Abstract
Aneurysmal subarachnoid hemorrhage (aSAH) is a devastating form of hemorrhagic stroke with 30-day mortality between 33 and 45%. Delayed cerebral ischemia (DCI) is the chief cause of morbidity and mortality in patients who survive the initial aSAH. DCI accounts for almost 50% of deaths in patients surviving to treatment of the ruptured aneurysm. The mechanisms for brain injury after aSAH and the brain's response to this injury are not fully understood in humans. MicroRNAs (miRs) are 22- to 25-nucleotide single-stranded RNA molecules that inhibit the expression of specific messenger RNA targets. In this work, miR profiling of human cerebrospinal fluid from eight patients after aSAH was performed daily for 10 days with the goal of identifying changes in miR abundance. Using the nanoString nCounter Expression Assay, we identified two specific clusters of miR that were differentially regulated over time. Quantitative RT-PCR was performed on select miRs from each cluster. The first cluster contained miRs known to be present in blood and decreased in abundance over time. miRs in this group include miR-92a and let-7b. The second cluster contained several poorly characterized miRs that increased in abundance over time. miRs in this group included miR-491. This second cluster of miRs may be released into the CSF by the brain itself as a result of the initial SAH. Temporal changes in the abundance of specific miRs in human CSF after aSAH may provide novel insight into the role of miRs in brain injury and the brain's response.
Keywords: microRNA, stroke, subarachnoid hemorrhage
stroke is a significant public health problem and is a leading cause of death and disability in the United States (14). Aneurysmal subarachnoid hemorrhage (aSAH) accounts for ∼5% of all strokes and is frequently a devastating condition. The 30-day mortality rate following SAH ranges between 33 and 45%. Spontaneous SAH is most often due to the rupture of a berry aneurysm arising from a branch of the circle of Willis at the base of the brain (8). Acute treatment of these patients focuses on the treatment of the aneurysm to prevent rehemorrhage, which is associated with mortality as high as 70%. Treatment options include open craniotomy with aneurysm clipping or endovascular occlusion of the aneurysm with detachable platinum coils (8).
Delayed cerebral ischemia (DCI) is a chief cause of morbidity and mortality in patients who survive the initial aSAH. DCI accounts for almost 50% of deaths in patients surviving to treatment (clipping or coiling) after aSAH. Vasospasm has been thought to be the most significant cause of DCI and affects between 30 and 70% of patients following aSAH. Vasospasm has an onset between days 3 and 5 and peaks after 2 wk. An additional complication of aSAH is acute hydrocephalus, which occurs in 20–30% of patients and often requires placement of an external ventricular drain (EVD) (8). The presence of an EVD allows sampling of cerebrospinal fluid (CSF). CSF is contiguous with the interstitial fluid of the brain and may reflect its constituents on a more global scale than cerebral microdialysis. Protein biomarkers in CSF have been analyzed, revealing increased levels of glutamate and S100B in patients with DCI (9).
microRNAs (miRs) are 22- to 25-nucleotide single-stranded RNAs that inhibit the expression of specific mRNA targets (5, 29). miR profiling can discriminate between different forms of cardiovascular disease such as ischemic cardiomyopathy and heart failure (29). Because it bathes the central nervous system (CNS) and comes into contact with the injured tissue, CSF has been suggested to be an excellent source of miR expression changes related to CNS injury (7). miR profiling of CSF from patients with primary central nervous system lymphoma has been performed, revealing a statistically significant decrease in miR-21, -19b, and -92a compared with control patients (4). Additionally, in this study the authors showed that miRs are stable in CSF samples for up to 96 h at room temperature (4).
Little is known about the role of miRs in stroke pathogenesis (10, 21). Several groups have found changes in miR levels in humans after acute ischemic stroke both in blood (16, 31) and CSF (30), yet the impact of these changes on brain injury have not been characterized. Furthermore, changes in miRs following aSAH have not yet been described at all.
This work presents the first evidence on changes in the abundance of miR in human CSF after aSAH. Understanding the temporal changes in the abundance of different miR in human CSF after aSAH is likely to provide insight into the role of miRs in brain injury and help understand the brain's response to aSAH.
METHODS
Study Subjects and Experiment Design
The study protocol and material were approved by the Ohio State University Institution Review Board, and all subjects gave written informed consent prior to participation. We have analyzed the abundance of miRs in aSAH CSF in eight patients. Men and women 18–80 yr of age were eligible to participate in the study.
Inclusion criteria.
1) Onset of new neurological signs of subarachnoid hemorrhage within 72 h at the time of evaluation. 2) Clinical signs consistent with the diagnosis of subarachnoid hemorrhage including severe thunderclap headache, cranial nerve abnormalities, decreased level of consciousness, meningismus and focal neurological deficits. 3) Computed tomography demonstrates subarachnoid hemorrhage. 4) Cerebral angiography reveals the presence of saccular aneurysm(s) in a location that explains the subarachnoid hemorrhage. 5) Treatment of cerebral aneurysm must be carried within 72 h of symptom onset. 6) Accepted treatments of aneurysms include surgical clipping or endovascular embolization. 7) Presence of an external ventricular drain with position of the catheter tip within the lateral or third ventricle confirmed by computed tomography.
Exclusion criteria.
1) Time of symptom onset cannot be reliably assessed. 2) No demonstrable aneurysm by cerebral angiography. 3) Evidence of traumatic, mycotic, or fusiform aneurysm by cerebral angiography. 4) Hunt and Hess scale of 4 or greater and Fisher scale of 0–1. 5) Severe prior physical disability that precludes evaluation of clinical outcome measures. 6) Severe terminal disease with life expectancy < 6 mo. 7) No family available to give informed consent.
CSF Collection
CSF was collected via ventriculostomy from eight patients with aSAH daily from days 3–12 posthemorrhage. All patients had Fisher 3 and Hunt-Hess 2 or 3 aSAH as well as hydrocephalus requiring ventriculostomy placement.
Following collection, the CSF was immediately frozen at −70°C and was not subjected to centrifugation. Analyzing only the CSF supernatant for miR levels would confound trends over time, as cells (especially leukocytes and red blood cells) in the CSF are known to lyse and may release miRs.
miR Extraction and Purification
Total nucleic acid from CSF was extracted using the Circulating Nucleic Acid Extraction Kit (Qiagen, Valencia, CA), following the manufacturer's protocol designed for isolation of miR omitting carrier RNA. To remove any DNA contamination in the extracted RNA, we performed DNA digestion with Qiagen RNase-free DNase set. For concentration of miRNA from enzymatic reactions, the RNA was further cleaned up, with the manufacturer's modifications using Qiagen RNeasy MinElute Clean up Kit.
NanoString Assay
The multiplexed nanoString nCounter miR system (nanoString Technologies) was used for miR expression profiling (11). This assay was performed at The Ohio State University Comprehensive Cancer Center Nucleic Acid shared services facility. Total RNA (100 ng) was used as input material. Small RNA samples were prepared by ligating a specific DNA tag onto the 3′-end of each mature miR according to manufacturer's instruction (nanoString Technologies). These tags normalized the melting temperatures of the miRs and provided identification for each miR species in the sample. Excess tags were then removed, and the resulting material was hybridized with a panel of miR:tag-specific nCounter capture and barcoded reporter probes. Hybridization reactions were incubated at 64°C for 18 h. Hybridized probes were purified and immobilized on a streptavidin-coated cartridge using the nCounter Prep Station (nanoString Technologies). nCounter Digital Analyzer was used to count individual fluorescent barcodes and quantify target RNA molecules present in each sample. For each assay, a high-density scan (600 fields of view) was performed.
Data Normalization and Analysis
Quality control, normalization, and data analysis was performed using nSolver 2.0 Analysis Software (nanoString Technologies). Additional data analysis was performed using Genespring GX (Agilent Technologies, Santa Clara, CA). For data visualization, the miRNA expression data were subjected to hierarchical clustering using dChip (v 1.3) software. All differentially expressed miR were identified using one-way analysis of variance (ANOVA) assay with significance level was set at P < 0.05 and with correction for false discovery rate (22–26).
Real-time PCR
Isolated circulating miRNA underwent reverse transcription using miRCURY LNA Universal cDNA synthesis kit (Exiqon, Woburn, MA) according to manufacturer's recommended protocol. MiRs were quantified via real-time polymerase chain reaction using ExiLENT SYBR Green (Exiqon) on a Mx3000P qPCR platform (Agilent Technologies, Santa Clara, CA). SNORD44 (hsa) was use as a housekeeping reference. ΔΔCt comparative analysis was done was used to normalize each assayed miR sequence. The LNA PCR primers sets (Exiqon) used targeted the below sequences: hsa-miR-92a-3p: UAUUGCACUUGUCCCGGCCUGU, hsa-let-7b-3p: UGAGGUAGUAGGUUGUGUGGUU, hsa-miR-491-3p: CUUAUGCAAGAUUCCCUUCUAC.
RESULTS
Patient Data
A total of eight female patients were enrolled in this study (Table 1). They ranged in age from 30 to 74 yr old. All had a significant amount of blood in the subarachnoid space on computed tomography scan (Fisher grade 3) and had a good clinical grade (Hunt-Hess grades 2 or 3). There were three anterior communicating artery aneurysms, two posterior communicating artery aneurysms, one internal carotid artery aneurysm, one middle cerebral artery aneurysm, and one posterior inferior cerebellar artery aneurysm. Seven patients were treated with coil embolization, and one underwent craniotomy and clipping of her aneurysm. Four of the patients suffered a clinical decline due to DCI, and three had radiographic evidence of infarction due to DCI.
Table 1.
Patient demographics
| Patient No. | Age | Sex | Fisher Score | Hunt-Hess Grade | Aneurysm Location | Treatment | DCI |
|---|---|---|---|---|---|---|---|
| 1 | 49 | F | 3 | 3 | R PComm | coiled | no |
| 2 | 74 | F | 3 | 2 | AComm | coiled | yes |
| 3 | 40 | F | 3 | 3 | L PICA | coiled | no |
| 4 | 30 | F | 3 | 3 | L MCA | coiled | yes |
| 5 | 40 | F | 3 | 3 | L ICA | coiled | no |
| 6 | 66 | F | 3 | 2 | L PComm | coiled | no |
| 7 | 70 | F | 3 | 3 | AComm | clipped | yes |
| 8 | 71 | F | 3 | 3 | AComm | coiled | yes |
F, female; L, left; R, right; AComm, anterior communicating artery; ICA, internal carotid artery; MCA, middle cerebral artery; PICA, posterior inferior cerebellar artery; PComm, posterior communicating artery; DCI, delayed cerebral ischemia.
Hierarchical Clustering
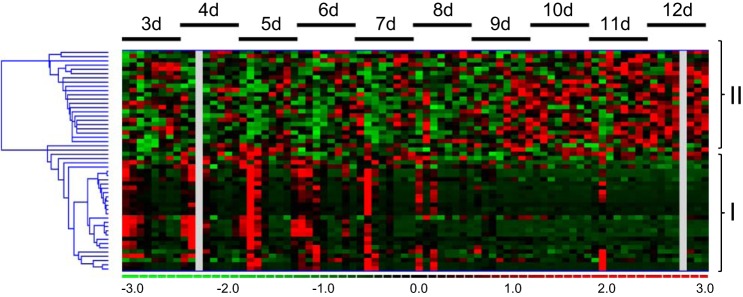
CSF was collected daily from day 3 to day 12 posthemorrhage. miRs were extracted and purified, and then miR expression profiling was run on an nCounter system using the nCounter Human v2 miR Expression Assay. Of 800 possible targets, 52 human miRs were detected in human CSF and fold expression relative to the first time point (day 3) characterized (Fig. 1). There were eight samples of aSAH CSF at each of the time points. Hierarchical clustering revealed two large clusters of miRs: one that showed decreased abundance over time compared with baseline (group 1) and a second that showed late increased abundance compared with baseline (group 2).
Fig. 1.
Hierarchical clustering of microRNA (miR) expression after hemorrhagic stroke. A: heat mapping (red, upregulated; green, downregulated) identification of 2 distinct clusters of miRs: group 1, decreased abundance over time; group 2, increased abundance over time. d, Day.
Analysis of NanoString and Quantitative RT-PCR Data
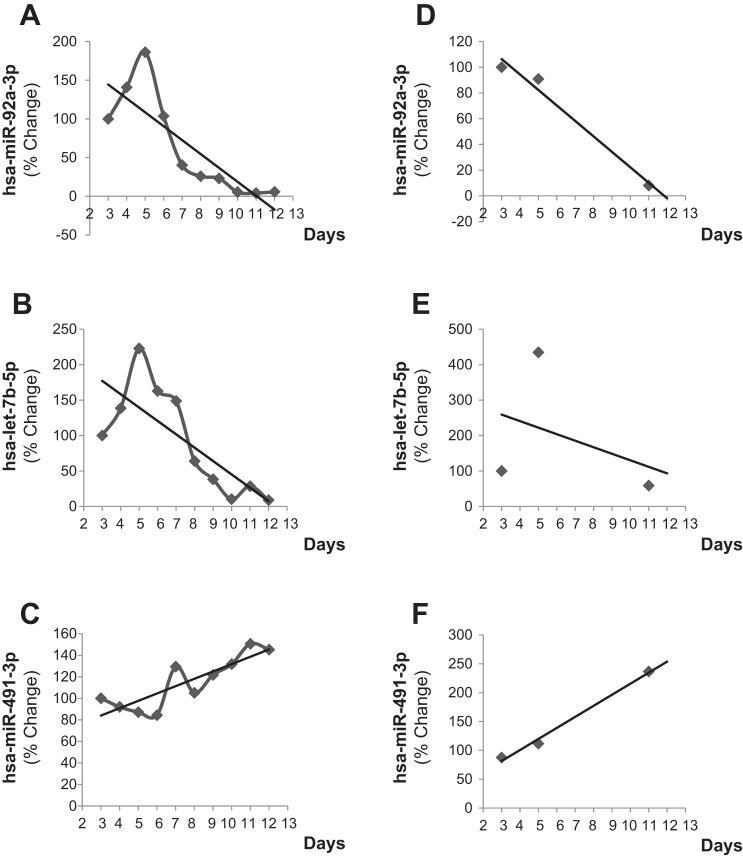
ANOVA with significance level was set at P < 0.05 and with correction for false discovery rate was performed of the nanoString data on each of the groups identified in the hierarchical clustering (Tables 2 and 3). In group 1, the overall downward trend was typified by the abundance of miR-92a and let-7b (Fig. 2, A and B). In group 2, the overall increase over time was typified by the abundance of miR-491 (Fig. 2C). These trends were confirmed with quantitative RT-PCR with samples collected on days 3, 5, and 11 (Fig. 2, D–F).
Table 2.
miRs with decreased abundance over time after aSAH (group 1)
| Fold Change |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | P Value | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | Day 10 | Day 11 | Day 12 |
| hsa-miR-410 | 3.17E-04 | 1 | 1.72 | 1.85 | 1.03 | −1.15 | −1.81 | −3.15 | −5.83 | −4.39 | −8.05 |
| hsa-miR-126-3p | 7.48E-04 | 1 | 1.63 | 2.14 | 2.17 | 1.67 | −1.05 | −3.72 | −4.02 | −3.28 | −5.31 |
| hsa-miR-331-3p | 0.001 | 1 | 1.08 | 1.66 | −1.18 | −1.20 | −2.41 | −3.49 | −9.72 | −10.73 | −3.56 |
| hsa-miR-142-3p | 0.002 | 1 | −1.50 | 1.15 | −1.12 | −1.61 | −2.68 | −3.98 | −7.21 | −5.41 | −6.96 |
| hsa-miR-20a/20b-5p | 0.002 | 1 | 1.31 | 1.75 | 1.34 | 1.09 | −1.25 | −3.44 | −11.06 | −3.83 | −8.47 |
| hsa-miR-486-3p | 0.002 | 1 | 2.42 | 1.36 | 1.02 | −1.22 | −1.20 | −2.41 | −12.96 | −3.38 | −6.16 |
| hsa-miR-92a-3p | 0.005 | 1 | 2.61 | 1.67 | 1.27 | −1.86 | −1.39 | −4.33 | −6.85 | −14.52 | −16.39 |
| hsa-miR-98 | 0.006 | 1 | 1.23 | −1.04 | −1.74 | −1.74 | −1.04 | −2.38 | −4.07 | −4.27 | −4.36 |
| hsa-miR-296-5p | 0.007 | 1 | 2.94 | 1.92 | 1.48 | −1.25 | −1.91 | −5.11 | −5.68 | −4.92 | −7.63 |
| hsa-miR-106b-5p | 0.008 | 1 | 1.45 | 2.29 | 1.69 | −1.07 | 1.55 | −3.91 | −4.67 | −1.64 | −2.91 |
| hsa-let-7b-5p | 0.012 | 1 | 3.18 | 2.37 | 1.90 | 1.14 | 1.07 | −3.10 | −4.68 | −5.05 | −10.02 |
ANOVA analysis of nanoString results of highly significant group 1 microRNAs (miRs) normalized to the first time point (day 3) following aneurysmal subarachnoid hemorrhage (aSAH).
Table 3.
miRs with increased abundance over time after aSAH (group 2)
| Fold Change |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | P Value | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | Day 10 | Day 11 | Day 12 |
| hsa-miR-1275 | 0.0031 | 1 | −1.08 | 1.07 | 1.11 | −1.46 | 1.03 | −1.19 | 3.39 | 2.16 | 1.80 |
| hsa-miR-491-3p | 0.0033 | 1 | −1.17 | −1.35 | −1.10 | 1.90 | 1.07 | 1.60 | 1.44 | 2.86 | 3.18 |
| hsa-miR-135a-5p | 0.0039 | 1 | 1.53 | 2.11 | 1.28 | 2.56 | 1.82 | 2.15 | 4.82 | 2.57 | 1.93 |
| hsa-miR-515-5p | 0.0073 | 1 | −1.81 | −1.21 | −2.18 | −1.80 | −1.72 | 1.44 | 1.21 | 1.14 | 2.33 |
| hsa-miR-1471 | 0.0078 | 1 | 2.77 | 1.15 | 2.78 | 1.60 | 2.99 | 1.08 | 2.44 | 1.16 | 2.86 |
| hsa-miR-585 | 0.0119 | 1 | −3.22 | −2.86 | −2.46 | −3.05 | −3.34 | −1.88 | −2.65 | −1.09 | 1.34 |
| hsa-miR-1287 | 0.0121 | 1 | −3.63 | −1.87 | −1.94 | −2.67 | −3.62 | −1.85 | −2.53 | −1.59 | 1.38 |
| hsa-miR-647 | 0.0159 | 1 | 1.18 | 1.41 | 1.24 | 1.04 | 1.59 | 2.55 | 1.87 | 2.88 | 3.32 |
| hsa-miR-552 | 0.0188 | 1 | 1.69 | 1.07 | 2.25 | 1.86 | 1.11 | 1.61 | 2.45 | 1.54 | 4.60 |
| hsa-miR-1255a | 0.0191 | 1 | −3.75 | −3.24 | −2.16 | −4.70 | −2.09 | −2.57 | −3.14 | −1.55 | −1.47 |
| hsa-miR-449b-5p | 0.0360 | 1 | 1.39 | 1.72 | 1.45 | 1.90 | 1.60 | 2.00 | 4.86 | 2.68 | 2.87 |
ANOVA analysis of nanoString results of highly significant group 2 miRs normalized to the first time point (day 3) following aSAH.
Fig. 2.
Temporal expression of pathologically relevant microRNA. A–C: mean percent change expression relative to the first time point [day 3 postaneurysmal subarachnoid hemorrhage (aSAH)] of hsa-miR-92a-3p, hsa-let-7b-3p (group 1), and has-miR-491-3p (group 2), respectively, as quantified via Nanostring nCounter Expression Assay. D–F: mean percent change expression validation of Nanostring miRs performed via quantitative RT-PCR of hsa-miR-92a-3p, hsa-let-7b-3p (group 1), and hsa-miR-491-3p (group 2), respectively.
DISCUSSION
Our understanding of brain injury and delayed cerebral ischemia after aSAH is limited. The working clinical cause is thought to be narrowing of large and small cerebral arteries that result in ischemia; however, while this is a clinically useful paradigm, it does not explain the cause for the vessel narrowing itself nor does it account for all cases of delayed cerebral ischemia (8). Because CSF is contiguous with the interstitial fluid of the brain, we believe that sampling of CSF may provide information on the molecular events in the extracellular milieu. Because aSAH results in the contamination of relatively acellular CSF with numerous circulating cells, it is quite likely that there are components in blood that contribute to brain injury and delayed cerebral ischemia after aSAH. However, by analyzing CSF over time, we have been able to identify temporally mediated changes that suggest the release of miRs into CSF days after the initial aSAH.
This is the first study to describe the temporally differentiated abundance of miRs in human CSF after aSAH. We hypothesize that the ability of a single miR to alter translation of numerous proteins makes them likely contributors to brain injury and the brain's response to injury. Recent studies have unveiled the role of miRs as key modulators of intracellular signaling mediators (2). Here we have identified two distinct clusters of miR abundance in human CSF in the days following SAH. Group 1 miRs decrease over time relative to the first time point and may be due to breakdown of miRs that were introduced into the CSF in the blood at the time of the initial hemorrhage (12, 13, 18). On the other hand, the increased abundance of group 2 miRs over time relative to the first time point may be due to the release of miRs into the CSF from the brain itself. The actual source of these group 2 miRs (ependymal cells, glial cells, or neurons) remains to be seen.
It is conceivable that some of the group 1 miRs contribute to brain injury following aSAH. The let-7 family of miRs is highly expressed in the cardiovascular system, and aberrant expression has been shown in cardiovascular diseases (3). Circulating let-7b has been shown to be decreased after acute myocardial infarction (15) and increased after some types of acute ischemic stroke (16). Additionally, in mice let-7b participates in angiogenesis by regulating the antiangiogenic factor tissue inhibitor of metalloproteinase 1 (19). However, the mechanism by which let-7b may contribute to brain injury following aSAH is undefined.
Perhaps the most compelling mechanism for an miR in CSF following aSAH contributing to brain injury is with miR-92a. The transcription factor Krüppel-like factor 2 (KLF2) plays an important role in endothelial cell function and is highly induced by atheroprotective flow patterns (i.e., shear stress with a significant forward direction) as well as by statins. KLF2 enhances vessel compliance and confers antithrombotic and anti-inflammatory effects on endothelial cells (1). miR-92a targets KLF2 and levels of miR-92a are reduced by atheroprotective flow patterns, resulting in increased levels of KLF2 messenger RNA (32). Overexpression of miR-92a in human umbilical vein endothelial cells suppressed the KLF2-regulated gene eNOS (6). It is possible that increased abundance of miR-92a following aSAH results in perturbation of endothelial function and an increased likelihood of secondary brain injury through delayed cerebral ischemia. This is supported by the finding that KLF2(−/−) mice suffer larger stroke burden and impairment in blood brain barrier function than control mice following transient middle cerebral artery occlusion (28).
It is also intriguing to hypothesize that the group 2 miRs play a role in the brain's response to injury as this group of miRs increases over time and is likely due to release from the brain into the CSF. In particular, miR-491 has been shown to be upregulated in human subjects who had committed suicide (27) and may regulate synaptic plasticity in the mouse amygdala (20). Cognitive impairment and behavioral difficulties are some of the most common symptoms in those who survive aSAH (8), but the mechanistic role of miR-491 in this problem, if any, is again undefined.
This study has several limitations that will be addressed in subsequent work. First, a larger sample size needs to be analyzed. In the current study a small sample size of only eight patients was included. Second, although nanoString analysis has been shown to provide excellent specificity and a low false positive rate (17), only a subset of the miRs were analyzed with quantitative RT-PCR. And third, any clinical relevance of our results will need to be explained in future studies.
Conclusions
Here we have identified a temporally differentiated abundance of miRs in human CSF after aSAH. Some decrease in abundance over time, while others increase in abundance over time. These miRs may have pleiotropic effects on brain injury and the brain's response to injury. Further research into CSF miRs may contribute to a better understanding of brain injury after aSAH and lead to the development of novel therapeutics.
GRANTS
Supported in part by National Institutes of Health Grants NS-42617, NS-085272, and GM-069589 and by a grant from the Brain Aneurysm Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: C.J.P. and C.K.S. conception and design of research; C.J.P., R.D., C.R., S.R., and C.K.S. interpreted results of experiments; C.J.P. drafted manuscript; C.J.P., R.D., S.W.Z., C.R., S.R., and C.K.S. edited and revised manuscript; C.J.P., R.D., C.R., S.R., and C.K.S. approved final version of manuscript; R.D. and C.R. performed experiments; R.D., C.R., S.R., and C.K.S. analyzed data; R.D. and S.R. prepared figures.
REFERENCES
- 1.Atkins GB, Jain MK. Role of Kruppel-like transcription factors in endothelial biology. Circ Res 100: 1686–1695, 2007. [DOI] [PubMed] [Google Scholar]
- 2.Avraham R, Yarden Y. Regulation of signalling by microRNAs. Biochem Soc Transact 40: 26–30, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y, Zhou HH. Let-7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int J Mol Sci 14: 23086–23102, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, Maghnouj A, Zollner H, Reinacher-Schick A, Schmiegel W, Hahn SA, Schroers R. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 117: 3140–3146, 2011. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, function. Cell 116: 281–297, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324: 1710–1713, 2009. [DOI] [PubMed] [Google Scholar]
- 7.Burgos KL, Javaherian A, Bomprezzi R, Ghaffari L, Rhodes S, Courtright A, Tembe W, Kim S, Metpally R, Van Keuren-Jensen K. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 19: 712–722, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly ES Jr, Rabinstein AA, Carhuapoma JR, Derdeyn CP, Dion J, Higashida RT, Hoh BL, Kirkness CJ, Naidech AM, Ogilvy CS, Patel AB, Thompson BG, Vespa P, American Heart Association Stroke Council, Council on Cardiovascular Radiology and Intervention, Council on Cardiovascular Nursing, Council on Cardiovascular Surgery and Anesthesia, Council on Clinical Cardiology. Guidelines for the management of aneurysmal subarachnoid hemorrhage: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke 43: 1711–1737, 2012. [DOI] [PubMed] [Google Scholar]
- 9.Jung CS, Lange B, Zimmermann M, Seifert V. CSF and serum biomarkers focusing on cerebral vasospasm and ischemia after subarachnoid hemorrhage. Stroke Res Treat 2013: 560305, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koutsis G, Siasos G, Spengos K. The emerging role of microRNA in stroke. Curr Top Med Chem 13: 1573–1588, 2013. [DOI] [PubMed] [Google Scholar]
- 11.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol 94: Unit25B 10.17, 2011. [DOI] [PubMed] [Google Scholar]
- 12.Lai CY, Wu YT, Yu SL, Yu YH, Lee SY, Liu CM, Hsieh WS, Hwu HG, Chen PC, Jeng SF, Chen WJ. Modulated expression of human peripheral blood microRNAs from infancy to adulthood and its role in aging. Aging Cell 13: 679–689, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Gao G, Yang C, Zhou K, Shen B, Liang H, Jiang X. The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int J Mol Sci 15: 10567–10577, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott M, Meigs J, Mozaffarian D, Nichol G, O'Donnell C, Roger V, Rosamond W, Sacco R, Sorlie P, Stafford R, Steinberger J, Thom T, Wasserthiel-Smoller S, Wong N, Wylie-Rosett J, Hong Y, American Heart Association Statistics Committee, Stroke Statistics Subcommittee. Heart disease and stroke statistics–2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 119: e21–e181, 2009. [DOI] [PubMed] [Google Scholar]
- 15.Long G, Wang F, Duan Q, Yang S, Chen F, Gong W, Yang X, Wang Y, Chen C, Wang DW. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One 7: e50926, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long G, Wang F, Li H, Yin Z, Sandip C, Lou Y, Wang Y, Chen C, Wang DW. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol 13: 178, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D'Andrade P, DeMayo M, Dennis L, Derveaux S, Feng Y, Fulmer-Smentek S, Gerstmayer B, Gouffon J, Grimley C, Lader E, Lee KY, Luo S, Mouritzen P, Narayanan A, Patel S, Peiffer S, Ruberg S, Schroth G, Schuster D, Shaffer JM, Shelton EJ, Silveria S, Ulmanella U, Veeramachaneni V, Staedtler F, Peters T, Guettouche T, Wong L, Vandesompele J. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Meth 11: 809–815, 2014. [DOI] [PubMed] [Google Scholar]
- 18.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differentiation 20: 1603–1614, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, Han J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest 118: 1944–1954, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrzykowski AZ, Spijker S. Impulsivity and comorbid traits: a multi-step approach for finding putative responsible microRNAs in the amygdala. Front Neurosci 8: 389, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rink C, Khanna S. MicroRNA in ischemic stroke etiology and pathology. Physiol Genomics 43: 521–528, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy S, Khanna S, Rink C, Biswas S, Sen CK. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics 34: 162–184, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy S, Khanna S, Shah H, Rink C, Phillips C, Preuss H, Subbaraju GV, Trimurtulu G, Krishnaraju AV, Bagchi M, Bagchi D, Sen CK. Human genome screen to identify the genetic basis of the anti-inflammatory effects of Boswellia in microvascular endothelial cells. DNA Cell Biol 24: 244–255, 2005. [DOI] [PubMed] [Google Scholar]
- 24.Roy S, Khanna S, Wallace WA, Lappalainen J, Rink C, Cardounel AJ, Zweier JL, Sen CK. Characterization of perceived hyperoxia in isolated primary cardiac fibroblasts and in the reoxygenated heart. J Biol Chem 278: 47129–47135, 2003. [DOI] [PubMed] [Google Scholar]
- 25.Roy S, Khanna S, Yeh PE, Rink C, Malarkey WB, Kiecolt-Glaser J, Laskowski B, Glaser R, Sen CK. Wound site neutrophil transcriptome in response to psychological stress in young men. Gene Express 12: 273–287, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy S, Patel D, Khanna S, Gordillo GM, Biswas S, Friedman A, Sen CK. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA 104: 14472–14477, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serafini G, Pompili M, Hansen KF, Obrietan K, Dwivedi Y, Shomron N, Girardi P. The involvement of microRNAs in major depression, suicidal behavior, and related disorders: a focus on miR-185 and miR-491-3p. Cell Mol Neurobiol 34: 17–30, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, Sheng B, Zhang F, Wu C, Zhang R, Zhu J, Xu K, Kuang Y, Jameson SC, Lin Z, Wang Y, Chen J, Jain MK, Atkins GB. Kruppel-like factor 2 protects against ischemic stroke by regulating endothelial blood brain barrier function. Am J Physiol Heart Circ Physiol 304: H796–H805, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 469: 336–342, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl Stroke Res 5: 711–718, 2014. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Sun G, Zhang L, Shi L, Zeng Y. Circulating microRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J Stroke Cerebrovasc Dis 23: 2607–2613, 2014. [DOI] [PubMed] [Google Scholar]
- 32.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G Jr, Wang KC, Geary GG, Zhang Y, Wang WC, Huang HD, Zhou J, Li YS, Chien S, Garcia-Cardena G, Shyy JY. Flow-dependent regulation of Kruppel-like factor 2 is mediated by microRNA-92a. Circulation 124: 633–641, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]