Abstract
HLDA10 collated a panel of monoclonal antibodies (mAbs) that primarily recognised molecules on human myeloid cell and dendritic cell (DC) populations. As part of the studies, we validated a backbone of mAbs to delineate monocyte and DC populations from peripheral blood. The mAb backbone allowed identification of monocyte and DC subsets using fluorochromes that were compatible with most ‘off the shelf' or routine flow cytometers. Three laboratories used this mAb backbone to assess the HLDA10 panel on blood monocytes and DCs. Each laboratory was provided with enough mAbs to perform five repeat experiments. The data were collated and analysed using Spanning-tree Progression Analysis of Density-normalised Events (SPADE). The data were interrogated for inter- and intra-laboratory variability. The results highlight the definition of DC populations using current readily available reagents. This collaborative process provides the broader scientific community with an invaluable data set that validates mAbs to leucocyte surface molecules.
Antigen-presenting cells process and present exogenous and endogenous antigens to T cells for specific, protective and homeostatic immune responses. Macrophages, monocytes, dendritic cells (DCs) and B cells all act as antigen-presenting cells but DCs have a specialist role, exemplified by their ability to process antigen, migrate to lymph nodes and initiate primary immune responses.1, 2, 3 DCs are distinguished morphologically, phenotypically and functionally from monocytes and macrophages. Practically, we define human monocytes as CD14pos cells and DCs as HLA-DR+ Lineage (CD3, CD19, CD56, CD14 (Lin)) negative (−) cells. DCs are derived from a common DC progenitor, which gives rise to the major plasmacytoid (pDC) and myeloid (mDC) subsets.4, 5 We divide the mDCs further into CD1c+ and CD141+ subsets.6, 7 Likewise, human monocytes are divided into functional subsets: the major CD14hiCD16− and a minor CD14loCD16+ subset, with a third CD14intCD16+ subset described in some studies.8, 9, 10 It has been noted that within the human HLA-DR+Lin− peripheral blood mononuclear cell (PBMC) gate there remains cells that lack the common mDC or pDC markers.6 A small subset of these cells includes CD34+ cells and this population has in fact been shown to be immunostimulatory.6, 11 To acknowledge the presence of CD34+ cells as including the haematopoietic stem cell population, we continue to analyse the HLA-DR+Lin−CD34+ population as a potentially important antigen-presenting cell population.
The focus of the Human Leucocyte Differentiation Antigen (HLDA) workshops is to establish ‘clusters of differentiation' (CD) that define monoclonal antibody (mAb) clones that have been demonstrated to recognise the same molecule targets. This is achieved by inter-laboratory testing using a variety of techniques and targets to validate each antibody for the benefit of all. The expression of the target antigen is thus determined for a large number of mAbs providing researchers and commercial interests with the assurance that those mAbs reported in the workshops have well-defined reactivity.
In humans, peripheral blood DCs are the most widely studied ex vivo population. The HLDA10 workshop provided the opportunity for inter-laboratory comparison of blood DCs defined by the same antibody backbone using flow cytometry. This enabled us to compare the robustness of gating strategies to define the commonly studied blood DC subsets. We used a mAb backbone that allowed us to analyse the blood DC and monocyte populations. To interrogate CD14+ and CD16+ monocyte populations alongside DC populations, the CD14 mAb was removed from the Lineage mix and CD14 and CD16 mAbs were used as independent parameters. We used the data to confirm the expression of antibodies that were validated through the HLDA10 blinded studies.
There are a number of computational programs available to analyse multi-parameter cytometry data.12 We report results analysed using Spanning-tree Progression Analysis of Density-normalised Events (SPADE), which is the first clustering program widely available for unbiased analysis of high-dimensional data.13 The advantage of SPADE is that it normalises the frequency of abundant and rare cell populations determined by all parameters in each sample in the data set. The result is global clustering of phenotypically similar cells into two dimensions for easy visualisation of global expression patterns from complex heterogeneous data. This is particularly useful in the analysis of flow cytometric data collected from multiple donors and across multiple sites. SPADE is available through Cytobank, which also hosts FlowRepositry, a web-based data sharing and repository function for community-based analysis of flow cytometry data.14
Results
Manual gating of blood DCs is consistent with unsupervised clustering by SPADE
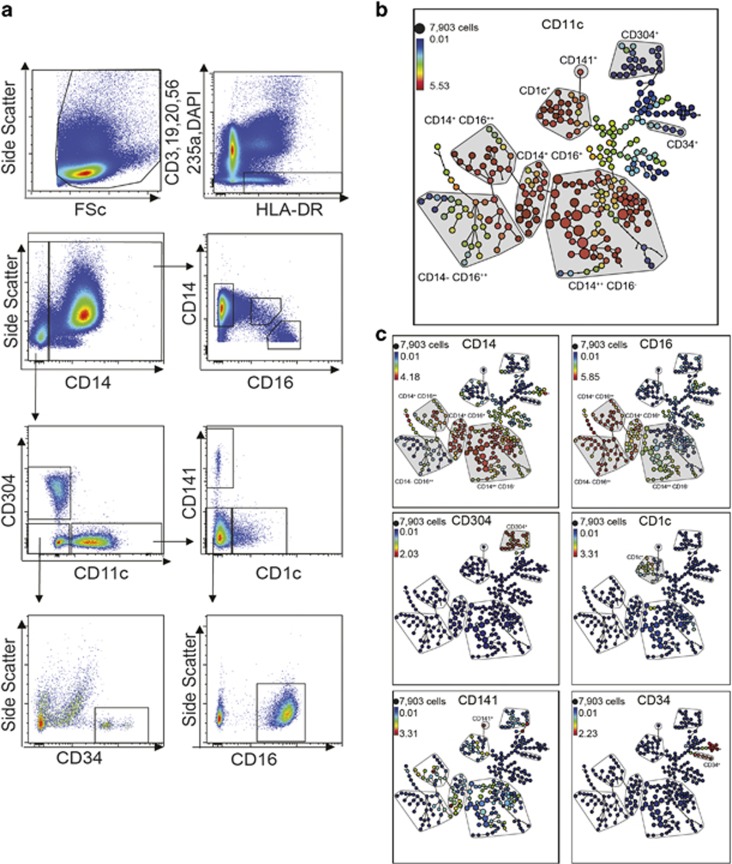
A backbone panel of 12 mAbs used to describe and delineate different myeloid subsets was assembled using commercially available conjugated mAbs to maximise compatibility on common instrumentation, while minimising fluorescence spill over (Table 1). We demonstrated specificity of the backbone for monocyte and DC subsets using the gating strategy shown in Figure 1a. Manual gating of viable, CD3−, CD20−, CD56− (Linneg), HLA-DR+ cells was performed to identify CD14+ monocytes and CD14− DCs. Monocytes were divided into the two main subsets, CD14++ CD16− ‘classical' and CD14+ CD16+ ‘non-classical' monocytes. CD14− cells were divided into CD304+ pDCs, CD11c+ mDCs and the remaining CD304− CD11c− cells. Myeloid DCs were divided into three mutually exclusive subsets based on the expression of CD1c+, CD141+ and CD16+. The CD304− CD11c− cells were further analysed for CD34+ cells.
Table 1. The mAb backbone panel for distinguishing monocyte and DC populations from peripheral blood.
| CD and fluorochrome | Clone details | Source |
|---|---|---|
| CD3 V450 | UCHT1 Ms IgG1, κ | 560365 BD Horizon |
| CD20 V450 | L27 Ms IgG1, κ | 642274 BD Horizon |
| CD19 V450 | HIB19 Ms IgG1, κ | 560353 BD Horizon |
| CD56 V450 | B159 Ms IgG1, κ | 560360 BD Horizon |
| CD16 V500 | 3G8 Ms IgG1, κ | 561394 BD Horizon |
| CD14 PerCPCy5.5 | M5E2 Ms IgG2a, κ | 550787 BD Pharmingen |
| CD11c AF700 | B-ly6 Ms IgG1, κ | 561352 BD Pharmingen |
| HLA-DR APC-H7 | L243 Ms IgG2a, κ | 641393 BD Horizon |
| CD304 APC | AD5-17F6 Ms IgG2a, κ | 130-090-900 Miltenyi Biotec |
| CD1c FITC | AD5-8E7 Ms IgG1 | 130-090-507 Miltenyi Biotec |
| CD141 PEVio770 | AD5-4H12 Ms IgG1 | 130-100-217 Miltenyi Biotec |
| CD34 PE-CF594 | 581 Ms IgG1, κ | 562383 BD Horizon |
Abbreviations: APC, allophycocyanin; CD, clusters of differentiation; DC, dendritic cell; FITC, fluorescein isothiocyanate; Ig, immunoglobulin; mAb, monoclonal antibody; PE, phycoerythrin; PerCP, peridinin chlorophyl protein. Of note, the CD141 mAb clone was found to give the best discrimination of the CD141+ population.
Figure 1.
Identification of human monocytes and DCs. (a) Events collected in each FCS file were first gated for consistent data collection using low trigger pulse width and Forward Scatter (FSc), for single cells using Side Scatter area and height and for leucocyte populations using Side Scatter and FSc. The viable myeloid and DC populations were manually gated as the viable (DAPI−), CD3, CD19, CD20, CD56, CD235a (Lin−) and HLA-DR+ cells. The Lin−HLA-DR+ population was gated into CD14+ monocytes and CD14− DCs. The CD14+ monocytes were divided into the CD14++ CD16− and CD16+ CD14+ cells. CD14− cells were divided into CD304+, CD11c+ and CD304− CD11c− cells. Remaining Lin−HLA-DR+ DCs were subsequently analysed for CD34+ cells. The CD11c+ DCs were divided into three mutually exclusive subsets by the expression of CD1c+, CD141+ or CD16+. (b) Semi-supervised clustering was performed on the Lin− HLA-DR+ cells, and SPADE analysis was performed on ArcSinH-transformed fluorescent parameters. SPADE clustering was performed from five healthy donors on the entire test mAb data set, including both PE- and FITC-conjugated test mAbs to generate unified SPADE trees. SPADE trees were generated based on the expression of CD11c and other subsets (c) CD14, CD16, CD141 and CD34. Monocyte and DC populations were manually annotated according to the same criteria as used for manual analysis. Heat maps represent the median ArcSinH-transformed fluorescence of the indicated markers and size of each node is representative of the number of cells.
We used this backbone panel to test the HLDA10 panel of 84 test mAbs in three laboratories. Each laboratory was provided with the same batch of commercial antibody and tested PBMC from three to five donors. We assessed the ability of the backbone to reliably define the expected populations by conventional gating. We compared our conventional gating with SPADE analysis to cluster Lin− HLA-DR+ PBMC. The analysis included the entire data set of all test mAbs from all donors allowing an unbiased picture of the resulting specificity. We mapped each of the backbone mAbs onto the resulting SPADE tree allowing each of the myeloid and DC cell types to be identified and also clearly demonstrated the continuity of expression of some backbone markers (for example, CD14 and CD16, Figure 1b) as well as discontinuity of others (CD304, CD34 and 10–66 (ILT-7)).
We used the profile of well-known mAbs included in the test panel to annotate our SPADE output for monocyte and DC subsets according to nomenclature outlined by the International Immunological Union8 and as we used in previous workshops.6, 15 The Lin−HLA-DR+ cells were divided on the basis of CD14 expression. The CD14+ monocytes were divided into CD14++ CD16− and CD16+ CD14+ cells. CD14− cells were subdivided into CD304+ pDCs, CD11c+ DCs and CD304− CD11c− cells. The CD304− CD11c− cells included a small CD34+ subset. The CD11c+ DCs were divided into three mutually exclusive subsets by the expression of CD1c+, CD141+ or CD16+ (Figure 1c). As expected, this analysis showed that DCs clustered quite separately from monocytes. Within the Lin− HLA-DR+ fraction of PBMC, there were still substantial populations of cells falling outside these discreet populations, many of which have been described in previous workshops.
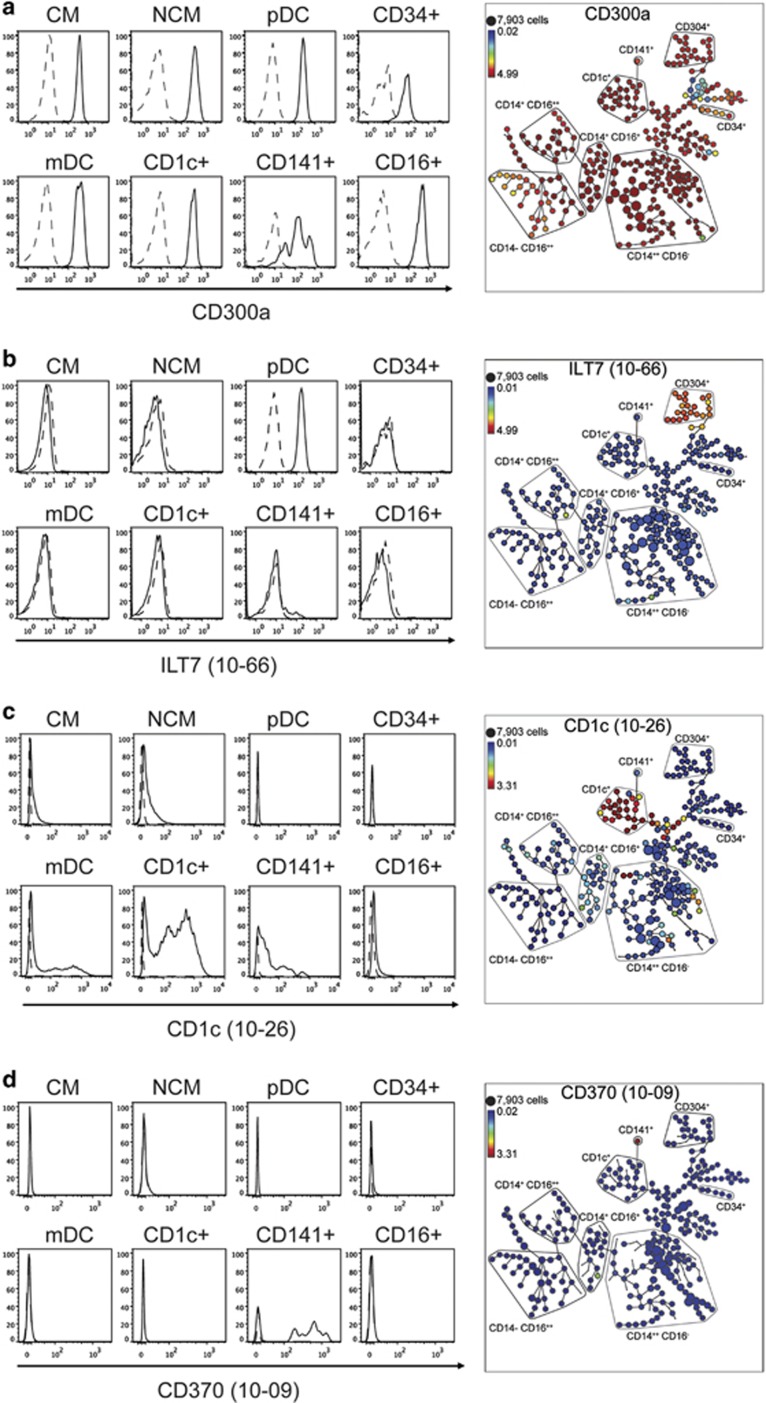
We validated the SPADE clustering by demonstrating that the expression of known DCs and myeloid subset-specific markers was consistent with the SPADE clustered populations (Figure 2). We compared the SPADE clusters with manually gated bivariate plots for binding of the pan leucocyte marker CD300a mAb clone MEM260, 17G10.2 (10–66) to the CD304+ pDC-specific ILT-7 molecule, L161 (10–26) to CD1c+ DCs and 9A11 (10-09) to CLEC9A on CD141+ mDCs. This was performed by backgating the expression of each mAb onto the delineated populations.
Figure 2.
Validation of SPADE clustering to identify discreet DC subpopulations in the combined analysis of purified and PE-conjugated test mAbs. Manually gated bivariate plots for submitted mAbs to well-described monocyte- and DC-specific molecules were compared with manually annotated SPADE trees generated using the DC backbone. Each SPADE tree was interrogated for binding of well-described monocyte or DC markers. (a) CD300a (clone MEM260) was used as a pan myeloid marker, (b) ILT7 (clone 17G10.2) identified pDCs co-expressing CD304 (clone 7G3), (c) CD1c (clone L161) identified DCs co-expressing BDCA-1 (clone AD5-8E7) and (d) CLEC9A (clone 9A11) identified DCs co-expressing CD141+ (clone AD54H12). CM,CD14++ CD16− classical monocytes; DCs; CD1c+ DCs; CD141+ DCs and CD16+ DCs are gated as the CD11c+ mDC subsets, mDC,CD11c+CD304− myeloid, NCM,CD14+ CD16++ non-classical monocytes, pDC,CD304+ plasmacytoid DCs.
Expression of C-type lectin (CLEC) molecules to myeloid and DC populations
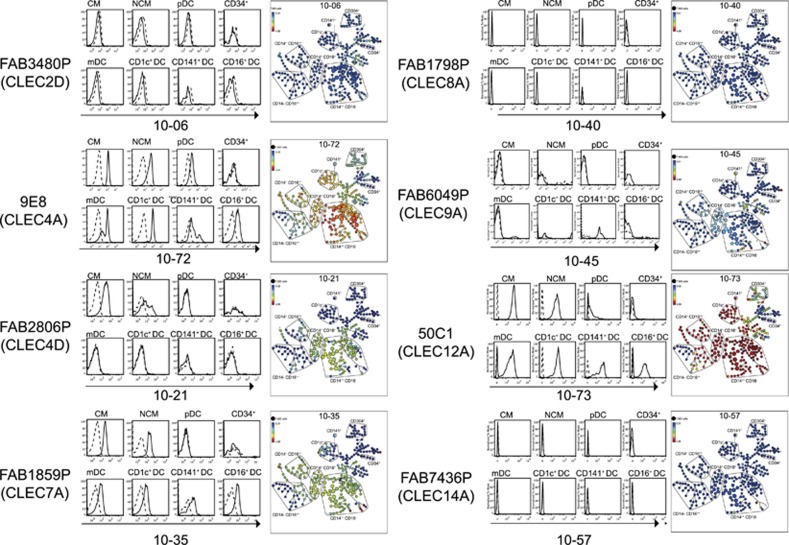
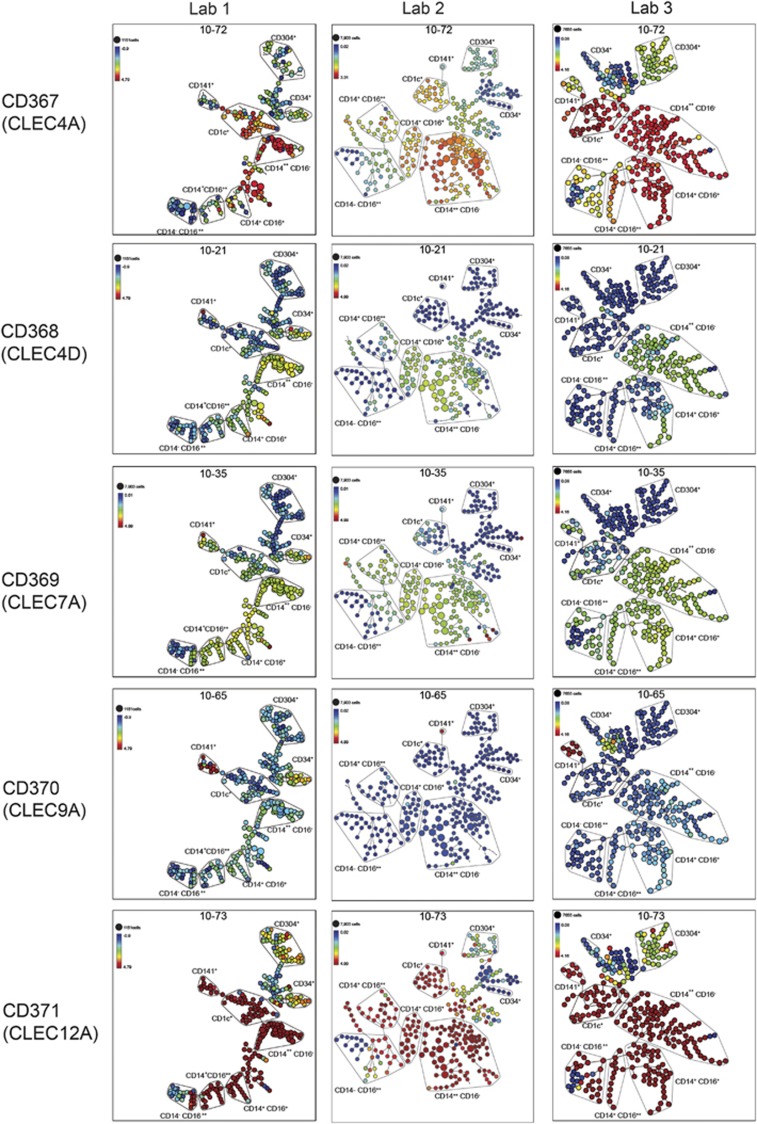
We used SPADE to analyse the binding of mAbs specific for different CLEC molecules to the myeloid and DC populations (Figure 3). There was little binding of mAbs to CLEC2D (10-06), CLEC8A (10-40) or CLEC14A (10-57) to any population; however, these mAbs showed little binding to cell lines and thus we did not confirm their activity.
Figure 3.
Comparison of the histograms showing expression of different CLECs on the gated populations with the SPADE analysis. CD1c+ DCs, CD141+ DCs and CD16+ DCs are gated as the CD11c+ mDC subsets. The mAb clone name and molecule is shown on the left of each histogram set and the HLDA10 workshop code is indicated on the x axis of the histogram and in the SPADE diagram. CM,CD14++ CD16− classical monocytes; mDC,CD11c+CD304− mDCs; NCM,CD14+ CD16++ non-classical monocytes; pDC,CD304+ plasmacytoid DCs.
The three antibodies to CLEC4A, 10-13 (not shown, data from all mAbs are available at http://HCDM.org), 10-71 (not shown) and 10-72 (Figure 3) showed similar specificity and bound strongly to the CD14++ CD16− and CD14+ CD16++ myeloid cells as well as CD11c+CD16+ and CD1c+ DC populations.
The three antibodies to CLEC7A, 10-01 (not shown), 10-35 (Figure 3) and 10-79 (not shown) showed similar specificity and bound strongly to the CD14++ CD16− and CD14+ CD16++ myeloid cells and CD11c+CD16+ DC but not to the CD1c+ DC population.
The three antibodies to CLEC12A, 10-07 (not shown), 10-51 (not shown) and 10-73 (Figure 3) all bound strongly to all monocyte and DC populations with the exception of pDCs.
The two antibodies to CLEC4D, 10-21 (Figure 3) and 10-78 (not shown) bound only to the CD14++ CD16− monocytes.
CLEC4D (10-21), CLEC7A (10-35) and CLEC12A (10-73) mAbs bound most strongly to the CD14++ CD16− and CD14+ CD16++ myeloid cells as well as cells that are CD11c+CD16+ and CD1c+ DC populations.
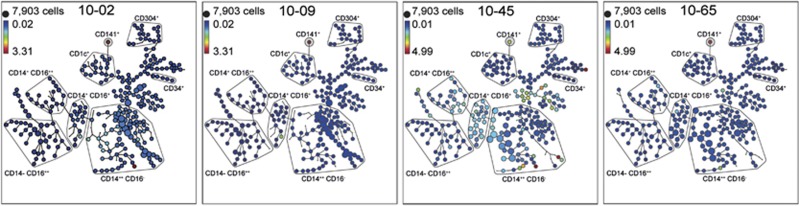
The HLDA10 test panel included three clones 10-02, 10-09, 10-45 and 10-65 to the CLEC9A molecule. One of these clones 8F9 (10-02 and 10-65) was obtained from two sources. The analysis of the data from five healthy donors by SPADE identified the most intense binding of each of these mAbs as being the small but distinct CD141+ DC population (Figure 4). The CLEC9A mAbs 10-02, 10-09 and 10-65 gave the most consistent results across the three laboratories (Figure 5). CD nomenclature was allocated to five CLEC molecules defined by mAb in HLDA10. These were CD367 (CLEC4A), 368 (CLEC4D), 369 (CLEC7A), 370 (CLEC9A) and 371 (CLEC12A). The mAbs to each of these molecules were assessed across the data from three different laboratories. The binding patterns were consistent demonstrating the robustness of the mAbs and the population discrimination (Figure 5).
Figure 4.
Comparison of antibodies directed against CLEC9a. Expression patterns of CLEC9a identified by submitted antibodies robustly identified the rare CD141+ DC subsets. Antibodies 10-02 and 10-65 were the same clone but labelled with different fluorophores.
Figure 5.
Using the DC backbone across laboratories and flow cytometers resulted in similar pattern of expression. Data from each of the three contributing laboratories for one mAb validated in the HLDA10 workshop that contributed to a new CD was analysed by SPADE. Each SPADE tree is generated from three to five donors.
Expression of other markers on DCs and monocytes
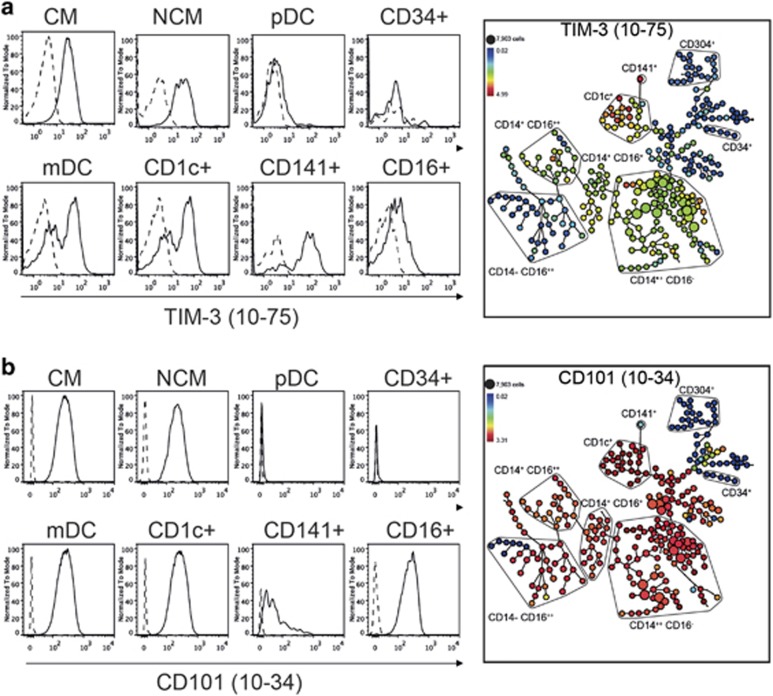
The two mAbs, 10-24 and 10-75, bind TIM-3 and were clustered as CD366. The analysis across the laboratories demonstrated that TIM-3 is expressed on CD14+ classical monocytes and CD1c+ and CD141+ mDCs. There was little-to-no expression on CD16+ cells or pDCs (Figure 6a). Similarly, the CD101 mAb 10-34 bound to all monocyte and DC populations with the exception of pDCs and CD34+ cells (Figure 6b).
Figure 6.
Binding of mAbs to TIM-3 and CD101 to DC subsets confirms their expression on these cells. (a) TIM-3 mAb 10-75 binds to CD141+ DCs and CD1c+ DCs. (b) CD101 mAb 10-34 binds CD1c+ and CD16+ DC populations and not to CD141+ DCs and pDCs.
SPADE clustering was consistent between laboratories
We further analysed the data from the three different laboratories by SPADE clustering. Of the test 84 mAbs, only the 34 mAbs directly conjugated to phycoerythrin (PE) were tested by all three groups, and this data was used in the inter-laboratory analysis. To reduce the size of the data set compensated, Lin− HLA-DR+ gates were exported to new FCS files for analysis. As a consequence of the different flow cytometers used in the three laboratories, FCS files from each laboratory had different parameter structures and dynamic ranges. Therefore, parameter readouts differed in intensities and were not combined into a single data set for clustering. Instead, each laboratory data set was clustered independently using the same SPADE parameters and manually annotated. As the SPADE analysis effectively reduces the multi-dimensional data set into two dimensions, it is expected that each of the resulting SPADE trees differ.13 When annotation of the clusters was performed by backgating with the same discriminatory markers used earlier, we could identify the same populations of myeloid cells and DCs (Figure 5).
Discussion
Whether a monocyte or DC is defined by ontogeny, location or function, the manner in which cells are currently investigated by flow cytometry is directed by the ability to have a mAb or other agent that can be used to define, isolate or probe the cell.16 Mouse models enable the additional use of molecular tools to tag molecules in genetically modified animals creating the opportunity to study ‘in vivo' the expression and function of gene products. However, this is much more difficult in human experimental systems.
The delineation of myeloid cell subsets and their distinction from DC populations is often directed by studies in the mouse.1 The emphasis on mouse models as inbred, pathogen free and a statistically reproducible source of data contrasts with the variation in human samples and the frequent lack of a corresponding mAb specific for human homologues. The value of the HLDA workshops has been to validate mAbs to human cell surface molecules and provide expression data using conventional flow cytometry on leucocyte subsets. We report here the phenotyping of human peripheral blood myeloid and DC populations completed by three laboratories using the same panel of mAbs and conventional flow cytometry. We have used SPADE analysis combined with traditional flow cytometry analysis techniques to demonstrate sensitivity and display global staining patterns of each submitted antibody.
The panel of mAbs to define the human myeloid and DC subsets included those used to identify monocyte populations combined with those used to identify DCs. In human studies, CD14 expression is the critical phenotypic marker on monocytes.10 All CD14− cells are removed routinely in the gating hierarchy. CD16 is then used as a marker to distinguish different CD14+ monocyte subsets. Similarly, DCs are generally seen as not expressing the lineage markers CD3, CD19, CD20, CD56, CD235 and CD14. Thus CD14 is generally included in the Lin+ cells to remove monocytes. In these HLDA10 studies, CD14 and CD16 mAbs were separated from the Lineage mAbs so that we could interrogate monocyte and DC subsets. This confirmed the presence of a population of CD14− cells that were CD16+ that would not be generally included in the monocyte gates and are likely to be another ‘DC subset' as described previously.6 Whether or not they derive from a monocyte population is under investigation.
The DC backbone panel contained 12 different antibodies with 8 fluorophores and generally kept the PE channel to be used for the test mAb. As not all mAbs were PE labelled, a second strategy used AlexaFluor 488-coupled secondary reagent to detect the test mAb. When this strategy was used, the CD1c marker was removed from the panel and the CD1c DC population was determined as the Lin−HLA-DR+ CD11c+ CD16− CD141− cells. Importantly, we included all the data from PE and purified antibodies in a unified analysis that omitted CD1c from the clustering algorithm. When we did this, CD1c+ DCs still formed a discreet myeloid cluster in the unified SPADE tree that could be identified robustly by backgating samples that did have CD1c in the panel demonstrating accurate analysis of these cells. The definition of this gating strategy was consistent with our previous studies6 and more recent panels described in the literature.17, 18
CLEC form a large family of pattern recognition receptors that are classically defined by their ability to bind carbohydrates.19 Already a number of CLECs are well described as markers of DCs. The use of unsupervised gating enabled us to identify critical expression patterns for some specific markers. The mAbs to CLEC9A (10-02, 10-09, 10-45, 10-65) were clearly most abundant on the CD141+ DCs, although there was consistent low-level expression detected with two mAbs on CD14+ monocytes.20 There have been suggestions that XCR1, the orphan chemokine receptor for lymphotactin, may be a superior marker for the CD141+ DC subset, but as yet no mAb to XCR1 is readily available.21, 22
The two CLEC12A mAbs (10-17, 10-51) bound consistently to most monocytes and DC populations with the clear exception of pDCs. This appears to be in contrast with the original description of 10-17 binding to pDCs, which were identified in these studies by conventional gating as CD303+ cells within CD14− cells.23 The lack of binding to pDCs by two different clones to CLEC12A across three laboratories confirms lack of expression.
CLEC4A was identified by three mAbs in the panel and all demonstrated specific binding to CD1c DCs and monocytes but not to pDCs or CD141+ DCs.
TIM-3 is generally regarded as a monocyte marker but mAb 10-75 showed that it was also present on both CD141+ DCs and CD1c+ DCs, with CD141+ DCs expressing the higher level.
CD101 has been described as having high-level expression on monocytes, granulocytes, DCs and activated T cells. We confirmed the binding of CD101 mAb 10-34 on all DC populations with the exception of pDC18 and CD141+ DCs.
Testing the panel of mAbs across three sites provided comparable results. Variability in peripheral blood samples from healthy human donors did not result in significant differences between the sites. The comparison of the data by unsupervised clustering showed remarkable similarity in the results. The different appearance of the SPADE trees generated using data from three independent laboratories is caused by the inherent stochastic nature of the downsampling and clustering used by the program. Importantly, the clustering algorithms generate the same general clusters when queried for the expression patterns of both backbone and test mAbs. In particular, mAbs to CD1c in both the backbone and test panel (10-26) allowed us to confirm the similarity of clusters.
The use of high dimensional clustering confirmed the distinction of blood monocyte and DC populations, highlighting the continuum of expression of many of the molecules used to distinguish them. It has highlighted a number of mAbs that will be able to be taken forward into further subset analysis using such technologies as mass cytometry CyTOF.24 The development of these technologies relies on the availability of well-validated mAbs that can be used in the large parameter experiments required to make the most of the big data analysis.
Methods
HLDA10 panel
Contributors to the HLDA10 supplied antibodies to the DCR group at the ANZAC Research Institute. Antibodies were diluted to 100 μg ml−1. Antibodies were used at saturating concentrations determined by information provided by supplier and titration on cell lines. The table of mAbs included in this panel is in the accompanying paper in this volume reviewing the workshop (http://www.nature.com/doifinder/10.1038/cti.2015.40).
DC backbone
An 8-colour panel of 12 mAbs used to identify monocyte and DC populations was assembled from commercially available sources and is listed in Table 1. We endeavoured to follow standard guidelines in panel design to maximise separation of dim antigens while using fluorophores that allowed the panel to be readily accessible across a number of research and diagnostic flow cytometry instruments.25 This and the limitations of the fluorophores available for many DC-specific reagents necessarily influenced the panel design. The relatively dim violet excited V450 fluorophore was used for lineage markers as cells stained with lineage mAbs were removed from the analysis. DAPI (4,6-diamidino-2-phenylindole) was detected using the same violet detector as the lineage markers. In one data set, DAPI was detected on a 355-nm laser and compensated out of the violet channels. Most mAbs submitted to HLDA10 were directly conjugated to PE, which was excited using either a 488- or a 561-nm laser line, and appropriate detectors were kept free for the test reagents. We specifically chose the AD5-4H12 clone to identify CD141 as we used this clone to define the CD141 subset.7 The AD5-17F6 and AD5-8E7 used to identify CD304+ pDCs and CD1c+ DCs, respectively, were limited by the availability of conjugated forms of these clones.
Fresh blood DC and monocyte populations
Venous blood from healthy donors and clinical samples was collected into citrate–phosphate–dextrose anticoagulant blood collection bags, with informed consent approved by the Sydney Local Health District Human Research Ethics Committee, Sydney Australia (HREC/11/CRGH61, HREC/12/CRGH/59 and HREC/07/RPAH/28) and the Universtiy of Auckland Human Participants Ethics Committee, New Zealand (Ethics approval 010558) consistent with the declaration of Helsinki. Mononuclear cells (PBMC) were isolated from healthy donor fresh venous blood using density-gradient centrifugation over Ficoll-Paque, (GE Healthcare, Little Chalfont, UK) or Lymphoprep (Axis-Shield, Dundee, Scotland) and Leucocep tubes (Greiner-Bio-One, Stonehouse, Gloucestershire, UK).
Flow cytometry analysis
Five million PBMC were incubated with the mAb backbone and the directly conjugated test mAb (PE or fluorescein isothiocyanate (FITC)) or a Streptavidin PE in the case of biotinylated test mAb. Where the test mAbs were purified or ascites, indirect staining was performed using a goat anti-mouse IgG AlexaFluor 488 (ThermoFisher, Melbourne, VIC, Australia) followed by blocking with 10% mouse serum. Where FITC-conjugated mAbs or indirect staining were used, CD1c was omitted from the backbone.
Flow cytometric analysis was performed using an FACS Aria II SORP (Auckland Laboratory) or an Influx (Sydney Laboratory) flow cytometer (all from BD Biosciences, San Jose, CA, USA). Initial data analysis was performed using FlowJo version 9 or X (Treestar, Ashland, OR, USA).
SPADE was performed using Cytobank (Cytobank Inc., Mountain View, CA, USA).14 FCS files were processed with FlowJo V9 to select the data in the single cell, DAPI− live cell, Lin−, HLA-DR+ gate before SPADE analysis. SPADE clustering was performed on each individual laboratory-generated data set on ArcSinH-transformed fluorescent parameters using 300 nodes and 10% down sampling from five donors to generate unified SPADE trees based on the expression of CD11c, CD14, CD16, CD1c, CD141 and CD34. Populations were manually annotated based upon expression backbone markers according to the same criteria as used for manual analysis. Heat maps are displayed showing the median ArcSinH-transformed fluorescence of the indicated markers, with the size of each node representing the number of cells.
Acknowledgments
This work was supported by funding from the Concord Cancer Center, University of Sydney and the Australasian College of Pathologists. We would like to acknowledge the generous support of BD Biosciences (Dr Robert Balderas) and Miltenyi Biotech (Dr Katharina Winnemöller) for the provision of mAb backbones for DC studies and to those who contributed mAbs to the HLDA10 panel. Details of the providers of antibodies are on the HLDA10 website at http://HCDM.org or in the following reference (http://www.nature.com/doifinder/10.1038/cti.2015.40).
The authors declare no conflict of interest.
References
- Reynolds G, Haniffa M. Human and mouse mononuclear phagocyte networks: a tale of two species? Front Immunol 2015; 6: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M, Bigley V, Collin M. Human mononuclear phagocyte system reunited. Semin Cell Dev Biol 2015; 41: 59–69. [DOI] [PubMed] [Google Scholar]
- Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology 2013; 140: 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 2013; 31: 563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Breton G, Oliveira TY, Zhou YJ, Aljoufi A, Puhr S et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med 2015; 212: 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald KPA, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DNJ. Characterization of human blood dendritic cell subsets. Blood 2002; 100: 4512–4520. [DOI] [PubMed] [Google Scholar]
- Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med 2010; 207: 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010; 116: e74–e80. [DOI] [PubMed] [Google Scholar]
- Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 2011; 118: e16–e31. [DOI] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L. Blood monocytes and their subsets: established features and open questions. Front Immunol 2015; 6: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner W, Hart DN. The phenotype of freshly isolated and cultured human bone marrow allostimulatory cells: possible heterogeneity in bone marrow dendritic cell populations. Immunology 1995; 85: 611–620. [PMC free article] [PubMed] [Google Scholar]
- Chester C, Maecker HT. Algorithmic tools for mining high-dimensional cytometry data. J Immunol 2015; 195: 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr, Bruggner RV, Linderman MD et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011; 29: 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom 2010; Chapter 10: Unit 10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ju X, Azlan M, Hart DN, Clark GJ. Screening of the HLDA9 panel on peripheral blood dendritic cell populations. Immunol Lett 2011; 134: 161–166. [DOI] [PubMed] [Google Scholar]
- Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol 2014; 14: 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autissier P, Soulas C, Burdo TH, Williams KC. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytometry A 2010; 77: 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung E, Esposito L, Todd JA, Wicker LS. Multiplexed immunophenotyping of human antigen-presenting cells in whole blood by polychromatic flow cytometry. Nat Protoc 2010; 5: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa N. Dendritic cell immunoreceptors: C-type lectin receptors for pattern-recognition and signaling on antigen-presenting cells. J Dermatol Sci 2007; 45: 77–86. [DOI] [PubMed] [Google Scholar]
- Sancho D, Mourao-Sa D, Joffre OP, Schulz O, Rogers NC, Pennington DJ et al. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J Clin Investig 2008; 118: 2098–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med 2010; 207: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Imai T, Kakizaki M, Nishimura M, Takagi S, Yoshie O. Identification of single C motif-1/lymphotactin receptor XCR1. J Biol Chem 1998; 273: 16551–16554. [DOI] [PubMed] [Google Scholar]
- Marshall AS, Willment JA, Pyz E, Dennehy KM, Reid DM, Dri P et al. Human MICL (CLEC12A) is differentially glycosylated and is down-regulated following cellular activation. Eur J Immunol 2006; 36: 2159–2169. [DOI] [PubMed] [Google Scholar]
- Diggins KE, Ferrell PB Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods 2015; 82: 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H, Trotter J. Selecting reagents for multicolor BD flow cytometry. Postepy Biochem 2009; 55: 461–467. [PubMed] [Google Scholar]