed (known) quantities are shaded. The segmentation algorithms aims to estimate , along with the segmentation of healthy tissue p(ki|y).
A. The Probabilistic Generative Model
Normal healthy tissue classes
We model the normal healthy tissue label ki of voxel i in the healthy part of the brain using a spatially varying probabilistic atlas, or prior p(ki = k) that is constructed from prior examples. At each voxel i ∈ {1,…, I}, this atlas indicates the probability of the tissue label ki to be equal to tissue class k ∈ {1,…, K} (Fig. 1, blue). The probability of observing tissue label k at voxel i is modeled through a categorical distribution
| (1) |
where for all i and for all i, k. The tissue label ki is shared among all C channels at voxel i. In our experiments we assume K = 3, representing gray matter (G), white matter (W) and cerebrospinal fluid (CSF), as illustrated in Fig. 2.
Fig. 2.
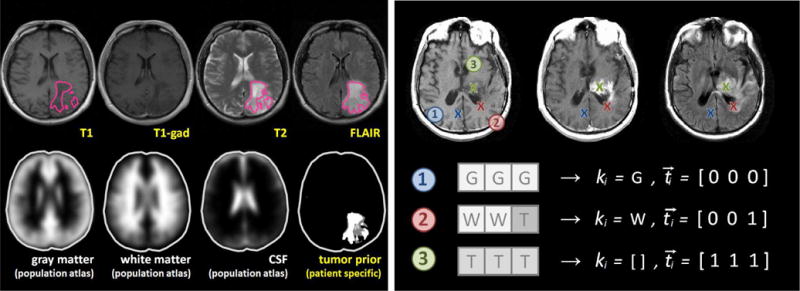
Illustration of the probabilistic model. The left panel shows images of a low-grade glioma patient with lesion segmentations in the different channels (outlined in magenta); in the bottom row it shows the probabilistic tissue atlases used in the analysis, and the patient-specific tumor prior inferred from the segmentations in the different channels. The right panel shows three voxels i with different labels in T1-, T1c- and FLAIR-MRI for a high-grade glioma patient. In voxel 1 all three channels show the characteristic image intensity of gray matter (G). In voxel 2 white matter (W) is visible in the first two channels, but the third channel contains a tumor-induced change (T), here due to edema or infiltration. In voxel 3 all channels exhibit gray values characteristic of tumor: a hypo-intense signal in T1, a hyper-intense gadolinium uptake in T1c – indicating the most active regions of tumor growth – and a hyper-intense signal in the FLAIR image. The initial tissue class ki remains unknown. Both ki and ti are to be estimated. Inference is done by introducing a transition process – with latent prior αi (Fig. 1) – which is assumed to have induced the channel-specific tissue changes implied by in the tumor label vector ti.
Tumor class
We model the tumor label using a spatially varying latent probabilistic atlas α [49], [50] that is specific to the given patient (Fig. 1, red). At each voxel i, this atlas contains a scalar parameter αi that defines the probability of observing a tumor at that voxel, forming the 3D parameter volume α. Parameter αi is unknown and is estimated as part of the segmentation process. We define a latent tumor label that indicates the presence or absence of tumor-induced changes in channel c ∈ [1,…, C] at voxel i, and model it as a Bernoulli random variable with parameter αi. We form a binary tumor label vector (where [·]T indicates the transpose of the vector) of the tumor labels in all C channels, that describes tumor presence in voxel i with probability
| (2) |
Here, we assume tumor occurrence to be independent from the type of the underlying healthy tissue. We will introduce conditional dependencies between the underlying tissue class and the likelihood of observing tumor-induced intensity modifications in Sec. II-C.
Observation model
The image observations are generated by Gaussian intensity distributions for each of the K tissue classes and the C channels, with mean and variance , respectively (Fig. 1, purple). In the tumor (i.e., if , the normal observations are replaced by intensities from another set of channel-specific Gaussian distributions with mean and variance representing the tumor class. Letting θ denote the set of mean and variance parameters for normal tissue and tumor classes, and denote the vector of the intensity observations at voxel i, we form the data likelihood:
| (3) |
where is the Gaussian distribution with mean μ and variance v.
Joint model
Finally, the joint probability of the atlas, the latent tumor class, and the observed variables is the product of the components defined in Eqs. (1–3):
| (4) |
We let Y denote the set of the C image volumes, T denote the corresponding C volumes of binary tumor labels, K denote the tissue labels, and α denote the parameter volume. We obtain the joint probability over all voxels i ∈ I by forming , assuming that all voxels represent independent observations of the model.
B. Maximum Likelihood Parameter Estimation
We derive an expectation-maximization scheme that jointly estimates the model parameters {θ, α} and the posterior distribution of tissue labels ki and tumor labels ti. We start by seeking maximum likelihood estimates of the model parameters {θ, α}:
| (5) |
| (6) |
and
| (7) |
| (8) |
where label vector indicates tumor in all channels with , and normal tissue for all other channels. As an example with three channels, illustrated in Fig. 2 (voxel 2), suppose ti = [0, 0, 1] and ki = W indicating tumor in channel 3 and image intensities relating to white matter in channels 1 and 2. This results in the tissue label vector si = [W, W, T].
E-step
In the E-Step of the algorithm, making use of given estimates of the model parameters , we compute the posterior probability of all K * 2C tissue label vectors si. Expanding Eq. (4), we use ti(si) and ki(si) that are corresponding to si to simplify notation:
| (9) |
and for all i. Using the tissue label vectors, we can calculate the probability that tumor is visible in channel c of voxel i by summing over all the configurations ti for which (or equivalently ):
| (10) |
where δ is the Kronecker delta that is equal to 1 for and 0 otherwise. In the same way we can estimate the probability for the healthy tissue classes k:
| (11) |
where indicates that one or more of the C channels of label vector si contain k.
M-step
In the M-Step of the algorithm, we update the parameter estimates using closed-form update expressions that guarantee increasingly better estimates of the model parameters [65]. The updates are intuitive: the latent tumor prior is an average of the corresponding posterior probability estimates
| (12) |
and the intensity parameters and are set to the weighted statistics of the data for the healthy tissues (k = 1, …,K)
| (13) |
| (14) |
Similarly, for the parameters of the tumor class (T), we obtain
| (15) |
| (16) |
We alternate between updating the parameters {θ, α} and the computation of the posteriors p(si|yi; θ, α) until convergence, which is typically reached after 10–15 iterations.
C. Enforcing Additional Biological Constraints
Expectation-maximization is a local optimizer. To overcome problems with initialization, we enforce desired properties of the solution by replacing the exact computation with an approximate solution that satisfied additional constraints. These constraints represent our prior knowledge about tumor structure, shape or growth behaviour1.
Conditional dependencies on tumor occurrence
A possible limitation in the generalization of our probabilistic model is the dimensionality of tissue label vector si that has K*2C possible combinations in Equation (9) and, hence, the computational demands and memory requirements that grow exponentially with the number of channels C in multimodal data sets. To this end, we may want to impose prior knowledge on p(ti|ki) and p(ti) by only considering label vectors si that are biologically plausible. First, instead of assuming independence between tissue class and tumor occurrence, we assume conditional dependencies, such as for all c. We impose this dependency by removing, in this example, all tumor label vectors containing both CSF and tumor from the list of vectors si that are included in Equation (9). Second, we can impose constraints on the co-occurrence of tumor-specific changes in the different image modalities (rather than assuming independence here as well), and exclude additional tumor label vectors. We consider, for example, that the edema visible in T2 will always coincide with the edema visible in FLAIR, or that lesions visible in T1 and T1c are always contained within lesions that are visible in T2 and FLAIR.
Together, these constraints reduce the total number of label vectors si to be computed in Equation (9), for a standard glioma imaging sequences with T1c, T1, T2, and FLAIR, from K * 2C = 3 * 24 = 48 to as few as ten vectors: three healthy vectors with t = [0, 0, 0, 0] (corresponding to [G, G, G, G], [W, W, W, W], and [CSF, CSF, CSF, CSF ]); edema with tumor-induced chances visible in FLAIR in the forth channel t = [0, 0, 0, 1] (with [W, W, W, T] and [G, G, G, T]); edema with tumor-induced changes visible in both FLAIR and in T2 t = [0, 0, 1, 1] (with [W, W, T, T] and [G, G, T, T]); the non-enhancing tumor core with changes in T1, T2, FLAIR, but without hyper-intensities in T1c t = [0, 1, 1, 1] ([W, T, T, T ] and [G, T, T, T]); the enhancing tumor core with hyper-intensities in T1c and additional changes in all other channels t = [1, 1, 1, 1] ([T, T, T, T]).
Hyper- and hypo-intense tumor structures
During the iterations of the EM algorithm we enforce that tumor voxels are hyper- or hypo-intense with respect to the current average image intensity of the white matter tissue (hypo-intense for T1, hyper-intense for T1c, T2, FLAIR). Similar to [51], we modify the probability that tumor is visible in channel c of voxel i by comparing the observed image intensity with the previously estimated prior to calculating updates for parameters θ (Eq. 16). We set the probability to zero if the intensity does not align with our expectations:
| (17) |
For hypo-intensity constraints we modify the posterior probability updates in the same way, using as a criterion.
Spatial regularity of the tumor prior
Little spatial context is used in the basic model, as we assume the tissue class si in each voxel to be independent from the class labels of other voxels. Atlas πk encourages spatially continuous classification for the healthy tissue classes by imposing similar priors in neighboring voxels. To encourage spatial regularity of the tumor labels, we extend the latent atlas α to include a Markov random field (MRF) prior:
| (18) |
Here, Ni denotes the set of the six nearest neighbors of voxel i, and β is a parameter governing how similar the tumor labels tend to be at the neighboring voxels. When β = 0, there is no interaction between voxels and the model reduces to the one described in Section II. By applying a mean-field approximation [66], we derive an efficient approximate algorithm. We let
| (19) |
denote the currently estimated “soft” count of neighbors that contain tumor in channel c. The mean-field approximation implies
| (20) |
where , replacing the previously defined Eq. (9), using a channel-specific as a modification of αi that features the desired spatial regularity.
III. Discriminative Extensions
High-level context at the organ or lesion level, as well as regional information, is not considered in the segmentation process of the generative model. Although we use different constraints to incorporate local neighbourhood information, the generative model treats each voxel as an independent observation and estimates class labels only from very local information. To evaluate global patterns, such as the presence of characteristic artifacts or tumor sub-structures of specific diagnostic interest, we present two alternative discriminative probabilistic methods that make use of both local and non-local image information. The first one, acting at the regional level, is improving the output of the generative model and maintaining its hyper- and hypo-intense lesion maps, while the second one, acting at the voxel level, is transforming the generative model output to any given set of biological tumor labels.
A. The Probabilistic Discriminative Model
We employ an algorithm that predicts the probability of label l ∈ L for a given observation j which is described by feature vector derived from the segmentations of the generative model. We seek to address two slightly different problems. In the first task, class labels L indicate whether a segmented region j is a result of a characteristic artifact rather than of tumor-induced tissue changes, essentially indicating false positive regions in the segmentations of the generative algorithm that should be removed from the output. In the second task, class labels L represent dense, voxel-wise labels with a semantic interpretation, for example structural attributes of the tumor that do not coincide with the hyper-and hyper-intense segmentations in the different channels, but labels such as “necrosis”, or “non-enhancing core”. We test both cases in the experimental evaluation, using on channel-wise tumor probabilities and on normalized intensities yi to derive input features for the discriminative algorithms.
To model relations between lj and xj for observation j ∈ N, we choose random forests, an ensemble of D randomized decision trees [67]. We use the random forest classifier as it is capable of handling irrelevant predictors and, to some degree, label noise. During training each tree uses a different set of samples . It consists of n randomly sampled observations Xn that only contain features from a random subspace of dimensionality m = log(P), where P is the number of features. We learn an ensemble of D different discriminative classifiers, indexed by d, that can be applied to new observations xj during testing, with each tree predicting the membership L(d)j. When averaging over all D predictions that we obtain for the individual observation, we obtain an estimate of p(lj|xj) = 1/DΣL(d)j. We choose logistic regression trees as discriminative base classifiers for our ensemble, as the resulting oblique random forests perform multivariate splits at each node and are, hence, better capable of dealing with the correlated predictors derived from a multi-modal image data set [68]. For both discriminative approaches we use an ensemble with D = 255 decision trees.
B. A Discriminative Approach Acting at the Regional Level
As many characteristic artifacts have, at the pixel level, a multimodal image intensity patterns that is similar to the one of a lesion, we design a discriminative probabilistic method that is postprocessing and “filtering” the basic output of the generative model. In addition to the pixel-wise intensity pattern, it evaluates regional statistics of each connected tumor area, such as volume, location, shape, signal intensities. It replaces commonly used postprocessing routines for quality control that evaluate hand-crafted rules on lesion size or shape and location by a discriminative probabilistic model, similar to [44].
Features and labels
The discriminative classifier acts at the regional level to remove those lesion areas from the output of the generative model that are not associated with tumor, but that stem from arbitrary biological or imaging variation of the voxel intensities. To this end we identify all R isolated regions in the binary tumor map of the FLAIR image (containing voxels i with . We choose FLAIR since it is the most inclusive image modality. As artifacts may be connected to lesion areas, we over-segment larger structures using a watershed algorithm, subdividing regions with connections that are less than 5mm in diameter to reduce the number mixed regions containing both tumor pixels and artifact patterns. For each individual region r ∈ 1 … R we calculate a feature vector xr that includes volume, surface area, surface-to-volume ratio, as well as regional statistics that are minimum, maximum, mean and median of the normalized image intensities in the four channels. We scale the image intensities for each channels linearly to match the distribution of intensities in a reference data set. We also determine the absolute and the relative number of voxels i with within region r, i.e., the volume of the active tumor. We calculate the linear dimensions of the region in axial, sagittal, and transversal direction, the maximal ratio between these three values indicating eccentricity, and the relative location of the region with respect to the center of the brain mask, as well as minimum, maximum, mean and median distances of the regions’s voxels from the skull, as a measure of centrality within the brain. We then determine the total number of FLAIR lesions for the given patient and assign this number as another feature to each lesion, together with its individual rank with respect to volume both in absolute numbers and as a normalized rank within [0, 1].
Overall, we construct P = 39 features for each region r (Fig. 6). To assign labels to each region, we inspect them visually and assign those overlapping well with a tumor area to the true positive “tumor” class Lr = 1, all other to the false positive “artifact” class Lr = 0. When labeling regions in the BRATS training data set (Sec. V), all regions labeled as true positives have at least 30% overlap with the “whole tumor” annotation of the experts.
Fig. 6.
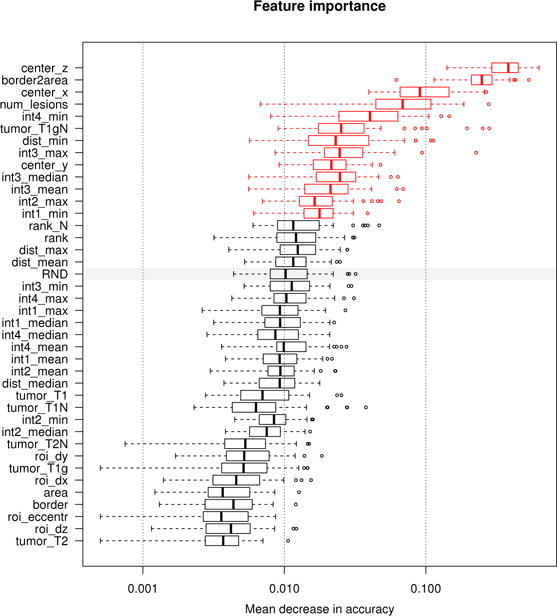
Measuring feature relevance of the discriminative model. Features relevant for discriminating between false positives and true positives regions. We evaluate the permutation importance [67] of each feature extracted for the FLAIR regions (see text for details). Boxplots show the decrease in accuracy for all 255 trees of the oblique random forest (boxes representing quartiles) with high values indicating high relevance. The gray bar indicates the performance of a random feature (“RND”) under this measure, features displayed in red perform significantly better (as indicated by a paired Cox-Wilcoxon test at 5% level). Location and shape of the regions are most discriminative, as well as the general number of lesions visible in the given FLAIR image, and selected image intensities.
C. A Discriminative Approach Acting at the Voxel Level
The generative model returns a set of probabilistic maps indicating the presence of hypo- or hyper-intense modifications of the tissue. In most applications and imaging protocols, however, it is necessary to localize arbitrary tumor structures – with biological interpretations and clinically relevant semantic labels, such as “edema”, “active tumor” or “necrotic core”. These structures do not correspond one-by-one to the hypo-and hyper-intense lesions, but have to be inferred by evaluating spatial context and tumor structure as well. We use the probabilistic output of the generative model, together with few secondary features that are derived from the same probabilistic maps and image intensity features, as input to a classifier predicting the posterior probability of the desired semantic labels. The discriminative classifier evaluates local and non-local features to map the output of the generative model to semantic tumor structure and to infer the most likely label L for each given voxel, similar to [32].
Features and labels
To predict a dense set of semantic labels L we extract the following set of features xj for each voxel j: the tissue prior probabilities p(kj) for the K = 3 tissue classes ; the tumor probability for all C = 4 channels , and the C = 4 image intensities after they have been scaled linearly to the intensities of a reference data set . From these data we derive two types of features. First, we construct the differences of local image intensities or probabilities for all three types of input features . This feature captures the difference between the image intensity or probability xj of voxel j and the corresponding image intensity or probability of another voxel k. For every voxel j in our volume we calculate these differences for 20 different directions, with spatial offsets in between 3mm to 3cm, i.e., distances that correspond to the extension of most relevant tumor structures. To reduce noise the subtracted values of xk are extracted after smoothing the image intensities locally around voxel k (using a Gaussian kernel with 3mm standard deviation). We calculate differences between tumor or tissue probability at a given voxel and the probability of the same location on the contralateral side. Second, we evaluate the geodesic distance between voxel j and specific image features that are of particular interest in the analysis. The path is constrained to areas that are most likely gray matter, white matter or tumor as predicted by the generative model. More specifically, we use the distance of of voxel j to the boundary of the the brain tissue (the interface of white and gray matter with CSF), the distance to the boundary of the T2 lesion representing the approximate location of the edema. This latter distance is calculated independently for voxels outside and inside the edema. In the same way, we calculate and representing the inner and outer distance to the next T1c hyper-intensity. We calculate the number of voxels that are labeled as “edema” or “active tumor” among the direct neighbours of voxel , and determine the x-y-z location of the voxel in the co-registered NMI space
Overall, we construct P = 651 image features for each voxel j and, when adapted to the BRATS training data set (Sec. V), five labels Lj as provided by clinical experts.
IV. Experiment 1: Properties and Performance of the Generative Model
In a first experiment, we evaluate the relevance of different components and parameters of the probabilistic model, compare it with related generative approaches,and evaluate the performance on the public BRATS glioma dataset, and test the generalization in a transfer to a related application dealing with stroke lesion segmentation.
A. Data and Evaluation
Glioma data
We use the public BRATS 2012–2013 dataset that provides a total of 45 annotated multimodal glioma image volumes [1]. Training datasets consist of 10/20 low/high-grade cases with native T1, Gadolinium-enhanced T1 (T1c), T2 and FLAIR MR image volumes. The test dataset contains no labels, but can be evaluated by uploading image segmentations to a server; it includes 4/11 low-grade/high-grade cases. Experts have delineated tumor edema, Gadolinium-enhancing “active” core, non-enhancing solid core, cystic/necrotic core. We co-register the probabilistic MNI tissue atlas of SPM99 with the T1 image of each dataset using the FSL software, and sampled to 1mm3 isotropic voxel resolution. We perform a bias field correction using a polynomial spline model (degree 3) together with a multivariate tissue segmentation using an EM segmenter that is robust against lesions2 [51]. Image intensities of each channel in each volume are scaled linearly to match the histogram of a reference.
Stroke data
Images are acquired in patients with acute and subacute ischemic stroke. About half of the 18 datasets comprise T1, T2, T1c and FLAIR images in patients in the sub-acute phase, acquired about one or two days after the event; another half comprises T1, T1c, T2 base diffusion and mean diffusivity (MD) images acquired in acute stroke patients within the first few hours after the onset of symptoms. For both groups the imaging sequences return tissue contrasts of normal tissues and lesion areas that are similar to hyper- and hypo-intensities expected in glioma sequences; stroke lesions are characterised here by T1 hypo-, T1c hyper-, T2 hyper-and FLAIR /MD hyper-intense changes. For the quantitative evaluation of the algorithm, we delineate the lesion in every 10th axial, sagittal, and coronal slice, in each of the four modalities. In addition, we annotate about 10% of the 2D slices twice to estimate variability. We register the probabilistic atlas and perform a model based bias field correction as for the glioma data. Image intensities are scaled to the same reference as for the glioma cases.
Evaluation
To measure segmentation performance in the experiments with this dataset, we combine the set of four tumor labels (edema and the three tumor core subtypes) to one binary “complete lesion” label map. We compare this map with the hyper-intense lesion as segmented in T2 and FLAIR. Separately, we compare the “enhancing core” label map with the hyper-intense lesion as segmented in T1c MRI. Quantitatively, we calculate volume overlap between expert annotation A and predicted segmentation B using the Dice score . We compute Dice scores for whole brain when testing performances on the BRATS data set. We also calculate Dice score within a 3cm distance from the lesion to measure local differences in lesion segmentation rather than in global detection performances.
B. Model Properties and Evaluation on the BRATS data set
Comparison of generative modelling approaches
We compare the proposed generative model against related generative tissue segmentations models and evaluate the relevance of individual components of our approach on the BRATS training data set. We calculate Dice scores in the area containing the lesion and the 3cm margin.
Figure 3A illustrates the benefit of the proposed multivariate tumor and tissue segmentation over a univariate segmentations that treat tumor voxels as intensity outliers similar to Van Leemput’s EM segmentation approach for white matter lesion [51]. On the given data this baseline approach leads to a high number of false positives, either requiring stronger spatial regularization or a more adaptive tuning of the outlier threshold. Figure 3B reports the benefit of enforcing intensity constraints within the proposed generative model. While the benefit for the large hyper-intense regions visible in T2 and FLAIR is minor, the difference for segmenting the enhancing tumor core visible in T1c in high-grade patients is striking: the constraint disambiguates tumor-related hypo-intensities – similar to those visible in native T1, for example, from edema – from hyper-intensities induced by the contrast agent in the active rim. Figure 3C reports a comparison between our approach and Prastawa’s classic tumor EM segmentation approach [22] that models lesions as an additional class with a “flat” global atlas prior. We test different values for the tumor prior α in Eq. 2, evaluating result for αflat ∈ [.005, .01, .02, .04, .1, .2, .4]. We find that every channel and every segmentation task has a different optimal αflat. However, each optimally tuned generative model with flat prior is still outperformed by the proposed generative model.
Fig. 3.
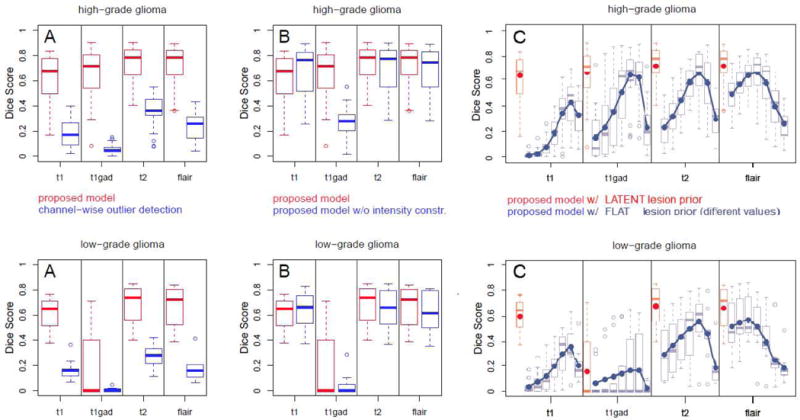
Evaluation of the generative model and comparison against alternative generative modeling approaches: high-grade (top) and low-grade cases (bottom). Reported are Dice scores for channel-specific segmentations for both low- and high-grade cases in the BRATS training set calculated in the lesion area. Boxplots indicate quartiles, circles indicate outliers. Results of the proposed model are shown in red, while results for related but different generative segmentation methods are shown in blue. Figure A reports performances of univariate tumor segmentations similar to [51]. Figure B: performances of our algorithm with and without constraints on the expected tumor intensities indicating their relevance. Figure C: performance of a generative model with “flat” global tumor prior αflat – i.e., the model of the standard EM segmenter – and evaluating seven different values αflat ∈ [.005, …, .4]. Blue lines and dots in C indicate average Dice scores. The proposed model outperforms all tested alternatives.
Enforcing spatial regularity
Our model has a single parameter that has to be set which is the regularization parameter β coupling segmentations of neighbouring voxels. Based on our previous study [55], we performed all experiments reported in Figure 3 with weak spatial regularization (β = .3). To confirm these preliminary results we test different regularization settings with β ∈ [0, 2−3, 2−2,…, 23], now also evaluating channel-specific performance in the lesion area (Figure 7 in the online Supplementary Materials). We find a strong regularization to be optimal for the large hyper-intense lesions in FLAIR β ≥ .5, suppressing small spurious structures, while little or no regularization is best for the hardly visible hypo-intense structures visible in T1 (β < .5). Both T2 and T1c are rather insensitive to regularization. We find the previous value of β = .3 to work well, but choose β = .5 for both low- and high-grade tumors in further experiments, somewhat better echoing the relevance of FLAIR.
Evaluation on the BRATS test set
We apply our segmentation algorithm to the BRATS test sets that have been used for the comparison of twenty glioma segmentation methods in the BRATS evaluation [1]. We identify the segmentations in FLAIR with the “whole tumor” region of the BRATS evaluation protocol, and the T1c enhancing regions with the “active tumor” region. We evaluate two sets of segmentations: segmentations that are obtained by thresholding the corresponding probabilities at 0.5, and the same segmentations after removing all regions that are smaller than 500mm2 in the FLAIR volume. This latter postprocessing approach was motivated by our observation that smaller regions correspond to false positives in almost all cases. We calculate Dice scores for the whole brain.
Table I reports Dice scores for the BRATS test sets with results of about .60 for the whole tumor and about .50 for the active tumor region (‘raw’). As visible from Figure 4, results are heavily affected by false positive regions that have intensity profiles similar to those of the tumor lesions. Applying the basic, size-based postprocessing rule improves results in most cases (‘postproc.’). Most false positives are spatially separated from the real lesion and when calculating Dice scores from a region that contains the FLAIR lesion and a 3cm margin only, results improve drastically to average values of .78 (±.09 std.) for the whole tumor to and .55 (±.27 std) for the active region (not shown in the table) which aligns well with results obtained for T2 and T1c on the training set (Fig. 3).
Table I.
Dice scores on the test sets used in this study, for the two tasks of segmenting the whole lesion (top) and the Gadolinium enhancing structures (bottom). BRATS results are calculated on the whole brain, stroke results in the lesion area. Inter-rater represents the overlap over multiple segmentations of the corresponding task and datasets done by human raters [1]. Reported are mean with standard deviation and median with median absolute deviance.
| Task: complete lesion (FLAIR) | mean (±std) | median (±MAD) |
|---|---|---|
|
| ||
| BRATS glioma – generative (raw) | .58 (±.22) | .67 (±.11) |
| BRATS glioma – gener. (postproc.) | .62 (±.21) | .72 (±.11) |
| BRATS glioma – gener.-discr. (region) | .69 (±.24) | .79 (±.06) |
| BRATS glioma – gener.-discr. (pixel) | .78 (±.13) | .83 (±.05) |
| INTER-RATER (4 raters) | .86 (±.06) | .87 (±.06) |
|
| ||
| Zurich stroke | .78 (±.11) | .79 (±.07) |
| INTER-RATER (2 raters) | .79 (±.11) | .80 (±.12) |
| Task: enhancing core (T1c) | mean (±std) | median (±MAD) |
|---|---|---|
|
| ||
| BRATS glioma – generative (raw) | .46 (±.26) | .60 (±.15) |
| BRATS glioma – gener. (postproc.) | .51 (±.27) | .64 (±.15) |
| BRATS glioma – gener.-discr. (region) | .53 (±.27) | .66 (±.14) |
| BRATS glioma – gener.-discr. (pixel) | .54 (±.29) | .66 (±.15) |
| INTER-RATER (4 raters) | .76 (±.10) | .78 (±.08) |
|
| ||
| Zurich stroke | .45 (±.33) | .64 (±.18) |
| INTER-RATER (2 raters) | .82 (±.08) | .83 (±.05) |
Fig. 4.
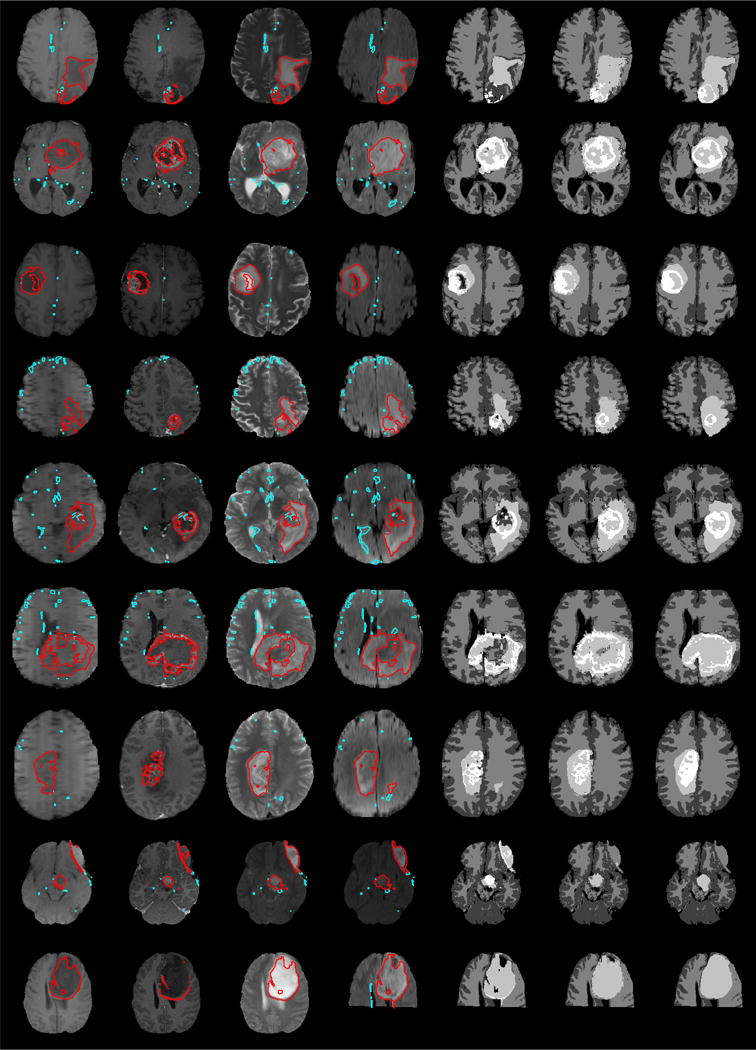
Exemplary BRATS test sets, with results for generative and generative discriminative models. Shown are axial views through the tumor center for T1, T1c, T2 and FLAIR image (columns from left to right) and the segmented hypo- or hyper-intense areas (red and cyan). Regions outlined in red have been identified as “true positive” regions by the regional discriminative classifier and the resulting tumor labels are shown in column 5 with edema (bright gray) and active tumor region (white). Column 6 shows results of the voxel-wise generative-discriminative classifier, and column 7 the expert’s annotation. Gray and white matter segmentations displayed in the last three columns are obtained by the generative model.
C. Generalization Performance and Transfer to the Stroke Data Set
We test the generalization performance of the generative model by using it for delineating ischemic stroke lesions that are similar in terms of lesion size and clinical image information. We apply the generative model as optimized for the BRATS dataset to the stroke images. As a single modification we allow T1c lesions to be both inside the FLAIR and T2 enhancing area and outside, as bleeding (which leads to the T1c hyper-intensities) may not coincide with the local edema. Stroke images contain cases with both active and chronic lesions with significantly different lesion patterns.
Although datasets, imaging protocol, and even major acquisition parameters differ, we obtain results that are comparable to the tumor data. We calculate segmentations accuracies in the lesion area and observe good agreement between manual delineation and automatic segmentation in all four modalities (Fig. 5). We also observe false positives at the white matter–gray matter interfaces, similar to those we observed for the glioma tests data (Fig. 4). Most false positive regions are disconnected from the lesion and could be removed with little user interaction or postprocessing. Inter-rater differences and performance of the algorithm are comparable to those from the glioma test set, with Dice scores close to .80 for segmenting the edema and around .50–.60 for T1c enhancing structures (Table I). Results on the stroke data underline the versatility of the generative lesion segmentation model and its good generalization performance not only across different imaging sequences, but also across applications. To the best of our knowledge this is one of the first attempts to automatically segment ischemic stroke lesion in multimodal images using a generative model.
Fig. 5.
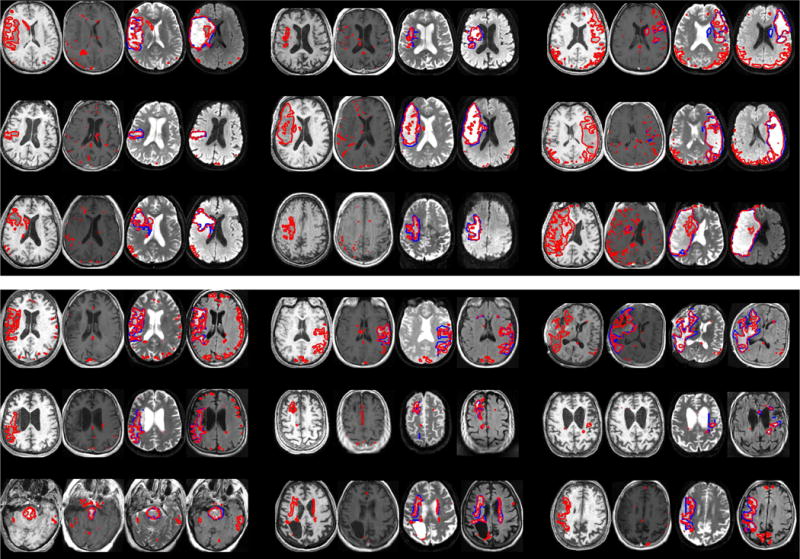
Generalization to ischemic stroke cases (showing acute stroke: rows 1–3; subacute stroke: rows 4–6) with T1, T1c, FLAIR/MD, T2/MAD images of each patient (from left to right, three patients per row). Automatic segmentations are delineated in red, lesions in manually segmented volumes are shown in blue (typically beneath a red line); T1 and T1c lesions were only visible for some cases.
V. Experiment 2: Properties and Performances of the Generative-Discriminative Model Extensions
Results of the generative model show its robustness and accuracy for delineating lesion structures. Still, it also shows to be sensitive to artifacts that cannot be recognized by evaluating the multimodal intensity pattern at the voxel level, and hypo- and hyper-intense structures can only be matched with selected tumor labels. To this end, we evaluate the two discriminative modeling strategies that are considering non-local features as input and arbitrary labels as output. We first evaluate model properties on the BRATS training set and then compare performances to results of other state-of-the art tumor segmentation algorithms on the BRATS test set.
A. Relevant Features and Information used by the Discriminative Models
The random forest classifier handles learning tasks with small sample sizes in high dimensional spaces by only relying on few “strong” variables and ignoring irrelevant features [69]. Still, in order to understand the information used when modeling the class probabilities, we can visualize the importance of the input features used. To this end we evaluate the relevance of the individual features using Breiman’s feature permutation test [67] that compares the test error with the error obtained after the values of a given feature have been randomly permuted throughout all test samples. The resulting decrease in test accuracy, or increase in test error, indicates how relevant the chosen feature is for the overall classification task. Repeated for each feature of all trees in the decision forest, this measure helps to rank the features and to compare the relevance as shown in Figure 6. In our test we augment the dataset with a random feature (random samples from a Gaussian distribution with mean 0 and standard deviation of 1) to establish a lower baseline of the relevance score. For each feature we compare the distribution of changes in classification error against the changes of this random feature in a paired Cox-Wilcoxon test. We analyze feature relevance in a cross-validation on the BRATS training set.
Results for the first discriminative model acting at the regional level are shown in Fig. 6. We find plausible features to be relevant: the relative location of the region with respect to the center of the brain (indicated as center_x, center_y, center_z in the figure), the surface-to-volume ratio (border2area), the total number of lesions visible for the given patient (num_lesions), the ratio of segmented voxels in T1c (tumorT 1cN), and some descriptors of image intensities, such as the minimum in FLAIR (int4_min), the maximum, median and average of the T2 intensities (int3_*), as well as the maximum in T1c (int1_max) and the minimum in T1 (int2_min).
For the second discriminative model acting at the pixel level we find about 80% of the features to be relevant, with with some variation across the different classification tasks. The features that rank highest in all tests are those we derived from the probability maps of the generative model: the total number of local edema or active tumor voxels, the geodesic distance to the nearest edema or active tumor pixels, but also the relative anatomical location in the MNI space, and selected image intensities and intensity differences (such as the intensity values of T1 and FLAIR for edema and T1c for active core, and local differences in the T1 image intensities).
B. Performance on the BRATS Test Set
Figure 4 displays nine exemplary image volumes of the BRATS test set. Shown are the raw probability maps of the generative model (red and cyan; columns 1–4), those regions that are selected by the regional discriminative model (cyan) and the derived tumor segmentation (column 5), as well as the output of the voxel-level tumor classifier (column 6), together with an expert annotation (column 7).
Quantitative results are reported in Table I, and we find both discriminative models to improve results over those derived from the “raw” probability maps of the generative model. With few exceptions most “false positive” artifact regions are removed (Fig. 4). The voxel-level model shows to be more accurate than the regional-level model, also correcting for “false negative” areas in the center of the tumor (rows 1, 3, 6, and 7). In addition to whole tumor and active tumor areas, the second discriminative model is also predicting the location of necrotic and fluid filled structures, as well as the “tumor core” label (with a Dice score of .58; segmentations not shown in the figure). Sensitivities and specificities for this latter model are balanced with sensitivities of .75/.58/.63 for the three tumor regions (whole tumor/tumor core/active tumor) and a specificities of .86/.71/.56.
The BRATS challenge also allows us to compare the two generative-discriminative modeling approaches with eighteen other state of the art methods including inter-active ones, and we reproduce results of the challenge in Figure 8 in the online Supplementary Materials of this manuscript. The generative model with discriminative post-processing at the regional level (indicated by Menze (G)) performs comparable to most other approaches in terms of Dice score and robust Hausdorff distance for “whole tumor” and “active tumor”. However, it cannot model the “tumor core” segmentation task as this structure does not have a direct correspondence to any of the segmented hyper- and hypo-intensity regions and, hence, does does not provide competitive results for this tumor sub-structure. The voxel-level generative-discriminative approach (indicated by Menze (D)) is able to predict “tumor core” labels. It ranks first among the twenty evaluated methods in terms of average Hausdorff distances for both “tumor core” and in “whole tumor” segmentation, and it is the second best automatic method for the “active tumor” segmentation. In the evaluation of the average Dice scores it is second best for “whole tumor”, it is ranking third among the automated methods for the “tumor core” task, and its result are statistically indistinguishable from the inter-rater variation for “active tumor”. Most notably, the voxel-level generative-discriminative approach is outperforming all discriminative models that are similar in terms of random forest classifier and feature design [37], [32], [2], [34], but that do not rely on the input features derived from the probability maps of the generative model.
VI. Summary and Conclusions
In this paper, we extend the atlas-based EM segmenter by a latent atlas class that represents the probability of transition from any of the “healthy” tissues to a “lesion” class. In practice, the latent atlas serves as an adaptive prior that couples the probability of observing tumor-induced intensity changes across different imaging channels for the same voxel. Using the standard brain atlas for healthy tissues together with the highly specific multi-channel information provides us with segmentations of the healthy tissues surrounding the tumor, and enables us to automatically segment the images. The proposed generative algorithm produces outlines of the tumor-induced changes for each channel which makes it independent of the multimodal imaging protocol. We complement the basic probabilistic model with a discriminative model and test two different modeling strategies, both of them addressing shortcomings of the generative model, and find the resulting discriminative-generative model to define the state of the art in tumor segmentation on the BRATS data set [1].
The proposed generative algorithm generalizes the probabilistic model of the standard EM segmenter. As such, it can be improved by combining registration and segmentation [62], or by integrating empirical or physical bias-field correction models [15], [70]. The generative segmentation algorithm that we optimized for glioma images exhibits a good level of generalization when applied to multimodal images from patients with ischemic stroke. The method is likely to also work well for traumatic brain injury with similar hypo- and hyper-intensity patterns, and it can also be adapted to multimodal segmentation tasks beyond the brain. It may be interesting to evaluate relations to multi-channels approaches that do not rely on multiple physical channels, but high-dimensional sets of features extracted from one or few physical images [71]. Analyzing feature relevance indicated that the location of a voxel or region within the MNI space helped in removing false positives, as most of them appeared at white matter–gray matter interfaces or in areas that are often subject to B-field inhomogeneities. Extensions of the generative model may use a location prior that lowers the expectation of tumor occurrences in these areas. Moreover, preliminary findings suggest that results may improve by using non-Gaussian intensity models for the lesion classes.
Some tumor structures – such as necrotic or cystic regions, or the solid tumor core – cannot easily be associated with local channel-specific intensity modifications, but are rather identified based on the wider spatial context and their relation with other tumor compartments. We addressed the segmentation of such secondary structures by combining our generative model with discriminative model extensions evaluating additional non-local features. As an alternative, relations between visible tumor structures can be enforced locally using MRF as proposed by [35], or in a non-local fashing following the hierarchical approach following [54]. Future work may also aim at integrating image segmentation with tumor growth models enforcing spatial or temporal relations as in [53], [14]. Tumor growth models – often described through partial differential equations [72] – offer a formal description of the lesion evolution, and could be used to describe the propagation of channel-specific tumor outlines in longitudinal series [73], as well as a shape and location prior for various tumor structures [23]. This could also promote a deeper integration of underlying functional models of disease progression and formation of image patterns in the modalities that are used to monitor this process [74].
Supplementary Material
Acknowledgments
This research was supported by the The National Alliance for Medical Image Analysis (NIH NIBIB NAMIC U54-EB005149), The Neuroimaging Analysis Center (NIH NIBIB NAC P41EB015902), NIH NIBIB (R01EB013565), the Lundbeck Foundation (R141-2013-13117), the European Research Council through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images), the Swiss NSF project Computer Aided and Image Guided Medical Interventions (NCCR CO-ME), the German Academy of Sciences Leopoldina (Fellowship Programme LPDS 2009-10), the Technische Universität München – Institute for Advanced Study (funded by the German Excellence Initiative and the European Union Seventh Framework Programme under grant agreement n 291763), the Marie Curie COFUND program of the the European Union (Rudolf Mössbauer Tenure-Track Professorship to BHM).
Footnotes
An implementation of the generative tumor segmentation algorithm in Python is available from http://ibbm.in.tum.de.
available from http://www.medicalimagecomputing.com/downloads/ems.php
To support the further use and analysis of our generative segmentation algorithm, we make an implementation available in Python from http://ibbm.in.tum.de, also illustrating its use on reference data from the BRATS challenge.
References
- 1.Menze B, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) IEEE Transactions on Medical Imaging. 2014:33. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer S, Wiest R, Nolte L-P, Reyes M. A survey of MRI-based medical image analysis for brain tumor studies. Phys Med Biol. 2013 Jul;58(13):R97–R129. doi: 10.1088/0031-9155/58/13/R97. [DOI] [PubMed] [Google Scholar]
- 3.Angelini ED, Clatz O, Mandonnet E, Konukoglu E, Capelle L, Duffau H. Glioma dynamics and computational models: A review of segmentation, registration, and in silico growth algorithms and their clinical applications. Curr Medl Imaging Rev. 2007;3:262–276. [Google Scholar]
- 4.Kaus M, Warfield SK, Nabavi A, Chatzidakis E, Black PM, Jolesz FA, Kikinis R. Segmentation of meningiomas and low grade gliomas in MRI. Proc MICCAI. 1999:1–10. [Google Scholar]
- 5.Tsai Y-F, Chiang I-J, Lee Y-C, Liao C-C, Wang K-L. Automatic MRI meningioma segmentation using estimation maximization. Proc IEEE Eng Med Biol Soc. 2005;3:3074–3077. doi: 10.1109/IEMBS.2005.1617124. [DOI] [PubMed] [Google Scholar]
- 6.Konukoglu E, Wells WM, Novellas S, Ayache N, Kikinis R, B PM, Pohl KM. Monitoring slowly EVolving tumors. Proc ISBI. 2008:1–4. doi: 10.1109/ISBI.2008.4541120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuadra BBach, Pollo C, Bardera A, Cuisenaire O, Thiran JP. Atlas-based segmentation of pathological brain MR images using a model of lesion growth. IEEE T Med Imag. 2004;23:1301–14. doi: 10.1109/TMI.2004.834618. [DOI] [PubMed] [Google Scholar]
- 8.Styner M, Lee J, Chin B, Chin M, Commowick O, Tran H, Markovic-Plese S, Jewells V, Warfield S. 3D segmentation in the clinic: A grand challenge ii: MS lesion segmentation. MIDAS Journal. 2008:1–5. [Google Scholar]
- 9.Irimia A, Chambers MC, Alger JR, Filippou M, Prastawa MW, Wang B, Hovda DA, Gerig G, Toga AW, Kikinis R, Vespa PM, Van Horn JD. Comparison of acute and chronic traumatic brain injury using semi-automatic multimodal segmentation of MR volumes. J Neurotrauma. 2011;28:2287–2306. doi: 10.1089/neu.2011.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenton M, Hamoda H, Schneiderman J, Bouix S, Pasternak O, Rathi Y, Vu M-A, Purohit M, Helmer K, Koerte I, et al. A review of magnetic resonance imaging and diffusion tensor imaging findings in mild traumatic brain injury. Brain imaging and behavior. 2012;6:137–192. doi: 10.1007/s11682-012-9156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farr TD, Wegener S. Use of magnetic resonance imaging to predict outcome after stroke: a review of experimental and clinical evidence. J Cerebr Blood Flow Metab. 2010;30:703–717. doi: 10.1038/jcbfm.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rekik I, Allassonnière S, Carpenter TK, Wardlaw JM. Medical image analysis methods in MR/CT-imaged acute-subacute ischemic stroke lesion: Segmentation, prediction and insights into dynamic evolution simulation models. a critical appraisal. NeuroImage: Clinical. 2012 doi: 10.1016/j.nicl.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarabalka Y, Charpiat G, Brucker L, Menze BH. Enforcing monotonous shape growth or shrinkage in video segmentation. Proc BMCV (British Machine Vision Conference) 2013 [Google Scholar]
- 14.Alberts E, Charpiat G, Tarabalka Y, Weber MA, Zimmer C, M BH. Proc MICCAI Brain Lesions Workshop (BrainLes) Springer; 2015. A nonparametric growth model for estimating tumor growth in longitudinal image sequences. (LNCS). [Google Scholar]
- 15.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE T Med Imaging. 1999;18:885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- 16.Prastawa M, Bullitt E, Ho S, Gerig G. A brain tumor segmentation framework based on outlier detection. Med Image Anal. 2004;8:275–283. doi: 10.1016/j.media.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Pohl KM, Fisher J, Levitt JJ, Shenton ME, Kikinis R, Grimson WEL, Wells WM. A unifying approach to registration, segmentation, and intensity correction. LNCS 3750, Proc MICCAI. 2005:310–318. doi: 10.1007/11566465_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharaki EI, Shen D, Davatzikos C. ORBIT: A multiresolution framework for deformable registration of brain tumor images. IEEE T Med Imag. 2008;27:1003–17. doi: 10.1109/TMI.2008.916954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuadra B Bach, Pollo C, Bardera A, Cuisenaire O, Thiran JP. Atlas-based segmentation of pathological brain MR images using a model of lesion growth. IEEE T Med Imag. 2004;23:1301–14. doi: 10.1109/TMI.2004.834618. [DOI] [PubMed] [Google Scholar]
- 20.Gering D, Grimson W, Kikinis R. Recognizing deviations from normalcy for brain tumor segmentation. Lecture Notes In Computer Science. 2002 Sep;2488:388–395. [Google Scholar]
- 21.Moon N, Bullitt E, Van Leemput K, Gerig G. Model-based brain and tumor segmentation. Proc ICPR. 2002:528–31. [Google Scholar]
- 22.Prastawa M, Bullitt E, Moon N, Leemput KV, Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Acad Radiol. 2003;10:1341–48. doi: 10.1016/s1076-6332(03)00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gooya A, Pohl KM, Bilello M, Cirillo L, Biros G, Melhem ER, Davatzikos C. GLISTR: glioma image segmentation and registration. IEEE Trans Med Imag. 2012;31:1941–1954. doi: 10.1109/TMI.2012.2210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parisot S, Duffau H, Chemouny S, Paragios N. Joint tumor segmentation and dense deformable registration of brain MR images. Proc MICCAI. 2012:651–658. doi: 10.1007/978-3-642-33418-4_80. [DOI] [PubMed] [Google Scholar]
- 25.Gooya A, Biros G, Davatzikos C. Deformable registration of glioma images using em algorithm and diffusion reaction modeling. IEEE Trans Med Imag. 2011;30:375–390. doi: 10.1109/TMI.2010.2078833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenzi M, Menze Bjoern H, Niethammer M, Ayache N, Pennec X. Sparse Scale-Space Decomposition of Volume Changes in Deformations Fields. Proc MICCAI. 2013;8150:328–335. doi: 10.1007/978-3-642-40763-5_41. [DOI] [PubMed] [Google Scholar]
- 27.Cobzas D, Birkbeck N, Schmidt M, Jagersand M, Murtha A. 3D variational brain tumor segmentation using a high dimensional feature set. Proc ICCV. 2007:1–8. [Google Scholar]
- 28.Lefohn A, Cates J, Whitaker R. Interactive, GPU-based level sets for 3D brain tumor segmentation. Proc MICCAI. 2003:564–572. [Google Scholar]
- 29.Gorlitz L, Menze BH, Weber M-A, Kelm BM, Hamprecht FA. Proc DAGM. 2007. Semi-supervised tumor detection in magnetic resonance spectroscopic images using discriminative random fields; pp. 224–233. (LNCS). [Google Scholar]
- 30.Lee C, Wang S, Murtha A, Greiner R. Segmenting brain tumors using pseudo conditional random fields. LNCS 5242, Proc MICCAI. 2008:359–66. [PubMed] [Google Scholar]
- 31.Wels M, Carneiro G, Aplas A, Huber M, Hornegger J, Comaniciu D. A discriminative model-constrained graph cuts approach to fully automated pediatric brain tumor segmentation in 3D MRI. LNCS 5241, Proc MICCAI. 2008:67–75. doi: 10.1007/978-3-540-85988-8_9. [DOI] [PubMed] [Google Scholar]
- 32.Zikic D, Glocker B, Konukoglu E, Criminisi A, Demiralp C, Shotton J, Thomas OM, Das T, Jena R, P SJ. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. Proc MICCAI. 2012 doi: 10.1007/978-3-642-33454-2_46. [DOI] [PubMed] [Google Scholar]
- 33.Geremia E, Clatz O, Menze BH, Konukoglu E, Criminisi A, Ayache N. Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. Neuroimage. 2011;57:378–90. doi: 10.1016/j.neuroimage.2011.03.080. [DOI] [PubMed] [Google Scholar]
- 34.Geremia E, Menze BH, Ayache N. Spatially adaptive random forests. Proc IEEE ISBI. 2013 [Google Scholar]
- 35.Bauer S, May C, Dionysiou D, Stamatakos GS, Büchler P, Reyes M. Multi-Scale Modeling for Image Analysis of Brain Tumor Studies. IEEE Trans Bio-Med Eng. 2011 Aug; doi: 10.1109/TBME.2011.2163406. [DOI] [PubMed] [Google Scholar]
- 36.Wu W, Chen AY, Zhao L, Corso JJ. Brain tumor detection and segmentation in a conditional random fields framework with pixel-pairwise affinity and superpixel-level features. International journal of computer assisted radiology and surgery. 2013:1–13. doi: 10.1007/s11548-013-0922-7. [DOI] [PubMed] [Google Scholar]
- 37.Tustison NJ, Shrinidhi KL, Wintermark M, Durst CR, Kandel BM, Gee JC, Grossman MC, Avants BB. Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with antsr. Neuroinformatics. 2015 Apr;13(2):209–225. doi: 10.1007/s12021-014-9245-2. [Online]. Available: http://dx.doi.org/10.1007/s12021-014-9245-2. [DOI] [PubMed] [Google Scholar]
- 38.Dvorak P, Menze BH. Structured prediction with convolutional neural networks for multimodal brain tumor segmentation. Proc MICCAI MCV (Medical Computer Vision Workshop) 2015 [Google Scholar]
- 39.Urban G, Bendszus M, Hamprecht FA, Kleesiek J. Multi-modal Brain Tumor Segmentation using Deep Convolutional Neural Networks. Proc MICCAI BRATS (Brain Tumor Segmentation Challenge) 2014:31–35. [Google Scholar]
- 40.Iglesias JE, Konukoglu E, Zikic D, Glocker B, Van Leemput K, Fischl B. Is synthesizing MRI contrast useful for inter-modality analysis? Proc MICCAI. 2013 doi: 10.1007/978-3-642-40811-3_79. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy S, Carass A, Prince J. A compressed sensing approach for MR tissue contrast synthesis. Proc IPMI. 2011:371–383. doi: 10.1007/978-3-642-22092-0_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy S, Carass A, Shiee N, Pham DL, Calabresi P, Reich D, Prince JL. Longitudinal intensity normalization in the presence of multiple sclerosis lesions. Proc ISBI. 2013 doi: 10.1109/ISBI.2013.6556791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batmanghelich NK, Taskar B, Davatzikos C. Generative-discriminative basis learning for medical imaging. IEEE Trans Med Imaging. 2012 Jan;31(1):51–69. doi: 10.1109/TMI.2011.2162961. [Online]. Available: http://dx.doi.org/10.1109/TMI.2011.2162961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menze BH, Kelm BM, Bachert P, Weber M-A, Hamprecht FA. Mimicking the human expert: Pattern recognition for an automated assessment of data quality in MR spectroscopic images. Magn Reson Med. 2008;1466:1457–66. doi: 10.1002/mrm.21519. [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed/18421692. [DOI] [PubMed] [Google Scholar]
- 45.Criminisi A, Juluru K, Pathak S. A discriminative-generative model for detecting intravenous contrast in ct images. Med Image Comput Comput Assist Interv. 2011;14(Pt 3):49–57. doi: 10.1007/978-3-642-23626-6_7. [DOI] [PubMed] [Google Scholar]
- 46.Tu Z, Narr KL, Dollar P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans Med Imaging. 2008 Apr;27(4):495–508. doi: 10.1109/TMI.2007.908121. [Online]. Available: http://dx.doi.org/10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iglesias JE, Liu C-Y, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging. 2011 Sep;30(9):1617–1634. doi: 10.1109/TMI.2011.2138152. [Online]. Available: http://dx.doi.org/10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- 48.Speier W, Iglesias JE, El-Kara L, Tu Z, Arnold C. Robust skull stripping of clinical glioblastoma multiforme data. Med Image Comput Comput Assist Interv. 2011;14(Pt 3):659–666. doi: 10.1007/978-3-642-23626-6_81. [DOI] [PubMed] [Google Scholar]
- 49.Riklin-Raviv T, Menze BH, Van Leemput K, Stieltjes B, Weber MA, Ayache N, Wells WM, Golland P. Joint segmentation via patient-specific latent anatomy model. Proc MICCAI-PMMIA (Workshop on Probabilistic Models for Medical Image Analysis) 2009:244–255. [Google Scholar]
- 50.Riklin-Raviv T, Van Leemput K, Menze BH, Wells WM, 3rd, Golland P. Segmentation of image ensembles via latent atlases. Med Image Anal. 2010;14:654–665. doi: 10.1016/j.media.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Leemput K, Maes F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE T Med Imaging. 2001;20:677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 52.Bauer S, Tessier J, Krieter O, Nolte L, Reyes M. Integrated spatio-temporal segmentation of longitudinal brain tumor imaging studies. Proc MICCAI-MCV, Springer LNCS. 2013 [Google Scholar]
- 53.Tarabalka Y, Charpiat G, Brucker L, Menze BH. Spatio-temporal video segmentation with shape growth or shrinkage constraint. IEEE Transactions in Image Processing. 2014;23:3829–3840. doi: 10.1109/TIP.2014.2336544. [DOI] [PubMed] [Google Scholar]
- 54.Corso JJ, Sharon E, Dube S, El-Saden S, Sinha U, Yuille A. Efficient multilevel brain tumor segmentation with integrated Bayesian model classification. IEEE T Med Imag. 2008;9:629–40. doi: 10.1109/TMI.2007.912817. [DOI] [PubMed] [Google Scholar]
- 55.Menze BH, Van Leemput K, Lashkari D, Weber M-A, Ayache N, Golland P. Proc MICCAI. 2010. A generative model for brain tumor segmentation in multi-modal images; pp. 151–159. (LNCS 751). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menze BH, Van Leemput K, Lashkari D, Weber M-A, Ayache N, Golland P. Segmenting glioma in multi-modal images using a generative model for brain lesion segmentation. Proc MICCAI-BRATS (Multimodal Brain Tumor Segmentation Challenge) 2012:7p. doi: 10.1007/978-3-642-15745-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menze BH, Geremia E, Ayache N, Szekely G. Segmenting glioma in multi-modal images using a generative-discriminative model for brain lesion segmentation. Proc MICCAI-BRATS (Multimodal Brain Tumor Segmentation Challenge) 2012:8. [Google Scholar]
- 58.Wells W, Grimson W, Kikinis R, Jolesz F. Adaptive segmentation of MRI data. Proc Computer Vision, Virtual Reality and Robotics in Medicine. 1995:57–69. [Google Scholar]
- 59.Wells WM, Grimson WEL, Kikinis R, Jolesz FA. Adaptive segmentation of MRI data. IEEE T Med Imaging. 1996;15:429–442. doi: 10.1109/42.511747. [DOI] [PubMed] [Google Scholar]
- 60.Ashburner J, Friston K. Multimodal image coregistration and partitioning–a unified framework. Neuroimage. 1997 Oct;6(3):209–217. doi: 10.1006/nimg.1997.0290. [Online]. Available: http://dx.doi.org/10.1006/nimg.1997.0290. [DOI] [PubMed] [Google Scholar]
- 61.Pohl KM, Fisher J, Grimson W, Kikinis R, Wells W. A Bayesian model for joint segmentation and registration. Neuroimage. 2006;31:228–239. doi: 10.1016/j.neuroimage.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 62.Pohl KM, Warfield SK, Kikinis R, Grimson WEL, Wells WM. Coupling statistical segmentation and -PCA- shape modeling. Proc MICCAI. 2004:151–159. doi: 10.1007/978-3-540-30135-6_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Leemput K. Encoding probabilistic brain atlases using Bayesian inference. IEEE TMI. 2009;28:822–837. doi: 10.1109/TMI.2008.2010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iglesias JE, Sabuncu MR, Leemput KV. Improved inference in Bayesian segmentation using Monte Carlo sampling: Application to hippocampal subfield volumetry. Med Image Anal. 2013 doi: 10.1016/j.media.2013.04.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dempster A, et al. Maximum likelihood from incomplete data via the em algorithm. Journal of the Royal Statistical Society. 1977;39:1–38. [Google Scholar]
- 66.Jordan MI, Gaharamani Z, Jaakola TS, Saul LK. An introduction to variational methods for graphical models. Machine Learning. 1999;37:183–233. [Google Scholar]
- 67.Breiman L. Random forests. Mach Learn J. 2001;45:5–32. [Google Scholar]
- 68.Menze BH, Kelm BM, Splitthoff DN, Koethe U, Hamprecht FA. On oblique random forests. Proc ECML (European Conference on Machine Learning) 2011 [Google Scholar]
- 69.Breiman L. Consistency for a simple model of random forests. UC Berkeley, Tech Rep. 2004:670. [Google Scholar]
- 70.Poynton C, Jenkinson M, Wells W., III Atlas-based improved prediction of magnetic field inhomogeneity for distortion correction of EPI data. Proc MICCAI. 2009:951–959. doi: 10.1007/978-3-642-04271-3_115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toews M, Zollei L, W W., III Invariant feature-based alignment of volumetric multi-modal images. Proc IPMI. 2013 doi: 10.1007/978-3-642-38868-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Menze BH, Stretton E, Konukoglu E, Ayache N. Image-based modeling of tumor growth in patients with glioma. In: Garbe CS, Rannacher R, Platt U, Wagner T, editors. Optimal control in image processing. Springer; Heidelberg/Germany: 2011. [Google Scholar]
- 73.Konukoglu E, Clatz O, Menze BH, Stieltjes B, Weber M-A, Mandonnet E, Delingette H, Ayache N. Image guided personalization of reaction-diffusion type tumor growth models using modified anisotropic eikonal equations. IEEE T Med Imag. 2010;29:77–95. doi: 10.1109/TMI.2009.2026413. [DOI] [PubMed] [Google Scholar]
- 74.Menze BH, Van Leemput K, Honkela A, Konukoglu E, Weber MA, Ayache N, Golland P. A generative approach for image-based modeling of tumor growth. Proc IPMI (Inform Proc Med Imag) 2011 doi: 10.1007/978-3-642-22092-0_60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


