Abstract
Syndecan-4 (Syn4), a single-pass transmembrane heparin sulphate proteoglycan (HSPG), plays significant role in the formation of focal adhesions and interacts with many growth factors to regulate cell migration and neural induction. Here, we show the new roles of syndecan-4(syn4) in zebrafish embryonic neurogenesis. Syn4 is broadly and dynamically expressed throughout the early stages of embryonic development. Knockdown of syn4 increases the expression of the marker genes of multiple types of neural cells. The increased expression of the marker genes is resulted from excessive proliferation of the neural cells. In addition, disrupting syn4 expression results in truncated and multiple aberrant branching of caudal primary (CaP) axons. Collectively, these data indicate that Syn4 suppresses the cellular proliferation during neurogenesis and is crucial for the formation of CaP axons during zebrafish embryogenesis.
Proteoglycans (PGs) are extracellular glycoproteins that contain sulphated glycosaminoglycan chains (GAG)1. Several studies have indicated that PGs play remarkable roles in regulating the interactions of cell surface molecules, such as the molecules mediating the cell-matrix, ligand-receptor and cell-cell interactions. They also function as co-regulators of many growth factors that are associated with neural fate, including FGF, HGF, Wnt, TGFβ and BMP2,3,4,5, indicating PGs act as important modulators during neural development. Syndecan-4 (Syn4) is a heparan sulphate PG in the Syndecan family6,7 and is composed of a variable extracellular domain, a single-span transmembrane domain and cytoplasmic domains that are highly conserved among the Syndecans6,8. As a unique family member, Syn4 is able to bind protein kinase Cα (PKCα) and phosphatidylinositol 4,5-bisphosphate (PIP2)8,9,10. Moreover, Syn4 interacts with integrin to be involved in the formation of focal adhesions and stress fibers11 and bind to chemokines12,13 to modulate the planar cell polarity3,14. During Xenopus and zebrafish embryonic development, syn4 is expressed in the migrating neural crest and modulates the formation of polarized cell protrusion to control the directional migration3. Recent observations demonstrate that syn4 is required for neural induction involving in FGF/ERK and PKCγ/Rac/JNK pathway15 and Wnt/PCP pathway to control the neural tube closure in mammalian embryos14. All these data show the important roles of syn4 in embryonic neurogenesis. However, potential functions of syn4 in the development of neural cells themselves have not been fully addressed.
In the present study, the roles of syn4 during zebrafish embryonic neural development were addressed. The results showed that syn4 was dynamically expressed throughout the early stages of zebrafish embryonic development. Knockdown of syn4 promoted the generation of neural cells by enhancing the proliferation of neural cells. Furthermore, the length of CaP axon was severely reduced and the number of axonal branches was significantly increased as the expression of syn4 was down-regulated. Taken together, these results indicate that syn4 suppresses the proliferation of neural cells and is crucial for the formation of CaP axons.
Results
Spatio-temporal expressing patterns of syn4 during zebrafish embryonic development
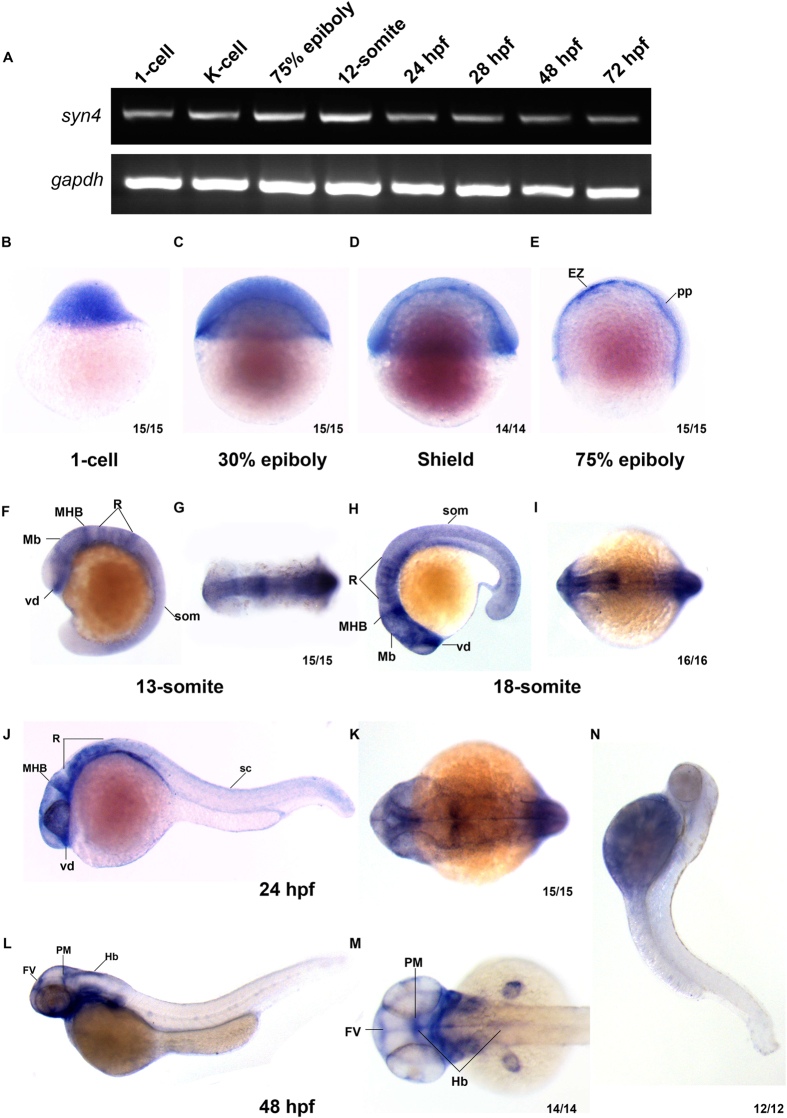
The zebrafish was used as a model organism to investigate the function of syn4 in early embryonic development. The semi-quantitative reverse transcription-PCR (sqRT- PCR) and whole mount in situ hybridization (WISH) showed that syn4 was expressed from 1-cell stage to 72 hours post fertilization (hpf). Notably, the syn4 expressing level was higher at 12-somite stage. These results demonstrate that syn4 is both maternally and zygotically expressed during zebrafish embryogenesis (Fig. 1A).
Figure 1. Syn4 expression pattern analysis in zebrafish.
(A) sqRT-PCR reveals that syn4 is expressed throughout embryonic developmental stages. (B–M) WISH shows the spatiotemporal expression pattern of syn4. (B–D) Syn4 is ubiquitously distributed before the gastrulation. (E) In 75% epiboly, syn4 is expressed in the thin evacuation zone and prechordal plate. (F–K) Syn4 is expressed in ventral diencephalon, midbrain, hind brain and neural crest during Segmentation Period. (L,M) In 48 hpf, syn4 is restricted to the ventricular zone in the forebrain, posterior midbrain and hindbrain. (N) Negative control of syn4 at 48 hpf. EZ: thin evacuation zone, FV: ventricular zone in the forebrain, Hb: hindbrain, Mb: midbrain, MHB: mid-hindbrain boundary, pp: prechordal plate, PM: posterior midbrain, R: rhombomeres, som: somites, sc: spinal cord, vd: ventral diencephalon. Lateral view, dorsal to the right in (B–E), dorsal view, anterior to the left in (G,I,K,M) lateral view, dorsal to the right and anterior to the top in (F), lateral view, dorsal to the top and anterior to the left in (H,J,M).
WISH analysis revealed that the spatial expression of syn4 was dynamic during embryonic development. The syn4 was ubiquitously distributed before the gastrulation (Fig. 1B,C). Then zygotic syn4 mRNA highly expressed in the shield (Fig. 1D) and enriched in the thin evacuation zone and prechordal plate from the late gastrulation stage onward (Fig. 1E). Since the segmentation stage, syn4 was predominantly expressed in rhombomere, ventral diencephalon, midbrain, hindbrain and neural crest till 24 hpf (Fig. 1F–K). Then syn4 mRNA was restricted to the ventricular zone in the forebrain, posterior midbrain and hindbrain at 48 hpf (Fig. 1L,M). The detailed expressing patterns of syn4 are consistent with previous observations in Xenopus embryos and zebrafish embryonic brains3,14,16. The results indicate that syn4 has a highly dynamic expressing pattern and may play a role in the nervous development.
Syn4 regulates the migration of neural crest and muscle patterning
In the view of specific distribution of syn4 in the neural tissues, we tried to investigate the function of syn4 during zebrafish neural development. Syn4 antisense morpholino oligonucleotide (syn4 MO), which was previously reported to block the syn4 translation17, was injected into embryos at the one-cell stage. As syn4 is involved in neural crest (NC) migration3, we assessed whether the syn4 MO we used were able to induce defective NC migration by analyzing the expression of the NC markers crestin and sox10 and observed a strong inhibition of trunk NC migration in syn4 MO-injected embryos at 18 somite stage (18hpf) (supplemental Fig. S1C–F). The results indicate that syn4 MO is able to induce the defective NC migration as described t3,14. Given the role of Syn4 in muscle development18,19, we also performed the WISH to detect the expressing pattern of myod, which is a transcription factor that controls skeletal myogenesis20. The results showed that the expression of myod appeared abnormal curvature in the morphants (supplemental Fig. S2A–E). Fast and slow muscles in the trunk are stained with phalloidin, which recognizes actin fibers of both muscle types, at 18 somite stage (supplemental Fig. S3A–C”) and 26hpf (supplemental Fig. S3D–E”). Injection of syn4 MO severely altered the morphology of muscle such that it appeared abnormally loose and wavy. Consistent with the previous observations18,19, the results indicate that syn4 is involved in muscle development.
To test the efficiency and specificity of syn4 MO, we co-injected the syn4 MO with the pEGFP-N1-syn4 recombinant plasmid that contained the syn4 MO binding 5′-UTR sequences and evaluated the efficiency by analyzing the fluorescence intensity of syn4-GFP fusing protein. As expected, the green fluorescence intensity was severely reduced in the co-injected embryos (supplemental Fig. S1A,B). Co-injection of syn4 mRNA without 5′-UTR syn4 MO binding sequences was able to rescue the defective NC migration and muscle patterning (supplemental Fig. S1C–F). Altogether, these data demonstrate that the syn4 MO is able efficiently to block syn4 translation as described previously.
To further confirm the MO specificity, we design a series of truncated mutants based on a previous observations21 (supplemental Fig. S4A). All of the mutant mRNAs were injected into zebrafish embryos. The results demonstrated the mutant lacking the C2 domain in plasma membranes (syn4-ΔC2) showed the phenotypes highly similar to the ones induced by the syn4 MO (supplemental Fig. S4B–J). The results clearly indicate that the ectopic expression syn4 ΔC2 mRNA is able to disrupt the syn4 functions in zebrafish embryos and the syn4-ΔC2 is disrupts syn4 function by acting as a dominant negative. Altogether, the results indicate the syn4 MO and the mutant syn4-ΔC2 are able to reveal the specific roles of syn4 involved in zebrafish embryonic development. The following results were mostly obtained from injection of syn4 MO in zebrafish embryos. The mutant syn4-ΔC2 was used to confirm the specificity of the phenotypes caused by syn4 MO in the following experiments.
Knockdown of syn4 expression increases neural progenitors
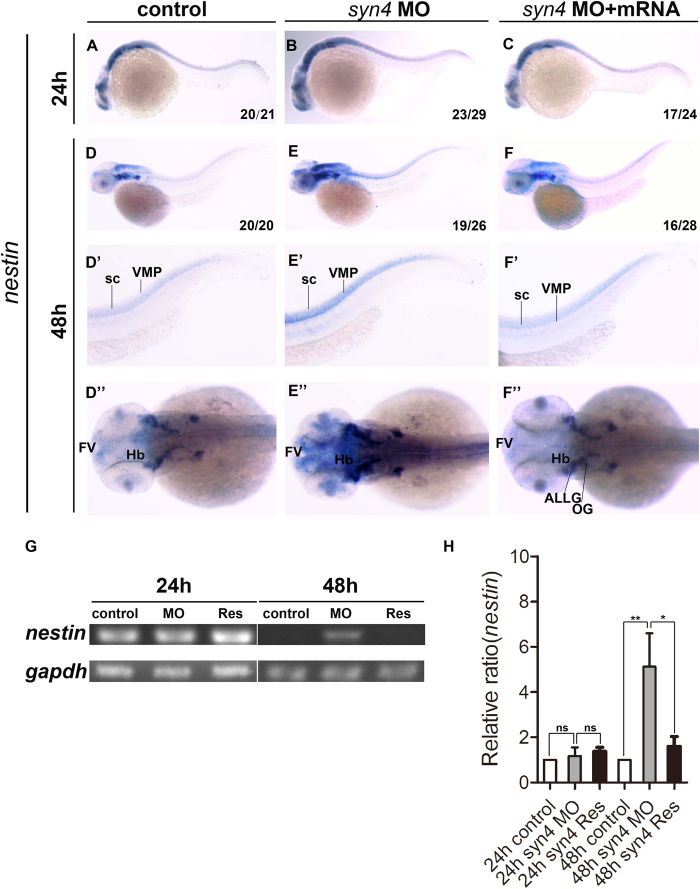
Since syn4 was expressed in neural tissues and required for neural crest migration, we then investigated whether syn4 was involved in the embryonic neurogenesis. The effect of down-regulated syn4 expression on neural stem cells and progenitor cells were examined by WISH with nestin22. The results showed that nestin expressing pattern was identical between the syn4 MO-injected and the control embryos at 24 hpf (Fig. 2A,B). However, the expression level was significantly increased in the syn4 morphants at 48 hpf (Fig. 2D–E”). Accordingly, sqRT-PCR showed the similar results (Fig. 2G,H). The syn4 mRNA was able to rescue the defective phenotypes (Fig. 2F–F”) caused by the syn4 MO and the dominant negative mutant syn4-ΔC2 mRNA (supplemental Fig. S4B–G”). The results show that syn4 negatively regulates nestin expression during embryonic neural development.
Figure 2. Knockdown of syn4 expression up-regulated the expression of nestin.
(A–C) Expression of nestin at 24 hpf, lateral view. (D–F) Expression of nestin at 48 hpf, lateral view. (E–E”) Nestin transcript level was increased significantly in syn4 morphants, dorsal view. (F–F”) The defective phenotypes were rescued by co-injecting syn4 mRNA. (G) sqRT-PCR showed the similar results. (H) Densitometric quantification of sqRT-PCR. The relative nestin expressing level is significantly higher in syn4 morphants (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student’s unpaired t-test). ALLG: anterior lateral line ganglion, FV: ventricular zone in forebrain, Hb: hindbrain, OG: octaval ganglion, sc: spinal cord, VMP: ventral motoneuron precursors.
Loss of syn4 results in the increase of neuroglial cells
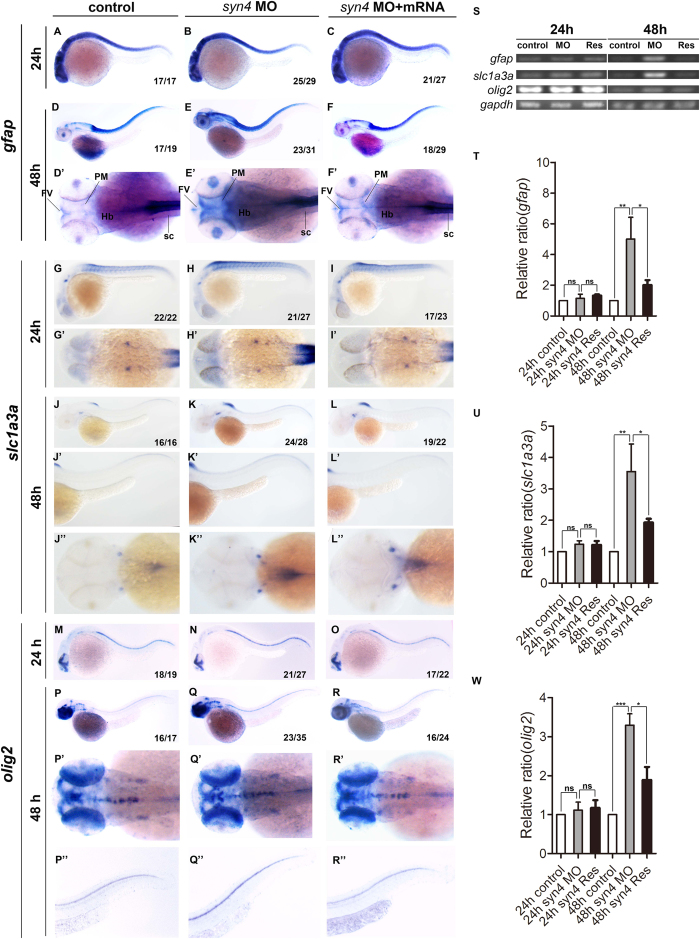
Since the neural stem cells/progenitor cells were regulated by Syn4, we were curious whether other neural cell types such as neuroglial cells were modulated by Syn4 as well. The expression of glial fibrillary acidic protein (gfap), an astrocyte marker, was not altered in the morphants and controls at 24 hpf (Fig. 3A,B), but up-regulated in the ventricular zone, especially in posterior midbrain and the spinal cord in the syn4 morphants by 48 hpf (Fig. 3D–E’). The syn4 mRNA was able to abrogate the increased gfap expression (Fig. 3F,F’). Next, we tested the expression of slc1a3a, also known as glast that expresses in radial glia–astrocyte lineage23,24, in embryonic neural tissues. The results showed that there was little difference between the controls and the syn4 morphants at 24 hpf (Fig. 3G–I’). The expressing levels of slc1a3a were obviously increased in dorsal spinal cord in the morphants at 48 hpf (Fig. 3J–K”). We then examined the expression of oligodendrocyte transcription factor 2 (olig2) in zebrafish embryos. Olig2 is a marker of oligodendrocytes and motor neuron (MN) precursors25,26. Not surprisingly, the olig2 expression was increased in the spinal cord in the syn4 morphants at 48 hpf (Fig. 3P–R”). The sqRT-PCR analysis confirmed the altering patterns of the expressing levels of gfap, slc1a3a, and olig2 in zebrafish embryos (Fig. 3S–W). Altogether, these data indicate that the neuroglial cells are increased when syn4 expression is down-regulated.
Figure 3. Loss of syn4 increased neuroglial cells.
(A–R”) WISH results of gfap (A–F”), slc1a3a (G–L”) and olig2 (M–R”) at 24 hpf and 48 hpf in zebrafish. (D–F’) The expression of gfap, (J–L”) slc1a3a, (Q–R”) olig2 are higher in the syn4 morphants than controls at 48 hpf. (S) sqRT-PCR analyzes of the transcript level of gfap, slc1a3a and olig2. (T–W) Densitometric quantification of sqRT-PCR. The syn4 morphants shown a higher expression level of gfap, slc1a3a and olig2 (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student’s unpaired t-test). FV: ventricular zone in forebrain, Hb: hindbrain, PM: posterior midbrain, sc: spinal cord. Lateral views, dorsal to the top and anterior to the left in (A–C,D–F,G–I,J–L’,M–O,P–R,P”–R”), Dorsal view, anterior to the left in (D’–F’,G’–I’,J”–L”,P’–R’).
Knockdown of syn4 expression promotes the generation of neurons
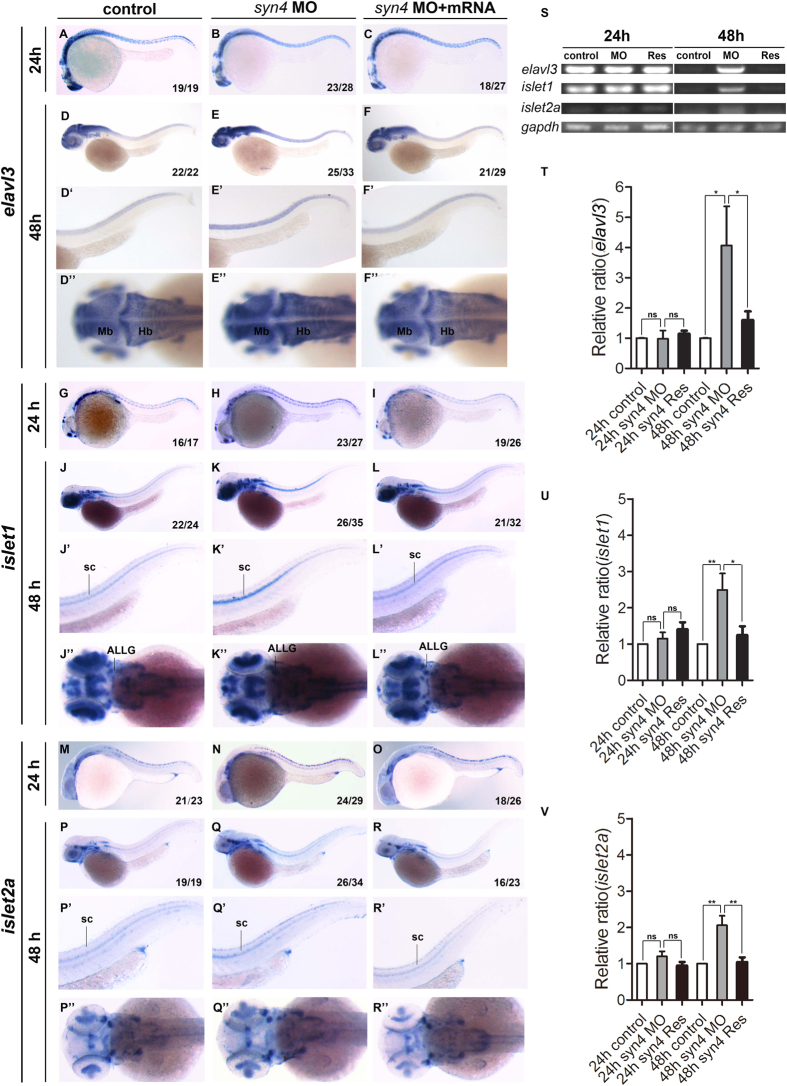
Since syn4 modulated the number of neural progenitors and neuroglial cells, we further tested whether syn4 was involved in neuron specification. Thus, we examined the expression of elavl3, the earliest marker of pan-neuronal cells27, in zebrafish embryos. Knockdown of syn4 didn’t alter the expression of elavl3 at 24 hpf (Fig. 4A,B). However, injection of syn4 MO resulted in a dramatic increased expressing levels of elavl3 in spinal cord compared with the controls at 48 hpf (Fig. 4D–E”).
Figure 4. Knockdown of syn4 expression promotes the generation of neurons.
(A–R”) WISH results of elavl3, islet1, and islet2a at 24 hpf and 48 hpf in zebrafish. (E–E”) The expression of elavl3, (K–K”) islet1, (Q–Q”) islet2a in spinal cord were increased in syn4 morphants at 48 hpf. co-injecting syn4 mRNA was able to rescue the defects. (S) sqRT-PCR analyzes of the transcript level of elavl3, islet1 and islet2a. (T–V) Densitometric quantification of sqRT-PCR. The syn4 morphants shown a higher expression level of elavl3, islet1 and islet2a (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student’s unpaired t-test). ALLG: anterior lateral line ganglion, Hb: hindbrain, Mb: midbrain, sc: spinal cord. Lateral views, dorsal to the top and anterior to the left in (A–F’,G–L’,M–R’). Dorsal view, anterior to the left in (D”–F”,J”–L”,P”–R”).
To test the formation of motor neurons in syn4-depleted embryos, we analyzed the expression of islet1 and islet2a, which label primary motoneurons (PMNs), sensory Rohon-Beard neurons, and retina neurons. Rostral primary (RoP) and middle primary (MiP) MNs express islet1 but not islet2a, while CaP and variable primary (VaP) MNs express islet228. The islet1 and islet2a expression were both increased in ventral spinal cord in the morphants at 48 hpf (Fig. 4J–K”). Co-injection of syn4 mRNA was able to rescue the enhancement of islet1 and islet2a expression induced by the syn4 MO (Fig. 4J–L”). The statistical results of sqRT-PCR were consistent with the results of WISH (Fig. 4S–V).
To gain further insight into the functions of syn4, we also analyzed another member of Syndecan family in zebrafish, syndecan2 (syn2) and found that syn2 mRNA was unable to rescue the phenotypes caused by the syn4 MO (supplemental Fig. S5A–F’). Taken together, the observations demonstrate that Syn4 modulates the embryonic development of neural stem cell/progenitors, neuronal glial cells, and neurons.
The proliferation of neural cells is increased by down-regulation of the syn4 expression
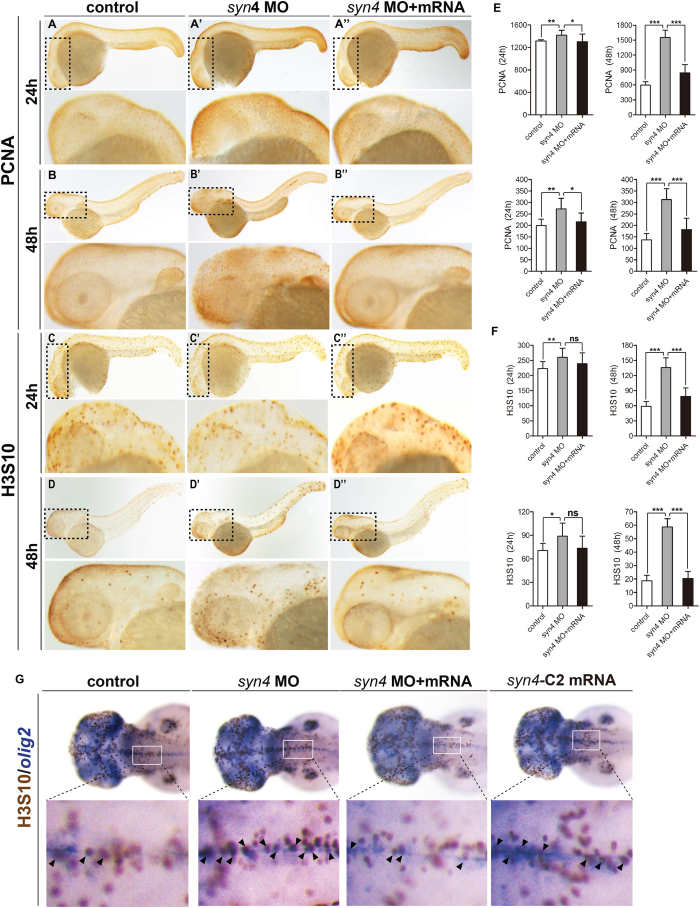
To reveal how the neural cells were regulated by the syn4 expression, we performed immunohistochemistry staining of whole-mount embryos to detect the proliferating cell nuclear antigen (PCNA), which is a marker of cells with proliferative potential29. Statistical results revealed that the numbers of PCNA positive cells were increased in the syn4 morphants at 48 hpf. The phenotypes were fully rescued by the syn4 mRNA (Fig. 5A–B”). We also counted phospho-Histone H3 (Ser10), which is another marker to label the proliferative cells, positive cells in zebrafish embryos. The results showed that the number of phospho-Histone H3 (Ser10) positive cells was increased in the morphants at 48 hpf (Fig. 5C–D”). The increased staining cells were more obvious in zebrafish embryonic brains (Fig. 5E,F, supplemental Fig. S6A–C’). Examination of the embryo sections also showed that the phospho-Histone H3 (Ser10) positive cells were increased in the 48hpf zebrafish embryos that were disrupted the Syn4 function (supplemental Fig. S6D–G). In addition, we also performed Two-Color Whole-Mount Staining with DAB and BCIP/NBT to identify whether neural cells, including neuroglial cells and motor neurons, carried increasing signal of phospho-histone H3 in zebrafish embryos after disruption of syn4 function. The results showed that proliferation of all tested types of neural cells was up-regulated in the morphants at 48 hpf (Fig. 5G and data not shown). Taken together, our data indicate that Syn4 suppresses the proliferation of neural cells during zebrafish embryonic neurogenesis.
Figure 5. Whole-mount immunohistochemistry staining of PCNA and H3S10 in zebrafish embryos.
(A–D”) Immunostaining of PCNA and H3S10 in 24 hpf and 48 hpf embryos, the box regions are magnified. (E,F) Quantification of PCNA-positive cells (E) and H3S10 -positive cells (F) in different treatment groups. Both PCNA positive cells and H3S10 positive cells are considerably increased in syn4 morphants and the defects were partially rescued by co-injection with syn4 mRNA. For each group, 18 embryos are scored. (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student’s unpaired t-test). (G) The double WISH with H3S10 and olig2. Arrows label the H3S10 positive cells, the box regions are magnified. Lateral views, dorsal to the top and anterior to the left in (A–D”,G).
Syn4 is required for the formation of CaP axons
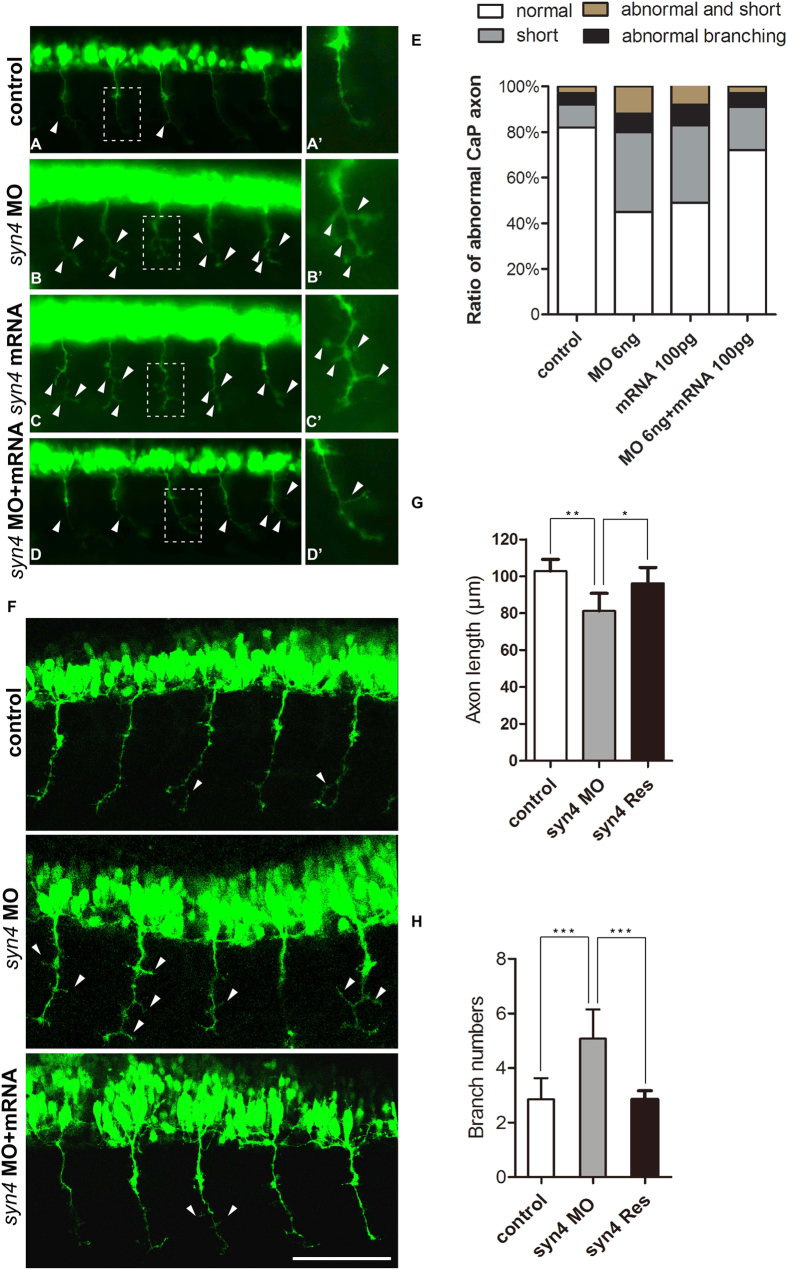
As the expression of several markers of motor neuron was altered in the syn4 MO injected embryos, we considered that syn4 might be associated with motor neuron development. There are two distinct classes of spinal motor neurons: PMNs and secondary motor neurons (SMNs) in zebrafish30. The PMNs are classified into CaP, MiP, and RoP MNs31. These PMNs can be identified by their stereotypical cell body positions and axon projection patterns. Specially, the CaP MNs is easy to be observed as their locates in the middle of each spinal cord and axonal projection extends ventrally32. Therefore, we selected CaP MNs as the target objects in Tg [hb9: GFP]ml2 transgenic zebrafish. The the expression of islet1/2a was not altered before 48hpf (supplemental Fig. S2F–K), however, 56% syn4-MO injected embryos showed abnormal PMNs with multiple aberrant branching axons and truncated axons at 26 hpf of the zebrafish embryos (Fig. 6A–B’). To assess whether overexpression of syn4 was able to disturb the outgrowth of CaP axons, we injected syn4 mRNA at one-cell stage and quantitated the number of PMNs with defective phenotypes at 26 hpf. The result showed that 53% syn4 mRNA-injected embryos had shortened CaP axons and/or abnormally branched axons (Fig. 6C). The length of CaP axon outgrowth in the morphants averaged 79.9 um, which was significantly reduced, compared to the control embryos’s CaP axons that averaged 100.2 um (Fig. 6F,G). In addition, the averaged branch number of per CaP axon in syn4 MO-injected embryos was 4.71, while control embryos carried only 3.57 branches in average (Fig. 6F,H). The syn4 mRNA co-injection was able to rescue the defective CaP axon morphology (Fig. 6D). These results indicate that accurate syn4 expression is crucial.
Figure 6. Syn4 is required for the CaP axons formation.
(A–D’) Embryos at 26 hpf show PMNs in green fluorescence. (B,B’) Syn4 morphants show shortened and branching CaP axons, (C,C’) syn4 mRNA-injected embryos have same phenotypes as syn4 morphants. (D,D’) Co-injection of syn4 MO with syn4 mRNA partially rescues CaP axon defects. (E) Statistical results of CaP axon outgrowth are shown in (A–D’). For each group 60 axons from 12 embryos are scored. (F) The CaP axons of control embryos and syn4 morphants are photographed under confocal microscopy. (G) Statistical results of axon length in (F–G). The length of CaP axons is reduced in syn4 morphants (**P < 0.01, t-test). (H) the branch number of CaP axons is increased (***P < 0.001, t-test) in syn4 morphants. For each group 75 axons from 15 embryos are scored. The box regions are magnified and show in (A’–D’). Arrows indicate shortened CaP axons and abnormal branchs. Lateral views, dorsal to the top and anterior to the left in (A–D’,F). Scale bars: 100 μm.
Discussion
Previous observations have shown that Syn4 is an element of the PCP non-canonical Wnt signaling pathways to regulate the neural tube closure and neural crest migration during Xenopus and zebrafish embryonic development3,14. Syn4 also modulates the neural induction through the ERK or PKC-dependent signaling pathways15. Here, the new role of syn4 is revealed in our works. We identify that the neural cells are prominently increased after the inhibition of syn4 during the early stage of zebrafish embryonic neurogenesis. Further evidences reveal that syn4 inhibits the cell proliferation during the development of neural system and maintains motorneuron morphology.
The prior data have indicated that Syn4 has a positive effect on cell proliferation by regulating PKCα activation to mediate growth factor-stimulated proliferation in the wounded adult tissue and adult skeletal muscle cells2,33,34,35. Syn4 has negative effects on the cell proliferation in multiple tumor cell lines such as breast carcinoma and glioblastoma cells. In the tumor cells, the ability of Syn4 to bind to the fibronectin is competitively blocked by tenascin-C in integrin signaling, resulting in the proliferation of tumor cells36. Since Syn4 is indispensable to the cell spreading by interaction with the HepII site in fibronectin, disrupting the interaction between Syn4 and FNIII13 results in cell spreading obstacle37,38. The data suggest that there exists a mechanism that fibronectin signaling attenuates cell proliferation by Syn436. In consistent with the results obtained from the tumor cell lines, we find that Syn4 inhibits the proliferation of neural cells. Therefore, further work is required to determine whether fibronectin is involved in the inhibition of neural proliferation during embryonic neural development.
The extension of axons and dendrites to form stereotypic neuronal connections is crucial to neural development. It has been reported that the biosynthesis of heparan sulfate (HS) is required for normal axon elongation and branching in animals, including mouse, C. elegans and zebrafish. HSPGs, the carrier proteins of HS, are involved in properly axon architecture. Syndecan-3 and glypican-2 are shown to be required for axon guidance5,39. However, little is known about the role of syn4 in the architecture of axon. We identify that perturbed expression of syn4 results in aberrant elongation and abnormal additional branching of CaP axons. The C2 domain of Syn4 is essential for the function of syn4 in the formation of CaP axons. These evidences is similar to the phenotype that defects in the extension and targeting of axons caused by abnormal FGF signaling, suggesting that Syn4 may regulate the axon architecture via the FGF signaling in the zebrafish5,40,41. Future experiments are required to explore the factors that are combined with syn4 in regulating CaP axons formation.
Our results show that muscle patterning is altered and the morphology of muscle severely altered in the syn4 morphants since 18hpf. The axons of motor neuron show aberrant elongation and additional branching in the morphants at 26hpf. In view of the axon of primary motor neuron start to project to the myotomes at 18hpf, while the myotomes have formed before the time point42,43. Our results suggest that the early defective muscle patterning induced by knockdown of Syn4 functions may be independent of the proliferation of the neural cells. The results also suggest that the early defective muscle patterning may contribute to defective axon branching and length in the syn4 morphants. However, further experiments are required to address the question. Syn2 and syn4 have a high degree of structural similarity44. However, we identify that overexpression of syn2 is unable to rescue the phenotypes caused by knockdown of Syn4 functions in present works. The results indicate that Syn4 specifically modulate the proliferation in neural cells and axon patterning of motor neurons and are consistent with previous observations that Syn2 and Syn4 are engaged in a number of distinct developmental processes3,45,46.
Methods
Zebrafish maintenance and strains
All experiments performed on zebrafish (Danio rerio) were according to standard procedures47. Embryos were staged as described and raised at 28.5 °C in the same system as adults (Aquatic Ecosystems)48. The Tg[hb9:GFP]ml2 transgenic lines49 and AB strains were used in our studies.
All zebrafish experiments were performed in accordance with the guidelines of the animal ethical committee of West China Hospital. All experimental protocols were proved by the Animal Ethical Committee, West China Hospital, Sichuan University.
Syn2/4 cloning
Syn2/4 cDNA was amplified from zebrafish embryos using the primers shown in the supplemental Table S1 and subcloned into the pGEM-T easy vector to synthetize antisense probes for in situ hybridization. The coding region of syn2/4 was subcloned into the pcDNA3.1+ vector for mRNA synthesis. The syn4 primer1 resultant PCR products (with 5′-UTR sequence) were ligated into pEGFP-N1 vector.
Construction of syn4 mutants
Five mutants of zebrafish syn4 were designed based on a previous observations21 and generated by PCR. The primers were shown in the supplemental Table S1. The C-terminal domain deletion mutants included ΔC1, ΔC2, ΔV and ΔC1–2. The N-terminal domain deletion mutant ΔEc lacks the extracellular domain.
Total RNA isolation and Semi-quantitative Reverse transcription-PCR (sqRT-PCR) and Statistical analysis
Total RNAs of an approximate 100 embryos at different stages, including 1-cell, K-cell, 75% epiboly, 12-somite, 24 h, 28 h, 48 h, 72 h post-fertilization (hpf) were isolated. Each reaction used 20μl total RNA with the Prime Script Reverse-transcription PCR kit (TaKaRa DRR014A). The resultant cDNA were used as the templates for PCR. The primer pairs used for sqRT-PCR analysis were shown in the supplementary (supplemental Table S1). Statistical analysis was performed using Student’s unpaired t-test. Differences were considered significant for p < 0.05.
In vitro transcription of Antisense RNA probes and mRNAs
Antisense RNA probes for in situ hybridization were synthesized in vitro following manufacturer’s instructions of Riboprobe system kit (Promega). Plasmid was linearized and the probes syn4, elavl3, gfap, islet1, islet2a, olig2, slc1a3a and nestin were synthesized. Capped syn4 mutant mRNA and full-length mRNA that lacked the morpholino sequence were synthesized using the T7 Mmessagem MACHINE kit (Ambion).
Antisense morpholino and mRNA or plasmid injections
Antisense morpholino oligonucleotides (MO) against syn4: TGAGGTAAACTTTCAACAT CTTCTC were purchased from Gene Tools (Philomath, OR, USA) and used as previously described17. The mRNAs and MO were injected into the yolk and the plasmid was injected into the cell at the one cell stage. Unless stated otherwise, a volume of 1 nl was injected into embryos with the concentration of 6 ng/nl of MO, 100–150 ng/ul of mRNA and 100–200 ng/ul of plasmid.
Whole-mount in situ hybridization and histology
Whole-mount in situ hybridization (WISH) or the double WISH with DAB and NBT/BCIP was carried out as previously described50. The antisense RNA probe was synthesized from the relative cDNA with a digoxygenin (DIG) RNA labeling kit (Roche). In brief, embryos were permeabilized with Proteinase K (10 mg/mL, Promega) for 2 minutes/24 hpf or 30 minutes/48 hpf. Hybridization was done overnight at 65 °C with the DIG-labeled antisense probes. After washed at 65 °C with formamide and SSCT concentration gradient (50% formamide, 2*SSCT to 0.2*SSCT), restore to the room temperature and washed again (0.1*SSCT to 0.05*SSCT), DAB staining followed NBT/BCIP (Roche) staining was performed according to the manufacturer’s instructions. The following probes were used: syn4, crestin, elavl3, slc1a3a, gfap, islet1, islet2a, olig2, sox10 and nestin.
Immunohistochemistry Staining of Whole-mount Embryos and Statistical analysis
Immunohistochemistry was performed using these sections as previously described51, the following primary antibodies were used: mouse anti-PCNA (Calbiochem 1:500) and rabbit anti-phospho-Histone H3 (Ser10) (Millipore 1:500). We counted the positive cells at the brain and the whole body of each fish, and calculate the average to compare the difference among groups. Statistical analysis was performed using Student’s unpaired t-test. Differences were considered significant for p < 0.05.
Immunofluorescence staining
Immunofluorescence staining was performed as previously described52 using the following antibodies: rabbit anti-phospho-Histone H3 (Ser10) (Millipore 1:200), phalloidin (Sigma 1:1000). In brief, Zebrafish embryos were fixed in 4% paraformaldehyde at 4 °C overnight, washed three times in PBS for 5 min each wash, then incubated in acetone at −20 °C for 7 min and washed three times in PBS for 5 min each wash. Blocking the embryos with 5% BSA and dilute the antibodies with blocking solution used for immunostaining at 4 °C overnight.
For cross-section samples, embryos were equilibrated in 30% sucrose prepared in PBS and embedded in optimal cutting temperature compound at −20 °C. Sections with a thickness of 8–10 um.
Additional Information
How to cite this article: Luo, N. et al. Syndecan-4 modulates the proliferation of neural cells and the formation of CaP axons during zebrafish embryonic neurogenesis. Sci. Rep. 6, 25300; doi: 10.1038/srep25300 (2016).
Supplementary Material
Acknowledgments
The authors thank the members of Dr. Mo’s laboratory for their critical discussion and review of the manuscript. This work was supported by the National Basic Research Program of China (to X.M. 2015CB942800), the Nature Science Foundation of China (to H.X. 31171384, to X.M. 81361120381).
Footnotes
Author Contributions N.L. and H.L. designed and performed experiments and wrote the manuscript. L.Q. and J.H. performed the molecular experiments. Y.L. performed the histological experiments. Y.L. and S.L. performed the microinjection. R.L. and W.M. performed WISH. B.X., Y.J., Y.W., H.X. and X.M. contributed to designing, interpretation of the experiments and completed the manuscript. X.M. supervised the whole experiments.
References
- Okina E., Manon-Jensen T., Whiteford J. R. & Couchman J. R. Syndecan proteoglycan contributions to cytoskeletal organization and contractility. Scand J Med Sci Sports 19, 479–489 (2009). [DOI] [PubMed] [Google Scholar]
- Bass M. D. & Humphries M. J. Cytoplasmic interactions of syndecan-4 orchestrate adhesion receptor and growth factor receptor signalling. Biochem J 368, 1–15 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews H. K. et al. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development 135, 1771–1780 (2008). [DOI] [PubMed] [Google Scholar]
- Kennea N. L. & Mehmet H. Neural stem cells. J Pathol 197, 536–550 (2002). [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y. Heparan sulfate proteoglycans in the nervous system: their diverse roles in neurogenesis, axon guidance, and synaptogenesis. Semin Cell Dev Biol 12, 99–106 (2001). [DOI] [PubMed] [Google Scholar]
- Multhaupt H. A. et al. Syndecan signaling: when, where and why? J Physiol Pharmacol 60 Suppl 4, 31–38 (2009). [PubMed] [Google Scholar]
- Partovian C., Ju R., Zhuang Z. W., Martin K. A. & Simons M. Syndecan-4 regulates subcellular localization of mTOR Complex2 and Akt activation in a PKCalpha-dependent manner in endothelial cells. Mol Cell 32, 140–149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambaerts K., Wilcox-Adelman S. A. & Zimmermann P. The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr Opin Cell Biol 21, 662–669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E. S., Woods A. & Couchman J. R. Multimerization of the cytoplasmic domain of syndecan-4 is required for its ability to activate protein kinase C. J Biol Chem 272, 11805–11811 (1997). [DOI] [PubMed] [Google Scholar]
- Oh E. S., Woods A. & Couchman J. R. Syndecan-4 proteoglycan regulates the distribution and activity of protein kinase C. J Biol Chem 272, 8133–8136 (1997). [DOI] [PubMed] [Google Scholar]
- Woods A. & Couchman J. R. Syndecan-4 and focal adhesion function. Curr Opin Cell Biol 13, 578–583 (2001). [DOI] [PubMed] [Google Scholar]
- Brule S. et al. The shedding of syndecan-4 and syndecan-1 from HeLa cells and human primary macrophages is accelerated by SDF-1/CXCL12 and mediated by the matrix metalloproteinase-9. Glycobiology 16, 488–501 (2006). [DOI] [PubMed] [Google Scholar]
- Charnaux N. et al. Syndecan-4 is a signaling molecule for stromal cell-derived factor-1 (SDF-1)/CXCL12. FEBS J 272, 1937–1951 (2005). [DOI] [PubMed] [Google Scholar]
- Munoz R., Moreno M., Oliva C., Orbenes C. & Larrain J. Syndecan-4 regulates non-canonical Wnt signalling and is essential for convergent and extension movements in Xenopus embryos. Nat Cell Biol 8, 492–500 (2006). [DOI] [PubMed] [Google Scholar]
- Kuriyama S. & Mayor R. A role for Syndecan-4 in neural induction involving ERK- and PKC-dependent pathways. Development 136, 575–584 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister W., Devine C. A. & Key B. Distinct expression patterns of syndecans in the embryonic zebrafish brain. Gene Expr Patterns 13, 126–132 (2013). [DOI] [PubMed] [Google Scholar]
- Lambaerts K. et al. Syntenin, a syndecan adaptor and an Arf6 phosphatidylinositol 4,5-bisphosphate effector, is essential for epiboly and gastrulation cell movements in zebrafish. J Cell Sci 125, 1129–1140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., McFarland D. C. & Velleman S. G. Migration of turkey muscle satellite cells is enhanced by the syndecan-4 cytoplasmic domain through the activation of RhoA. Mol Cell Biochem 375, 115–130 (2013). [DOI] [PubMed] [Google Scholar]
- Ronning S. B. et al. Syndecan-4 Regulates Muscle Differentiation and Is Internalized from the Plasma Membrane during Myogenesis. PLoS One 10, e0129288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y. et al. Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish. Dev Biol 358, 102–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase I., Osaki K., Tanaka G., Utani A. & Futaki S. Molecular interplays involved in the cellular uptake of octaarginine on cell surfaces and the importance of syndecan-4 cytoplasmic V domain for the activation of protein kinase Calpha. Biochem Biophys Res Commun 446, 857–862 (2014). [DOI] [PubMed] [Google Scholar]
- Mahler J. & Driever W. Expression of the zebrafish intermediate neurofilament Nestin in the developing nervous system and in neural proliferation zones at postembryonic stages. BMC Dev Biol 7, 89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T. et al. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci 17, 9212–9219 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storck T., Schulte S., Hofmann K. & Stoffel W. Structure, expression, and functional analysis of a Na(+)-dependent glutamate/aspartate transporter from rat brain. Proc Natl Acad Sci USA 89, 10955–10959 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. C., Mehta A., Richardson J. S. & Appel B. Olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol 248, 356–368 (2002). [DOI] [PubMed] [Google Scholar]
- Park H. C., Shin J., Roberts R. K. & Appel B. An olig2 reporter gene marks oligodendrocyte precursors in the postembryonic spinal cord of zebrafish. Dev Dyn 236, 3402–3407 (2007). [DOI] [PubMed] [Google Scholar]
- Kim C. H. et al. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett 216, 109–112 (1996). [DOI] [PubMed] [Google Scholar]
- Korzh V., Edlund T. & Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development 118, 417–425 (1993). [DOI] [PubMed] [Google Scholar]
- Lam C. S., Marz M. & Strahle U. gfap and nestin reporter lines reveal characteristics of neural progenitors in the adult zebrafish brain. Dev Dyn 238, 475–486 (2009). [DOI] [PubMed] [Google Scholar]
- Myers P. Z. Spinal motoneurons of the larval zebrafish. J Comp Neurol 236, 555–561 (1985). [DOI] [PubMed] [Google Scholar]
- Myers P. Z., Eisen J. S. & Westerfield M. Development and axonal outgrowth of identified motoneurons in the zebrafish. J Neurosci 6, 2278–2289 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Klapac L. R. & Beattie C. E. Zebrafish topped is required for ventral motor axon guidance. Dev Biol 273, 308–320 (2004). [DOI] [PubMed] [Google Scholar]
- Nikkari S. T., Jarvelainen H. T., Wight T. N., Ferguson M. & Clowes A. W. Smooth muscle cell expression of extracellular matrix genes after arterial injury. Am J Pathol 144, 1348–1356 (1994). [PMC free article] [PubMed] [Google Scholar]
- Cornelison D. D., Filla M. S., Stanley H. M., Rapraeger A. C. & Olwin B. B. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev Biol 239, 79–94 (2001). [DOI] [PubMed] [Google Scholar]
- Rauch B. H. et al. Syndecan-4 is required for thrombin-induced migration and proliferation in human vascular smooth muscle cells. J Biol Chem 280, 17507–17511 (2005). [DOI] [PubMed] [Google Scholar]
- Huang W., Chiquet-Ehrismann R., Moyano J. V., Garcia-Pardo A. & Orend G. Interference of tenascin-C with syndecan-4 binding to fibronectin blocks cell adhesion and stimulates tumor cell proliferation. Cancer Res 61, 8586–8594 (2001). [PubMed] [Google Scholar]
- Tumova S., Woods A. & Couchman J. R. Heparan sulfate chains from glypican and syndecans bind the Hep II domain of fibronectin similarly despite minor structural differences. J Biol Chem 275, 9410–9417 (2000). [DOI] [PubMed] [Google Scholar]
- Saoncella S. et al. Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc Natl Acad Sci USA 96, 2805–2810 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander A. D. & Selleck S. B. The elusive functions of proteoglycans: in vivo veritas. J Cell Biol 148, 227–232 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane S., Cornel E., Amaya E. & Holt C. E. Inhibition of FGF receptor activity in retinal ganglion cell axons causes errors in target recognition. Neuron 17, 245–254 (1996). [DOI] [PubMed] [Google Scholar]
- McFarlane S., McNeill L. & Holt C. E. FGF signaling and target recognition in the developing Xenopus visual system. Neuron 15, 1017–1028 (1995). [DOI] [PubMed] [Google Scholar]
- Panzer J. A. et al. Neuromuscular synaptogenesis in wild-type and mutant zebrafish. Dev Biol 285, 340–357 (2005). [DOI] [PubMed] [Google Scholar]
- Kotani Y. et al. Neuromuscular regulation in zebrafish by a large AAA+ ATPase/ubiquitin ligase, mysterin/RNF213. Sci Rep 5, 16161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E. S. & Couchman J. R. Syndecans-2 and -4; close cousins, but not identical twins. Mol Cells 17, 181–187 (2004). [PubMed] [Google Scholar]
- Arrington C. B. & Yost H. J. Extra-embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development 136, 3143–3152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E., Hermanson S. & Ekker S. C. Syndecan-2 is essential for angiogenic sprouting during zebrafish development. Blood 103, 1710–1719 (2004). [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). Neuron 35, 39–50 (1995). [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- Arkhipova V. et al. Characterization and regulation of the hb9/mnx1 beta-cell progenitor specific enhancer in zebrafish. Dev Biol 365, 290–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T. Double in situ Hybridization Techniques in Zebrafish. Methods 23, 345–358 (2001). [DOI] [PubMed] [Google Scholar]
- Le A. T. & Stainier D. Y. R. Fibronectin Regulates Epithelial Organization during Myocardial Migration in Zebrafish. Developmental Cell 6, 371–382 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. ApoA-II directs morphogenetic movements of zebrafish embryo by preventing chromosome fusion during nuclear division in yolk syncytial layer. J Biol Chem 286, 9514–9525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.