Summary
Every nucleosome across the genome must be disrupted and reformed when the replication fork passes, but how chromatin organization is re-established following replication is unknown. To address this problem, we have developed Mapping In vivo Nascent Chromatin with EdU and sequencing (MINCE-seq) to characterize the genome-wide location of nucleosomes and other chromatin proteins behind replication forks at high temporal and spatial resolution. We find that the characteristic chromatin landscape at Drosophila promoters and enhancers is lost upon replication. The most conspicuous changes are at promoters that have high levels of RNA polymerase II (RNAPII) stalling and DNA accessibility and show specific enrichment for the BRM remodeler. Enhancer chromatin is also disrupted during replication, suggesting a role for transcription factor (TF) competition in nucleosome re-establishment. Thus, the characteristic nucleosome landscape emerges from a uniformly packaged genome by the action of TFs, RNAPII and remodelers minutes after replication fork passage.
Introduction
The passage of the replication fork disrupts nucleosomes and DNA-binding proteins. Various classes of chromatin regulators, including remodelers, chaperones and modifying enzymes, have evolved to re-establish chromatin states after replication (MacAlpine and Almouzni, 2013; Ramachandran and Henikoff, 2015; Whitehouse and Smith, 2013). However, the cascade of events that establish the chromatin landscape is not known (Narlikar et al., 2013). Hence, mapping chromatin features immediately after the replication fork passes may provide insight into the processes that result in the characteristic chromatin landscape.
The importance of proper chromatin structure is underscored by the observation that many defects in replication-coupled chromatin assembly and maturation are lethal (Anderson et al., 2011; Klapholz et al., 2009; Moshkin et al., 2002). Disruption of chromatin assembly by reducing histone levels or by depleting assembly-associated histone chaperones Caf1 and Asf1 inhibits replication fork movement (Groth et al., 2007; Hoek and Stillman, 2003), increases susceptibility to DNA damage (Clemente-Ruiz et al., 2011; Prado et al., 2004) and, at least in Drosophila embryos, triggers a G2/M checkpoint and subsequent lethality (Gunesdogan et al., 2014). Both a histone H3 mutation that affects replication-coupled nucleosome assembly and the depletion of Caf1 alters neuronal development in C. elegans (Nakano et al., 2011).
The potential to reveal the determinants of the chromatin landscape motivate the mapping of the newly replicated and the maturing epigenome. Previous work has shown that well-positioned nucleosomes are deposited in the same position post-replication (Lucchini et al., 2001), leading to the assumption that nucleosome-depleted regions remain accessible for rapid re-binding of transcription factors (TFs) (Whitehouse and Smith, 2013). However, the generality of this observation has not been determined (Kaufman and Rando, 2010).
Genome-wide mapping of nucleosome assembly and chromatin maturation remains an outstanding challenge in the field of epigenome dynamics (Narlikar et al., 2013; Whitehouse and Smith, 2013). Current methods to map newly replicated DNA by labeling with nucleotide analogues are limited to the resolution of kilobases. This limitation precludes the mapping of nucleosomes, which requires resolution on the order of 10 bp. Furthermore, current methods require either cell sorting which does not generally provide enough material for routine epigenome mapping, or cell-cycle synchronization which can be technically challenging and not applicable to all cell lines and organisms. Mapping the newly replicated epigenome is further complicated by the high speed of the replication fork, measured at 2-4 kb/min (Blumenthal et al., 1974) necessitating labeling times of minutes to track events that occur within a few kilobases of replication fork passage.
To circumvent these issues and to map the newly replicated epigenome at base-pair resolution, we have developed a method, Mapping In vivo Nascent Chromatin with EdU and sequencing (MINCE-seq). In MINCE-seq, newly replicated DNA is labeled with the nucleotide analogue ethynyl deoxyuridine (EdU) (Salic and Mitchison, 2008), which is coupled with biotin ex vivo using click chemistry (Sirbu et al., 2011). Coupling with biotin ensures highly specific purification of newly replicated DNA from asynchronous cells, even if it is only a fraction of a percent of total DNA. Micrococcal nuclease (MNase) treatment recovers DNA fragments bound both by nucleosomes and non-histone DNA-binding proteins, which enables the mapping of newly replicated chromatin at near base-pair resolution (Henikoff et al., 2011). We find that the newly replicated chromatin landscape differs from the average landscape at active promoters and enhancers genome-wide. The functional features of promoters and enhancers correlate with the change in the chromatin landscape that replication brings about, which in turn reveals mechanisms that establish the characteristic chromatin landscapes at these genomic landmarks.
Results
MINCE-seq delineates chromatin maturation
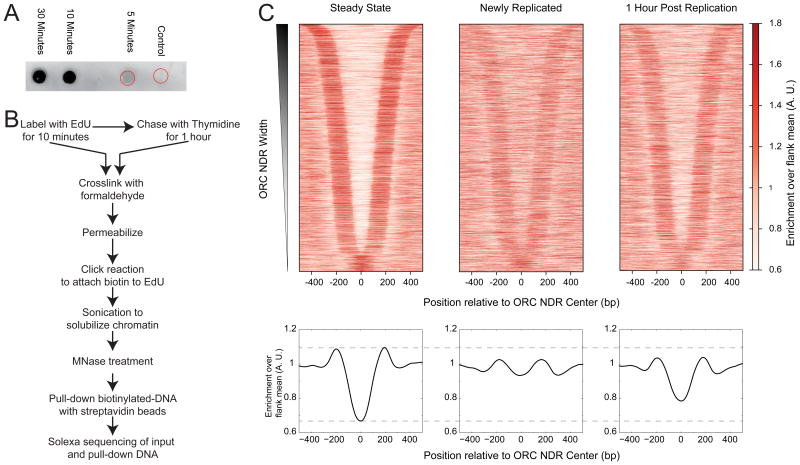
EdU has been used to visualize replication forks (Salic and Mitchison, 2008) and to isolate and identify proteins involved with newly replicated chromatin (Leung et al., 2013; Sirbu et al., 2011). Nucleosomes assemble rapidly behind a replication fork (McKnight and Miller, 1977), and the speed of the fork implies that the replication-coupled assembly machinery is associated with a given region of DNA for only a very short window of time. To capture the nucleosome landscape specifically set up by replication-coupled assembly, we first determined the shortest EdU labeling time that would lead to detectable newly replicated DNA in our chromatin preparation. In Drosophila S2 cells, we found that EdU that was tagged with biotin was undetectable after 5 minutes of labeling, but robustly detected after just 10 minutes of labeling (Figure 1A), which would result in labeling of at most 10-20 kb of newly replicated DNA behind each replication fork. Despite being a small fraction of total DNA, EdU pulse-labeled DNA is highly enriched by streptavidin pull-down, as indicated by using a spike-in control, which showed only very low levels of contaminating unlabeled DNA (Figure S1).
Figure 1. EdU labeling of newly replicated chromatin.
A) Dot blot of DNA purified from chromatin preparations of cells treated with EdU for indicated times. Control cells were not treated with EdU, but were processed the same way as other samples. Red circles indicate the location of sample application for control and 5 minute samples. The DNA applied to a nitrocellulose membrane was probed with IRDye 800CW streptavidin and detected using ODYSSEY infrared scanner. B) Schematic of the MINCE-seq protocol C) Nucleosome profiles of steady state, newly replicated and 1 hour post replication plotted as heatmaps (top) and average plots (bottom) around ORC nucleosome-depleted regions (NDRs; n=4230). NDRs were computationally identified around published ORC sites, and the center of the NDR was used as the reference point for plotting nucleosome profiles. All profiles are averages over a 50-bp sliding window. See also Figure S1 and Tables S1.
We performed replicate incorporations of i) a 10 minute pulse of EdU and ii) a 10 minute pulse of EdU followed by 1 hour chase with thymidine. After 10 minutes of labeling with EdU, we crosslinked and permeabilized cells, and then performed a “click” reaction to attach biotin tags to newly replicated DNA. We then treated chromatin with Micrococcal nuclease (MNase) followed by DNA isolation. Biotin-tagged DNA was then isolated using streptavidin beads (Figure 1B). The input DNA and the DNA pulled-down using streptavidin beads were sequenced with a library protocol that captures all fragments from 25 to 500 bp (Henikoff et al., 2011), enabling characterization of MNase protected fragments representing not only nucleosomes but also non-histone DNA-binding proteins. To verify that MINCE-seq was mapping newly replicated chromatin, we examined ORC binding sites, which have been mapped by the modENCODE project for S2 cells (Lubelsky et al., 2014). ORC binds during the G1 phase of the cell cycle in metazoans, after which the origins are licensed (Asano and Wharton, 1999; Bell and Dutta, 2002) to be replicated in the S-phase. Because a hallmark of ORC binding is nucleosome depletion and high turnover (Deal et al., 2010; MacAlpine et al., 2010), we can monitor the status of ORC sites after replication. In our input nucleosome maps, we observed a nucleosome depleted region (NDR) around ORC sites (Figure 1C, left) as expected from published results. In our MINCE-seq map, we observed dramatically higher nucleosome occupancy around ORC sites immediately after replication (Figure 1C, center). The nucleosome depletion around ORC sites was partially regained after a 1 hour chase, indicating that the maturation of chromatin creates a landscape amenable to ORC binding later (Figure 1C, right). Our data suggest that replication-coupled nucleosome assembly resets the chromatin landscape of ORC sites. Thus, in contrast to previous observations (Lucchini et al., 2001), we find the newly replicated chromatin landscape to be distinct from the average landscape. Thus, MINCE-seq is able to capture transient molecular events happening just behind the replication fork at replication origins.
Replication resets the promoter nucleosome landscape
We next examined the newly replicated nucleosome landscape at promoters. Promoters of expressed genes have a characteristic nucleosome landscape: A nucleosome-depleted region (NDR, Figure S2A, left) is flanked by the genic nucleosome array, starting with the +1 nucleosome position, and the upstream nucleosome array, starting with the -1 nucleosome position (Yuan et al., 2005). The transcription preinitiation complex (PIC) forms at the NDR, and elongating RNAPII stalls predominantly at the entry site of the +1 nucleosome position (Weber et al., 2014). In our input MNase-seq dataset, we observed a peak of <50-bp protected fragments at the NDRs of expressed genes (Figure S2B, left) whereas at non-expressed genes, the NDR, the nucleosome array and the peak of <50-bp particles were absent (Figure S2A, B, right), as seen previously (Teves and Henikoff, 2011; Weber et al., 2010).
To confirm that the 147-bp protected fragments represent nucleosomes, we compared nucleosome profiles from an input MINCE-seq dataset to those obtained by H2A.Z ChIP-seq (Weber et al., 2014). We saw clear concordance of nucleosome positions (Figure S2C). Likewise, to confirm that the <50-bp protected fragments represent transcription factors (TFs), we plotted enrichment of <50 bp protection at the well-established binding site of GAGA factor (GAF) at the Hsp70bB promoter. We observed a clear peak over the known GAGA factor binding site (Figure S2D). We also observed peaks of <50 bp fragments from the MINCE-seq input dataset at GAF binding sites genome-wide (Orsi et al., 2014) (Figure S2E). These comparisons to published data indicate that the ∼147 bp fragments and the <50 bp fragments predominantly represent nucleosomes and TFs respectively as has been observed in previous studies (Carone et al., 2014; Teves and Henikoff, 2011; Weber et al., 2010). This enables us to track nucleosome and TF profiles simultaneously in the same experimental dataset.
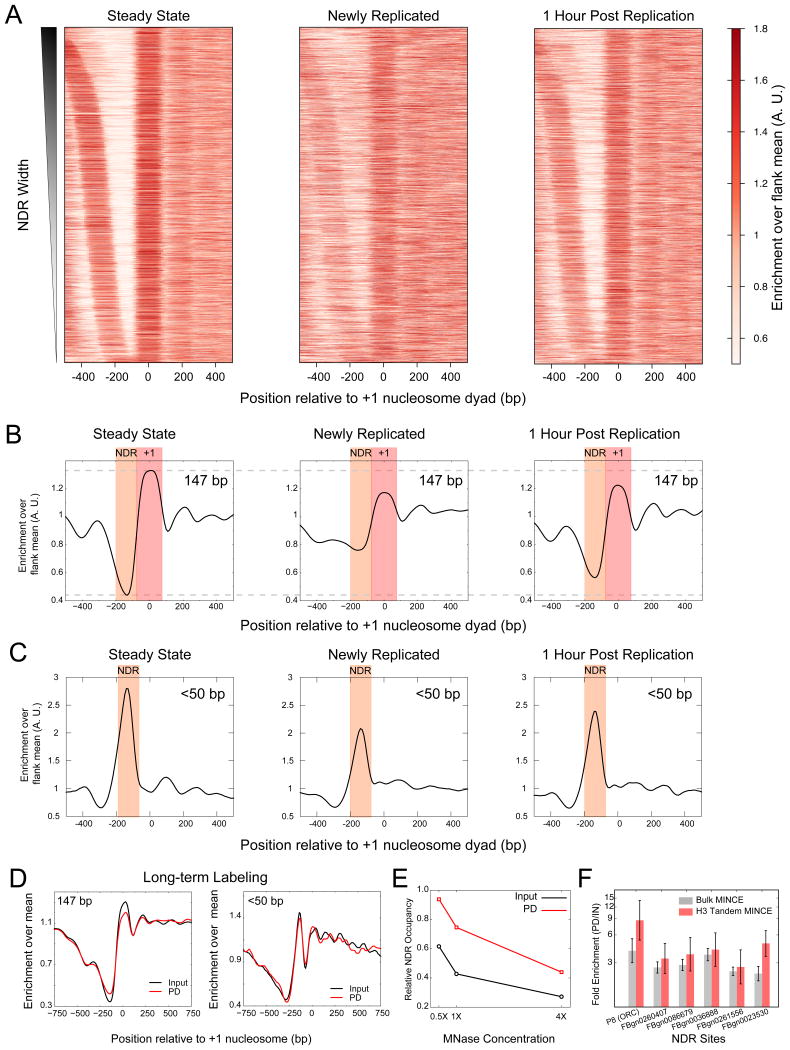
A nucleosome might be reassembled at the same position, or it might be reassembled at a different position and then translocated to its average steady-state position when chromatin matures. In newly replicated chromatin, we observed a higher signal at the NDR and a lower signal at ±1 positions, which indicates that after replication, nucleosomes fill NDRs in patterns distinct from their average positions (Figure 2A, B, S3A). Furthermore, we observed weaker upstream and downstream nucleosome arrays in newly replicated DNA. In contrast to the nucleosome gain at the NDR just after replication, we observed a conspicuous decrease in the <50-bp protection at the NDR, indicating loss of DNA-binding proteins and the PIC (Figure 2C). An hour after replication the nucleosome landscape resembles the steady state landscape with a clear NDR and strong +1 and -1 nucleosomes, indicating that the nucleosome gain at the NDR after replication is a dynamic process. The relative nucleosome gain at the NDR and loss at ±1 positions were also clearly observed when we plotted the difference heatmap of newly replicated compared to steady-state chromatin (Figure S3B). Nucleosome gains at the NDR and losses at ±1 positions were also evident when we quantified MINCE-seq and input datasets for each gene and generated distributions at the NDR, +1 and -1 positions (Figure S3C-F).
Figure 2. Replication resets promoter nucleosome landscapes.
A) Nucleosome profile (∼147 bp protections) of expressed genes (n=5113) plotted as a heatmap, where genes are ordered by NDR width. B) Average plots of nucleosome profiles relative to the +1 dyad positions for the same genes plotted in (A). C) Average profiles of <50-bp protected fragments representing DNA-binding proteins, plotted relative to the +1 dyad positions. D) Nucleosome profiles (left) and profiles for <50 bp fragments averaged over expressed genes plotted for input and EdU pull-down after 24 hour labeling of cells with EdU. Profiles are averages over a 50-bp sliding window. E) Nucleosomal enrichment at NDRs averaged over expressed genes (n=5113) for steady state (input) and newly replicated (EdU pull-down) plotted for different MINCE-seq experiments with increasing MNase concentrations. F) qPCR enrichment of nucleosomes over NDR regions in newly replicated chromatin (EdU pull-down) compared to steady state (input) plotted for MINCE and tandem H3-MINCE ChIP (N=6, 2 biological replicates with 3 technical replicates each). Data are presented as mean +/- SEM. See also Figure S2 and Table S2.
To rule out the possibility that nucleosome gains at NDRs are due to differential EdU labeling, we performed long-term EdU labeling. We found no significant differences in the nucleosome and TF landscapes compared to steady state (input) data, indicating that EdU labeling is uniform and that the changes in the chromatin landscape we observed during short-term labeling are due to dynamics of chromatin assembly and maturation (Figure 2D). The similarity between long-term labeling nucleosome and TF occupancy profiles and input profiles also implies that EdU incorporation does not affect nucleosome or TF sensitivity to MNase digestion. To directly exclude this possibility, we performed MINCE-seq on aliquots of the same starting material treated with three different MNase concentrations over an 8-fold range (Figure S3G). We observed nucleosome gain at promoters for newly replicated DNA at all MNase concentrations (Figure 2E, S3H). With increasing MNase concentration, we observed lower signal at NDRs and upstream regions relative to genic regions for both newly replicated and steady state chromatin, as expected (Weiner et al., 2010). Furthermore, the rate of loss of signal at NDRs is similar for both newly replicated chromatin and steady state chromatin (Figure 2E), implying that at the concentrations we use, the MNase sensitivity of EdU-labeled chromatin is similar to that of bulk chromatin. We conclude that the nucleosome gains we observe are not due to differential MNase sensitivity of EdU-labeled chromatin compared to bulk chromatin.
To confirm that the particles observed at NDRs behind the replication fork correspond to nucleosomes, we performed qPCR of tandem H3-MINCE ChIP at NDRs at an ORC binding site and at five promoters. These sites were chosen because they showed significant nucleosome gains after replication in our MINCE-seq datasets. We first labeled cells with EdU and performed a click reaction to couple biotin to EdU. We next performed H3-ChIP on soluble chromatin followed by DNA isolation. Isolated DNA was subjected to streptavidin pull-down. We estimated the enrichment of biotinylated DNA in tandem H3-ChIP-MINCE by qPCR. With this tandem H3-ChIP-MINCE-PCR protocol, we mapped newly replicated DNA that is specifically bound by histone H3. Using this protocol, we observed higher enrichment of nucleosomes at NDRs compared to a control +1 nucleosome, similar to bulk MINCE-PCR (Figure 2F), confirming that the 147-bp particles we observed over NDRs behind the replication fork are nucleosomes. Thus, after replication, nucleosomes are assembled at promoters, displacing DNA-binding proteins. NDRs form again at the promoter during the maturation of newly assembled chromatin.
Cell-type specific transcription differentiates promoter landscape resetting during replication
To ask if resetting of the promoter landscape during replication is a general phenomenon, we performed MINCE-seq with BG3 cells, which is a cell type distinct from S2 cells: BG3 cells have been derived from the central nervous system of 3rd instar larvae whereas S2 cells have been derived from embryos. Using MINCE-seq on BG3 cells, we observed nucleosome gains over NDRs upstream of +1 nucleosomes at expressed genes (Figure S4A), similar to what was observed in S2 cells. In addition to confirming results with S2 cells, MINCE-seq of BG3 cells provides profiles for a set of differentially expressed genes, where we can examine the resetting of nucleosome landscapes at promoters in a cell type-specific manner. To compare across the two cell types, we generated MINCE-seq difference profiles of relative nucleosome gain or loss for genes that are expressed in BG3 cells but not in S2 cells (Figure 3A), and for genes that are expressed in S2 cells but not in BG3 cells (Figure 3B). We observed nucleosome gains at NDRs only in the cell type where genes are expressed (Figures 3A, B). Hence, even with the same underlying sequence, promoter nucleosome resetting during replication reflects cell type-specific transcription programs. In other words, starting from nucleosome-containing promoters after replication, maturation of promoter nucleosome landscapes reflects cell type-specific expression patterns.
Figure 3. Promoter nucleosome resetting during replication is cell-type specific and requires replication-coupled assembly.
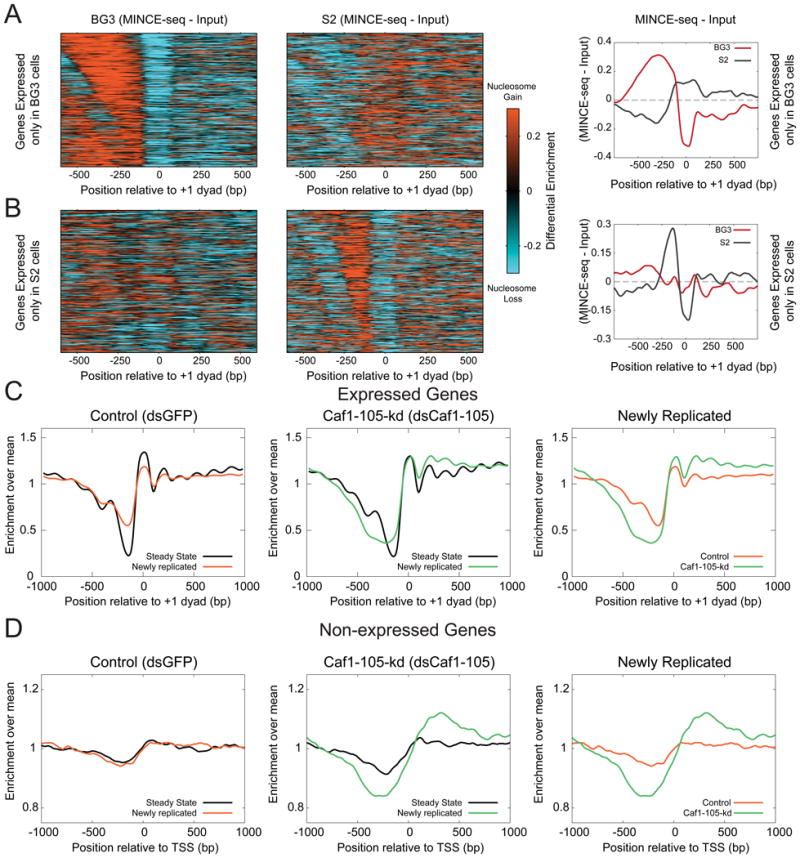
A) Difference between the newly replicated nucleosome landscape and the steady state landscape (input) is plotted as a heatmap for genes expressed in BG3 cells but not in S2 cells (N=683, left). Average plot of the difference between newly replicated and steady state nucleosome landscapes for the same genes plotted on the right. B) Same as (A) but for genes expressed in S2 cells and not in BG3 cells (N=728). C) Nucleosome profiles averaged over expressed genes (n=5113), plotted relative to the +1 dyad positions for the input (steady state) and pull-down (newly replicated) of MINCE-seq assays after depletion of mRNAs for GFP control and Caf1-105-kd proteins. D) Nucleosome profiles averaged over non-expressed genes (n=5202), plotted relative to transcription start sites (TSS). See also Figure S3 and S4.
Nucleosome assembly at promoters requires the replication-coupled nucleosome assembly pathway
To ask if nucleosome gain at promoters is due to replication-coupled nucleosome assembly, we knocked down the major subunit of the Caf1 complex, Caf1-105, achieving 90% protein reduction as quantified from immunoblotting (Figure S4B). The average nucleosome positioning around promoters is not altered by knockdown of Caf1-105, but nucleosome positions in newly replicated chromatin are dramatically affected (Figure 3C). At expressed genes, we observed the NDR to become more nucleosome-depleted in the Caf1-105 knockdown (Caf1-105-kd) compared to control, and the nucleosomes at the +1 position and downstream to increase in occupancy (Figure S4C). These results indicate that the nucleosome gains at promoters during replication that we observed is due to replication-coupled nucleosome assembly. As the replication-coupled nucleosome assembly pathway acts genome-wide, the Caf1-105-kd should have an effect not only at expressed genes but also at non-expressed genes. The nucleosome landscape of non-expressed promoters is featureless in both steady-state and newly replicated chromatin in the control (Figure 3D, left). When Caf1-105 was knocked down, we observed broad NDR formation upstream of the TSS and increased nucleosome density downstream (Figure 3D middle). This shows that chromatin is dynamic even at inactive promoters during replication, even though no apparent change is observed between the steady state and the newly replicated landscapes in control cells (Figure 3D, left). These changes in the newly replicated chromatin landscape with reduction in Caf1 imply that the replication-coupled nucleosome assembly pathway is required to reset the chromatin landscape around both active and inactive promoters during replication.
Promoters with high RNAPII enrichment show the largest gains in nucleosome occupancy upon replication
To understand the mechanisms behind establishment of the average nucleosome landscape starting from the newly replicated chromatin landscape, we devised a strategy to classify genes according to the patterns of successive changes in the nucleosome landscape upon replication. To identify genes with the largest changes in the nucleosome landscape upon replication, we assigned to each gene either nucleosome gain or nucleosome loss at its -1 and +1 positions and its NDR and looked for different patterns of nucleosome change at promoters (see Experimental Procedures). We observed distinct patterns of nucleosomes around promoters after replication (Figure 4A). The largest group (G1, n=3089) featured a nucleosome profile that is similar to the average MINCE-seq profile seen in Figure 1, with losses at ±1 positions and gains at NDRs (Figure 4B). Group 2 (n=149) featured NDRs that are more nucleosome-depleted upon replication compared to input. Group 3 (n=1390) featured a significant increase in nucleosome occupancy at NDRs, with no significant changes in occupancy at ±1 positions. The newly replicated nucleosome profile for Group 3 showed a shift in the +1 position towards the NDR and broad nucleosome occupancy at NDRs. This group represents the largest increase in nucleosome occupancy at NDRs. Groups 4 (n=98) and 6 (n=121) featured a strong -1 position, while groups 5 (n=106) and 7 (n=143) featured a strong +1 position. Furthermore, in groups with the strongest changes at the +1 position post-replication, the gains and losses were dependent on Caf1 function.
Figure 4. Patterns of changes in promoter nucleosome landscapes upon replication.
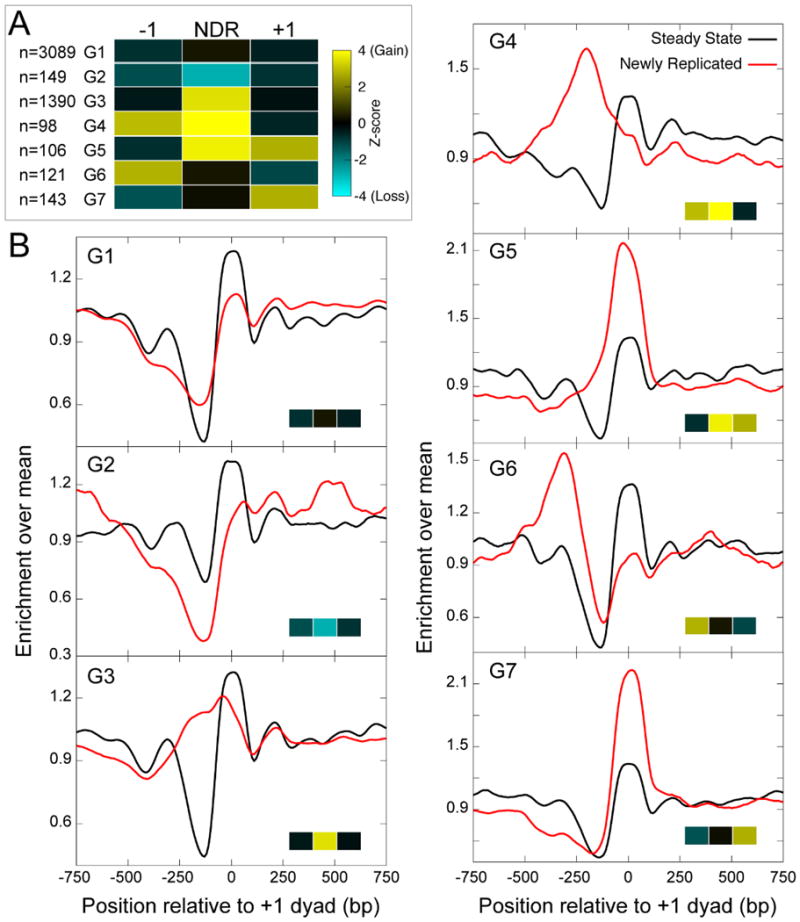
A) Heatmap of average Z-score values of ±1 positions and the NDR of genes classified according to the patterns of nucleosome gain/loss after replication. The Z-score is calculated by comparing the change in nucleosome enrichment upon replication to a control distribution. B) Nucleosome profiles averaged over the genes making up each of the promoter groups, plotted as a running average over a 50-bp window, relative to the +1 nucleosome dyad position.
To understand the functional differences between promoters grouped by their patterns of nucleosomes post-replication, we systematically analyzed which of the chromatin processes differentiated the promoter groups. We first analyzed the promoter enrichment of RNAPII transcription at steady state using 3′NT (Weber et al., 2014), which produces a base-pair resolution map of elongating and stalled RNAPII. We plotted the enrichment of RNAPII at promoters compared to gene bodies for each of the promoter groups (Figure 5A). We observed that the groups that changed the most in nucleosome occupancy at NDRs upon replication, namely groups 4, 5 and 3, featured the highest levels of RNAPII at promoters, whereas the groups with the smallest change in nucleosome occupancy at NDRs, groups 2, 6, 7 and 1 featured lower levels of RNAPII at promoters. Thus, highly active promoters showed the largest changes in nucleosome occupancy upon replication, while less active promoters featured only modest changes in nucleosome occupancy upon replication. Hierarchical ordering of the RNAPII enrichment profiles revealed that genes with the most RNAPII showed the greatest NDR nucleosome gains, and genes with the least RNAPII showed the least gains (Figure 5A).
Figure 5. Promoter features correlate with nucleosome changes during replication.
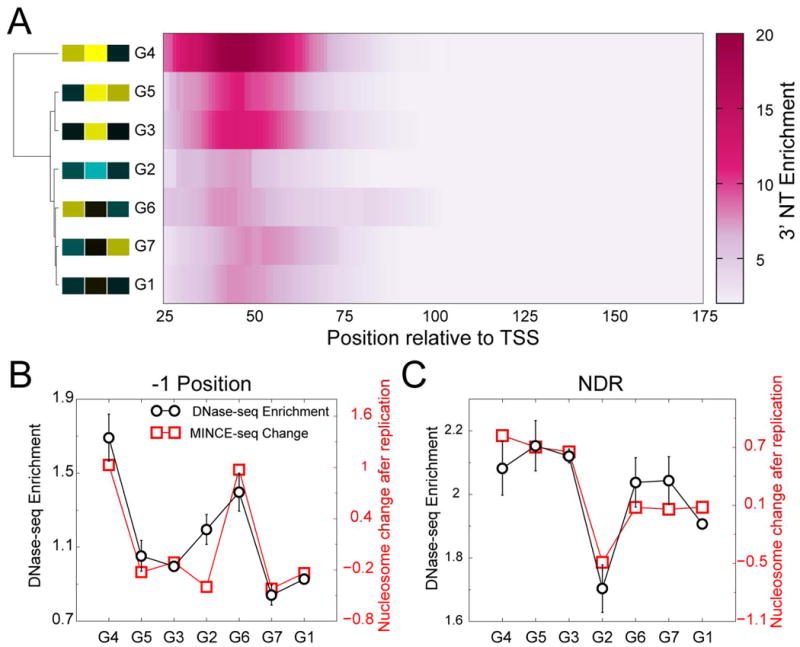
A) Heatmap of RNAPII enrichment relative to the TSS plotted for each promoter group. The rows of the heatmap are hierarchically ordered based on RNAPII enrichment profile, and the resulting dendogram is displayed on the left. The heatmap of ±1 and NDR Z-scores of the promoter groups are displayed next to the group label. B) Plots that compare DNase-seq enrichment and nucleosome gain/loss upon replication at the -1 position, averaged over genes making up each of the seven groups. C) Same as (B) for NDRs. Data are presented as mean +/- SEM.
Patterns of TF and remodeler binding at promoters correlate with nucleosome gains during replication
To further investigate the promoter groups, we determined the levels of non-histone protein binding at steady state by using modENCODE DNase-I mapping datasets (Kharchenko et al., 2011). DNase-I mapping broadly identifies nucleosome-depleted regions bound by transcription factors, and DNase-I enrichment can be used as a measure of global non-histone protein binding to DNA (Hesselberth et al., 2009). The DNase-seq enrichment of the promoter groups closely correlated with nucleosome gain at -1 positions and NDRs (Figure 5B, C). This strongly suggests that TFs could be driving promoter nucleosome loss during chromatin maturation after replication.
Because many TFs are known to recruit remodelers, we asked if remodelers are enriched at any of the promoter groups. We analyzed the profiles of the four major remodeler families in Drosophila: NURD, (P)BAP, INO80 and ISWI at the different promoter groups using data from published ChIP-chip experiments (Moshkin et al., 2012). For each remodeler, we found peaks in ∼1000 genes out of the ∼5000 expressed genes in S2 cells. When we plotted the profiles of the remodelers over these enriched genes in each of the 7 promoter groups, we observed similar profiles for NURD, INO80 and ISWI complexes (Figure 6A, B, C). However, we observed that BRM – a component of (P)BAP complexes – is differentially distributed in the promoter groups (Figure 6D). Groups 4 and 6, which showed -1 nucleosome gains during replication, featured higher BRM levels upstream of the gene, whereas group 5, which showed nucleosome gains at the NDR and +1 position during replication, featured the highest BRM levels in the gene body at steady state. This correspondence between BRM and post-replication chromatin changes suggests that BRM action at groups 4, 5 and 6 transforms their newly replicated landscapes back to their steady-state nucleosome landscapes.
Figure 6. Promoter groups feature distinct BRM profiles.
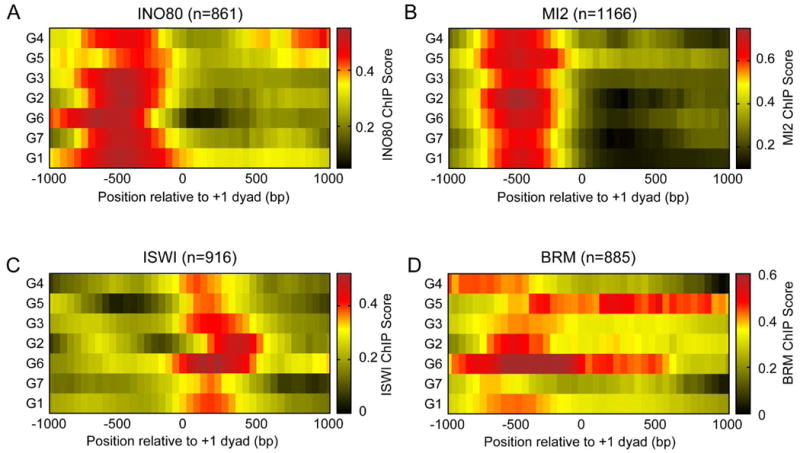
INO80 (A), MI2 (B), ISWI (C) and BRM (D) ChIP-chip scores averaged over genes making up each of the promoter groups plotted as averages over a 50-bp sliding window, relative to the +1 nucleosome position.
Replication resets cell-type specific enhancer chromatin
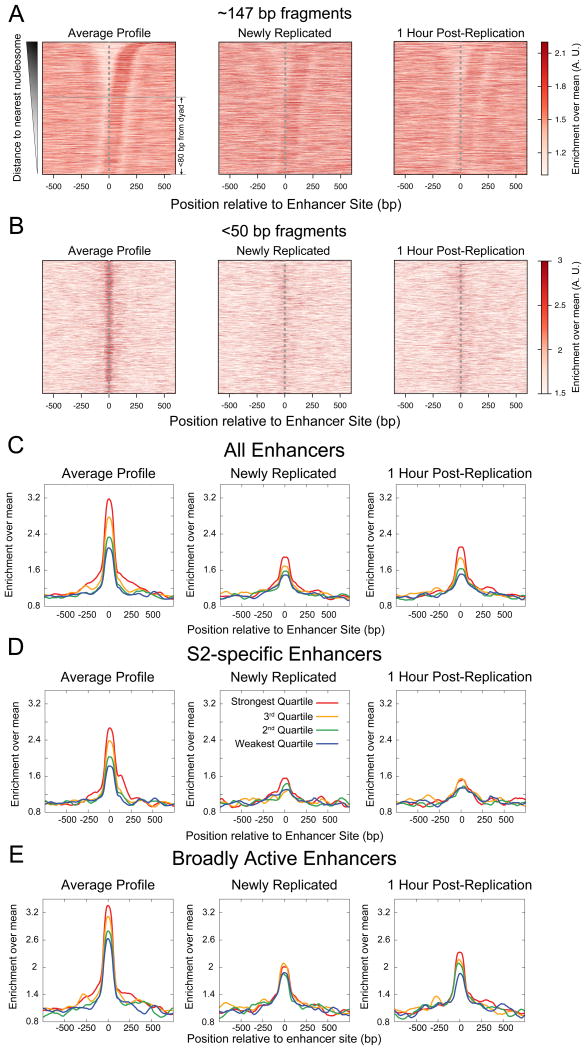
We wondered whether the resetting of the chromatin landscape that we observed at promoters also occurs in other regions of the genome, such as enhancers. We analyzed input and MINCE-seq datasets around enhancers in S2 cells identified by STARR-seq, a genome-wide reporter screen for enhancers (Arnold et al., 2013). We found that enhancers, unlike promoters, do not show uniform nucleosome depletion, but rather a majority of the enhancer sites were within 80 bp of the nearest nucleosome dyad position (Figure 7A, left). Regardless of the nucleosome position, we observed a striking enrichment of <50-bp protected fragments at enhancer sites (Figure 7B, left). The enrichment of <50-bp protection at enhancers strongly correlated with quartiles of enhancer strength as measured using STARR-seq (Figure 7C, left). This correlation was observed for both in enhancers specific to S2 cells and in broadly active enhancers that are active in multiple cell types (Figures 7D, E and S5A, B). However, no such correlation was observed for <50-bp protected fragments in S2 cells at enhancers specific to other cell types (Figure S5C, D). This cell type-specific enrichment of <50-bp protected fragments at enhancers implies that these changes reflect the functional status of enhancer sites. Thus, our input maps suggest enhancers as sites of competition between TFs and nucleosomes, where the extent of protection by TFs correlates with functional enhancer strength.
Figure 7. Replication resets cell-type specific enhancer chromatin.
A) Heatmap showing enrichment of ∼147 bp protection from MNase over enhancer sites, ordered by the distance to the nearest nucleosome and plotted in the direction of the nearest nucleosome for the input dataset (left), MINCE-seq dataset (middle) and MINCE-seq 1 hour post-replication dataset (right). B) Same as (A), but for <50-bp protected fragments. C) Enrichment of <50-bp protected fragments averaged over quartiles of enhancer strength, plotted relative to the center of enhancer sites at steady state (left), newly replicated (center) and 1 hour post-replication (right). Each quartile consists of 1374 enhancer sites. D) Enrichment of <50-bp protected fragments averaged over quartiles of enhancer strength for enhancers that are specific to S2 cells. The strongest, third, second and weakest quartiles consist of 363, 642, 834, 918 enhancer sites respectively. E) Enrichment of <50-bp protection averaged over quartiles of enhancer strength for broad enhancers, which are active in S2 cells and at least one other cell type (BG3 and OSC). The strongest, third, second and weakest quartiles consist of 1012, 733, 541 and 456 enhancer sites respectively. All profiles are averages over a 100-bp sliding window. See also Figure S5.
We next used MINCE-seq to ask what happens to <50-bp protected fragments at enhancers during replication fork passage. We observed a conspicuous weakening of the enrichment of these fragments after replication, both in broadly active and S2-specific enhancers (Figure 7B, C, D and E, middle). One hour post-replication, the broadly active enhancers strengthened significantly (Figure S5E) and we could observe ordering of enrichment according to quartiles of enhancer strength (Figure 7E, right). However, cell type-specific enhancers remained weak one hour post-replication (Figure S5E) and did not show any trend with respect to enhancer strength (Figure 7D, right). Our data indicate that replication-coupled nucleosome assembly efficiently displaces TFs from enhancers. Broadly active enhancers recover TFs faster than cell type-specific enhancers after replication, implying robust mechanisms for re-establishing the chromatin landscape of broadly active enhancers.
Discussion
Eukaryotic replication occurs over the course of S-phase, during which multiple forks replicate different regions of the genome. Variation in origin firing between different cells in a population (Kaykov and Nurse, 2015) means that even perfectly synchronized cells will not be replicating the same regions of the genome simultaneously. Furthermore, the speed of the replication fork implies that a given nucleosome is replicated within a few seconds, and so the newly replicated chromatin landscape cannot be mapped at nucleosome resolution using cell-cycle synchronization or after capturing S-phase cells by FACS. MINCE-seq, which is performed without sorting or synchronization, is able to capture chromatin dynamics behind replication forks in asynchronous cells, because it directly captures newly replicated DNA throughout the genome.
The passage of a replication fork removes all protein components of chromatin including nucleosomes, TFs and remodelers. However, it is not known how these components reassemble on the genome. Using MINCE-seq, we find that the characteristic rugged nucleosome landscape is flattened genome-wide during replication, which would include both nucleosome gain at NDRs and weakening of strong nucleosome positions. Considering that replication forks pass through a nucleosome in seconds, and we are profiling the chromatin landscape after 10 minutes of EdU labeling, it is reasonable to suppose that immediately after replication, the landscape is even flatter and the nucleosome distribution within a population is nearly uniform. This flat landscape post-replication also excludes all other DNA-binding proteins and remodelers. Thus, replication-coupled nucleosome assembly is so efficient that it outcompetes TFs to form a featureless landscape post-replication. Maturation of the newly replicated landscape involves rearrangement of nucleosomes by TFs to expose binding sites, form the NDR and achieve the characteristic positions of nucleosomes.
With MINCE-seq maps, we discovered that promoters, which are usually nucleosome depleted, gain nucleosomes during replication. Gene bodies, which usually have ordered nucleosome arrays, feature a uniform nucleosome density during replication. When we disrupted the replication-coupled nucleosome assembly pathway, we observed loss of the uniform nucleosome distribution during replication, both at expressed and non-expressed genes, confirming that MINCE-seq captures the newly replicated chromatin landscape. In vitro, it has been shown that RNAPII cannot initiate from a promoter that contains a nucleosome (Lorch et al., 1987). Promoters gain nucleosomes right after replication, and hence, RNAPII initiation would be inhibited. Nucleosome gain at promoters during replication correlates with promoter enrichment of RNAPII, suggesting that maturation of highly active promoters requires a greater extent of nucleosome depletion from the newly replicated state. Nucleosome gain during replication also correlates with promoter enrichment of TFs. TFs can displace nucleosomes by directly binding motifs in nucleosomal DNA and recruiting remodelers that displace nucleosomes (Tsukiyama et al., 1994). When we grouped promoters by patterns of nucleosome gain during replication, we observed differential profiles of BRM, the ATP-dependent nucleosome remodeler associated with gene activation (Armstrong et al., 2002). Thus, maturation of the newly replicated promoter landscape could involve the concerted action of TFs and remodelers specific to the cell type to expose the promoter DNA of active genes for binding of basal TFs to reform the pre-initiation complex and recruit RNAPII. In this way, replication-coupled nucleosome assembly and maturation could maintain cell type-specific transcription programs in spite of the disruption of chromatin during replication.
Our experiments also provide insights into the dynamics of factor binding at enhancers. The steady-state MINCE-seq profile shows highly specific short fragment (<50-bp) protection at enhancers whose enrichment correlates with enhancer activity. This indicates that we can quantitatively connect protein binding with high resolution at enhancer sites to the functional activity of the enhancers genome-wide. We observed dramatic weakening of short protected fragments at enhancers after replication, which implies that DNA-binding proteins are displaced by passage of the replication fork. Strikingly, broadly active enhancers, which are sites active in more than one cell type, regain DNA-binding proteins after replication at a faster rate compared to the cell-type specific enhancers. Previous work has shown that broadly active enhancers feature dinucleotide repeat motifs (DRMs) whereas cell type-specific enhancers feature motifs for specific DNA-binding proteins (Yanez-Cuna et al., 2014). The DRMs are nucleosome destabilizing and hence, faster recovery of broadly active enhancers might be due to intrinsic destabilization of nucleosomes. Cell type-specific enhancers have motifs for TFs that are at low abundance and these TFs may be less efficient in their competition with newly assembled nucleosomes, resulting in slower recovery of the <50-bp protection at cell-type specific enhancers post-replication. Thus, enhancers, similar to promoters, show replacement of TFs by nucleosomes behind the replication fork, and the characteristic enhancer chromatin would be regained through competition of TFs with the newly deposited nucleosomes.
Our findings might seem to contradict the observation that nucleosomes are deposited at the same positions within seconds post-replication (Lucchini et al., 2001). It is important to realize, however, that this study was limited to yeast rDNA repeat units, which is notable for having well-positioned nucleosomes that are completely lost upon transcription (Dammann et al., 1993). In yeast, many of the rDNA repeat units remain silent (Dammann et al., 1993), and so we presume that it is the silent nucleosome-containing subset of rDNA repeats that were profiled post-replication. As we also observed little difference between newly replicated and steady-state nucleosome profiles at non-expressed genes, our results provide confirmation of this original study, although the dynamic behavior that we see at active promoters and enhancers challenges the widely held assumption that nucleosomes pose no obstacle to TF rebinding post-replication (Annunziato, 2015). Rather, our results demonstrate that nucleosomes occupy active regulatory regions immediately post-replication, likely necessitating competition by TFs and BRM-class remodelers to clear them out.
In conclusion, MINCE-seq maps of both newly replicated and maturing chromatin reveal the genome-wide resetting of the nucleosome landscape during replication-coupled nucleosome assembly. The nucleosome landscape is also defined by histone variants and histone post-translational modifications (PTMs), the levels of which are significantly altered in newly replicated chromatin (Alabert et al., 2015). Patterns of variants and histone-PTMs on newly replicated DNA might be essential for maintenance of epigenomic states through the cell cycle. Future extensions of MINCE-seq have the potential to map these patterns genome-wide during replication at high spatial and temporal resolution.
Experimental Procedures
MINCE-seq
The MINCE protocol was adapted from i) a protocol to isolate proteins associated with newly replicated DNA (Sirbu et al., 2012) and ii) a crosslinking-MNase-seq protocol (Skene and Henikoff, 2015). Drosophila S2 cells were grown in HyClone SFX-Insect media supplemented with 18 mM L-Glutamine. To increase the fraction of cells in S-phase, cells were grown to saturation and split into fresh media at a density of 2×106 cells/ml 20 hours before starting the MINCE protocol (Rizzino and Blumenthal, 1978). During the time of harvest, ∼80% of cells are expected to be in S-phase (Rizzino and Blumenthal, 1978). Cells were labeled with 10 μM EdU for 10 minutes and rapidly harvested and washed with ice cold PBS. For a thymidine chase, cells were spun down and resuspended in fresh media with 20 μM thymidine for 1 hour. Cells were crosslinked with 1% formaldehyde for 15 minutes, quenched with 250 mM glycine for 5 minutes and washed three times with PBS. Cells were then permeabilized with 0.25% Triton X-100 in PBS for 30 minutes at room temperature, washed once with 0.5% BSA in PBS and resuspended in PBS. Click reagents were added in the following order to the indicated final concentration: biotin TEG-azide (100 μM), sodium ascorbate (10 mM) and CuSO4 (2 mM), and incubated for 1 hour at room temperature. Cells were washed three times with PBS, resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1), and incubated on ice for 10 minutes. Dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.1, 3 mM CaCl2) at 10 times the volume of lysis buffer was added and mixed well. The lysed cell suspension was sonicated with a Branson digital sonifier with following settings: total on time: 40s; power: 30%; pulse: 2.5s on; 5s off. The lysate was incubated at 37°C for 5 minutes, and then MNase was added to the lysate. MNase treatment concentrations and time were selected to preserve di- and tri-nucleosomes as visualized by electrophoresing the purified DNA in a 2% agarose gel. MNase reactions were stopped by addition of EGTA to a final concentration of 2 mM, of SDS to a final concentration of 0.9%, and 20 μg of proteinase K (Thermo) per ml of the reaction. The lysate was incubated at 65°C for 12 hours to reverse cross-links and degrade proteins. After incubation, phenol-chloroform-isoamyl alcohol (PCI) extraction was performed and RNAse-A was added to the aqueous phase and incubated at 37°C for 30 minutes. PCI extraction was performed again, and the DNA in the aqueous phase was precipitated using 70% isopropanol, 30 mM sodium acetate and glycogen at -80°C for at least 30 minutes followed by centrifugation at 4°C, 16,000 rcf for 30 minutes. The DNA pellet was washed with 75% ethanol and resuspended in 0.1x TE. After reserving an aliquot for input, biotinylated DNA was pulled down using M-280 streptavidin dynabeads (Life Technologies) following the manufacturer's protocol. After washes, the beads were resuspended in 0.1X TE followed by addition of SDS to a final concentration of 1% and 20 μg of proteinase K (Thermo) per ml of the reaction. The beads were incubated at 65°C for 15 minutes followed by PCI extraction and DNA precipitation as above. H3-ChIP for tandem H3-MINCE-ChIP was performed as published (Skene and Henikoff, 2015). Detailed protocols for H3 ChIP, sequencing and analysis are provided in the Supplemental Information.
BG3 cells were obtained from the Drosophila Genomics Resource Center (DGRC Stock #68) and grown in Shields and Sang M3 Insect media supplemented with 10% FBS, 2.5 g/l Bacto Peptone, 1 g/l yeast extract and 10 μg/ml insulin. MINCE-seq was performed exactly as above for BG3 cells except that cells in mid-log phase were used for EdU labeling.
To estimate the level of purification of small amounts of biotinylated DNA streptavidin pull-down, we performed a spike-in experiment. We spiked in yeast nucleosomal DNA before the streptavidin pull-down and estimated the depletion of yeast DNA in the pull-down relative to the input. The primers used to track yeast DNA are listed in Table S1.
RNAi
For dsRNA treatment, Drosophila S2-DRSC cells (DGRC Stock #181) were used (Weber et al., 2014). Published dsRNA amplicons for Caf1-105 (Chen et al., 2013) and control GFP (Hamada et al., 2005) were used. dsRNA was added to a final concentration of 10 μg/ 1×106 cells in serum-free media, mixed well and incubated for 30 minutes at room temperature, followed by supplementation with an equal volume of fresh media and FBS to a final concentration of 10%. After 4 days of dsRNA treatment, cells were treated with 10 μM EdU for 10 minutes and the MINCE protocol was performed as above. Sequencing and analysis methods are described in the Supplemental Information.
Classifying promoter nucleosome gain patterns upon replication
We generated a control distribution of MNase-seq differences at ±1 positions and NDRs by comparing the input datasets from newly replicated and 1 hour chase experiments (Figure S2, black lines). We determined the mean and standard deviation of the control distribution after fitting a normal curve to it. Using this mean and standard deviation, we calculated the Z-score of MINCE-seq signals at ±1 and NDR positions for every gene. A Z-score higher than +2 was marked as a gain and less than -2 was marked as a loss. These stringent criteria enabled identification of the most deviant patterns of nucleosome gain/loss at ±1 positions and NDRs. For each gene, the ±1 positions and the NDR were each coded as loss, gain or same and the genes were sorted to obtain the most significant patterns (Figure 4).
External datasets
Lists of ORC peaks (Lubelsky et al., 2014) and DNase-seq (Kharchenko et al., 2011) datasets used here were part of the modENCODE project. Enrichment profiles of RNAPII were calculated using published 3′NT data (Weber et al., 2014), by normalizing the 3′NT reads of each gene by the gene-body signal (3′NT reads between TSS+251 and TSS+2000 (or until TES if it occurs before TSS+2000)). Remodeller ChIP-chip data (Moshkin et al., 2012) was obtained from the Gene Expression Omnibus (GEO) databank under the accession number GSE32404. For enhnancer sites, we started with published STARR-seq summit positions (Arnold et al., 2013; Yanez-Cuna et al., 2014) of width 500 bp downloaded from http://starklab.org/data/yanez-cuna_genomeRes_2014/. Within each 500 bp STARR-seq summit, an enhancer site was defined as the peak position of the <50-bp size-class in our combined input datasets. If no peak was present in the 500 bp STARR-seq summit, the enhancer site was defined as the center of the 500 bp window of the summit.
Supplementary Material
Acknowledgments
We thank Christine Codomo for sequencing library preparation, Jorja Henikoff for help with data analysis and the FHCRC Genomics Shared Resource for Illumina sequencing. We also thank Paul Talbert and Kami Ahmad for critical comments on the manuscript.
Footnotes
Accession numbers: Reads from all samples have been deposited at the NCBI GEO database under the accession number: GSE76120.
Author contributions: S.R. and S.H. conceived and designed the experiments. S.R. performed the experiments and data analysis. S.R. and S.H. wrote the manuscript.
References
- Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AE, Karandikar UC, Pepple KL, Chen Z, Bergmann A, Mardon G. The enhancer of trithorax and polycomb gene Caf1/p55 is essential for cell survival and patterning in Drosophila development. Development. 2011;138:1957–1966. doi: 10.1242/dev.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT. The Fork in the Road: Histone Partitioning During DNA Replication. Genes (Basel) 2015;6:353–371. doi: 10.3390/genes6020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, Scott MP, Tamkun JW. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 2002;21:5245–5254. doi: 10.1093/emboj/cdf517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Asano M, Wharton RP. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 1999;18:2435–2448. doi: 10.1093/emboj/18.9.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annual review of biochemistry. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013;23:341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M, Gonzalez-Prieto R, Prado F. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS genetics. 2011;7:e1002376. doi: 10.1371/journal.pgen.1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Gunesdogan U, Jackle H, Herzig A. Histone supply regulates S phase timing and cell cycle progression. Elife. 2014;3:e02443. doi: 10.7554/eLife.02443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci U S A. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol. 2010;22:284–290. doi: 10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykov A, Nurse P. The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 2015;25:391–401. doi: 10.1101/gr.180372.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz B, Dietrich BH, Schaffner C, Heredia F, Quivy JP, Almouzni G, Dostatni N. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma. 2009;118:235–248. doi: 10.1007/s00412-008-0192-2. [DOI] [PubMed] [Google Scholar]
- Leung KH, Abou El Hassan M, Bremner R. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques. 2013;55:204–206. doi: 10.2144/000114089. [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 2014;24:1102–1114. doi: 10.1101/gr.160010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R, Wellinger RE, Sogo JM. Nucleosome positioning at the replication fork. EMBO J. 2001;20:7294–7302. doi: 10.1093/emboj/20.24.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome research. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL, Miller OL., Jr Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977;12:795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–2626. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, Dekkers DH, Demmers JA, Travers AA, Verrijzer CP. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol. 2012;32:675–688. doi: 10.1128/MCB.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Stillman B, Horvitz HR. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell. 2011;147:1525–1536. doi: 10.1016/j.cell.2011.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi GA, Kasinathan S, Hughes KT, Saminadin-Peter S, Henikoff S, Ahmad K. High-resolution mapping defines the cooperative architecture of Polycomb response elements. Genome Res. 2014;24:809–820. doi: 10.1101/gr.163642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Cortes-Ledesma F, Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO reports. 2004;5:497–502. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Henikoff S. Replicating Nucleosomes. Sci Adv. 2015;1:e1500587. doi: 10.1126/sciadv.1500587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzino A, Blumenthal AB. Synchronization of Drosophila cells in culture. In Vitro. 1978;14:437–442. doi: 10.1007/BF02616105. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. A simple method for generating high-resolution maps of genome-wide protein binding. Elife. 2015;4:e09225. doi: 10.7554/eLife.09225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev. 2011;25:2387–2397. doi: 10.1101/gad.177675.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Smith DJ. Chromatin dynamics at the replication fork: there's more to life than histones. Curr Opin Genet Dev. 2013;23:140–146. doi: 10.1016/j.gde.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, Arnold CD, Stampfel G, Boryn LM, Gerlach D, Rath M, Stark A. Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res. 2014;24:1147–1156. doi: 10.1101/gr.169243.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.