Abstract
The transcription factor NKX6.1 (NK6 homeobox 1) is important in the development of pancreatic β-cells and neurons. Although recent publications show that NKX6.1 is hypermethylated and downregulated during tumorigenesis, the function of NKX6.1 in carcinogenesis remains elusive. Here, we address the metastasis suppressor function of human NKX6.1 using cell, animal and clinical analyses. Our data show that NKX6.1 represses tumor formation and metastatic ability both in vitro and in vivo. Mechanistically, NKX6.1 suppresses cell invasion by inhibiting the epithelial-to-mesenchymal transition (EMT). NKX6.1 directly enhances the mRNA level of E-cadherin by recruiting BAF155 coactivator and represses that of vimentin and N-cadherin by recruiting RBBP7 (retinoblastoma binding protein 7) corepressor. Clinical cancer tumors with metastasis show low NKX6.1 protein expression coinciding with low E-cadherin and high vimentin expression. Our results demonstrate that NKX6.1 functions as an EMT suppressor by interacting with different epigenetic modifiers, making it a potential novel therapeutic option.
Introduction
Nkx6.1 (NK6 homeobox 1), first identified in Drosophila pancreatic β-cells,1 encodes a homeobox domain-containing protein that acts as a bifunctional transcription factor. NKX6.1 regulates insulin-secreting β-cell differentiation by binding to and activating the promoter of its own gene while simultaneously broadly repressing the activity of other genes, such as insulin.2 In addition, numerous reports have demonstrated essential functions of NKX6.1 in regional patterning and neuronal fate determination.3 Hypermethylation of the NKX6.1 gene promoter in acute lymphoblastic leukemia patients compared with controls has been verified.4 Moreover, hypermethylation of the NKX6.1 promoter is frequently detected in cervical cancer cell lines and squamous cell carcinoma tissues but not in normal cervical tissues.5 NKX6.1 has also been identified as a potential biomarker in cervical cancer screens.6 Accumulating evidence, including our own research, has shown that tumor suppressor gene inactivation is attributable to promoter hypermethylation in many types of cancer cells.7, 8, 9 Nevertheless, the fundamental biological role of NKX6.1 in carcinogenesis or cell metastasis remains elusive.
The epithelial-to-mesenchymal transition (EMT) has been well documented as a constitutive step in embryogenesis that is critical for organ development and differentiation.10 The importance of EMT in the pathogenesis of human diseases and cancers, through its involvement in organ fibrosis,11 therapeutic resistance12 and metastatic dissemination,13 has been increasingly appreciated. Growing evidence supports a complex multistep tumor metastasis process that includes the detachment of tumor cells from the basal membrane through EMT and proceeds to invasion, intravasation, circulation into blood vessels, extravasation and ultimately localization to a distant secondary organ to form a metastasis.14 EMT in carcinoma cells is defined as shedding of the differentiated epithelial phenotype, including cell–cell adhesion, apical–basal polarity and lack of motility, as well as transition to mesenchymal characteristics, including motility, invasiveness, resistance to apoptosis and, importantly, many features of tumor-initiating cells.15 Therefore, this cellular biological program, EMT, is an early and indispensable process for tumor cell dissemination and progression.
During the transition, the loss of epithelial markers, such as E-cadherin, or the acquisition of mesenchymal markers, such as vimentin or N-cadherin, is considered a vital event. Several EMT regulators, including SNAIL, SLUG, ZEB1 and TWIST, can repress E-cadherin directly or indirectly.13, 16 A set of EMT regulators become expressed and functionally activated in response to contextual oncogenic signaling cascades, such as hypoxia,17 and signaling occurring through a number of intracellular pathways, including transforming growth factor-β,18 Wnt,19 Notch,20 Hedgehog21 and epidermal growth factor receptor.22 Despite this growing awareness of the molecular players involved, the details of the mechanism that coordinately regulates epithelial genes and mesenchymal genes under the EMT program in human cancer remain poorly defined.
Here, we present data demonstrating that NKX6.1 acts as a metastasis suppressor in vitro and in vivo. Furthermore, we address how NKX6.1 inhibits cancer invasion and metastasis. Our results provide important evidence that NKX6.1 suppresses tumor metastasis through the epigenetic regulation of EMT. These findings suggest that NKX6.1 may serve as a potential therapeutic target in aggressive tumors.
Results
Ectopic expression of NKX6.1 suppresses the transformation and invasive ability of cancer cells
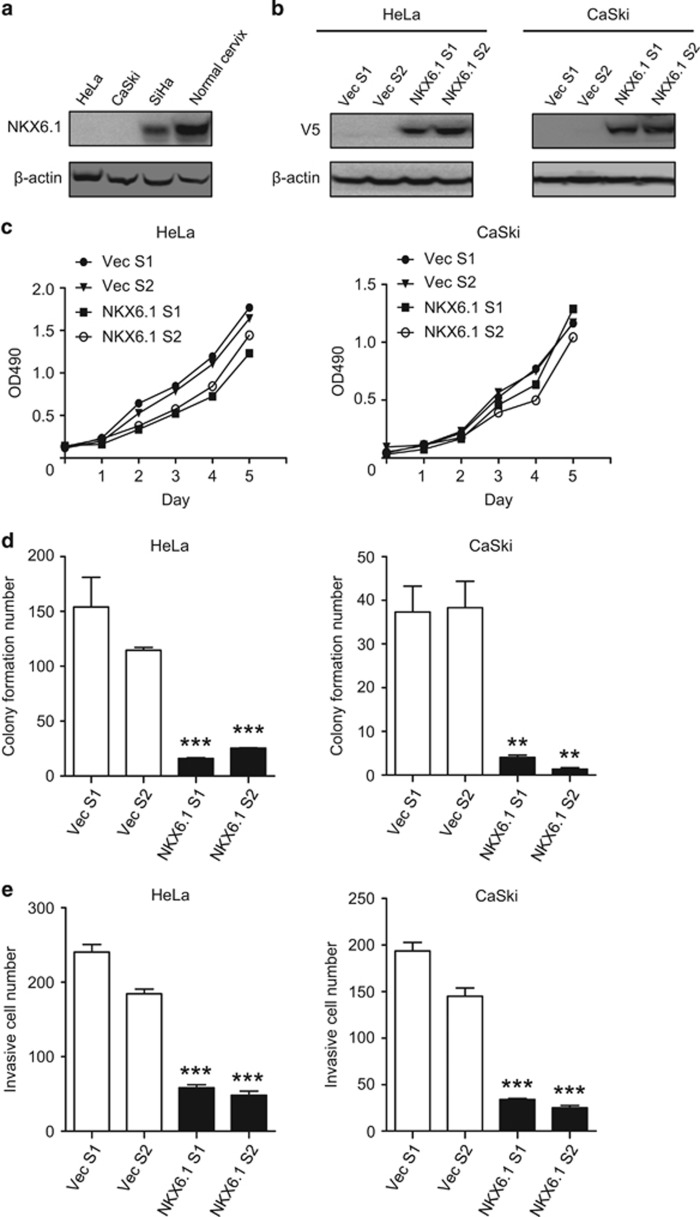
Promoter hypermethylation of NKX6.1 has been reported in cervical cancer cells but not in normal cervical cells.5 We first evaluated the protein expression of NKX6.1 in three cervical cancer cell lines and in normal cervixes. NKX6.1 protein expression was downregulated in cancer cell lines compared with normal cervixes (Figure 1a). To investigate the biological function of NKX6.1 in cancer cells in vitro, we stably overexpressed NKX6.1 in HeLa cells and CaSki cells, both of which normally show undetectable NKX6.1 expression (Figure 1b and Supplementary Figure 1a). The overexpression of NKX6.1 did not significantly affect cell viability (Figure 1c), but it did strongly reduce the colony formation ability, which indicates a suppressed cell transforming ability (Figure 1d). The stable overexpression of NKX6.1 also decreased the number of invading cells (Figure 1e). Representative images of anchorage-independent growth and invasion assays are shown in Supplementary Figures 1b and 1c.
Figure 1.
Ectopic expression of NKX6.1 suppresses the transformation and invasive ability of cancer cells in a constitutive expression system. (a) NKX6.1 expression was analyzed by western blotting in human cervical cancer cell lines and normal cervixes using an anti-NKX6.1 antibody. β-Actin was used as an internal control. (b) Expression of NKX6.1 in HeLa and CaSki cell lines stably transfected with NKX6.1 (NKX6.1 S1 and S2) or the empty vector (Vec S1 and S2) was analyzed by western blot analysis using an anti-V5 antibody. β-Actin was used as an internal control. (c) Cell proliferation (MTS) assays were performed using HeLa and CaSki cells expressing NKX6.1 or the empty vector. The absorbance values are presented as the mean±s.e. from four independent experiments. (d) Anchorage-independent growth assays were performed using HeLa and CaSki cells expressing NKX6.1 or the empty vector. (e) Matrigel invasion assays were performed using HeLa and CaSki cells expressing NKX6.1 or the empty vector. These results are presented as the mean±s.e. from three independent experiments in triplicates (analyzed by unpaired two-tailed t-test). **P<0.01 and ***P<0.001.
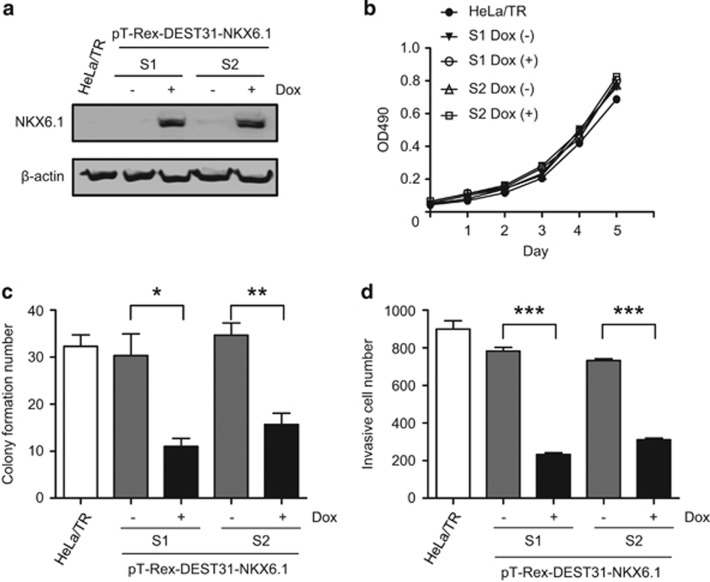
To further evaluate the suppressive effects of NKX6.1 on tumorigenesis, we established an inducible expression system in HeLa cells (termed HeLa-TR), in which NKX6.1 expression was induced by treating the cells with 1 μg/ml doxycycline (Dox). Western blotting and quantitative reverse-transcription-PCR (qRT–PCR) analyses confirmed that Dox treatment increased the NKX6.1 mRNA and protein levels (Figure 2a and Supplementary Figure 2a). We tested the effects of NKX6.1 overexpression on cell viability, transformation and cell invasion after induction for 7 days. Although the viability of NKX6.1-overexpressing cells was not significantly different from that of the controls (Figure 2b), NKX6.1 overexpression suppressed colony formation (Figure 2c and Supplementary Figure 2b) and invasion (Figure 2d and Supplementary Figure 2c). These data were consistent with the results obtained using cells stably expressing NKX6.1 and demonstrated that NKX6.1 reduces cell transformation and invasive ability.
Figure 2.
NKX6.1 suppresses the transformation and invasive ability of cancer cells in an inducible expression system. (a) Dox (1 ng/μl)-inducible NKX6.1 expression was established in HeLa cells (HeLa-TR) and was assessed by western blot analysis. β-Actin was used as an internal control. (b–d) Cell proliferation (MTS) (b), colony formation (c) and invasion (d) were performed using HeLa-TR and HeLa-TR-NKX6.1 cells. The results are presented as the mean±s.e. from three independent experiments in triplicates (analyzed by unpaired two-tailed t-test). *P<0.05, **P<0.01 and ***P<0.001.
Knockdown of NKX6.1 enhances the transformation and invasive ability of cancer cells
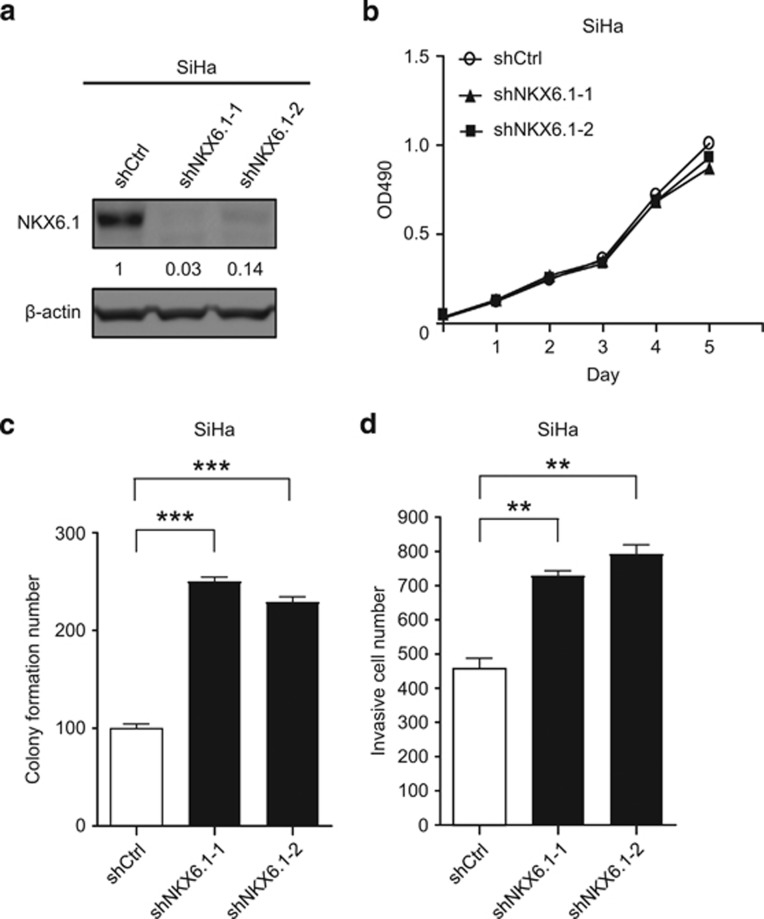
We next sought to evaluate whether the knockdown of NKX6.1 affects the malignant features of cancer cells. We infected SiHa cervical cancer cells, which show moderate NKX6.1 expression levels, with lentiviruses harboring two small hairpin RNAs (shRNAs) targeting distinct coding sequences of NKX6.1 (shNKX6.1-1 and shNKX6.1-2) or a control, nontargeting, shRNA (shCtrl). The two NKX6.1-shRNA transfectants exhibited suppressed NKX6.1 expression at both the mRNA and protein levels compared with the controls in SiHa cells (Figure 3a and Supplementary Figure 3a). NKX6.1 knockdown did not affect cell viability in SiHa cells (Figure 3b), but it did significantly enhance the cell transformation ability of SiHa cells (Figure 3c and Supplementary Figure 3b). Inhibition of NKX6.1 also enhanced the invasiveness of SiHa cells (Figure 3d and Supplementary Figure 3c). Furthermore, we also knocked down NKX6.1 in the HPV-immortalized cervical epithelial cells, Z172 cells,23 which normally show moderate NKX6.1 expression levels (Supplementary Figure 3d). NKX6.1 knockdown did not affect cell viability in Z172 cells (Supplementary Figure 3e), but it did significantly enhance the invasiveness of Z172 cells (Supplementary Figure 3f). Collectively, these results indicated that the loss of NKX6.1 increases the transformation and invasion abilities of cancer cells in vitro.
Figure 3.
Knockdown of NKX6.1 enhances transformation and invasive ability of cancer cells. (a) Expression of NKX6.1 in SiHa cells infected with lentiviruses harboring control shRNA (shCtrl) or NKX6.1 shRNA (shNKX6.1-1, shNKX6.1-2) was analyzed by western blot analysis. β-Actin was used as an internal control. The numbers in the western blots represent the ratios of targets to the internal control. (b–d) Cell proliferation (MTS assay) (b), colony formation (c) and invasion (Matrigel assay) (d) were assessed using SiHa cells. The data are presented as the mean±s.e. from three independent experiments in triplicates (analyzed by unpaired two-tailed t-test). **P<0.01 and ***P<0.001.
NKX6.1 suppresses tumorigenicity and metastasis in xenograft models
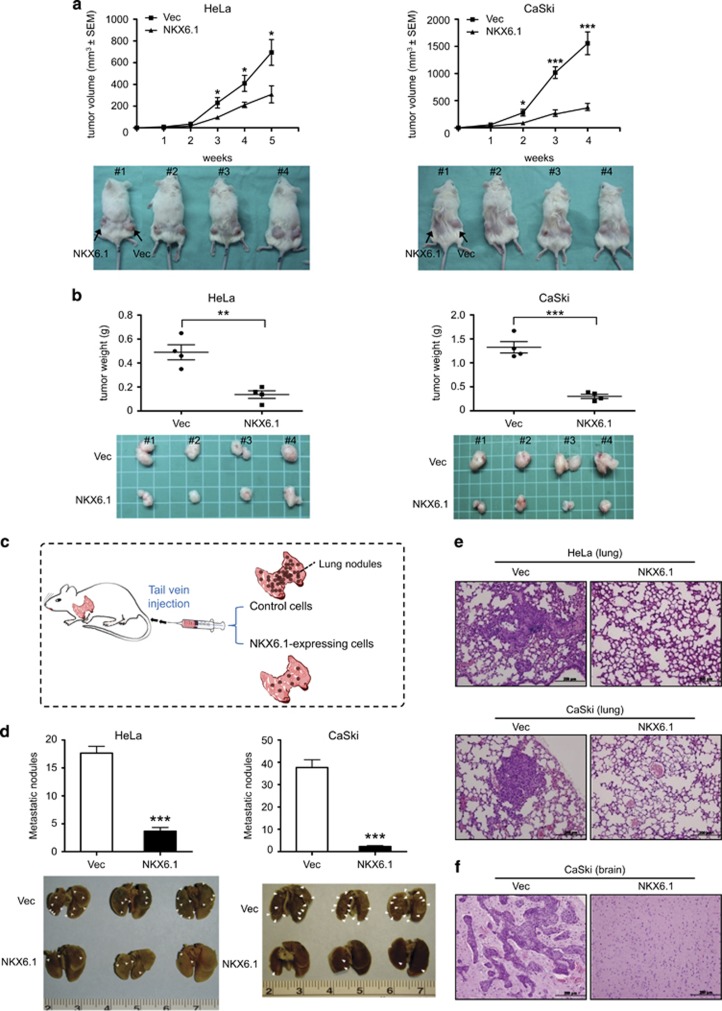
To explore the effect of NKX6.1 on tumorigenicity in vivo, we subcutaneously injected 106 HeLa or CaSki cells expressing NKX6.1 or their corresponding controls into both flanks of NOD-SCID (non-obese diabetic severe-combined immunodeficiency) mice. The tumor growth rate was slower and the tumor mass was smaller with NKX6.1-expressing HeLa or CaSki cells compared with the corresponding controls (Figures 4a and b).
Figure 4.
NKX6.1 suppresses tumor formation and metastasis in vivo. HeLa and CaSki cells transfected with the empty vector (right) or an NKX6.1 expression plasmid (left) were subcutaneously injected into NOD-SCID mice. The tumor growth curve (a) and tumor weights (b) of the NKX6.1-expressing cells were compared with that of cells harboring the vector only. (c) Schema for representing the metastasis assay model in vivo. (d) Control or NKX6.1-expressing HeLa or CaSki cells were injected into NOD-SCID mice via the tail vein. The lower images depict lungs excised from the mice; the arrows indicate lung nodules. (e and f) Hematoxylin and eosin staining of lungs (e) and brains (f) (original magnification, × 200) excised from the mice shown in (d). Scale bar=200 μm. The values are expressed as the mean±s.e. from three or four independent experiments (analyzed by unpaired two-tailed t-test). *P<0.05, **P<0.01 and ***P<0.001.
To further investigate the role of NKX6.1 in metastasis, we injected NKX6.1-expressing cells and their corresponding controls or NKX6.1 knockdown SiHa cells and their corresponding controls into mice via the tail vein. Two months after injection, there were fewer metastatic lung nodules and no brain metastases in the NKX6.1-expressing HeLa or CaSki cells compared with that in the controls (Figures 4c–f). In contrast, there were more metastatic lung nodules in NKX6.1 knockdown SiHa cells compared with that in the controls (Supplementary Figures 4a and c). Taken together, these data demonstrated that NKX6.1 has a critical role in suppressing tumor formation and metastatic behavior in xenograft models.
NKX6.1 represses cancer invasion by regulating EMT
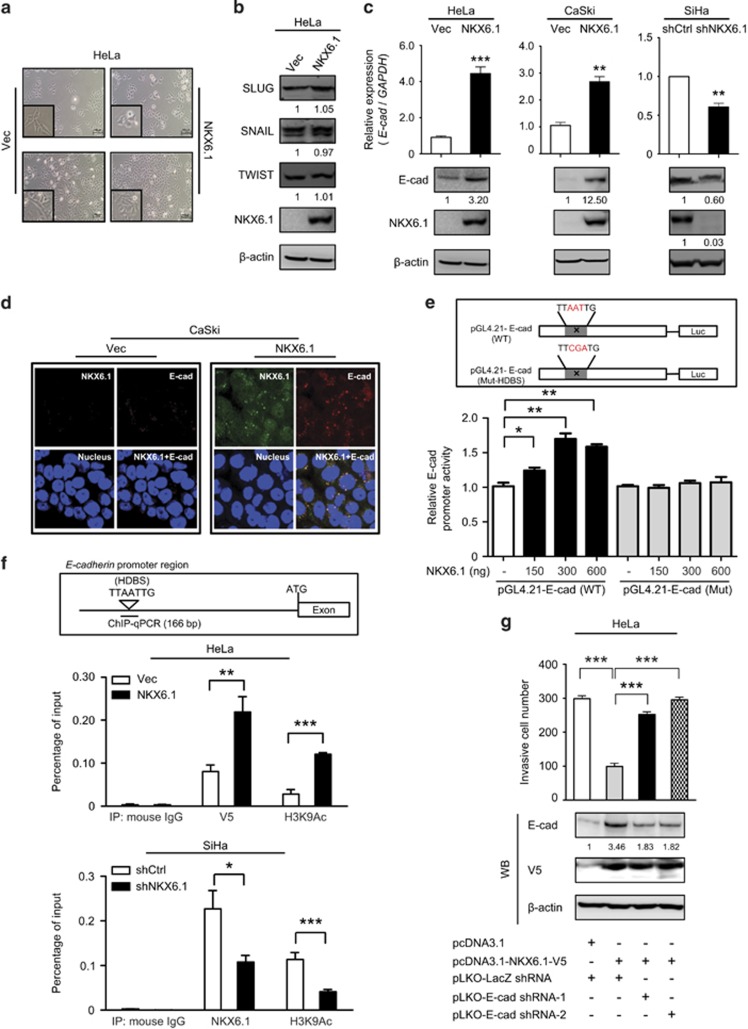
Furthermore, we sought to determine how NKX6.1 inhibits cancer invasion and metastasis. Our preliminary results showed that NKX6.1 overexpression enhanced E-cadherin expression and repressed vimentin expression. NKX6.1-expressing HeLa cells exhibited a more epithelial-like phenotype compared with the control cells, which displayed fibroblast-like mesenchymal features (Figure 5a). EMT in carcinoma cells is defined as a shedding of the differentiated epithelial phenotype and the transition to mesenchymal characteristics.10 We first analyzed the expression of SNAIL, TWIST and SLUG to assess the effect of NKX6.1 on EMT regulators. The ectopic expression of NKX6.1 did not significantly affect the expression of SNAIL, TWIST and SLUG in HeLa cells (Figure 5b), but it did increase E-cadherin at both the transcriptional and protein levels in HeLa or CaSki cells (Figure 5c). Moreover, knockdown of NKX6.1 in SiHa cells decreased E-cadherin expression at the mRNA and protein levels (Figure 5c). A similar regulation of E-cadherin expression by NKX6.1 was observed in CaSki cells using immunofluorescence (IF) (Figure 5d). These results showed that NKX6.1 regulates E-cadherin at the transcriptional level.
Figure 5.
NKX6.1 represses cancer invasion by increasing the expression of epithelial markers. (a) Representative images of the morphology of HeLa cells expressing NKX6.1 or the vector only at low (upper image) and high (lower image) cell density (original magnification, × 100). (b) The expression of EMT regulators was detected by western blotting in NKX6.1-expressing HeLa cells. (c) The expression of E-cadherin was analyzed in NKX6.1-expressing cells and NKX6.1-shRNA-knockdown cells by qRT–PCR (bar graphs) and western blotting (below). (d) IF assays were used to detect E-cadherin expression in NKX6.1-expressing CaSki cells. (e) The upper scheme depicts WT (pGL4.21-E-cadherin) and HDBS-mutated (Mut; pGL4.21-E-cadherin) E-cadherin promoters. The activity of different E-cadherin promoter constructs in HeLa cells was analyzed by a luciferase reporter assay. (f) Chromatin from HeLa cells expressing NKX6.1 or SiHa cells expressing NKX6.1 shRNAs was immunoprecipitated with indicated antibodies and then analyzed by quantitative PCR using E-cadherin-specific primers. The upper scheme depicts the position of the HDBS within the promoter region of E-cadherin. (g) HeLa cells were transfected with the indicated combinations of vectors, and Matrigel invasion assays were used to analyze the effects on cancer invasion. Two specific E-cadherin shRNAs (nos 1 and 2) were used. The numbers in the western blots represent the ratios of targets to the internal control. The data are presented as the mean±s.e. from three independent experiments in triplicates (analyzed by unpaired two-tailed t-test). *P<0.05, **P<0.01 and ***P<0.001.
In support of this finding, a putative NKX6.1 homeodomain-binding site (designated HDBS)2, 24 has been identified at position −1276 bp relative to the transcription start site of the E-cadherin locus. On the basis of this information, we hypothesized that NKX6.1 directly regulates E-cadherin transcription. Using promoter-luciferase reporter assays, we showed that the overexpression of NKX6.1 enhanced E-cadherin promoter activity in a dose-dependent manner. Moreover, mutation of the HDBS25 in the E-cadherin promoter abolished the observed activation by NKX6.1 (Figure 5e). Electrophoretic mobility shift assays (EMSAs) revealed that a wild-type (WT) HDBS containing the E-cadherin oligonucleotide probe interacted with NKX6.1-containing nuclear extracts and that this interaction was abolished by the addition of excess unlabeled competitor in HeLa cells (Supplementary Figure 5a). In contrast, an HDBS E-cadherin probe containing a mutated NKX6.1 HDBS was unable to interact with NKX6.1-containing nuclear extracts (Supplementary Figure 5a, lane 6).
To confirm that NKX6.1 binds to the endogenous E-cadherin promoter, we performed quantitative chromatin immunoprecipitation (qChIP) analyses. Our data demonstrated that NKX6.1 directly binds to the HDBS within the E-cadherin promoter. Notably, this binding was accompanied by an increase in H3K9 acetylation, a marker of transcriptionally active chromatin, in NKX6.1-expressing HeLa and CaSki cells, and this binding was accompanied by a decrease in H3K9 acetylation in NKX6.1 knockdown SiHa cells (Figure 5f and Supplementary Figures 5b and c). Thus, our data confirmed that NKX6.1 directly binds to the E-cadherin promoter through the HDBS. To further examine if E-cadherin mediates NKX6.1-induced suppression of the invasive property, we silenced E-cadherin expression using two shRNAs (Supplementary Figure 5d) and found that E-cadherin knockdown restores invasiveness in NKX6.1-overexpressing HeLa (Figure 5g and Supplementary Figure 5e) and CaSki (Supplementary Figure 5f) cells. These data indicated that NKX6.1 suppresses cancer invasion by directly binding to the E-cadherin promoter and activating its expression.
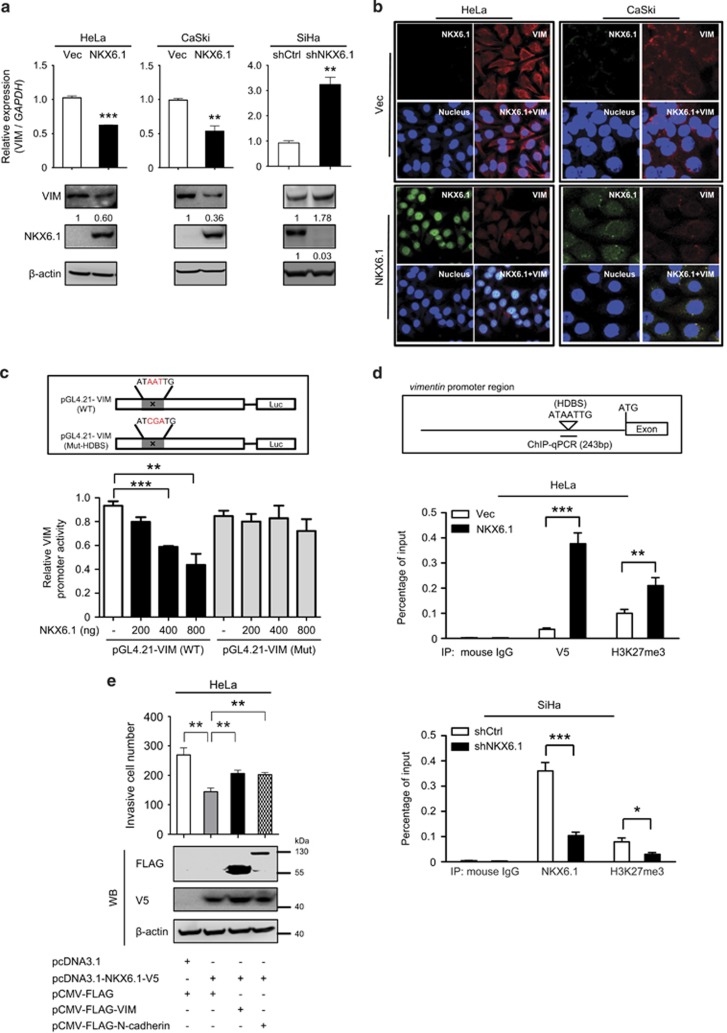
The gain of mesenchymal markers is another hallmark of the EMT process. Thus, we next sought to determine whether vimentin or N-cadherin is involved in the NKX6.1-mediated suppression of cell invasion. In contrast to the effects of NKX6.1 overexpression on E-cadherin, the overexpression of NKX6.1 in either HeLa or CaSki cells induced a decrease in the vimentin and N-cadherin mRNA and protein levels that was similar in both cell lines, as shown by qRT–PCR, western blotting and IF assays (Figures 6a and b and Supplementary Figure 6a). Conversely, knockdown of NKX6.1 in SiHa cells enhanced vimentin expression at both the mRNA and protein levels (Figure 6a). A putative NKX6.1 HDBS in the proximal promoter of the vimentin gene was detected at position −315 bp relative to the transcription start site. The overexpression of NKX6.1 decreased vimentin promoter activity, and site-directed mutagenesis of the putative HDBS within the vimentin promoter prevented this repression in HeLa cells (Figure 6c). EMSAs further showed that a WT HDBS-containing vimentin oligonucleotide probe interacted with NKX6.1-containing nuclear extracts and that this interaction was abolished by the addition of excess unlabeled competitor in HeLa cells (Supplementary Figure 6b). In contrast, a vimentin probe containing a mutated NKX6.1 HDBS was unable to interact with these NKX6.1-containing nuclear extracts (Supplementary Figure 6b, lane 5).
Figure 6.
NKX6.1 represses cancer invasion by decreasing the expression of mesenchymal markers. (a) The expression of vimentin in NKX6.1-expressing cells and NKX6.1-shRNA-knockdown cells was assessed by qRT–PCR (bar graphs) and western blotting (below). The numbers in the western blots represent the ratios of targets to the internal control. (b) IF assays were used to detect vimentin expression in NKX6.1-expressing HeLa and CaSki cells. (c) The upper scheme depicts WT (pGL4.21-VIM) and HDBS-mutated (Mut; pGL4.21-VIM) promoters of the vimentin gene. The activity of different vimentin promoter constructs in HeLa cells was analyzed by a luciferase reporter assay. (d) Chromatin from HeLa cells expressing NKX6.1 or SiHa cells expressing NKX6.1 shRNAs was immunoprecipitated with indicated antibodies and then analyzed by quantitative PCR using vimentin-specific primers. The upper scheme depicts the position of the HDBS within the promoter region of vimentin. (e) HeLa cells were transfected with the indicated combination of vectors, and Matrigel invasion assays were used to analyze the effects on cancer invasion. The data are presented as the mean±s.e. from three independent experiments in triplicates (analyzed by unpaired two-tailed t-test). *P<0.05, **P<0.01 and ***P<0.001.
Subsequent qChIP assays revealed that NKX6.1 overexpression in HeLa and CaSki cells increased the binding of NKX6.1 to the HDBS within the endogenous vimentin locus in association with an increase in the levels of H3K27me3, a marker of transcriptionally inactive chromatin. Conversely, knockdown of NKX6.1 in SiHa cells attenuated NKX6.1 binding and reduced the H3K27me3 levels (Figure 6d and Supplementary Figures 6c and d). A similar effect of NKX6.1 directly binding to the N-cadherin promoter through an HDBS (position −1835 bp relative to the transcription start site of the N-cadherin locus) was observed by qChIP assays in SiHa cells (Supplementary Figure 6e). Notably, the overexpression of vimentin or N-cadherin in NKX6.1-expressing HeLa and CaSki cells reversed the conversion to a mesenchymal phenotype (Figure 6e and Supplementary Figures 6f and h). Collectively, these data indicated that the mesenchymal genes, vimentin and N-cadherin, are both direct targets of NKX6.1, and these data suggested that decreases in the expression of these genes contribute to the suppressive effect of NKX6.1 on cancer invasion.
NKX6.1 suppresses EMT by interacting with the BAF155 (coactivator) and RBBP7 (corepressor) epigenetic modifiers
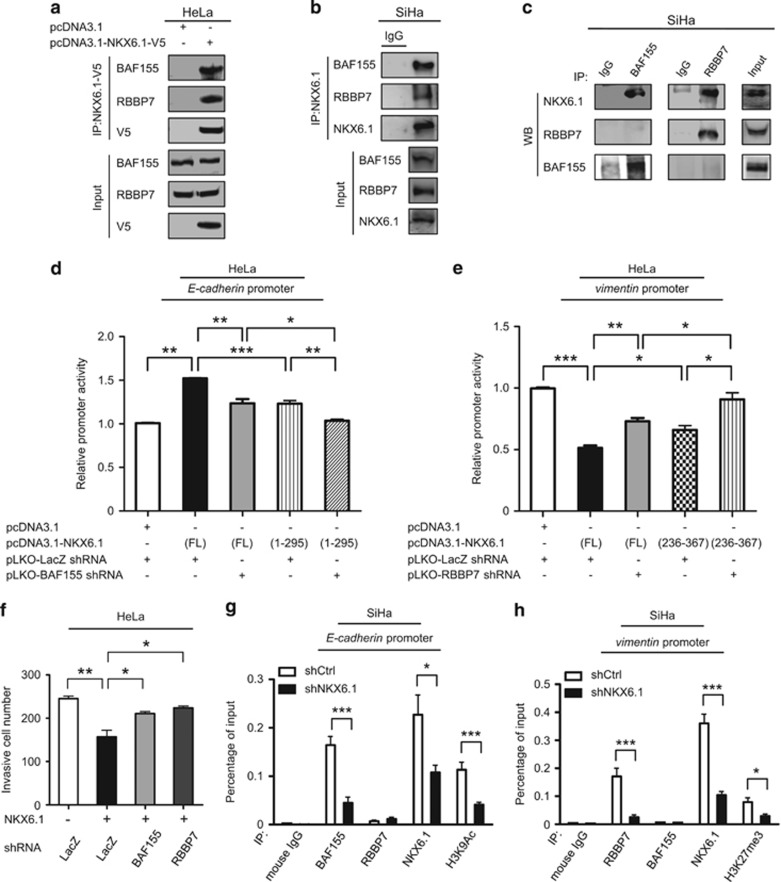
To test our hypothesis that NKX6.1 may interact with different cofactors to function as an activator or repressor involved in EMT regulation, we identified NKX6.1 interaction partners. First, we used an anti-V5 agarose affinity gel to pull down NKX6.1-associated proteins in V5-tagged NKX6.1-expressing HeLa cells and then performed liquid chromatography coupled with tandem mass spectrometry (Supplementary Figure 7a). A gene ontology analysis of candidate proteins using PANTHER (http://www.pantherdb.org/)26 showed that many of these proteins are involved in binding ability (Supplementary Figure 7b, left) with a portion of the subcategory being related to chromatin binding (Supplementary Figure 7b, right). These proteins included two chromatin modifiers as follows: SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily c, member 1 (SMARCC1 or BAF155) and retinoblastoma binding protein 7 (RBBP7).
BAF155 is a member of the SWI/SNF family, and members of this family display ATPase activities and are thought to regulate the transcription of certain genes by altering the chromatin structure around those genes.27 RBBP7 is a ubiquitously expressed nuclear protein that belongs to the polycomb repressive complex 2 and possesses HMT activity with specificity for Lys 9 (K9) and Lys 27 (K27) of histone H3.28 Because we were interested in the epigenetic mechanisms connected to NKX6.1, we further validated that NKX6.1 interacts with BAF155 or RBBP7 by using the V5 antibody to pull down endogenous BAF155 or RBBP7 in V5-tagged NKX6.1-expressing HeLa cells in coimmunoprecipitation (co-IP) assays (Figure 7a). Moreover, using an anti-FLAG antibody to pull down exogenous NKX6.1 in HeLa cells coexpressing FLAG-tagged BAF155 or RBBP7 and V5-tagged NKX6.1, we revealed the association of NKX6.1/BAF155 or NKX6.1/RBBP7, respectively (Supplementary Figures 7c and d). We also verified the interaction of NKX6.1 with BAF155 or RBBP7 at the endogenous level in SiHa cells (Figure 7b). Interestingly, the co-IP data showed that BAF155 and RBBP7 were mutually exclusive (Figure 7c). To determine whether the observed influence of BAF155 or RBBP7 on the transcriptional activity of NKX6.1 depends on its activation or repression domain, V5-tagged NKX6.1 full-length (FL)1 and two truncated NKX6.1 constructs, namely NKX6.1 (amino acids (aa) 1–295; lacking activation domain) and NKX6.1 (aa 236–367; lacking repression domain), were generated and expressed in HeLa cells (Supplementary Figure 7e). Depletion of BAF155 or overexpression of NKX6.1 (aa 1–295) reversed the E-cadherin promoter activity enhanced by the overexpression of NKX6.1. As expected, depletion of BAF155 and overexpression of NKX6.1 (aa 1–295) together showed a stronger suppressive effect on E-cadherin promoter activity in HeLa cells (Figure 7d). In contrast, the depletion of RBBP7 did not affect promoter activity (Supplementary Figure 7f, left). In addition, knockdown of RBBP7 or overexpression of NKX6.1 (aa 236–367) rescued the vimentin promoter activity repressed by NKX6.1 overexpression. Both knockdown of RBBP7 and overexpression of NKX6.1 (aa 236–367) had strong effects (Figure 7e), whereas knockdown of BAF155 did not alter promoter activity in HeLa cells (Supplementary Figure 7f, right).
Figure 7.
NKX6.1 suppresses EMT by interacting with the BAF155 (coactivator) and RBBP7 (corepressor) epigenetic modifiers. (a) Immunoprecipitates were prepared from HeLa cells after transfection with V5-tagged NKX6.1. (b and c) To identify interactions between endogenous NKX6.1 with BAF155 or RBBP7, immunoprecipitates were prepared from SiHa cells (no transfection). Cell lysates and immunoprecipitates obtained with anti-NKX6.1 (IP: NKX6.1) antibodies were blotted with anti-BAF155, anti-RBBP7 and anti-NKX6.1 antibodies (b); those obtained with anti-BAF155 (IP: BAF155) or anti-RBBP7 (IP: RBBP7) antibodies were blotted using anti-BAF155, RBBP7 and NKX6.1 antibodies (c). (d and e) HeLa cells transfected with the E-cadherin luciferase construct (d) or vimentin luciferase construct (e), and the indicated expression vectors were analyzed by a luciferase reporter assay. (f) HeLa cells were transfected with the indicated combinations of vectors, and Matrigel invasion assays were used to analyze the effects on cancer invasion. (g and h) Chromatin from SiHa cells expressing NKX6.1 shRNA was immunoprecipitated with indicated antibodies and then analyzed by quantitative PCR using E-cadherin-specific primers (g) or vimentin-specific primers (h). The data are presented as the mean±s.e. from three independent experiments in triplicates (analyzed by unpaired two-tailed t-test). *P<0.05, **P<0.01 and ***P<0.001.
When we functionally analyzed the NKX6.1 interaction with BAF155 or RBBP7, we found that knockdown of BAF155 or RBBP7 rescued the invasive ability repressed by NKX6.1 overexpression in HeLa cells (Figure 7f and Supplementary Figure 7g). Subsequently, we performed ChIP assays using an antibody specific for BAF155 and found a reduction in BAF155 binding to the HDBS within the E-cadherin promoter in NKX6.1 knockdown SiHa cells, and this reduction was accompanied by a decrease in the H3K9 acetylation level. Importantly, RBBP7 did not bind to the HDBS within the E-cadherin promoter (Figure 7g). Moreover, we also found that a reduction in RBBP7 binding to the HDBS within the vimentin promoter in NKX6.1 knockdown SiHa cells was accompanied by a decrease in the H3K27me3 level. Notably, BAF155 did not bind to the HDBS within the vimentin promoter (Figure 7h). The ChIP data showed that BAF155 and RBBP7 are mutually exclusive binding patterns. However, the loss of binding was not because of a decrease in BAF155 or RBBP7 expression in the NKX6.1 knockdown SiHa cells (Supplementary Figure 7h). These data indicated that NKX6.1 may interact with the BAF155 coactivator to function as an activator and that NKX6.1 may interact with the RBBP7 corepressor to function as a repressor in the regulation of EMT-related markers.
Clinical correlation of NKX6.1 and E-cadherin or vimentin expression
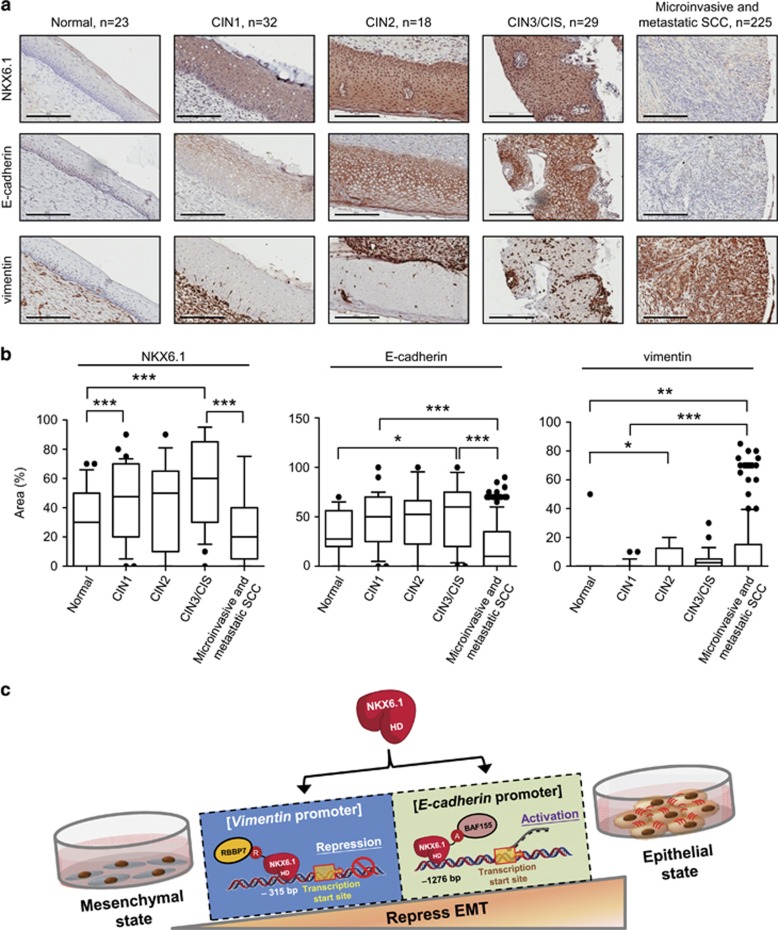
To assess the correlation of NKX6.1 and E-cadherin or vimentin expression in cancer specimens, we performed immunohistochemical staining of tissue microarrays of cervical cancer samples representing various stages of cancer. Immunohistochemical staining showed that NKX6.1 was expressed at low levels in normal epithelium, and the expression of NKX6.1 gradually increased from normal squamous epithelium to cervical intraepithelial neoplasia (CIN)-1, CIN2 and CIN3/CIS (carcinoma in situ) but decreased in microinvasive and metastatic lesions (Figures 8a and b and Supplementary Figure 8). These findings indicated a strong correlation between the loss of NKX6.1 expression and metastasis in cancer patients.
Figure 8.
Clinical correlation of NKX6.1 and E-cadherin or vimentin expression. (a) Representative results for NKX6.1 (upper), E-cadherin (middle) and vimentin (lower) immunohistochemical staining in samples representing a full spectrum of cervical cancer. Scale bar=200 μm. (b) Immunohistochemical staining data are expressed as the area percentage of positive staining of NKX6.1, E-cadherin and vimentin. The median for each group is indicated by a horizontal bar (*P<0.05, **P<0.01 and ***P<0.001, Mann–Whitney U-test). The details of grading are described in the Materials and methods section. (c) A proposed model of NKX6.1-suppressed EMT. NKX6.1 can bind to the promoter of E-cadherin and functions as an activator via its activating domain to interact with the coactivator BAF155. NKX6.1 also binds to the promoter of vimentin and functions as a repressor via its repression domain to interact with corepressor RBBP7. NKX6.1 is a bifunctional transcription factor in EMT regulation. A, activation domain; HD, homeodomain; R, repression domain.
E-cadherin was more weakly expressed in microinvasive and metastatic lesions compared with that in the CIN group, whereas vimentin had much higher expression levels in cases of malignant lesions compared with that in the CIN group (Figures 8a and b and Supplementary Figure 8). Compared with NKX6.1 expression in cervical cancer patients, NKX6.1 high expression cases showed significantly higher E-cadherin expression (47.8%) compared with NKX6.1 low expression cases (15.7% P<0.001) (Supplementary Table 1), revealing a positive correlation. In contrast, NKX6.1 low expression cases showed higher vimentin expression (23.2%) compared with that in cases with NKX6.1 high expression (13% P=0.05) (Supplementary Table 2), displaying a negative correlation. These clinical data suggested that NKX6.1 enhances E-cadherin expression and represses vimentin expression to inhibit cancer cell metastatic behavior.
Discussion
Mammalian NKX6.1 is a transcription factor that has an important role in pancreatic and neural development by regulating downstream targets. However, whether NKX6.1 is important in cancer and EMT-related processes has not been reported previously. Here, we provided evidence from cervical cancer cells, animal samples and clinical samples that showed that NKX6.1 is a bona fide metastasis suppressor that regulates EMT. Taking our findings together, we demonstrated that NKX6.1 activates epithelial gene expression and represses mesenchymal gene expression at the transcriptional level by interacting with different cofactors, namely BAF155 and RBBP7. A proposed model of NKX6.1-suppressed EMT based on our findings is presented in Figure 8c.
Our tissue microarray results showed that NKX6.1 expression is higher in precancerous lesions compared with that in normal epithelium. Therefore, we hypothesized that increased NKX6.1 expression in the early stages of tumor development may reflect a response to oncogenic signaling stress, similar to that of LMX1A, in cervical cancer.7, 29 Specificity protein 1 (Sp1) belongs to the Kruppel-like family of transcription factors, which bind to GC-rich promoter elements and contribute to tumorigenesis by regulating gene transcription related to proliferation or metastasis, and Sp1 protein expression is similar to that of the NKX6.1 protein in the development of squamous cell carcinoma.30 Moreover, Sp1 downregulation caused by protein instability leads to the loss of the transcriptional activation of E-cadherin, and consequently increases the amount of nuclear β-catenin for gene transcription related to metastasis in highly invasive lung tumor cells.31 Our preliminary data showed that Sp1 can enhance NKX6.1 promoter activity (data not shown). Taken together, these data suggest that Sp1 may be an activator of NKX6.1 in the early stages of tumor development.
It has been reported that the direct binding of NKX6.1 to DNA via its NH2-terminal domain is involved in Groucho-mediated transcriptional repression during neural patterning in the ventral neural tube and in pancreatic β-cell differentiation.2, 32 Moreover, NKX6.1 is capable of activating NKX6.1 gene transcription through interactions involving its COOH-terminal acidic domain. Nkx6.1 has been shown to mediate this feedback activation by recruiting transcription factor IIB and transcription factor IID to its promoter.2 Drosophila Nkx6.1 has also been reported to both activate and repress downstream target gene expression in the appropriate context.33 Taken together, these data suggest that Nkx6.1 likely has a bifunctional role in embryos and during β-cell differentiation. Our data showed that NKX6.1 functions as a bifunctional transcription factor in suppressing the EMT process in cancer cells by reciprocally modulating the expression of the epithelial marker, E-cadherin, and the mesenchymal marker, vimentin. In addition, our ChIP assays showed higher levels of K9-acetylated histone H3, a marker of transcriptionally active chromatin, at the E-cadherin promoter in NKX6.1-overexpressing cells compared with the levels observed in control cells (Figure 5f). Conversely, we found that the levels of K27-trimethylated histone H3, a marker of transcriptionally silent chromatin, were clearly higher at the vimentin promoter in NKX6.1-overexpressing cells compared with that in control cells (Figure 6d). These results suggested that EMT-related markers are modulated by NKX6.1 in a context-dependent manner and that NKX6.1 interacts with different modifiers of histone and chromatin by associating with either the repression domain or the activation domain of NKX6.1.
Several transcription factors have been known to function as both a repressor and an activator by interacting with different cofactors, such as nuclear receptors.34 To clarify this phenomenon, we selected two chromatin-remodeling cofactors, namely BAF155 and RBBP7, as potential candidates based on our liquid chromatography coupled with tandem mass spectrometry data. BAF155 contributes to the transcriptional activation of the nuclear glucocorticoid receptor target,35 and the re-expression of BAF155 in ovarian and colon cancer cell lines leads to reduced transformation ability, which implies that it is a tumor suppressor gene.36 Additionally, TWIST has been reported to suppress E-cadherin by interacting with several components of the nucleosome remodeling complex, including RBBP7, and recruiting them to proximal regions of the E-cadherin promoter for transcriptional repression.37 However, the functional interaction of NKX6.1 and BAF155 or RBBP7 in the regulation of EMT remains unclear.
As expected, we demonstrated that NKX6.1 directly activates E-cadherin by interacting with the BAF155 coactivator, and this activation is accompanied by an increase in H3K9 acetylation levels. Simultaneously, NKX6.1 directly represses vimentin by interacting with the RBBP7 corepressor, and this repression is accompanied by an increase in the H3K27me3 levels. Some EMT regulators, such as SNAIL, also mediate their repression function on the E-cadherin gene by recruiting histone modifiers, including histone deacetylase-1,38 lysine-specific demethylase-1,39 G9a methyltransferase40 and suppressor of variegation 3-9 homolog 1,41 as well as polycomb repressive complex 2 components, such as Suz12 and EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit).42 TWIST, another EMT regulator, has been reported to bind to the E-cadherin promoter by physically interacting with the BMI1 (polycomb repressive complex 1 subunit) and EZH2 (polycomb repressive complex 2 subunit) chromatin-modifying factors and forming a repressive complex in head and neck squamous cell carcinoma.43 Such chromatin changes are concordant with a programmed epigenetic switch related to EMT.44 Collectively, these studies indicate that histone modifiers are required for the bifunctional properties of NKX6.1.
The current study demonstrated that NKX6.1 functions as an EMT suppressor, thereby suppressing the metastatic behavior of cancer cells. Considered together, our findings suggest a therapeutic strategy for suppressing EMT based on re-expressing NKX6.1 or the use of epigenetic drugs. Thus, our results shed light on potential future therapeutic approaches for changing this phenotype, which is relevant to cancer metastasis.
Materials and methods
Cell culture
All the human cervical cancer cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were used within 6 months of thawing. HPV16 DNA-immortalized human cervical epithelial cells (Z172)23 were obtained from Dr CY Chou (National Cheng Kung University, Tainan, Taiwan). Detailed descriptions were conducted as described previously.29
Plasmids, shRNA clones and constructs
Detailed descriptions are available in Supplementary Materials and methods and Methods. All clones were verified by sequencing.
Generation of cells overexpressing stable or inducible transfects
Assays for generation of cells overexpressing stable or inducible transfects were conducted as described previously.9
Lentiviruses production, infection and gene silencing
Assays for lentiviruses production, infection and gene silencing were conducted as described previously.44
Real-time PCR and immunoblotting
Assays for real-time PCR and immunoblotting were conducted as described previously.7, 9 These primers used in real-time PCR were shown in Supplementary Table 3. The following primary antibodies were used in immunoblotting assay: anti-V5 (MCA1360; AbD Serotec, Raleigh, NC, USA), anti-NKX6.1 (generated by Yao-Hong Biotechnology Inc., Taipei, Taiwan), anti-E-cadherin (610404; BD Biosciences, San Jose, CA, USA), anti-vimentin (sc-6260; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-N-cadherin (610920; BD Biosciences), anti-SNAIL (GTX100754; GeneTex, Irvine, CA, USA), anti-TWIST (GTX127310; GeneTex) and anti-SLUG (sc-10436; Santa Cruz Biotechnology), anti-BAF155 (GTX114777; GeneTex) and anti-RBBP7 (sc-8272; Santa Cruz Biotechnology). Horseradish peroxidase-conjugated rabbit anti-mouse or goat anti-rabbit secondary antibodies (Santa Cruz Biotechnology) were used as appropriate.
Assays for cell viability, anchorage-independent growth and invasion assay
Assays for cell viability by MTS(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, Inner Salt), anchorage-independent growth and invasion were conducted as described previously.7
In vivo tumor xenograft and metastasis model
Six-week-old NOD-SCID female mice were used in the tumorigenicity and metastasis analysis. All animal studies were approved by the Institutional Animal Care and Use Committee of the National Defense Medical Center (Taipei, Taiwan). Detailed tumor xenograft and metastasis analyses were conducted as described previously.7, 45
Immunofluorescence
Assays for IF were conducted as described previously.9 The following antibodies were used in IF assay: anti-E-cadherin-eFluor (eBioscience, San Diego, CA, USA), vimentin-eFluor (eBioscience) or V5-FITC (AbD Serotec).
Site-directed mutagenesis and luciferase reporter assay
Assays for site-directed mutagenesis and luciferase reporter assay were conducted as described previously.29 The primers used to mutate homeodomain-binding site for site-directed mutagenesis assay were listed in Supplementary Table 3.
Electrophoretic mobility shift assay
The EMSA assay was performed by EMSA Gel-Shift Kit (Panomics, Inc., Santa Clara, CA, USA) according to manufacturer's protocol. WT and mutant probes of E-cadherin and vimentin were shown in Supplementary Table 3. An anti-V5 antibody (MCA1360; AbD Serotec) was added in supershift experiments.
ChIP assays
ChIP assays were performed using an EZ-Magna ChIP G Kit (Millipore, Billerica, MA, USA) according to the manufacturer's protocol. Detailed descriptions are available in Supplementary Materials and methods.
Immunoprecipitation
IP assays were conducted as described previously.9 Detailed descriptions are available in Supplementary Materials and methods.
Protein identification by mass spectrometry
The evidence of NKX6.1 bound to BAF155 or RBBP7 was performed by liquid chromatography coupled with tandem mass spectrometry according to the manufacturer's protocol. Shotgun proteomic identification by a nanoLC−nanoESi-MS/MS analysis was performed using a nanoAcquity system (Waters, Milford, MA, USA) connected to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nanospray interface (Proxeon, Odense, Denmark).
Tissue microarrays and Immunohistochemistry
Tissue microarrays were obtained from commercial sources (US Biomax, Rockville, MD, USA; Pantomics). A total of 23 normal cervix samples, 79 samples of CIN and 225 microinvasive and metastatic cervical squamous carcinomas were tested. Among CIN samples, 32, 18 and 29 were graded I, II and III, respectively. Detailed descriptions are available in Supplementary Materials and methods.
Statistical analysis
In vitro and in vivo experimental results were analyzed by unpaired two-tailed Student's t-test. P-values <0.05 were considered significant and the results are presented as the mean±s.e. from three independent experiments in triplicates. A Mann–Whitney U-test was used to evaluate the statistical significance of the IHC results and Fisher's exact test was used to compare the altered frequency of NKX6.1 and E-cadherin or vimentin between cervical cancer samples representing various stages of cancer. The statistical analyses were performed using GraphPad Prism software, version 5.0 (GraphPad Software Inc., San Diego, CA, USA).
Acknowledgments
We thank Yong-Jang Chen for technical assistance. RNA interference reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, supported by the National Research Program for Genomic Medicine Grants of NSC (NSC 97-3112-B-001-016). This work was supported by the following grants: NSC 100-2314-B-016-021, NSC 102-2320-B-016-016-MY3, MOST 103-2314-B-016-008 and MOST 103-2321-B-016-007 from the Ministry of Science and Technology, Taiwan, Republic of China, and the Liver Disease Prevention and Treatment Research Foundation, Taiwan, Republic of China.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Rudnick A, Ling TY, Odagiri H, Rutter WJ, German MS. Pancreatic beta cells express a diverse set of homeobox genes. Proc Natl Acad Sci USA 1994; 91: 12203–12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iype T, Taylor DG, Ziesmann SM, Garmey JC, Watada H, Mirmira RG. The transcriptional repressor Nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic beta-cell differentiation: evidence for feedback activation of the nkx6.1 gene by Nkx6.1. Mol Endocrinol 2004; 18: 1363–1375. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 2000; 101: 435–445. [DOI] [PubMed] [Google Scholar]
- Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H et al. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res 2007; 67: 2617–2625. [DOI] [PubMed] [Google Scholar]
- Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer 2008; 123: 161–167. [DOI] [PubMed] [Google Scholar]
- Lai HC, Lin YW, Huang RL, Chung MT, Wang HC, Liao YP et al. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer 2010; 116: 4266–4274. [DOI] [PubMed] [Google Scholar]
- Liu CY, Chao TK, Su PH, Lee HY, Shih YL, Su HY et al. Characterization of LMX-1 A as a metastasis suppressor in cervical cancer. J Pathol 2009; 219: 222–231. [DOI] [PubMed] [Google Scholar]
- Su HY, Lai HC, Lin YW, Liu CY, Chen CK, Chou YC et al. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer 2010; 127: 555–567. [DOI] [PubMed] [Google Scholar]
- Tsao CM, Yan MD, Shih YL, Yu PN, Kuo CC, Lin WC et al. SOX1 functions as a tumor suppressor by antagonizing the WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Hepatology 2012; 56: 2277–2287. [DOI] [PubMed] [Google Scholar]
- Micalizzi DS, Ford HL. Epithelial–mesenchymal transition in development and cancer. Fut Oncol 2009; 5: 1129–1143. [DOI] [PubMed] [Google Scholar]
- Duffield JS, Lupher M, Thannickal VJ, Wynn TA. Host responses in tissue repair and fibrosis. Annu Rev Pathol 2013; 8: 241–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave B, Mittal V, Tan NM, Chang JC. Epithelial–mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res 2012; 14: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110. [DOI] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA. Cancer stem cells and epithelial–mesenchymal transition: concepts and molecular links. Semin Cancer Biol 2012; 22: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9: 265–273. [DOI] [PubMed] [Google Scholar]
- Fan F, Samuel S, Evans KW, Lu J, Xia L, Zhou Y et al. Overexpression of Snail induces epithelial–mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med 2012; 1: 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang G, Li X, Zhang Y, Jiang Y, Shen J et al. Hypoxia induces epithelial–mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor -1alpha in hepatocellular carcinoma. BMC Cancer 2013; 13: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol 2012; 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vahdat LT, Wong S, Chang JC, Mittal V. Microenvironmental regulation of epithelial–mesenchymal transitions in cancer. Cancer Res 2012; 72: 4883–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno Y, Coelho AL, Jarai G, Westwick J, Hogaboam CM. Notch signaling mediates TGF-beta1-induced epithelial–mesenchymal transition through the induction of Snai1. Int J Biochem Cell Biol 2012; 44: 776–789. [DOI] [PubMed] [Google Scholar]
- Ohta H, Aoyagi K, Fukaya M, Danjoh I, Ohta A, Isohata N et al. Cross talk between hedgehog and epithelial–mesenchymal transition pathways in gastric pit cells and in diffuse-type gastric cancers. Br J Cancer 2009; 100: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Zhang X, Paladino D, Sengupta B, Ahmad S, Holloway RW et al. Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene 2012; 31: 2309–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecoraro G, Lee M, Morgan D, Defendi V. Evolution of in vitro transformation and tumorigenesis of HPV16 and HPV18 immortalized primary cervical epithelial cells. Am J Pathol 1991; 138: 1–8. [PMC free article] [PubMed] [Google Scholar]
- Jorgensen MC, Vestergard Petersen H, Ericson J, Madsen OD, Serup P. Cloning and DNA-binding properties of the rat pancreatic beta-cell-specific factor Nkx6.1. FEBS Lett 1999; 461: 287–294. [DOI] [PubMed] [Google Scholar]
- Donelan W, Koya V, Li SW, Yang LJ. Distinct regulation of hepatic nuclear factor 1alpha by NKX6.1 in pancreatic beta cells. J Biol Chem 2010; 285: 12181–12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2013; 41: D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev 2002; 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Yan MD, Yu PN, Li HJ, Kuo CC, Hsu CL et al. The role of Sp1 and EZH2 in the regulation of LMX1A in cervical cancer cells. Biochim Biophys Acta 2013; 1833: 3206–3217. [DOI] [PubMed] [Google Scholar]
- Wang YT, Chuang JY, Shen MR, Yang WB, Chang WC, Hung JJ. Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process. J Mol Biol 2008; 380: 869–885. [DOI] [PubMed] [Google Scholar]
- Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC et al. Sp1 expression regulates lung tumor progression. Oncogene 2012; 31: 3973–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 2001; 104: 861–873. [DOI] [PubMed] [Google Scholar]
- Syu LJ, Uhler J, Zhang H, Mellerick DM. The Drosophila Nkx6 homeodomain protein has both activation and repression domains and can activate target gene expression. Brain Res 2009; 1266: 8–17. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 2002; 108: 465–474. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 1998; 393: 88–91. [DOI] [PubMed] [Google Scholar]
- DelBove J, Rosson G, Strobeck M, Chen J, Archer TK, Wang W et al. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics 2011; 6: 1444–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J et al. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 2011; 21: 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 2009; 137: 371 e361–365. [DOI] [PubMed] [Google Scholar]
- Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial–mesenchymal transition. Oncogene 2010; 29: 4896–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 2012; 122: 1469–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wu Y, Wang Y, Wang C, Kang T, Rychahou PG et al. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene 2013; 32: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008; 28: 4772–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH et al. Bmi1 is essential in Twist1-induced epithelial–mesenchymal transition. Nat Cell Biol 2010; 12: 982–992. [DOI] [PubMed] [Google Scholar]
- Wu MZ, Tsai YP, Yang MH, Huang CH, Chang SY, Chang CC et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell 2011; 43: 811–822. [DOI] [PubMed] [Google Scholar]
- Lin YW, Tsao CM, Yu PN, Shih YL, Lin CH, Yan MD. SOX1 suppresses cell growth and invasion in cervical cancer. Gynecol Oncol 2013; 131: 174–181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.