Abstract
Background
We investigated the whole genome sequence (WGS) of a carbapenem-resistant Acinetobacter baumannii isolate belonging to the global clone 2 (GC2) and predicted resistance islands using a software tool.
Methods
A. baumannii strain YU-R612 was isolated from the sputum of a 61-yr-old man with sepsis. The WGS of the YU-R612 strain was obtained by using the PacBio RS II Sequencing System (Pacific Biosciences Inc., USA). Antimicrobial resistance genes and resistance islands were analyzed by using ResFinder and Genomic Island Prediction software (GIPSy), respectively.
Results
The YU-R612 genome consisted of a circular chromosome (ca. 4,075 kb) and two plasmids (ca. 74 kb and 5 kb). Its sequence type (ST) under the Oxford scheme was ST191, consistent with assignment to GC2. ResFinder analysis showed that YU-R612 possessed the following resistance genes: four β-lactamase genes blaADC-30, blaOXA-66, blaOXA-23, and blaTEM-1; armA, aadA1, and aacA4 as aminoglycoside resistance-encoding genes; aac(6')Ib-cr for fluoroquinolone resistance; msr(E) for macrolide, lincosamide, and streptogramin B resistance; catB8 for phenicol resistance; and sul1 for sulfonamide resistance. By GIPSy analysis, six putative resistant islands (PRIs) were determined on the YU-R612 chromosome. Among them, PRI1 possessed two copies of Tn2009 carrying blaOXA-23, and PRI5 carried two copies of a class I integron carrying sul1 and armA genes.
Conclusions
By prediction of resistance islands in the carbapenem-resistant A. baumannii YU-R612 GC2 strain isolated in Korea, PRIs were detected on the chromosome that possessed Tn2009 and class I integrons. The prediction of resistance islands using software tools was useful for analysis of the WGS.
Keywords: Acinetobacter baumannii, Whole genome sequence, GIPSy, Global clone 2
INTRODUCTION
Acinetobacter baumannii is an opportunistic pathogen that causes hospital-acquired infections, especially in intensive care units [1,2,3]. The incidence and worldwide dissemination of carbapenem-resistant A. baumannii isolates named global clone (GC)1 and GC2 have increased [4,5,6]. Carbapenem resistance in A. baumannii has mainly been ascribed to degradation by carbapenem-hydrolyzing oxacillinases including OXA-23, OXA-24, OXA-58, OXA-51, OXA-143, and their variants [2,7].
Large DNA sequences (6-200 kb) are termed genomic islands when they are acquired from other organisms through plasmids, transposons, and other mobile genetic elements [8,9]. Among the genomic islands of A. baumannii, multiple antibiotic resistance regions that have inserted into the target ATPase gene (comM) are termed resistance islands (AbaR) [10]. There are several known AbaRs, which differ according to the clone type [11,12]. The AbaR3-type resistance islands are the major resistance islands of GC1 isolates, and the AbaR4-type islands are associated with GC2 isolates [13,14,15]. Some AbaR4-type islands of A. baumannii harbor transposons such as Tn2009, Tn2006, and Tn6008 carrying the blaOXA-23 gene [16].
Because in vitro approaches to investigate genomic islands are time-consuming and expensive, several software tools for identifying putative genomic islands have been developed. Some useful software tools and resources for identification of antimicrobial resistance genes have also been reported, such as ResFinder from the Center for Genomic Epidemiology and the Comprehensive Antibiotic Research Database (CARD) [17,18]. Recently, an island prediction software tool (Genomic Island Prediction software [GIPSy]) was introduced for the prediction of genomic islands in bacteria [9]. Here, we investigated the whole genome sequence (WGS) of a carbapenem-resistant A. baumannii isolate belonging to GC2 and predicted the resistance islands using software tools.
METHODS
1. Bacterial strains and antimicrobial susceptibility tests
The A. baumannii YU-R612 strain was isolated from the sputum of a 61-yr-old man with sepsis in Severance Hospital, Seoul, Korea. The species was identified by conventional methods and the VITEK 32 GN system (bioMérieux, Marcy l'Etoile, France). Antimicrobial susceptibility testing was performed by using the VITEK II N211 system (bioMérieux). The YU-R612 strain was resistant to multiple antibiotics including piperacillin (minimum inhibitory concentration [MIC] ≥128 µg/mL), ceftazidime (≥64 µg/mL), cefotaxime (≥64 µg/mL), cefepime (≥64 µg/mL), imipenem (≥16 µg/mL), meropenem (≥16 µg/mL), ampicillin/sulbactam (≥32 µg/mL), ciprofloxacin (≥4 µg/mL), and gentamicin (≥16 µg/mL); however, it was susceptible to minocycline (≤1 µg/mL) and colistin (≤0.5 µg/mL). Modified Hodge tests and EDTA plus sodium mercaptoacetic acid (SMA) double-potentiation tests were carried out to screen carbapenemase and metallo-β-lactamase activity, respectively. Both tests showed negative results, which suggested the absence of metallo-β-lactamase genes in this strain.
2. Whole genome sequencing and PCR experiments
The WGS of the YU-R612 strain was obtained by using the PacBio RS II sequencing system (Pacific Biosciences Inc. Menlo Park, CA, USA) in a commercial laboratory (Macrogen, Seoul, Korea). PCR was performed by using in-house primers (Table 1).
Table 1. Nucleotide sequences of primers designed for this study.
| Primer | Sequence 5'-3' |
|---|---|
| P1 | ACCCCTTCATCATCTTCTGCA |
| P2 | TGAACAATCTGACTCGGGGT |
| P3 | GTCAGTTGCACTTGGTCGAA |
| P4 | GGCGTTCTTTGGAGTGGAAG |
| P5 | GGAATCGTTGCTGTTGGTCA |
| P6 | TGCAAAGTTAGCGATGAGGC |
| P7 | TCCAAGAGCAACGTACGACT |
| P8 | GGCCATTGACCTTCACGTAG |
3. Identification of the resistome and resistance islands
Antimicrobial resistance genes were analyzed by using ResFinder, the resources from the Center for Genomic Epidemiology (www.genomicepidemiology.org). A ResFinder threshold of ID=98% was selected. Genomic islands were determined by using resources from Genomic Island Prediction software (GIPSy) as previously described [9].
RESULTS
The A. baumannii strain YU-R612 genome consisted of a circular chromosome (ca. 4,075 kb) and two plasmids (ca. 74 kb and 5 kb). WGS analysis showed that the sequence type (ST) under the Oxford scheme was ST191 (1-3-3-2-2-94-3) and under the Institute Pasteur Multi Locus Sequence Typing (MLST) scheme was ST2, which was consistent with the assignment of this strain to GC2. ResFinder analysis showed that the YU-R612 strain possessed the following resistance genes on its chromosome: four β-lactamase genes, namely blaADC-30 (identity, 100%), blaOXA-66 (100%), blaOXA-23 (100%), and blaTEM-1 (100%); armA (100%), aadA1 (100%), and aacA4 (99.6%) as aminoglycoside resistance-encoding genes; aac(6')Ib-cr (99.2%) for fluoroquinolone resistance; msr(E) (100%) for macrolide, lincosamide, and streptogramin B resistance; catB8 (100%) for phenicol resistance; and sul1 (100%) for sulfonamide resistance.
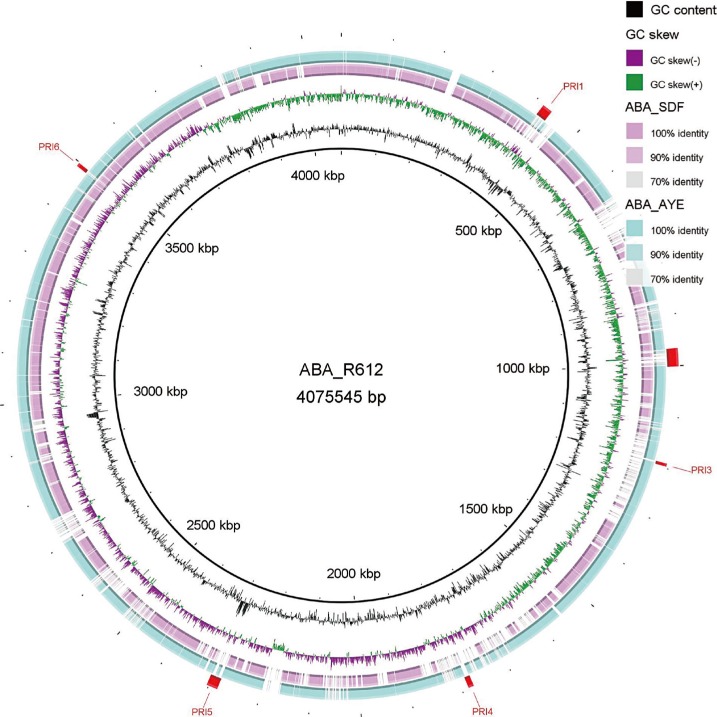
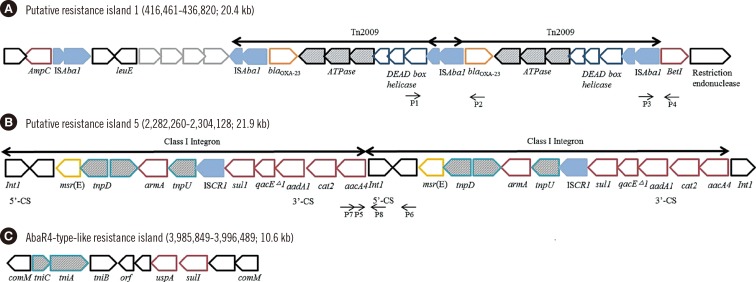
Six putative resistant islands (PRIs) were determined on the chromosome of the YU-R612 strain by using the GIPSy tool (Fig. 1). PRI1 possessed two copies of Tn2009 carrying the blaOXA-23 gene (Fig. 2A). PRI5 carried two copies of a class I integron including 5' conserved sequence (CS) and 3'CS, and their gene cassettes possessed sul1 and armA resistance genes (Fig. 2B). PRI2, PRI3, PRI4, and PRI6 carried several hypothetical proteins that could not be further annotated. Antimicrobial resistance genes detected by ResFinder were not found on PRI2, PRI3, PRI4, or PRI6.
Fig. 1. Circular genome comparison showing putative resistance islands predicted for Acinetobacter baumannii strain YU-R612 by GIPSy. The figure was plotted by using A. baumannii AYE strain as a reference and generated by using the Basic Local Alignment Search Tool (BLAST) Ring Image Generator (BRIG) software.
Abbreviations: PRI, putative resistance island; GIPSy, Genomic Island Prediction software.
Fig. 2. Structures of putative resistance island 1 (A) and putative resistance island 5 (B) identified by using GIPSy and an AbaR4-like resistance island (C) in the YU-R612 strain. The short arrows indicate the primers (P1 to P8).
Interestingly, in addition to the PRIs, a resistance island was identified that was inserted in the comM gene on the YU-R612 chromosome. The resistance island was 10.6 kb in size and very similar to AbaR4-type resistance islands, especially Tn6022, which consists of the tniC-tniA-tniB-tinE-orf-uspA-sul1 genes bracketed by a partial comM gene (Fig. 2C). There were two plasmids in the YU-R612 strain, which did not harbor PRIs or antimicrobial resistance genes.
This draft genome sequencing project has been deposited at the GenBank nucleotide database (GenBank nos. CP014215, CP014216, and CP014217) under BioProject ID PRJNA309091.
DISCUSSION
Strain YU-R612 is a multiple antibiotic-resistant isolate recovered from a patient with pneumonia. Resistance to multiple antibiotics correlated with the presence of antimicrobial resistance genes detected by ResFinder: blaOXA-23 for resistance to imipenem and meropenem, armA for gentamicin, and aac(6')Ib-cr for ciprofloxacin, which are major resistance genes for A. baumannii in Korea [19,20].
Under the Oxford scheme, the YU-R612 strain was shown to be ST191, indicating that it is a member of the GC2, which accounts for the majority of isolates recovered in Korea [19]. GC2, which includes clonal complex 92 in the MLST Oxford scheme, has also been identified among carbapenem-resistant A. baumannii strains in Asian and European countries [19]. Among the resistance islands of A. baumannii, AbaR4-type resistance islands are common in GC2 isolates [12]. Clonal dissemination and diversification of GC92 and the predominance of AbaR4-type resistance islands among these isolates have been reported in Korea [15].
AbaR4-type islands that shared a core structure similar to AbaR4 appeared to be distributed in GC2 isolates via integration into a distinct genomic site at comM. The backbone of AbaR4 comprised five open reading frames constituting the so-called transposition module with two other genes encoding the universal stress protein (uspA) and sulfate permease (sul1) [21]. In this study, a 10.6-kb AbaR4-type resistance island was detected in the YU-R612 strain. While some AbaR4-type resistance islands possess transposons such as Tn2006 and Tn1213 carrying antibiotic resistance genes such as blaOXA-23, the YU-Y612 strain did not carry any transposons or antibiotic resistance genes on its AbaR4-type-like resistance island. However, the blaOXA-23 gene on the Tn2009 of YU-R612 strain was detected in PRI1.
In this study, six PRIs were identified on the chromosome of the YU-R612 strain by using the GIPSy tool. Among them, two PRIs (PRI1 and PRI5) possessed transposons and class I integrons that carried important resistance genes: blaOXA-23, armA, and sul1. The duplication of Tn2009 in PRI1 and class I integrons in PRI5 was confirmed by using PCR to supplement a limitation of deep sequencing for the replication.
The GIPSy software facilitated the prediction of resistance islands and resistance genes in the WGS of this strain. Although some PRIs detected by software tools are different from known resistance islands, this software may be useful to identify novel genomic islands carrying clusters of resistance genes. Software tools provide a convenient way of identifying PRIs as well as acquired antimicrobial resistance genes, and they may simplify the complex steps in analyzing the WGS of bacteria in clinical microbiology laboratories.
In conclusion, the prediction of resistance islands in the WGS of a carbapenem-resistant A. baumannii strain belonging to GC2 in Korea revealed PRIs on the chromosome that carried Tn2009 and class I integrons. The predicted resistance islands using software tools helped in the analysis of the WGS in this strain.
Acknowledgments
This work was partly supported by a grant from the Korea Healthcare Technology R&D Project of the Ministry of Health, Welfare, and Family Affairs (HI12C1251-00013) and partly by a faculty research grant of Yonsei University College of Medicine for 2014 (6-2014-0039). This research was also supported by the research fund of Hanyang University (HY-201400000002367).
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee K, Yong D, Jeong SH, Chong Y. Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med J. 2011;52:879–891. doi: 10.3349/ymj.2011.52.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung HS, Lee Y, Park ES, Lee DS, Ha EJ, Kim M, et al. Characterization of the multidrug-resistant Acinetobacter species causing a nosocomial outbreak at intensive care units in a Korean teaching hospital: suggesting the correlations with the clinical and environmental samples, including respiratory tract-related instruments. Ann Clin Microbiol. 2014;17:29–34. [Google Scholar]
- 4.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 5.Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–762. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 6.Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 7.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 2014;38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 9.Soares SC, Geyik H, Ramos RT, de Sá PH, Barbosa EG, Baumbach J, et al. GIPSy: Genomic island prediction software. J Biotechnol. 2015:pii: S0168-1656(15)30115-2. doi: 10.1016/j.jbiotec.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Turton JF, Baddal B, Perry C. Use of the accessory genome for characterization and typing of Acinetobacter baumannii. J Clin Microbiol. 2011;49:1260–1266. doi: 10.1128/JCM.02335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krizova L, Dijkshoorn L, Nemec A. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob Agents Chemother. 2011;55:3201–3206. doi: 10.1128/AAC.00221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DH, Choi JY, Kim HW, Kim SH, Chung DR, Peck KR, et al. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob Agents Chemother. 2013;57:5239–5246. doi: 10.1128/AAC.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W. Comment on: AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother. 2012;67:512, author reply 513-4. doi: 10.1093/jac/dkr475. [DOI] [PubMed] [Google Scholar]
- 14.Hamidian M, Hall RM. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother. 2011;66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 15.Kim DH, Park YK, Ko KS. Variations of AbaR4-type resistance islands in Acinetobacter baumannii isolates from South Korea. Antimicrob Agents Chemother. 2012;56:4544–4547. doi: 10.1128/AAC.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nigro S, Hall RM. Distribution of the blaOXA-23-containing transposons Tn2006 and Tn2008 in Australian carbapenem-resistant Acinetobacter baumannii isolates. J Antimicrob Chemother. 2015;70:2409–2411. doi: 10.1093/jac/dkv102. [DOI] [PubMed] [Google Scholar]
- 17.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Lee J, Jeong SH, Lee J, Bae IK, Lee K. Carbapenem-non-susceptible Acinetobacter baumannii of sequence type 92 or its single-locus variants with a G428T substitution in zone 2 of the rpoB gene. J Antimicrob Chemother. 2011;66:66–72. doi: 10.1093/jac/dkq402. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Bae IK, Kim J, Jeong SH, Lee K. Dissemination of ceftazidime-resistant Acinetobacter baumannii clonal complex 92 in Korea. J Appl Microbiol. 2012;112:1207–1211. doi: 10.1111/j.1365-2672.2012.05283.x. [DOI] [PubMed] [Google Scholar]
- 21.Seputiene V, Povilonis J, Suziedeliene E. Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob Agents Chemother. 2012;56:1969–1973. doi: 10.1128/AAC.05678-11. [DOI] [PMC free article] [PubMed] [Google Scholar]