Abstract
Background
Penicillium expansum is a destructive phytopathogen that causes decay in deciduous fruits during postharvest handling and storage. During colonization the fungus secretes D-gluconic acid (GLA), which modulates environmental pH and regulates mycotoxin accumulation in colonized tissue. Till now no transcriptomic analysis has addressed the specific contribution of the pathogen's pH regulation to the P. expansum colonization process. For this purpose total RNA from the leading edge of P. expansum-colonized apple tissue of cv. 'Golden Delicious' and from fungal cultures grown under pH 4 or 7 were sequenced and their gene expression patterns were compared.
Results
We present a large-scale analysis of the transcriptome data of P. expansum and apple response to fungal colonization. The fungal analysis revealed nine different clusters of gene expression patterns that were divided among three major groups in which the colonized tissue showed, respectively: (i) differing transcript expression patterns between mycelial growth at pH 4 and pH 7; (ii) similar transcript expression patterns of mycelial growth at pH 4; and (iii) similar transcript expression patterns of mycelial growth at pH 7. Each group was functionally characterized in order to decipher genes that are important for pH regulation and also for colonization of apple fruits by Penicillium. Furthermore, comparison of gene expression of healthy apple tissue with that of colonized tissue showed that differentially expressed genes revealed up-regulation of the jasmonic acid and mevalonate pathways, and also down-regulation of the glycogen and starch biosynthesis pathways.
Conclusions
Overall, we identified important genes and functionalities of P. expansum that were controlled by the environmental pH. Differential expression patterns of genes belonging to the same gene family suggest that genes were selectively activated according to their optimal environmental conditions (pH, in vitro or in vivo) to enable the fungus to cope with varying conditions and to make optimal use of available enzymes. Comparison between the activation of the colonized host's gene responses by alkalizing Colletotrichum gloeosporioides and acidifying P. expansum pathogens indicated similar gene response patterns, but stronger responses to P. expansum, suggesting the importance of acidification by P. expansum as a factor in its increased aggressiveness.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2665-7) contains supplementary material, which is available to authorized users.
Keywords: Transcription profiling, Penicillium expansum, RNA-seq, Gene expression, pH regulation, Pathogenicity, pH-regulated genes, Fungal genes regulated by pH, Apple genes regulated by pH
Background
The genus Penicillium comprises a group of anamorphic fungi in the division Ascomycota [1]. Some Penicillium species are of economic importance because they are postharvest pathogens that cause spoilage in tropical and deciduous fruits. Penicillium expansum is the causal agent of blue mold, which is considered one of the most important postharvest pathogens [2] and that causes decay in deciduous fruits during postharvest handling and storage.
Penicillium expansum causes extensive maceration of the infected tissue by means of a common mechanism of tissue acidification [3]; it acidifies its host by activation of glucose oxidase 2 (gox2) and the catalyzed oxidation of glucose resulting in secretion of small molecules such as D-gluconic acid (GLA) [3, 4], which modulate the environmental pH and thereby activate several polygalacturonases that contribute to pectin depolymerization and consequently tissue maceration, at pH 3.5-4 [3, 5, 6]. Functional analysis of glucose oxidase 2-RNAi mutants showed a strong effect on pathogen interactions with their host: the greater the down-regulation of gox2, the stronger the impairment of GLA production, medium acidification, and apple fruit colonization [7]. More recently, Barad et al. [8] reported that P. expansum secreted not only GLA but also ammonia at the leading edge of colonization in apple fruits. Growth of the fungus in high-sucrose media induced rapid metabolism of sugar and increased GLA accumulation, and the decrease in available carbon reserves resulted in enhanced ammonia accumulation, probably because of amino acid catabolism, such as occurs with other pathogens [9, 10]. These results indicated that bidirectional pH regulation occurs in P. expansum-probably dependent on nutrient availability. The question concerns which metabolic processes are modulated by the pH level during colonization of apple fruits.
PacC is a transcription factor that activates genes of Aspergillus nidulans during the alkalization of the environment [11]. In a recent report [8], it was suggested that P. expansum's pacC gene was active also under acidic conditions, and that ammonia produced at the leading edge of the decaying tissue under low-pH conditions further contributed to the activation of pacC responsiveness. This may indicate that P. expansum uses ammonia accumulation during nutritional modification of the ambient environmental pH as a regulatory cue for activation of pacC, by signaling and activating alkaline-induced genes that contribute to pathogenicity and accumulation of secondary metabolites such as patulin [8]. This pH modulation under the influence of GLA and ammonia accumulation is important for understanding the fungal response to differential gene regulation by pH; however, the overall response remains poorly understood.
The diversity of recently described ecological interactions of Penicillium with fruits [3, 4] and with other resident microorganisms [5] have suggested a wide variety of genes that may contribute to the pathogenicity of this pathogen. However, the work of Barad et al. [8] has attributed to the pH regulation of the host by P. expansum specific importance for pathogenicity. In the present study we used a transcriptomic approach to analyze the effect of pH on fungal gene regulation and host responses to various pH-modulating pathogens. The effect of the fungal response was observed by comparing the responses of the apple to colonization by P. expansum as driven by expression of the fungal gene at pH 4 or 7. Our RNA-Seq data of fungal responses revealed nine differentially co-expressed gene clusters; they showed alterations of expression patterns that were associated with differential pH responses, and also with genes related to P. expansum colonization. The apple response to pH was elucidated by using qRT-PCR to analyze expressions of selected genes in response to acidifying and alkalinizing pathogens: P. expansum and Colletotrichum gloeosporioides, respectively. This analysis indicated similar patterns of host response to the respective pathogens, but the apple genes showed an extensive multifold greater response to infection by its natural pathogen P. expansum than to that by C. gloeosporioides. This may indicate the importance of the acidifying modulation of pH by Penicillium as a factor for greater aggressiveness of P. expansum.
Results and Discussion
Profiling the expression of P. expansum genes during colonization and growth at pH 4 and 7
Genomes of P. expansum [7] were sequenced by using paired-end reads by means of Illumina Hiseq 2500 [12]. For analysis of the P. expansum transcriptome we downloaded the reference draft genome accession JQFX00000000.1 from the Genbank site [13]. Ten libraries of single-end RNAseq (deposited in Genbank under accession SRP071104) were mapped to the reference genome by using the Bowtie2 software [14]. The 10 libraries contained the following features: (i) P. expansum grown in culture media at pH 4 for 3 h as well as a pooled sample collected at several time points-0.5, 1, 3, 10, and 24 h-with two replicates at each point; (ii) P. expansum grown in culture media at pH 7 for 3 h, as well as a pool collected at several time points-0.5, 1, 3, 10, and 24 h-with two replicates at each point; and (iii) the leading edge, comprising 1–2 mm of apple cv. 'Golden Delicious' tissue, colonized by P. expansum. For in vitro experiments, 106 spores of P. expansum were inoculated into germinating minimal medium (GMM) at pH 4.5 as described by Barad et al. [7] and transferred 2 days later to buffered inducing media at pH 4 or 7. At several different time points after transfer to inducing secondary media (SM), fungal mycelia were sampled and pooled for analysis. Pooled samples of RNA were compared with independent samples 3 h after transfer to inducing media.
Principal component analysis (PCA) of all expressed genes of P. expansum indicated that the replicates were very close to each other, and that the expression patterns of fungal genes exposed to the respective pH conditions for 3 h were almost identical to those of pooled samples of mycelia exposed for various intervals (Fig. 1a). Moreover, the general pattern of gene expression in decayed apple tissue was far different from those of cultures grown under different pH conditions, as could be expected (Fig. 1a). Hierarchical clustering analysis indicated differing patterns of gene expression between the in vivo and in vitro samples at both pH levels; and the colonized apple tissue contained more down- than up-regulated genes (Fig. 1b).
Fig. 1.
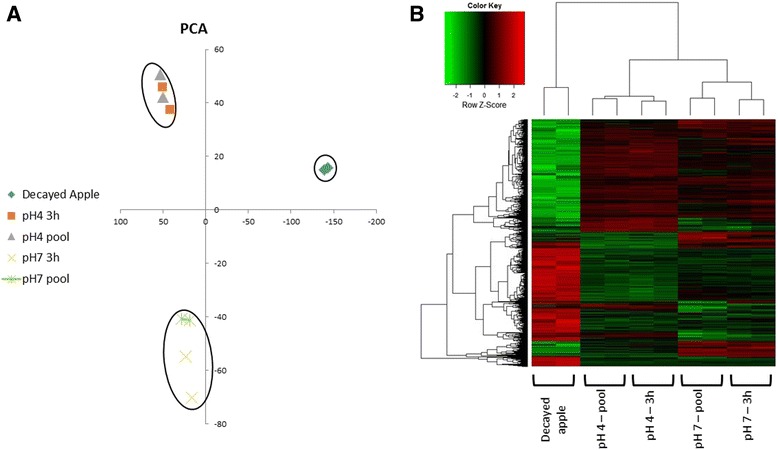
Principal component analysis (PCA) and heat map analysis of all the samples: duplicates of leading edge of inoculated apple; duplicates of pooled samples from all time points treated in vitro at pH 4.0; two replicates of P. expansum mycelia treated in vitro at pH 4.0 for 3 h; duplicates of a pool of samples from all time points treated in vitro at pH 7.0; and two replicates of P. expansum mycelia treated in vitro at pH 7.0 for 3 h. a PCA based on 5,646 differentially expressed genes. The results are plotted as: (♦) decayed apple; (■) in vitro at pH 4 for 3 h; (▲) pooled samples at pH 4; (X) in vitro at pH 7 for 3 h; (ӿ) pooled samples at pH 7. Circles are drawn around clustered points. b Expression heat map of differentially expressed P. expansum genes in the various samples
Out of 11,019 annotated genes [12], 5,646 genes were differentially expressed according to an false discovery rate (FDR, [15]) threshold < 0.05 and the log fold change was greater than 1 or smaller than −1 implying at least 2 fold change in expression scale (Fig. 1b). We sorted the differentially expressed gene patterns into nine different co-expressed gene clusters, which fell into three major groups:
Group I comprised clusters of genes whose expression patterns in the colonized tissue differed with respect to growth at pH 4 or pH 7. This group comprised clusters 1, 2, 3, and 6, which contained genes that are important for P. expansum colonization within apple fruits.
Group II comprised clusters in which expression patterns of the colonized tissue were similar to those of the pathogen's growth in culture at pH 4, and differed from those of fungal growth at pH 7. This group includes clusters numbers 5, 7, and 8.
Group III comprised gene clusters in which expression patterns of the colonized tissue were similar to those expressed by the pathogen grown in culture at pH 7. This group includes clusters numbers 4 and 9 (Fig. 2).
Fig. 2.

Expression patterns of 9 clusters of co-expressed differential genes. The normalized expression pattern (log 2-transformed, median centered) of 9 clusters that were derived from the hierarchical clustering algorithm by use of the hclust function algorithm in R [70, 71]. Each gene is plotted in gray, in addition to the mean expression profile for that cluster (blue). The figure was generated by using perl script define_clusters_by_cutting_tree.pl of the Trinity software package [67]
Group I includes clusters of expressed genes, whose level of expression under in vitro conditions differed from that exhibited during colonization. This group comprised clusters 1 and 2, with 2,248 and 233 genes, respectively, which exhibited higher expression under in vitro growth conditions at pH 4 or 7 than that manifested as down-regulation in the colonized apple tissue (Fig. 2). The genes expressed under both high and low pH are probably important for growth under in vitro conditions but not for pathogenicity. Group I also included clusters 3 and 6, with 2,220 and 72 genes, respectively, which comprise genes that are up-regulated in apple tissue colonization but down-regulated in vitro at both at pH 4 and pH 7. All the genes in this group probably are involved in growth in either nutritional environments or liquids, or both, that differ substantially from those within the apple tissue.
Group II comprised clusters 5, 7, and 8, in which the gene expression patterns in the colonized tissue were similar to those in vitro at pH 4 but different from those in vitro at pH 7 (Fig. 2). Cluster 7, with a gene count of 67 transcripts, showed similar overall expression levels during colonization of apple tissue and during in vitro growth at pH 4, which suggests that this cluster contained genes that are activated at pH 4, i.e., the usual fruit pH, therefore we consider that they may contribute to colonization at pH 4. Another two clusters present in this group were 5 and 8, with gene counts of 482 and 58, respectively; they include genes that were down-regulated both in colonized apple tissue and in vitro at pH 4, but maintained high expression levels in vitro at pH 7. These genes are probably necessary for growth under alkaline conditions.
Group III comprises gene clusters that showed similar expression levels in colonized apple tissue and during in vitro growth fungal at pH 7, but showed altered gene expression at pH 4 (Fig. 2). This group included clusters 9 and 4, with 24 and 242 transcript counts, respectively. Genes in cluster 9 showed similar high expression levels in colonized fruit and in vitro at pH 7, but low levels in vitro at pH 4; these genes probably are expressed in fruits under induced alkaline conditions or during fungal ammonia accumulation [8]. Cluster 4 included genes that exhibited low expression levels both in colonized fruit and in vitro at pH 7; we hypothesize that these genes are repressed under alkaline conditions or during ammonia secretion [8].
Gene ontology analysis of decayed apple tissue compared with in vitro conditions
We analyzed functional enrichment in each cluster using blast2go software [16] and Fisher’s Exact Test [17].
Clusters with different gene expression levels under colonization than under in vitro conditions
Higher expression levels of fungal genes when grown in culture than in colonized tissue
Genes in clusters 1 and 2 showed higher expression levels under in vitro conditions than in colonized tissue. Functionality of these genes showed involvement in respiratory activity, in light of their easier growth in culture media than in colonized apple tissue. GO-enriched terms in clusters 1 and 2 are depicted in Additional file 1: Figure S1 and Additional file 2: Figure S2, respectively.
Lower expression levels of fungal genes when grown in culture than in colonized tissue
Genes in clusters 3 and 6 showed lower expression levels when grown in vitro at pH 4 or pH 7 than when grown in colonized tissue (Fig. 2). These clusters included genes encoding for vesicle transport, which might indicate activation of secretion process(es); probably of mycotoxin(s) and effectors (Additional file 3: Figure S3 and Additional file 4: Figure S6 respectively). Cluster 3 included most of the 15 genes involved in patulin biosynthesis-patB, D, E, F, H, I, J, K, L, and O. Unexpectedly, these genes were not highly expressed in vitro at either pH 4 or pH 7, which supports a previous report by Li et al. [18], which suggested the importance of specific nutritional growth conditions for induced expression of the patulin biosynthesis gene cluster (PatA-PatO) in P. expansum. However, at the same time, the question arises of the specific mechanism(s)-possibly nutritional and possibly others-that modulate the conditions for expression of the mycotoxin cluster in vivo [18–20]. Cluster 3 also showed genes such as acetamidase, amidase-family proteins, and fatty-acid amide hydrolase, encoding for amidase activity (Additional file 3: Figure S3) these may indicate a possible source of NH4+ production during periods of limited sucrose levels and pathogenicity of P. expansum, which would support earlier suggestions by Barad et al. [8] that local pH modulation might result from ammonia accumulation in the leading edge during pathogenicity of P. expansum.
Cluster 6 shows enriched expression of genes associated with host-cell-wall degradation that are important for the virulence of P. expansum [5] (Additional file 4: Figure. S6). These genes have previously known functionalities associated with pathogenicity of fungi; these genes and activities include: chitinase-associated genes, pectin lyase, and polygalacturonase (PG) activities. Among these, PG was shown to be involved in maceration of apples by P. expansum, particularly in diseases characterized by tissue maceration or soft rot [21]; and pectin lyases contribute to the pathogenicity of many pathogens by degrading pectin polymers directly by means of a β-elimination mechanism that results in formation of 4,5-unsaturated oligo-galacturonides [22]. In addition, cluster 6 shows over-representation of genes encoding for aspartic endopeptidase-pep1-which is associated with pH modulation and pathogenicity of P. digitatum [23]. Aspartic endopeptidase catalyzes hydrolysis of elastin and collagen, the major structural proteins of basement membranes, and plays a significant role in virulence of P. digitatum on citrus fruits [23]. Aspartic endopeptidase was up-regulated during infection of citrus fruits, and contributed to fungal colonization, either by degradation of plant cell-wall components to provide a nitrogen supply, or even by inactivating defense proteins [24]. This type of response at low pH is probably a result of accumulation of GLA during the pathogenicity process of P. expansum, in order to ensure that secreted enzymes and metabolites are produced at the optimal pH to fully facilitate their physiological functions [3].
Clusters with similar expression levels in colonized apple and in vitro at pH 4
High expression level of fungal genes in colonized apple tissue and in samples grown in culture at pH 4
A single cluster-cluster 7-showed enhanced expression of genes both in colonized apple tissue and in vitro at pH 4 (Fig. 2). This cluster was enriched in genes involved in the glutamate metabolic process (Additional file 5: Figure S7); a process that includes high expression of glutamate decarboxylase (Table 1), which is required for normal oxidative-stress tolerance in Saccharomyces cerevisiae. γ-Aminobutyric acid (GABA) largely originates from decarboxylation of L-glutamate; it is associated with sporulation/spore metabolism [25] and in most fungi it serves as a carbon and nitrogen source. Also, γ-aminobutyric acid metabolism promotes asexual sporulation in Saccharomyces nodorum [26]. Moreover, in plants and fungi GABA synthesis has been associated with acidic pH, either in response to cytosolic acidification-probably as a pH-regulatory mechanism-or during growth under acidic conditions [25]. Since acidification of the host environment usually occurs during apple colonization by P. expansum, it is possible that γ-aminobutyric acid synthesis might be one of the mechanisms employed by the fungus to cope with this more acidic pH of the environment [6].
Table 1.
Genes involved in each process of cluster 7
| GO Term | Gene id | Genbank transcript id | Description |
|---|---|---|---|
| Nucleobase transport | PEXP_081890 | KGO47390 | Permease, cytosine/purine, uracil, thiamine, allantoin |
| PEXP_004400 | KGO37467 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| PEXP_081870 | KGO47388 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| Taurine metabolic process | PEXP_099510 | KGO46394 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 |
| PEXP_039450 | KGO48081 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| Beta-alanine metabolic process | PEXP_099510 | KGO46394 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 |
| PEXP_039450 | KGO48081 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| Transmembrane transport | PEXP_023020 | KGO42773 | Major facilitator superfamily domain, general substrate transporter |
| PEXP_104460 | KGO36460 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_019050 | KGO45782 | C4-dicarboxylate transporter/malic acid transport protein | |
| PEXP_083120 | KGO38619 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_105230 | KGO41447 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_012370 | KGO49188 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_060340 | KGO45245 | Iron permease FTR1 | |
| PEXP_059590 | KGO45345 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_094700 | KGO43568 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_025230 | KGO42652 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_030850 | KGO40270 | ABC transporter, integral membrane type 1 | |
| PEXP_078840 | KGO37868 | Major facilitator superfamily domain, general substrate transporter | |
| Glutamate metabolic process | PEXP_099510 | KGO46394 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 |
| PEXP_039450 | KGO48081 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| Glutamate decarboxylase activity | PEXP_099510 | KGO46394 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 |
| PEXP_039450 | KGO48081 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| Nucleobase transmembrane transporter activity | PEXP_081890 | KGO47390 | Permease, cytosine/purine, uracil, thiamine, allantoin |
| PEXP_004400 | KGO37467 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| PEXP_081870 | KGO47388 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| Cofactor binding | PEXP_107000 | KGO41320 | Aldolase-type TIM barrel |
| PEXP_068570 | KGO46687 | Aldolase-type TIM barrel | |
| PEXP_099510 | KGO46394 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| PEXP_080260 | KGO38010 | Pyridoxal phosphate-dependent transferase, major region, subdomain 2 | |
| PEXP_043260 | KGO39485 | Thiamine pyrophosphate enzyme, C-terminal TPP-binding | |
| PEXP_076130 | KGO36094 | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding | |
| PEXP_001130 | KGO44422 | Glyceraldehyde/Erythrose phosphate dehydrogenase family | |
| PEXP_039450 | KGO48081 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| Glucosylceramidase activity | PEXP_081260 | KGO47327 | Glycoside hydrolase, family 30 |
| 3-isopropylmalate dehydratase activity | PEXP_074470 | KGO48717 | Aconitase/3-isopropylmalate dehydratase large subunit, alpha/beta/alpha, subdomain 1/3 |
| N-acylphosphatidylethanolamine-specific phospholipase D activity | PEXP_024950 | KGO42624 | N-acyl-phosphatidylethanolamine-hydrolysing phospholipase D |
| Transmembrane transporter activity | PEXP_104460 | KGO36460 | Major facilitator superfamily domain, general substrate transporter |
| PEXP_034210 | KGO40038 | CDR ABC transporter | |
| PEXP_019050 | KGO45782 | C4-dicarboxylate transporter/malic acid transport protein | |
| PEXP_105230 | KGO41447 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_081890 | KGO47390 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| PEXP_004400 | KGO37467 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| PEXP_030850 | KGO40270 | ABC transporter, integral membrane type 1 | |
| PEXP_081870 | KGO47388 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| Integral component of membrane | PEXP_023020 | KGO42773 | Major facilitator superfamily domain, general substrate transporter |
| PEXP_104460 | KGO36460 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_034210 | KGO40038 | CDR ABC transporter | |
| PEXP_019050 | KGO45782 | C4-dicarboxylate transporter/malic acid transport protein | |
| PEXP_083120 | KGO38619 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_054930 | KGO44009 | Major intrinsic protein | |
| PEXP_105230 | KGO41447 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_012370 | KGO49188 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_059590 | KGO45345 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_094700 | KGO43568 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_027630 | KGO43014 | Amino acid transporter, transmembrane | |
| PEXP_025230 | KGO42652 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_030850 | KGO40270 | ABC transporter, integral membrane type 1 | |
| PEXP_038200 | KGO47945 | Mitochondrial substrate/solute carrier | |
| PEXP_078840 | KGO37868 | Major facilitator superfamily domain, general substrate transporter | |
| Lysosome | PEXP_081260 | KGO47327 | Glycoside hydrolase, family 30 |
| 3-Isopropylmalate dehydratase complex | PEXP_074470 | KGO48717 | Aconitase/3-isopropylmalate dehydratase large subunit, alpha/beta/alpha, subdomain 1/3 |
Low expression level of fungal genes in colonized apple and in samples grown in vitro at pH 4
In contrast to the single up-regulated cluster 7, two clusters-5 and 8-showed repressed expression of genes both in colonized apple tissue and in vitro at pH 4 (Fig. 2). Cluster 5 showed enrichment of genes that are involved in fungal pathogenicity and that are known to be active under alkaline conditions; they exhibit, for example, pectate lyase and hydrolase activities [27] (Additional file 6: Figure S5) and the transcription factor PacC (Table 2), which suggests that the acidic pH-repressed alkaline-expressed genes are involved in pathogenicity. It is likely that P. expansum adopted its acidifying life pattern characterized by production of organic acids-mainly gluconic acid secretion [7]-in order to optimize the pH of the media. These present findings support a previous publication by Barad et al. [28] which indicated that whereas pacC expression was inhibited under acidic conditions during the accumulation of gluconic acid, it might be activated in the leading edge of colonized tissue where ammonia is accumulated at pH 4 [8]. Part of the localized pH modulation may be regulated by the Na+ and Ca2+ transporters that are present in cluster 5, that modulate the internal equilibrium of hydrogen ions and contribute to cytoplasmic pH (Table 2).
Table 2.
Genes involved in each process of cluster 5
| GO Term | Gene id | Genbank transcript id | Description |
|---|---|---|---|
| Regulation of pH | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_043720 | KGO39291 | Alkali metal cation/H+ antiporter Nha1, C-terminal | |
| PEXP_009670 | KGO49300 | Cation/H+ exchanger | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| L-Arabinose metabolic process | PEXP_023030 | KGO42774 | Glycoside hydrolase, superfamily |
| PEXP_071150 | KGO40611 | Alcohol dehydrogenase superfamily, zinc-type | |
| PEXP_089320 | KGO41720 | Glycoside hydrolase, superfamily | |
| Calcium ion transport | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| Kynurenine metabolic process | PEXP_039010 | KGO48037 | putative cyclase |
| PEXP_045070 | KGO39237 | putative cyclase | |
| PEXP_009350 | KGO49268 | Pyridoxal phosphate-dependent transferase, major region, subdomain 2 | |
| Glyoxylate cycle | PEXP_011610 | KGO49112 | Malate synthase A |
| PEXP_077890 | KGO37773 | Pyruvate/Phosphoenolpyruvate kinase | |
| Tryptophan catabolic process | PEXP_039010 | KGO48037 | Putative cyclase |
| PEXP_045070 | KGO39237 | Putative cyclase | |
| PEXP_009350 | KGO49268 | Pyridoxal phosphate-dependent transferase, major region, subdomain 2 | |
| Inorganic ion transmembrane transport | PEXP_059490 | KGO45335 | Sulfate anion transporter |
| PEXP_043720 | KGO39291 | Alkali metal cation/H+ antiporter Nha1, C-terminal | |
| PEXP_048620 | KGO39686 | Nickel/cobalt transporter, high-affinity | |
| PEXP_009670 | KGO49300 | Cation/H+ exchanger | |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_094650 | KGO43563 | Ammonium transporter | |
| Disaccharide metabolic process | PEXP_052570 | KGO42305 | Glycosyl transferase, family 20 |
| PEXP_011870 | KGO49138 | Pectinesterase, catalytic | |
| PEXP_069330 | KGO46763 | Glycoside hydrolase, family 61 | |
| PEXP_011880 | KGO49139 | Glycoside hydrolase, family 28 | |
| PEXP_066270 | KGO46936 | Hexokinase, N-terminal | |
| PEXP_053240 | KGO42372 | Glycoside hydrolase, family 61 | |
| PEXP_107940 | KGO41788 | Alpha-amylase, C-terminal all beta | |
| Substrate-specific transmembrane transporter activity | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_059490 | KGO45335 | Sulfate anion transporter | |
| PEXP_016910 | KGO46049 | Xanthine/uracil/vitamin C permease | |
| PEXP_043720 | KGO39291 | Alkali metal cation/H+ antiporter Nha1, C-terminal | |
| PEXP_052330 | KGO42281 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_033600 | KGO39977 | Amino acid/polyamine transporter I | |
| PEXP_094860 | KGO43584 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_068270 | KGO46657 | Nucleobase cation symporter-1, NCS1 | |
| PEXP_006100 | KGO36811 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_086200 | KGO40808 | Amino acid/polyamine transporter I | |
| PEXP_003610 | KGO37353 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_043410 | KGO39260 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_048620 | KGO39686 | Nickel/cobalt transporter, high-affinity | |
| PEXP_006360 | KGO36837 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_000260 | KGO44335 | Amino acid/polyamine transporter I | |
| PEXP_072400 | KGO48510 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_009030 | KGO48810 | Na dependent nucleoside transporter | |
| PEXP_068910 | KGO46721 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_031270 | KGO40312 | Permease, cytosine/purine, uracil, thiamine, allantoin | |
| PEXP_009670 | KGO49300 | Cation/H+ exchanger | |
| PEXP_110420 | KGO38764 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_095790 | KGO43677 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_048590 | KGO39683 | Amino acid/polyamine transporter I | |
| PEXP_104220 | KGO36561 | Cation/H+ exchanger | |
| PEXP_019490 | KGO45826 | C4-dicarboxylate transporter/malic acid transport protein | |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_076320 | KGO37552 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_001370 | KGO37112 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_084160 | KGO41053 | Amino acid/polyamine transporter I | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_062420 | KGO45644 | General substrate transporter | |
| PEXP_022750 | KGO42746 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_068690 | KGO46699 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_034320 | KGO40048 | Arsenical pump membrane protein, ArsB | |
| PEXP_074940 | KGO36159 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_094650 | KGO43563 | Ammonium transporter | |
| Solute:proton antiporter activity | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_043720 | KGO39291 | Alkali metal cation/H+ antiporter Nha1, C-terminal | |
| PEXP_009670 | KGO49300 | Cation/H+ exchanger | |
| PEXP_104220 | KGO36561 | Cation/H+ exchanger | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| Cation:cation antiporter activity | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_043720 | KGO39291 | Alkali metal cation/H+ antiporter Nha1, C-terminal | |
| PEXP_104220 | KGO36561 | Cation/H+ exchanger | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| Metal ion binding | PEXP_046510 | KGO39177 | Cytochrome P450, E-class, CYP52 |
| PEXP_036890 | KGO35901 | Transcription factor, fungi | |
| PEXP_084560 | KGO41093 | 4-Hydroxyphenylpyruvate dioxygenase | |
| PEXP_055320 | KGO44077 | Polyketide synthase, enoylreductase | |
| PEXP_042410 | KGO39400 | Manganese/iron superoxide dismutase, C-terminal | |
| PEXP_072700 | KGO48540 | Annexin | |
| PEXP_089160 | KGO41704 | Terpenoid synthase | |
| PEXP_102680 | KGO36423 | Transcription factor, fungi | |
| PEXP_042010 | KGO48399 | Protein of unknown function DUF3468 | |
| PEXP_011320 | KGO49083 | Zinc finger, C2H2 | |
| PEXP_095680 | KGO43666 | Protein of unknown function DUF3468 | |
| PEXP_050580 | KGO42108 | Cytochrome P450 | |
| PEXP_059140 | KGO45293 | Polyketide synthase, enoylreductase | |
| PEXP_100050 | KGO46448 | Heavy metal-associated domain, HMA | |
| PEXP_103630 | KGO36296 | Urease accessory protein UreD | |
| PEXP_083550 | KGO40992 | Thiamine pyrophosphate enzyme, C-terminal TPP-binding | |
| PEXP_070880 | KGO40584 | Polyketide synthase, enoylreductase | |
| PEXP_027120 | KGO42963 | hypothetical protein | |
| PEXP_040610 | KGO48229 | Transcription factor, fungi | |
| PEXP_075350 | KGO36016 | Alpha-actinin | |
| PEXP_011040 | KGO49055 | Cytochrome P450, E-class, group I | |
| PEXP_005060 | KGO36707 | Transcription factor, fungi | |
| PEXP_070870 | KGO40583 | Aldehyde dehydrogenase, C-terminal | |
| PEXP_046450 | KGO39171 | Amidohydrolase 1 | |
| PEXP_008360 | KGO37036 | 4-Hydroxyphenylpyruvate dioxygenase | |
| PEXP_048620 | KGO39686 | Nickel/cobalt transporter, high-affinity | |
| PEXP_101160 | KGO38158 | Pectin lyase fold/virulence factor | |
| PEXP_022330 | KGO42704 | Transcription factor, fungi | |
| PEXP_058230 | KGO38945 | Alcohol dehydrogenase superfamily, zinc-type | |
| PEXP_053390 | KGO42387 | Polyketide synthase, enoylreductase | |
| PEXP_017020 | KGO46060 | Cytochrome P450 | |
| PEXP_021500 | KGO44639 | Transcription factor, fungi | |
| PEXP_043440 | KGO39263 | Polyketide synthase, enoylreductase | |
| PEXP_102420 | KGO36222 | Protein of unknown function DUF3468 | |
| PEXP_108970 | KGO41891 | Polyketide synthase, enoylreductase | |
| PEXP_095570 | KGO43655 | Transcription factor, fungi | |
| PEXP_088350 | KGO41611 | Transcription factor, fungi | |
| PEXP_056220 | KGO38423 | Cytochrome P450 | |
| PEXP_099120 | KGO46300 | Hypothetical protein | |
| PEXP_028450 | KGO43127 | Transcription factor, fungi | |
| PEXP_110420 | KGO38764 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_050400 | KGO42090 | Zinc finger, C2H2 | |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_085230 | KGO41160 | Hypothetical protein | |
| PEXP_000560 | KGO44365 | Class II aldolase/adducin N-terminal | |
| PEXP_071150 | KGO40611 | Alcohol dehydrogenase superfamily, zinc-type | |
| PEXP_090470 | KGO45017 | Pyruvate carboxyltransferase | |
| PEXP_080790 | KGO38063 | hypothetical protein | |
| PEXP_026670 | KGO42918 | Transcription factor, fungi | |
| PEXP_084100 | KGO41047 | Transcription factor, fungi | |
| PEXP_102160 | KGO38273 | Casein kinase II, regulatory subunit | |
| PEXP_036970 | KGO35909 | Cytochrome P450, E-class, group I | |
| PEXP_032030 | KGO39788 | Hypothetical protein | |
| PEXP_089350 | KGO41723 | Transcription factor, fungi | |
| PEXP_005230 | KGO36724 | Transcription factor, fungi | |
| PEXP_057180 | KGO44225 | ATP adenylyltransferase, C-terminal | |
| PEXP_002560 | KGO37249 | Hypothetical protein | |
| PEXP_081210 | KGO47322 | ATP-grasp fold, subdomain 1 | |
| PEXP_071070 | KGO40603 | Molybdenum cofactor synthesis C-terminal | |
| PEXP_095390 | KGO43637 | Transcription factor, fungi | |
| PEXP_054870 | KGO44003 | Transcription factor, fungi | |
| PEXP_042700 | KGO39429 | Hypothetical protein | |
| PEXP_036990 | KGO35911 | Ureohydrolase | |
| PEXP_041200 | KGO48300 | Transcription factor, fungi | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_048130 | KGO39637 | Transcription factor, fungi | |
| PEXP_010010 | KGO48952 | Transcription factor, fungi | |
| PEXP_085290 | KGO41166 | Transcription factor, fungi | |
| PEXP_079380 | KGO37922 | Zinc finger, C2H2 | |
| PEXP_057190 | KGO44226 | Thiamine pyrophosphate enzyme, C-terminal TPP-binding | |
| PEXP_069040 | KGO46734 | Cytochrome P450 | |
| PEXP_007560 | KGO36956 | Zinc finger, C2H2 | |
| PEXP_081220 | KGO47323 | Transcription factor, fungi | |
| PEXP_087170 | KGO40934 | Transcription factor, fungi | |
| PEXP_105830 | KGO41509 | Transcription factor, fungi | |
| PEXP_043140 | KGO39473 | Cytochrome P450 | |
| PEXP_067370 | KGO46568 | hypothetical protein | |
| PEXP_073070 | KGO48577 | Exonuclease, RNase T/DNA polymerase III | |
| PEXP_051020 | KGO42152 | Amidohydrolase 1 | |
| PEXP_061400 | KGO45512 | Hypothetical protein | |
| PEXP_101010 | KGO38143 | Glycoside hydrolase, family 71 | |
| PEXP_043150 | KGO39474 | Terpenoid synthase | |
| PEXP_015320 | KGO47645 | ATP-grasp fold, subdomain 1 | |
| Pectate lyase activity | PEXP_101160 | KGO38158 | Pectin lyase fold/virulence factor |
| PEXP_080220 | KGO38006 | Pectate lyase, catalytic | |
| Calcium ion transmembrane transporter activity | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| Sequence-specific DNA binding RNA polymerase II transcription factor activity | PEXP_102680 | KGO36423 | Transcription factor, fungi |
| PEXP_042010 | KGO48399 | Protein of unknown function DUF3468 | |
| PEXP_095680 | KGO43666 | Protein of unknown function DUF3468 | |
| PEXP_040610 | KGO48229 | Transcription factor, fungi | |
| PEXP_070870 | KGO40583 | Aldehyde dehydrogenase, C-terminal | |
| PEXP_021500 | KGO44639 | Transcription factor, fungi | |
| PEXP_102420 | KGO36222 | Protein of unknown function DUF3468 | |
| PEXP_088350 | KGO41611 | Transcription factor, fungi | |
| PEXP_099120 | KGO46300 | Hypothetical protein | |
| PEXP_085230 | KGO41160 | Hypothetical protein | |
| PEXP_080790 | KGO38063 | Hypothetical protein | |
| PEXP_102160 | KGO38273 | Casein kinase II, regulatory subunit | |
| PEXP_032030 | KGO39788 | Hypothetical protein | |
| PEXP_005230 | KGO36724 | Transcription factor, fungi | |
| PEXP_057180 | KGO44225 | ATP adenylyltransferase, C-terminal | |
| PEXP_002560 | KGO37249 | Hypothetical protein | |
| PEXP_095390 | KGO43637 | Transcription factor, fungi | |
| PEXP_042700 | KGO39429 | Hypothetical protein | |
| PEXP_048130 | KGO39637 | Transcription factor, fungi | |
| PEXP_010010 | KGO48952 | Transcription factor, fungi | |
| PEXP_085290 | KGO41166 | Transcription factor, fungi | |
| PEXP_087170 | KGO40934 | Transcription factor, fungi | |
| PEXP_067370 | KGO46568 | Hypothetical protein | |
| PEXP_101010 | KGO38143 | Glycoside hydrolase, family 71 | |
| Hydrolase activity, hydrolyzing O-glycosyl compounds | PEXP_000940 | KGO44403 | Protein of unknown function DUF2985 |
| PEXP_044190 | KGO39338 | Aldolase-type TIM barrel | |
| PEXP_049330 | KGO39518 | Glycoside hydrolase, family 35 | |
| PEXP_023030 | KGO42774 | Glycoside hydrolase, superfamily | |
| PEXP_101680 | KGO38226 | Concanavalin A-like lectin/glucanases superfamily | |
| PEXP_023340 | KGO42463 | Glycoside hydrolase, family 43 | |
| PEXP_070780 | KGO40574 | Glycoside hydrolase, family 31 | |
| PEXP_072880 | KGO48558 | Glycoside hydrolase, superfamily | |
| PEXP_048880 | KGO39711 | Concanavalin A-like lectin/glucanases superfamily | |
| PEXP_069330 | KGO46763 | Glycoside hydrolase, family 61 | |
| PEXP_011880 | KGO49139 | Glycoside hydrolase, family 28 | |
| PEXP_089320 | KGO41720 | Glycoside hydrolase, superfamily | |
| PEXP_076310 | KGO37551 | Glycoside hydrolase, family 32 | |
| PEXP_026640 | KGO42915 | Glycoside hydrolase, family 16, CRH1, predicted | |
| PEXP_057570 | KGO44264 | Glycoside hydrolase family 3 | |
| PEXP_053240 | KGO42372 | Glycoside hydrolase, family 61 | |
| PEXP_042140 | KGO48412 | Concanavalin A-like lectin/glucanase, subgroup | |
| PEXP_107940 | KGO41788 | Alpha-amylase, C-terminal all beta | |
| ATPase activity, coupled to transmembrane movement of substances | PEXP_023060 | KGO42777 | CDR ABC transporter |
| PEXP_050880 | KGO42138 | ABC transporter, integral membrane type 1 | |
| PEXP_110420 | KGO38764 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_040720 | KGO48240 | ABC transporter, integral membrane type 1 | |
| PEXP_071990 | KGO40700 | ABC transporter, integral membrane type 1 | |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_104880 | KGO41412 | ABC transporter, integral membrane type 1 | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_077310 | KGO37715 | CDR ABC transporter | |
| PEXP_086490 | KGO40844 | ABC transporter, integral membrane type 1 | |
| PEXP_057530 | KGO44260 | ABC transporter, integral membrane type 1 | |
| 4-Hydroxyphenylpyruvate dioxygenase activity | PEXP_084560 | KGO41093 | 4-Hydroxyphenylpyruvate dioxygenase |
| PEXP_008360 | KGO37036 | 4-Hydroxyphenylpyruvate dioxygenase | |
| Integral component of membrane | PEXP_046990 | KGO39741 | Sodium/calcium exchanger membrane region |
| PEXP_024890 | KGO42618 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_041670 | KGO48366 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_059490 | KGO45335 | Sulfate anion transporter | |
| PEXP_043720 | KGO39291 | Alkali metal cation/H+ antiporter Nha1, C-terminal | |
| PEXP_052330 | KGO42281 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_037770 | KGO36126 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_102860 | KGO36441 | Mitochondrial carrier protein | |
| PEXP_023060 | KGO42777 | CDR ABC transporter | |
| PEXP_033600 | KGO39977 | Amino acid/polyamine transporter I | |
| PEXP_014370 | KGO47550 | Amino acid transporter, transmembrane | |
| PEXP_094860 | KGO43584 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_006100 | KGO36811 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_046430 | KGO39169 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_086200 | KGO40808 | Amino acid/polyamine transporter I | |
| PEXP_024440 | KGO42573 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_003610 | KGO37353 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_043410 | KGO39260 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_048620 | KGO39686 | Nickel/cobalt transporter, high-affinity | |
| PEXP_006360 | KGO36837 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_044620 | KGO39381 | Tetracycline resistance protein, TetA/multidrug resistance protein MdtG | |
| PEXP_072400 | KGO48510 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_028370 | KGO43119 | Amino acid transporter, transmembrane | |
| PEXP_009920 | KGO48943 | Peroxisomal biogenesis factor 11 | |
| PEXP_033410 | KGO39958 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_050880 | KGO42138 | ABC transporter, integral membrane type 1 | |
| PEXP_056980 | KGO44205 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_102420 | KGO36222 | Protein of unknown function DUF3468 | |
| PEXP_074410 | KGO48711 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_068910 | KGO46721 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_060610 | KGO45433 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_009670 | KGO49300 | Cation/H+ exchanger | |
| PEXP_017290 | KGO46087 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_110420 | KGO38764 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_059360 | KGO45322 | Mitochondrial carrier protein | |
| PEXP_095790 | KGO43677 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_040720 | KGO48240 | ABC transporter, integral membrane type 1 | |
| PEXP_071990 | KGO40700 | ABC transporter, integral membrane type 1 | |
| PEXP_048590 | KGO39683 | Amino acid/polyamine transporter I | |
| PEXP_104220 | KGO36561 | Cation/H+ exchanger | |
| PEXP_000120 | KGO44321 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_019490 | KGO45826 | C4-dicarboxylate transporter/malic acid transport protein | |
| PEXP_088420 | KGO41618 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_076320 | KGO37552 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_001370 | KGO37112 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_001360 | KGO37111 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_104880 | KGO41412 | ABC transporter, integral membrane type 1 | |
| PEXP_005240 | KGO36725 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_108540 | KGO41848 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_095400 | KGO43638 | Sodium/calcium exchanger membrane region | |
| PEXP_046610 | KGO39187 | Sodium/calcium exchanger membrane region | |
| PEXP_098820 | KGO46270 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_001950 | KGO37170 | Mitochondrial carrier protein | |
| PEXP_050270 | KGO42077 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter | |
| PEXP_023180 | KGO42789 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_010400 | KGO48991 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_098540 | KGO46242 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_066250 | KGO46934 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_062420 | KGO45644 | General substrate transporter | |
| PEXP_026170 | KGO42868 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_077310 | KGO37715 | CDR ABC transporter | |
| PEXP_022750 | KGO42746 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_068690 | KGO46699 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_066860 | KGO46517 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_086490 | KGO40844 | ABC transporter, integral membrane type 1 | |
| PEXP_067360 | KGO46567 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_057530 | KGO44260 | ABC transporter, integral membrane type 1 | |
| PEXP_034320 | KGO40048 | Arsenical pump membrane protein, ArsB | |
| PEXP_074940 | KGO36159 | Major facilitator superfamily domain, general substrate transporter |
A second cluster that was under-represented, both in colonized apple tissue and in vitro at pH 4 was cluster 8 (Fig. 2). This cluster mainly showed enrichment in carbon utilization and sugar-transport-related genes (Table 3, Additional file 7: Figure S8), which suggests that they contribute to a series of processes that induce carbon catabolism and thereby result in GLA accumulation [8]. This behavior may indicate that these processes are usually repressed during colonization unless there is a significant increase in ammonia accumulation.
Table 3.
Genes involved in each process of cluster 8
| GO Term | Gene id | Genbank transcript id | Description |
|---|---|---|---|
| Phosphate ion transport | PEXP_088430 | KGO41619 | Phosphate transporter |
| PEXP_030290 | KGO40453 | Phosphate transporter | |
| Carbon utilization | PEXP_105770 | KGO41503 | Ribose/galactose isomerase |
| PEXP_105760 | KGO41502 | Aldolase-type TIM barrel | |
| Transmembrane transport | PEXP_104850 | KGO41409 | Major facilitator superfamily domain, general substrate transporter |
| PEXP_048020 | KGO39626 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_070650 | KGO40561 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_011530 | KGO49104 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_033090 | KGO39926 | Amino acid/polyamine transporter I | |
| PEXP_078910 | KGO37875 | Cation efflux protein | |
| PEXP_049390 | KGO39524 | Cation/H+ exchanger | |
| PEXP_030290 | KGO40453 | Phosphate transporter | |
| PEXP_034270 | KGO40044 | Major facilitator superfamily domain, general substrate transporter | |
| Cation transport | PEXP_002160 | KGO37191 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter |
| PEXP_078910 | KGO37875 | Cation efflux protein | |
| PEXP_049390 | KGO39524 | Cation/H+ exchanger | |
| PEXP_004260 | KGO37441 | Sodium\x3aneurotransmitter symporter | |
| Neurotransmitter transport | PEXP_004260 | KGO37441 | Sodium\x3aneurotransmitter symporter |
| Triose-phosphate isomerase activity | PEXP_105760 | KGO41502 | Aldolase-type TIM barrel |
| Ribose-5-phosphate isomerase activity | PEXP_105770 | KGO41503 | Ribose/galactose isomerase |
| Cysteine dioxygenase activity | PEXP_086730 | KGO40868 | Cysteine dioxygenase type I |
| asparaginase activity | PEXP_042000 | KGO48398 | L-asparaginase, type II |
| Integral component of membrane | PEXP_002160 | KGO37191 | ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter |
| PEXP_104850 | KGO41409 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_025010 | KGO42630 | Hypothetical protein | |
| PEXP_083360 | KGO38643 | FAD-binding 8 | |
| PEXP_048020 | KGO39626 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_070650 | KGO40561 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_011530 | KGO49104 | Major facilitator superfamily domain, general substrate transporter | |
| PEXP_033090 | KGO39926 | Amino acid/polyamine transporter I | |
| PEXP_078910 | KGO37875 | Cation efflux protein | |
| PEXP_049390 | KGO39524 | Cation/H+ exchanger | |
| PEXP_004260 | KGO37441 | Sodium\x3aneurotransmitter symporter | |
| PEXP_034270 | KGO40044 | Major facilitator superfamily domain, general substrate transporter |
Clusters with similar expression levels in colonized apple tissue and in vitro at pH 7
High expression level of fungal genes in colonized apple tissue and in fungi grown in vitro at pH 7
One of the most important clusters that showed high expression levels both in colonized apple tissue and in vitro at pH 7 was number 9 (Fig. 2), which showed significant enrichment of cellular amino-acid metabolism (Table 4, Additional file 8: Figure S9). Ammonia accumulation is induced under the limited-nutrient conditions present at the leading edge of the decay, in order to enable the pathogen to use a battery of pectolytic enzymes for tissue maceration [8]. Among the genes involved in cellular amino-acid metabolism process is glucose-methanol-choline oxidoreductase (GMC) (Table 4), whose families are clusters of FAD flavoprotein oxidoreductases-highly complex genes that contribute to several oxidation and reduction processes [29]. These enzymes include a variety of proteins, not all of which were present in cluster 9, such as choline dehydrogenase (CHD), which was present in clusters 1, 3, and 9, methanol oxidase (MOX), and cellobiose dehydrogenase, which was present in cluster 3; these are proteins that share a number of homologous regions that show sequence similarities. Since ammonia accumulation at the leading edge of the Penicillium-colonized tissue did not increase local pH [8], it is possible that the accumulated ammonia may activate gene expression, either directly or indirectly by generation of hydrogen peroxide and activation of RBOH in the killed cell [30]. To elucidate the role of ammonia, we analyzed the relative expression levels of several genes-MepB, CuAC and ACC-from cluster 9 in Penicillium-colonized fruits exposed to 22 μM of NH4+, and found induction of their relative expression levels (Fig. 3), which indicates the capability of ammonia to activate this process.
Table 4.
Genes involved in each process of cluster 9
| GO Term | Gene id | Genbank transcript id | Description |
|---|---|---|---|
| Ammonium transmembrane transport | PEXP_025350 | KGO42664 | Ammonium transporter |
| Cellular amino acid metabolic process | PEXP_082650 | KGO38572 | Pyridoxal phosphate-dependent transferase, major region, subdomain 2 |
| PEXP_109810 | KGO38703 | Glucose-methanol-choline oxidoreductase | |
| PEXP_004920 | KGO36657 | Glutamate synthase, central-N | |
| PEXP_053110 | KGO42359 | Catalase-peroxidase heme | |
| PEXP_056170 | KGO38539 | 1-aminocyclopropane-1-carboxylate deaminase | |
| Amine metabolic process | PEXP_053110 | KGO42359 | Catalase-peroxidase heme |
| PEXP_056170 | KGO38539 | 1-aminocyclopropane-1-carboxylate deaminase | |
| PEXP_000620 | KGO44371 | Copper amine oxidase, N2-terminal | |
| Hydrogen peroxide catabolic process | PEXP_053110 | KGO42359 | Catalase-peroxidase heme |
Fig. 3.
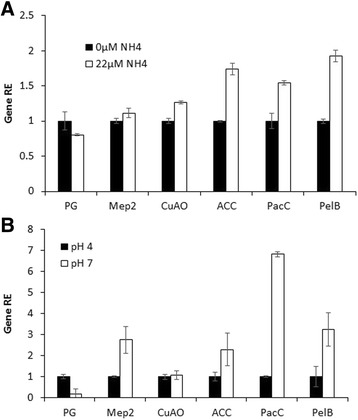
Effects of ammonia and pH on the relative expressions of genes involved in the pathogenicity process. Expressions were analyzed of: polygalacturonase (PG), Mep2, Copper amine oxidase (CuAO), ACC deaminase, PacC, and pectate lyase A (PelA). a Samples from the leading edge of infection in apples treated with exogenous NH4Cl at 0 and 22 μM, according to Barad et al. [8]; b Samples grown at pH 4 or pH 7, for 3 h in shaking secondary media (SM)
Another highly expressed family of genes in cluster 9 was the glucose-methanol-choline oxidoreductase family. Heat-map analysis of this family showed that genes with the same activity were differentially expressed between different clusters (Fig. 4). For example, 11 GMC oxidoreductase genes were detected in P. expansum: three in cluster 1, three in cluster 3, and one in cluster 9, whereas the remaining four were not differentially expressed. The GMC oxidoreductase family also exhibited close similarity to the glucose oxidase family. We were able to detect three glucose oxidase transcripts, as described by Ballester et al. [12], which showed differing expression patterns: gox1 of P. expansum was not differentially expressed under our conditions, gox2 was found in cluster 3, and gox3 was detected in cluster 1 (Fig. 4). Analysis of only the expression pattern at pH 4 compared with that at pH 7 showed that both gox2 and glucose oxidase 3 (gox3) were upregulated at pH 7 but not at pH 4 [7]. This differential expression pattern of the gox family suggests that genes were being selectively activated on the basis of their optimal conditions with respect to pH, in vitro, or in vivo, to enable the fungus to cope with varied conditions and to make optimal use of the inventory of available enzymes [31].
Fig. 4.
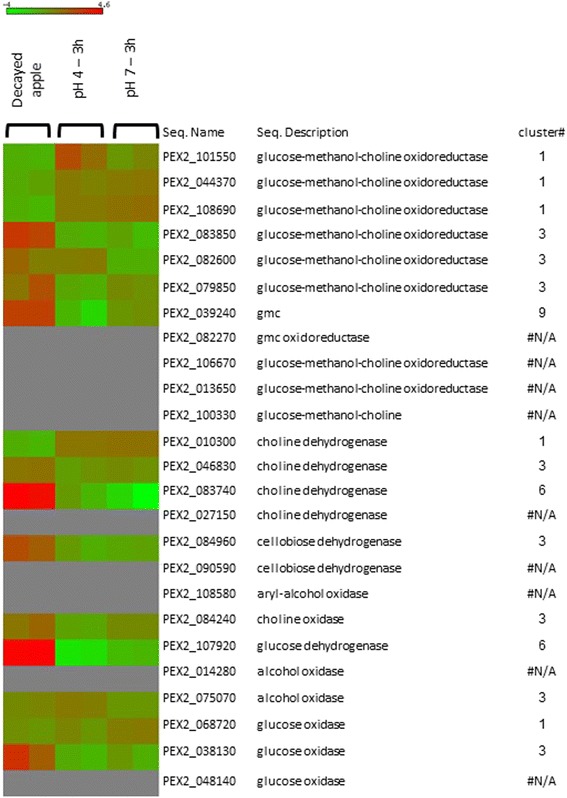
Heat map of the expression pattern of GMC oxidoreductase gene family. The expression patterns of genes belonging to the GMC oxidoreductase gene family are shown as a heat map obtained with matrix2png software [72]. Gene identity, gene description, and its location in the differentially expressed gene clusters are indicated
Another process that was enriched in this cluster was amine metabolism (Additional file 8: Figure S9), which involves the gene copper amine oxidase (CuAO) (Table 4). In plants, wounding of tissue usually results in an increase in the steady-state levels of copper amine oxidase expression and H2O2 accumulation [32]. Activation of CuAO during Penicillium attack also may lead to enhanced accumulation of H2O2 at the wound site, thereby contributing to extension of necrotic lesions and extensive plant-cell damage [32]. Also, copper amine oxidase may catalyze oxidation of the aliphatic diamines putrescine and cadaverine at their primary amino groups [33]. The products of putrescine oxidation by CuAO are H2O2, NH3, and Δ1-pyrroline; and Δ1-pyrroline is further catabolized to γ-aminobutyric acid, which subsequently is transaminated and oxidized to succinic acid. Thus, in Penicillium copper amine oxidase may contribute to the balance of reactive oxygen species (ROS) produced in the cell wall extracellular matrix [34]. This specific regulatory mechanism and the molecular signals inducing modulation of copper amine oxidase in Penicillium during decay highlight the relevance of these enzymes as an H2O2-delivering system in colonized tissue. The overrepresentation of glucose-methanol-choline oxidoreductase- and copper amine oxidase-activated genes in cluster 9 may indicate their contribution to the ROS and oxidoreductase process at the leading edge of the colony; also, in the same cluster there was activation of the H2O2 catabolic process (Additional file 8: Figure S9) through catalase peroxidase, which suggests a hitherto unknown mechanism of Penicillium survival under oxidative stress. Interestingly, similar up-regulation of catalase peroxidase under pathogenic conditions was reported to account for the survival of P. marneffei, an intracellular pathogen that causes common opportunistic infections in humans, and of P. simplicissimum, a plant pathogen in which strong expression of the catalase peroxidase transcripts may contribute to survival of this fungus in host cells [35]. Considered together, the relative expressions of the oxidoreductase genes and copper amine oxidase under fruit pH levels ranging from 3.7 to 4.2, again may indicate the importance of local ammonification, as reported by Barad et al. [8], as a mechanism to induce activation of genes usually overexpressed at pH 7 (Fig. 3).
The overrepresentation in cluster 9, of several nitrogen-metabolism-regulating genes, such as glutamate synthase and MepB (Table 4) also is important for nitrogen metabolism in fungi. The ammonia transporter encoded by mepB can lead to an internal/external modulation of ammonia in the hypha, activation of pacC, and high-pH induced genes [36]. Accumulation of glutamate, followed by its transformation to glutamine by glutamate synthase activity may be the basis for accumulation of several amino acids, and both stages are activated by ammonia accumulation (Fig. 3). Overall, the nitrogen mobilization promoted by infection could be considered as part of a "slash-and-burn" strategy that deprives the pathogen of nutrients and modulates alkaline-expressed genes. Such nutritional and metabolic changes might occur as a differential-attack mechanism promoting pathogen development [37].
One interesting gene also activated in cluster 9 in cellular amino acid metabolism (Additional file 8: Figure S9) is the gene encoding for aminocyclopropane-1-carboxylate deaminase (ACC) (Table 4). The ACC functions as a deaminase, degrading aminocyclopropane-1-carboxylate deaminase to 2-oxobutyrate and ammonia, which is a precursor of the plant hormone ethylene. A similar reaction process catalyzing ACC synthase was found in Penicillium digitatum attacking citrus fruits [38, 39], which suggests that colonization may be associated with ethylene production and induced tissue senescence. Taken together these results indicate that ROS production by the pathogen may be accompanied by host tissue induced senescence.
Low expression level of fungal genes in colonized apple tissue and in fungus grown in vitro at pH 7
Group III also includes cluster 4, in which colonized apple tissue showed under-representation of transcripts similar to that in vitro at pH 7 (Fig. 2). This cluster shows enrichment of genes involved in the oxido-reduction process, such as cysteine dioxygenase (Table 5, Additional file 9: Figure S4). Among representatives of this group are the cytochrome P-450 (CytP) monooxygenases, which are enzymes that: catalyze conversion of hydrophobic intermediates of primary and secondary metabolic pathways; detoxify natural and environmental pollutants that are accumulated during colonization as phytochemical molecules belonging to the polyphenols; and enable fungal growth under varied colonizing conditions [40]. They do this by inserting one oxygen atom into the aliphatic position of an organic substrate, while the other oxygen atom is reduced to water.
Table 5.
Genes involved in each process of cluster 4
| GO Term | Gene id | genbank transcript id | Description |
|---|---|---|---|
| Oxidation-reduction process | PEXP_035940 | KGO40210 | Cytochrome P450, E-class, CYP52 |
| PEXP_094610 | KGO43560 | Multicopper oxidase, type 1 | |
| PEXP_069560 | KGO47265 | Cytochrome P450 | |
| PEXP_047900 | KGO39614 | Cytochrome P450 | |
| PEXP_074010 | KGO48671 | Cytochrome P450 | |
| PEXP_065530 | KGO46863 | Short-chain dehydrogenase/reductase SDR | |
| PEXP_073620 | KGO48632 | Polyketide synthase, enoylreductase | |
| PEXP_079160 | KGO37900 | FAD-linked oxidase, N-terminal | |
| PEXP_057820 | KGO44289 | Cysteine dioxygenase type I | |
| PEXP_043230 | KGO39482 | Isocitrate and isopropylmalate dehydrogenases family | |
| PEXP_101170 | KGO38159 | Oxoglutarate/iron-dependent dioxygenase | |
| PEXP_060240 | KGO45235 | Short-chain dehydrogenase/reductase SDR | |
| PEXP_034900 | KGO40106 | Aldolase-type TIM barrel | |
| PEXP_106700 | KGO41290 | Isocitrate and isopropylmalate dehydrogenases family | |
| PEXP_026690 | KGO42920 | Short-chain dehydrogenase/reductase SDR | |
| PEXP_069740 | KGO47756 | Redoxin | |
| PEXP_103230 | KGO36363 | Cytochrome P450 | |
| PEXP_091400 | KGO45110 | hypothetical protein | |
| PEXP_027810 | KGO43163 | Polyketide synthase, enoylreductase | |
| PEXP_023550 | KGO42484 | Indoleamine 2,3-dioxygenase | |
| PEXP_077820 | KGO37766 | Polyketide synthase, enoylreductase | |
| PEXP_043790 | KGO39298 | Dihydrolipoamide succinyltransferase | |
| PEXP_082800 | KGO38587 | Cytochrome P450 | |
| PEXP_043420 | KGO39261 | Polyketide synthase, enoylreductase | |
| PEXP_030200 | KGO40444 | Cytochrome P450 | |
| PEXP_061260 | KGO45498 | Short-chain dehydrogenase/reductase SDR | |
| PEXP_078860 | KGO37870 | Dimeric alpha-beta barrel | |
| PEXP_042100 | KGO48408 | NADPH-cytochrome p450 reductase, FAD-binding, alpha-helical domain-3 | |
| PEXP_037510 | KGO35963 | Oxoglutarate/iron-dependent dioxygenase | |
| PEXP_010380 | KGO48989 | Aldo/keto reductase | |
| PEXP_000410 | KGO44350 | Acyl transferase/acyl hydrolase/lysophospholipase | |
| PEXP_076770 | KGO37511 | 3-oxo-5-alpha-steroid 4-dehydrogenase, C-terminal | |
| PEXP_066880 | KGO46519 | FAD dependent oxidoreductase | |
| PEXP_008580 | KGO37058 | hypothetical protein | |
| PEXP_001620 | KGO37137 | Ubiquinone biosynthesis protein Coq7 | |
| PEXP_030860 | KGO40271 | NADPH-dependent FMN reductase | |
| PEXP_107740 | KGO41768 | Polyketide synthase, enoylreductase | |
| PEXP_049110 | KGO39496 | Monooxygenase, FAD-binding | |
| PEXP_017320 | KGO46090 | Polyketide synthase, enoylreductase | |
| Taurine metabolic process | PEXP_057820 | KGO44289 | Cysteine dioxygenase type I |
| PEXP_050220 | KGO42072 | Acetate/Proprionate kinase |
Carbohydrate active enzymes (CAZy) cluster distribution
The diverse complex carbohydrates that contribute to Penicillium maceration capabilities are controlled by a panel of enzymes involved in their assembly (glycosyltransferases) and breakdown (glycoside hydrolases, polysaccharide lyases, carbohydrate esterases), which collectively are designated as Carbohydrate-Active enZymes (CAZymes) [41]. In plant pathogens CAZymes promote synthesis, degradation, and modification of carbohydrates that play important roles in the breakdown of plant cell walls and in host/pathogen interactions [42]. Penicillium uses these enzymes to macerate the colonized tissue by degradation of complex carbohydrates of the hosts to simple monomers that can be utilized as nutrients [43, 44].
The CAZymes analysis toolkit (CAT) [45] was used to identify 771 putative CAZymes in P. expansum within the various clusters (Fig. 5). CAZymes formed 8.00 % of all the transcripts in cluster 1, with 15, 24, 19, 57, and 65 of CAZymes families including auxiliary activity (AA), carbohydrate-binding modules (CBM), carbohydrate esterases (CE), glycosyltransferases (GT), and glycoside hydrolases (GH), respectively. In cluster 2 they formed 5.57 % of all the transcripts with 2, 3, and 8 of CAZymes families including carbohydrate esterases, glycosyltransferases (GT), and glycoside hydrolases (GH), respectively. In cluster 3 they formed 17.92 % of all the transcripts, with 24, 74, 28, 149, and 123 of CAZymes families including auxiliary activity (AA), carbohydrate-binding modules (CBM), carbohydrate esterases (CE), glycosyltransferases (GT), and glycoside hydrolases (GH), respectively. In cluster 4 they formed 14.46 % of all transcripts, with 2, 3, 4, 15, and 11 of CAZymes families including including auxiliary activity (AA), carbohydrate-binding modules (CBM), carbohydrate esterases (CE), glycosyltransferases (GT), and glycoside hydrolases (GH), respectively. In cluster 5 they formed 22.61 % of all transcripts, with 7, 11, 11, 5, 23, and 52 of CAZymes families including auxiliary activity (AA), carbohydrate-binding modules (CBM), carbohydrate esterases (CE), polysaccharide lyases (PL), glycosyltransferases (GT), and glycoside hydrolases (GH), respectively. In cluster 6 they formed 29.16 % of all transcripts, with 2, 3, 2, 5, and 9 of CAZymes families including auxiliary activity (AA), carbohydrate-binding modules (CBM), PL, glycosyltransferases (GT), and glycoside hydrolases (GH), respectively. In cluster 7 they formed 22.61 % of all transcripts, with 4 of CAZymes families including glycosyl hydrolases. In cluster 8 they formed 12.06 % of all transcripts, with 2 and 5 of CAZymes families including carbohydrate esterases (CE) and glycoside hydrolases (GH), respectively. In cluster 9 they formed 16.67 % of all transcripts, with 2 of CAZymes families including each of auxiliary activity (AA) and glycoside hydrolases (GH) (Fig. 5).
Fig. 5.
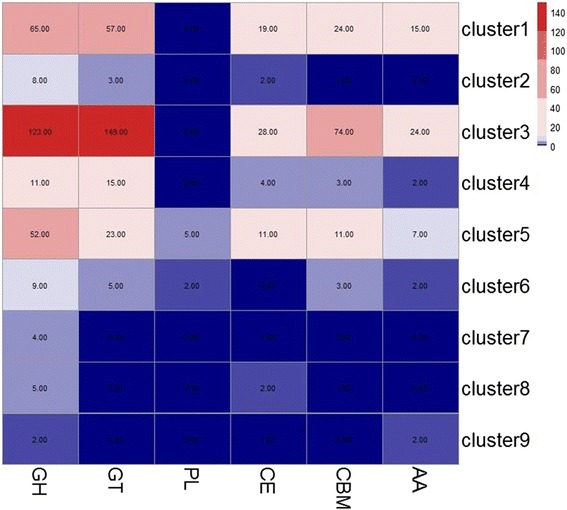
Comparison of the CAZyme repertories identified in each cluster of co-expressed genes. Enzyme families are represented by their class (GH-glycoside hydrolases; GT-glycosyltransferases; PL-polysaccharide lyases; CE-carbohydrate esterases; and CBM-chitin-binding modules) and family number according to the Carbohydrate-Active Enzyme Database. Abundance of the various enzymes within a family is represented on a color scale from 0 (dark blue) to 150 occurrences (dark red) per cluster
Carbohydrate esterases, glycoside hydrolases, and polysaccharide lyases are associated with the ability to utilize the diversity of carbohydrates present in the environment and within host fruits. Glycosyltransferases are mainly involved in the basal activities of fungal cells, e.g., fungal-cell-wall synthesis, glycogen cycle, and trehalose cycle [41, 44, 46]. The wide occurrence of CAZymes in the various Penicillium clusters, taken together with their importance in degradation of the plant cell wall, indicates their basic contribution to colonization of the host fruit.
Differentially expressed genes in Penicillium expansum- and Colletotrichum gloeosporioides-infected apple compared with healthy tissue
In order to analyze the effect of Penicillium expansum infection during acidification on the host (apple tissue) response, we used RNAseq to analyze the differentially expressed genes induced by P. expansum infection. Overall, in the P. expansum-infected apple tissue we found 4,292 differentially expressed genes with FDR threshold < 0.001 and with expression levels increasing or decreasing by a factor greater or less than 8, respectively, i.e., greater or less than +3 or −3, respectively, on a logarithmic (base 2) scale. We found 2,427 up-regulated and 1,865 down-regulated genes in colonized apple tissue. Analysis of the enriched pathways with MatGeneMap [47] revealed 21 significantly up-regulated pathways (Table 6) and 21 down-regulated ones (Table 7) with FDR < 0.05. Among the significantly up-regulated processes induced by Penicillium in apple tissue were: the jasmonic acid (JA), the mevalonate, and the flavonoid biosynthesis pathways, and the geranyl geranyldiphosphate biosynthesis I super pathway. Among the down-regulated pathways were the glycogen biosynthesis I and the starch biosynthesis pathways. Jasmonic acid is a lipid-derived signaling compound involved in regulation of diverse processes in plants such as fruit ripening, root growth, tendril coiling, senescence, and resistance to pathogens [48]; JA and related compounds are synthesized in plants via the octadecanoid pathway [49]. Biosynthesis of jasmonates starts with oxygenation of linolenic acid, which is thought to be released from membrane lipids through the action of a lipase, followed by several oxidation processes. This activation may indicate that there is a host response prior to fungal maceration induced by Penicillium, and this supposition is supported by findings that fruits pretreated with JA and related compounds showed enhanced resistance to pathogens [50].
Table 6.
Apple up-regulated pathways by infection with P. expansum
| Pathway name | p value (adjusted) |
|---|---|
| Jasmonic acid biosynthesis | 5.73E-11 |
| Mevalonate pathway | 9.12E-08 |
| Flavonoid biosynthesis | 7.62E-05 |
| Superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 7.62E-05 |
| Glutathione-mediated detoxification | 0.00171 |
| Trans,trans-farnesyl diphosphate biosynthesis | 0.0032 |
| Glutamate dependent acid resistance | 0.0032 |
| Chorismate biosynthesis | 0.00325 |
| Amygdalin and prunasin degradation | 0.00988 |
| Salicylate biosynthesis | 0.01194 |
| DIMBOA-glucoside degradation | 0.01415 |
| β-Alanine biosynthesis II | 0.01456 |
| Acetate formation from acetyl-CoA II | 0.01456 |
| Pyruvate fermentation to acetate III | 0.01456 |
| Glutamate degradation III (via 4-aminobutyrate) | 0.01693 |
| Superpathway of phenylalanine and tyrosine biosynthesis | 0.01693 |
| Phospholipid desaturation | 0.03878 |
| Glycolipid desaturation | 0.03878 |
| 13-LOX and 13-HPL pathway | 0.04864 |
| Divinyl ether biosynthesis II (13-LOX) | 0.04864 |
| Superpathway of phenylalanine, tyrosine, and tryptophan biosynthesis | 0.04864 |
The analysis was performed with MetGenMap
Table 7.
Apple down-regulated pathways by infection with P. expansum
| Pathway name | p value (adjusted) |
|---|---|
| Glycogen biosynthesis I (from ADP-D-Glucose) | 9.064E-08 |
| Starch biosynthesis | 6.194E-07 |
| C4 Photosynthetic carbon assimilation cycle | 0.0002403 |
| UDP-galactose biosynthesis (salvage pathway from galactose using UDP-glucose) | 0.0008945 |
| Lipoate biosynthesis and incorporation I | 0.0019429 |
| Methylerythritol phosphate pathway | 0.0031538 |
| Acyl carrier protein metabolism | 0.0034133 |
| Trans-lycopene biosynthesis | 0.0059809 |
| Fatty acid biosynthesis initiation I | 0.0070497 |
| Pyridoxal 5'-phosphate biosynthesis | 0.0087438 |
| Superpathway of pyridoxal 5'-phosphate biosynthesis and salvage | 0.0132283 |
| Xylitol degradation | 0.0161255 |
| Starch degradation | 0.0173149 |
| Heme biosynthesis I | 0.0178181 |
| Heme biosynthesis from uroporphyrinogen I | 0.0195451 |
| Colanic acid building blocks biosynthesis | 0.0199615 |
| Xylose degradation I | 0.0284483 |
| Sucrose biosynthesis | 0.0303525 |
| 5-Aminoimidazole ribonucleotide biosynthesis II | 0.0332884 |
| Biotin-carboxyl carrier protein | 0.0357646 |
| Methanol oxidation to formaldehyde | 0.0437164 |
The analysis was performed with MetGenMap
The second process that was activated was the mevalonate pathway, which produces isoprenoids that are vital for diverse cellular functions; these isoprenoids include sterols, carotenoids, chlorophyll, plant hormones, and defense isoprenoids. The penetration of Penicillium probably induces defense isoprenoids such as were found in damaged plant leaves [51]. These comprise a wide variety of defense-related genes, including those that activate biosynthesis of JA and ethylene, as a possible response to pathogen penetrations, as was reported for Botrytis cinerea [52]. qRT-PCR analysis of the ethylene-responsive transcription factor 2-like showed a significant induction of this gene in the apple tissue as a result of the Penicillium infection (Table 8).
Table 8.
Relative expression (Pe/Cg) of selected genes in apple tissue infected with P. expansum or C. gloeosporioides
| Apple down-regulated genes | Apple (Pe)/apple (Cg) | Apple up-regulated genes | Apple (Pe)/apple (Cg) |
|---|---|---|---|
| Programmed cell death protein 4-like | 0.025/0.112 = 0.223 | Histone deacetylase hdt3-like | 0.70/1.21 = 0.583 |
| Auxin-repressed kda isoform x1 | 0.002/0.009 = 0.222 | Phenylalanine ammonia-lyase 1 | 9.41/3.38 = 2.781 |
| ap2-like ethylene-responsive transcription factor at2g41710 isoform x2 | 0.015/0.063 = 0.238 | Chalcone synthase | 16.33/24.23 = 0.670 |
| udp-glucose:glycoprotein glucosyltransferase | 0.104/0.291 = 0.357 | Peroxidase 47 | 98.83/10.55 = 9.368 |
| Senescence-associated carboxylesterase 101-like | 0.079/0.192 = 0.411 | Respiratory burst oxidase homolog protein d-like | 185.14/24.15 = 7.664 |
| 1-Aminocyclopropane-1-carboxylate oxidase 1 | 0.034/0.064 = 0.544 | Lipoxygenase | 490.05/205.74 = 2.381 |
| Anthocyanidin 3-o-glucosyltransferase 5-like | 0.007/0.041 = 0.169 | Indole-3-acetic acid-induced protein arg2 | 9.97/6.49 = 1.535 |
| Expansin 1 | 0.002/0.040 = 0.067 | Zinc finger an1 domain-containing stress-associated protein 12-like | 17.66/17.78 = 0.993 |
| ap2-like ethylene-responsive transcription factor at2g41710 isoform x2 | 0.012/0.045 = 0.282 | NADPH--cytochrome p450 reductase isoform x2 | 8.18/4.04 = 2.022 |
| Lysine-specific histone demethylase 1 homolog 1-like | 0.075/0.192 = 0.394 | Ethylene-responsive transcription factor 2-like | 33.81/33.83 = 0.999 |
| Serine threonine-protein kinase-like protein ccr4 | 11.12/1.83 = 6.049 |
The relative expression of the apple genes was compared with healthy tissue
The third up-regulated process was related to biosynthesis of flavonoids, which are linked to fungal potential cytotoxicity and capacity to interact with enzymes through protein complexation [53]. Some flavonoids provide stress protection, for example, by acting as scavengers of free radicals such as reactive oxygen species (ROS), as reported by Falcone Ferreyra et al. [53]. During P. expansum pathogenicity, the fungus produces gluconic acid, with H2O2 as a by-product. The host reacts to ROS by activating the flavonoids biosynthesis pathway and thereby initiating a resistance response to fungal penetration. Production of secondary metabolites such as anthocyanins, isoflavonoids, and flavonol glycosides may contribute to this resistance. Plants such as Arabidopsis respond to the combination of biotic, i.e., bacterial, and abiotic, e.g., UV-B radiation, stresses through synthesis of defense-related compounds such as phytoalexins and lignin, which serve as structural barriers that restrict the spread of pathogens. These responses modify the expression of genes involved in the production of protective metabolites such as flavonols [53]. This behavior suggests that there is a significant apple response to Penicillium penetration under these susceptible conditions. This activation matches the fourth activated pathway of P. expansum colonization, in which the geranyl-geranyl diphosphate-mediated processes are activated, probably in biosynthesis of essential compounds such as chlorophylls, carotenoids, tocopherols, plastoquinones, and gibberellins, but mainly in production of a variety of secondary metabolites. All of which indicates the significant response of apple fruits in coping with the colonization process.
Two significantly down-regulated processes that modulate carbohydrate metabolism were found: glycogen biosynthesis I from ADP-D-glucose, and starch biosynthesis. Glycogen and starch are both multibranched polysaccharides of glucose that serve as a means of energy storage for fungi; their downregulation may indicate their importance as energy stores that rapidly can be mobilized from the cytosol/cytoplasm and that perform an important function during glucose consumption by Penicillium during the acidification process [7]. Down-regulation of these processes probably results from catabolism of the substrates during attack by P. expansum and strongly supports previous findings of Prusky et al. [3], Hadas et al. [4], and Barad et al. [7] that indicate acidification of the tissue as a factor in pathogenicity of P. expansum.
In order to analyze the responses of apple fruits to fungal acidification or alkalization, the differential expressions of specific genes induced during colonization by Penicillium were compared to those expressed during colonization of the same host by Colletotrichum. For that purpose, the relative expressions of 21 specific key genes, of which 10 were up-regulated and 11 down-regulated by Penicillium (Table 8), were compared in apple RNA extracted from the leading edges of tissue colonized by the two respective pathogens. Genes that were up-regulated in the fruit showed the same response pattern when fruits were colonized by either Penicillium or Colletotrichum, but the relative expression obtained in Penicillium-inoculated apples was always significantly higher than that in Colletotrichum-inoculated apples: the ratio of P. expansum: C. gloeosporioides relative expression responses ranged from 0.58 to 9.36. This indicates a stronger host response to Penicillium colonization than to Colletotrichum colonization. Analysis of the down-regulated genes also showed similar patterns of host responses to the two respective pathogens, but lower ratios of P. expansum: C. gloeosporioides relative expressions, ranging from 0.067 to 0.54, which indicates that, although the response patterns were similar, the alkalizing pathogen induced lower expression levels. These results indicate that host responses to Penicillium and to Colletotrichum attack did not show opposite senses of gene modulation, as might be expected from their contrasting pH modulation directions, but only reduced expression levels in the Colletotrichum-inoculated apples.
Conclusions
Overall, gene-expression profiling of pH-dependent genes of P. expansum during colonization of apple fruits revealed three major effects: (1) a pattern of high gene expression during colonization that had no connection with pH response; (2) gene expression patterns during colonization similar to those obtained under in vitro growth conditions at pH 4; and (3) gene expression patterns during colonization similar to those obtained under in vitro growth conditions at pH 7. These three main trends indicate the existence of pH-regulated genes, expressed at pH 4 and pH 7 that may contribute to P. expansum colonization. One of the key processes overrepresented in cluster 7 at pH 4 was high expression of glutamate decarboxylase (GAD). One of the key products of this enzyme-GABA-largely originates from decarboxylation of L-glutamate [54], and it had been found to function in communication between tomato (Lycopersicon esculentum var. commune Bailey) plants and the fungus Cladosporium fulvum [55]. Hyphae of C. fulvum are restricted to the apoplast, therefore the fungus is dependent on the contents of the apoplastic nutrients. During infection, the GABA concentration in the apoplast increased from about 0.8 mM to 2–3 mM; this can be attributed to stimulation of glutamate dehydrogenase activity by decreased pH and increased cytosolic calcium, which are associated with pathogen attack [56]. In the present study, similar conditions were present in the Penicillium/apple interactions, where acidification by GLA accumulation may enhance the consumption of this amino acid.
Accumulation of ammonia, induced under the limited-nutrient conditions present at the edge of the decaying tissue, may contribute to gene expression, either by alkalization or by direct induction of ammonia during gene activation [8]. Modulation of ammonia levels by MepB transporters and/or the amine metabolic process (both overrepresented at pH 7 and decayed tissue in cluster 9) may contribute to the alkalizing effects and pH increase. Under these pH conditions also an over-representation of glucose-methanol-choline (GMC) oxidoreductase was observed. GMC shows similar activity to the GOX of P. expansum that contributed to oxidation and reduction processes [29] by providing the H2O2 required by ligninolytic peroxidases in Pleurotus species [57]. This combination of GMC genes [58], described here for the first time in P. expansum under alkaline conditions, indicates activity of new undescribed mechanisms that contribute to cell-wall degradation by generation of H2O2 during fungal colonization at pH 7.
In addition, the copper amino oxidase (CuAO) contributes to the enhanced accumulation of H2O2 at the wound site which, in turn, contributes to the extended necrotic lesions and extensive plant cell damage [32]. The CuAO catalyzes oxidation of aliphatic diamines of the primary amino groups that contribute to H2O2 and NH3 accumulation, which emphasizes the relevance of the H2O2-delivering system in colonized tissue [33]. This over-representation of GMC and CuAO indicates the contribution to ROS production and oxidoreductase processes at the leading edge of the developing colony, and attributes the presence of fungal catalase peroxidases the need to protect colonizing hyphae.
These oxidative processes were concurrent with activation of Cyt P-450 monoxygenase-in group 3, cluster 4-which catalyzes the conversion of hydrophobic intermediates of primary and secondary metabolic pathways, thereby detoxifying natural and environmental pollutants and allowing fungi to grow under difficult oxidative conditions [40], which accounts for the activation of genes coding for proteases, cell-wall-related enzymes, redox homoeostasis, and detoxification processes that are expressed during the infection process.
Exploiting these strong oxidative conditions during its necrotrophic development, the fungus further advances the colonization process by activation of aminocyclopropane-1-carboxylate deaminase (ACC) (cluster 9), a precursor of the plant hormone ethylene. A similar reaction process catalyzing ACC synthase was found when P. digitatum attacked citrus fruits [38, 39], which indicates that the colonization process involves induction of senescence of the colonized tissue, in conjunction with the strong oxidative process.
Out of the 771 putative CAZymes identified in P. expansum, eight were found to be expressed in clusters 7 and 9, which were the most important clusters modulating pathogenicity (Fig. 5). Given the importance of CAZymes during cell-wall degradation, it is clear that they contribute strongly to the apple maceration process. The present transcriptome analysis showed induced ammonification, and strong oxidative and senescence processes, accompanied by strong activation of pectolytic enzymes, all of which indicate the pH dependence of the tools used by the pathogen to colonize the environment. All these induced metabolic changes indicate the significant role of pH modulation in the pathogenicity of P. expansum in fruits.
Analysis of the fruit transcriptome suggested that even if apple fruits are susceptible to Penicillium, the fungus activates significant gene processes related to fruit resistance. This may indicate that the necrotrophic maceration of the tissue occurs, not as a senescence response, but as an active fungal process that supports its pathogenicity. Also interesting is the stronger response of Penicillium than of Colletotrichum, which indicates that the pH adjustment effected by the fungus did not affect the pattern but the level of host response. At the same time, these findings indicate that the fungal matching to the host is the main factor activating the maceration process.
Methods
Fungal strains and culture conditions
The WT P. expansum isolate Pe-21 was obtained from decayed apples (Malus domestica cv. Golden Delicious) purchased from a local market in Israel [4]. Cultures were grown at 27 °C in the dark, and maintained on PDA plates (Difco, MD, USA) unless otherwise indicated. Conidia were harvested with 10 mL of sterile distilled water supplemented with 0.01 % (v/v) Tween 80 (Sigma-Aldrich, Copenhagen, Denmark). Cells were visualized with a model BX60F-3 microscope and a model SZ-60 stereoscope (both from Olympus America Inc., Melville, NY, USA) and counted with a hemocytometer (Brand, GMBH, Werheim, Germany).
Assay for colonization and disease development
'Golden Delicious' apples were inoculated by placing 5 μL of a conidial suspension containing 106 spores mL−1 on each of six 2-mm-deep, 2-mm-diameter wounds spaced evenly in a circle around the upper part of the stem end of the fruits. Following inoculation, the fruits were incubated for 5 days at 25 °C in covered plastic containers containing wet paper towels. Samples of healthy tissue and of the leading edge of each wound were collected and frozen with liquid nitrogen for subsequent RNA analysis.
In vitro procedure of P. expansum
Fungal spores were inoculated at 106 spores mL−1 into 40 mL of a primary medium, i.e., glucose minimal medium, in 125-mL flasks containing (per liter) 10 g sucrose, 5 g yeast extract (Difco Laboratories, MD, USA), 50 mL nitrate salts, and 1 mL trace elements, at pH 4.5. The cultures were incubated at 25 °C with shaking at 150 rpm for 48 h. Cultures were harvested by vacuum filtration through a sterile Büchner funnel fitted with a Whatman number 1 filter paper, and the remaining mycelia were washed twice with 50 mL of sterile distilled water. The washed mycelia were resuspended in 50 mL of 0.2 M phthalate-buffered or phosphate-buffered liquid secondary medium (SM) at pH 4 or pH 7, respectively. This SM contained (per liter) 60 g sucrose, 7 g NaNO3, 3 g tryptone (Difco Laboratories, MD, USA), 1 g KH2PO4, 0.5 g MgSO4 · 7H2O, and 0.5 g KCl. The cultures were incubated at 25 °C on a rotating shaker at 150 rpm for 0.5, 1, 3, 10, or 24 h. A sample of mycelia from the cultures was collected into a 1.5-mL Eppendorf at each time point, and the mycelia were frozen with liquid nitrogen for RNA extraction.
RNA extraction
RNA was extracted from the in vitro samples with the SV Total RNA Isolation kit (Promega, Madison,WI, USA). Purity of the extracted RNA was assayed with an ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA), and the extracts were stored at −80 °C pending further analysis.
Total RNA of apple fruits was extracted according to Yang et al. [59], with minor changes: aliquots were taken from pooled samples from the leading edges of the six inoculation areas of each apple. The samples were ground to a fine powder in liquid nitrogen and transferred into 50-mL centrifuge tubes with 10 mL of CTAB RNA extraction buffer comprising 100 mM Tris-borate pH 8, 2 M NaCl, 25 mM ethylenediaminetetraacetic acid (EDTA) pH 8, 2 % (w/v) CTAB, 2 % (w/v) polyvinylpolypyrrolidone, and 2 % (v/v) β-mercaptoethanol. The mixture was shaken for 3 min and then incubated at 65 °C for 15 min. Samples were extracted twice with an equal volume of 24:1 (v:v) chloroform:isoamyl alcohol, and the phases were separated by centrifugation at 10,000 × g for 10 min. Following centrifugation, LiCl was added to a final concentration of 2.5 M and RNA was allowed to precipitate overnight at 4 °C. The precipitated RNA was pelleted at 4 °C for 30 min at 10,000 × g, washed with 70 % ethanol, and resuspended at 65 °C for 3 min in SSTE buffer comprising 10 mM Tris pH 8, 1 M NaCl, 1 mM EDTA pH 8, and 0.5 % (w/v) SDS. Samples were extracted with equal volumes of 24:1 (v:v) chloroform:isoamyl alcohol, and with equal volumes of 24:1:25 (v:v:v) chloroform:isoamyl alcohol:water-saturated phenol, and the phases were separated by centrifugation at 10,000 × g for 10 min. The RNA was ethanol-precipitated overnight, and resuspended in diethyl-pyrocarbonate-treated water. The RNA was further treated with Turbo DNAse (Ambion, Austin, TX, USA).
Preparations of libraries
A 500-ng of total RNA from 11 samples was processed with the TruSeq RNA Sample Preparation Kit v2 (RS-122-2001) (Illumina, San Diego, CA, USA). Libraries were evaluated with the Qubit and TapeStation software (Agilent Technologies, Palo Alto, CA, USA), and sequencing libraries were constructed with barcodes to enable multiplexing of pools of the eight in vitro samples-duplicates of pH 4 3 h, pool pH 4, pH 7 3 h and pool pH 7-in one lane. The results showed 14.5–32.1 million single-end 50-bp reads.
Three individual lanes were used for the in vivo samples and yielded 199.5–253.3 million single-end, 100-bp reads from the duplicates of the leading edges of infected apple tissue and 31.7 million single-end 100-bp reads from healthy apple tissue. All samples were sequenced on a HiSeq 2500 The transcriptome of the healthy apple tissue and the leading edge of infected tissue were sequenced with Trueseq protocols at the Genome Center of the Life Sciences and Engineering Faculty, Technion-Israel Inst. Technology, Haifa, Israel.
Bioinformatic analysis of RNAseq Data
Three libraries with total single-end, 100-nucleotides-long RNA-seq reads were generated from the in vivo samples and eight libraries with 50-nucleotides-long total single-end RNA-seq reads were generated from the in vitro samples. The libraries contained the following sequences: 1) duplicates of in-vitro P. expansum mycelia in pH 4 for 3 h with 21,506,394 and 26,786,825 reads, respectively; 2) duplicates of in-vitro P. expansum mycelia in pH 7 for 3 h with 16,745,224 and 32,103,824 reads, respectively; 3) duplicates of pooled samples from all time points during in vitro exposure in medium at pH 4, with 19,665,360 and 14,581,483 reads, respectively; (4) duplicates of pooled samples from all time points during in vitro exposure in medium at pH 7, with 17,324,922 and 19,406,152 reads, respectively; (5) duplicates of leading edge samples of inoculated apple tissue with 253,299,784 and 199,419,262 reads; (6) one sample of healthy apple cv. 'Golden Delicious' tissue, with 31,719,018 reads.
The datasets are available at the NCBI Sequence Read Archive (SRA) under accession number SRP071104.
The Bowtie2 softweare [14] was used to align the RNA-seq outputs against the transcriptome of P. expansum. The libraries were aligned against the P. expansum genome (downloaded from NCBI Accession no. JQFX00000000.1). The apple samples were also aligned against the Malus × domestica. Whole Genome v1.0 that was downloaded from GDR (Genome Database for Rosaceae) [60]. RSEM software [61] was used for transcript quantification of the RNAseq data and then the edgeR package [62] was used to calculate differentially expressed genes.
The genes of P. expansum were annotated by using BLASTx [63] against the non-redundant NCBI protein database, after which their GO term [64] was assigned by combining BLASTx data and interproscan analysis [65] by means of the BLAST2go software pipeline [16]. GO-enrichment was analysed by using Fisher’s Exact Test with multiple testing correction of FDR [15]. Heatmap and clustering of the genes were visualized by using the R software ggplots2 package [66]. We used a threshold of FDR < 0.05 [15] and the criterion that expression level increased or decreased by a factor greater or less than 2, respectively, i.e., greater than or less than +1 or −1, respectively, on a logarithmic (base 2) scale. The normalized expression value was centered and log 2 transformed for visualization purposes with a script taken from trinity pipeline [67]. The CAZY analysis was done with the CAZymes Analysis Toolkit (CAT) [45]. A heat map of Cazymes in each cluster was plotted by using the R package pheatmap [68].
Ammonia atmosphere analysis
To create an ammonia vapor atmosphere, 250-mL aliquots of 5 N NaOH with 0 or 50 μM of NH4Cl were placed in closed containers for 24 h before use, after which, five apple fruits were placed inside each container for 5 days. Concentrations of 0 and 50 μM NH4Cl in NaOH solutions resulted in detected ammonia concentrations of 0 and 22 μM, respectively. All presented gene-expression results are relative to 0 μM NH4Cl in 5 N NaOH.
Gene-expression analysis by qRT-PCR
Real-time qPCR was performed with the StepOnePlus System (Applied Biosystems, Grand Island, New York, USA). PCR amplification was performed with 3.4 μL of cDNA template in 10 μL of reaction mixture containing 6.6 μL of the mix from the SYBR Green Amplification Kit (ABgene, Surrey, UK) and 300 nM of primers; Additional file 10: Tables S1 and Additional file 11: Table S2 (see Supporting Information) list the forward and reverse primers for each of the indicated genes. The PCR was carried out as follows: 10 min at 94 °C, and 40 cycles of 94 °C for 10 s, 60 °C for 15 s, and 72 °C for 20 s. The samples were subjected to melting-curve analysis: efficiencies were close to 100 % for all primer pairs, and all products showed the expected sizes of 70 to 100 bp. All of the samples were normalized to 28S expression levels, and the values were expressed as the change (increasing or decreasing) of levels relative to a calibrator sample. Results were analyzed with the StepOnePlus software v.2.2.2 (Applied Biosystems, Grand Island, New York, USA). Relative quantification was performed by the ∆∆CT method [69]. The ∆CT value was determined by subtracting the CT results for the target gene from those for the endogenous control gene-18S for apple analysis and 28S for P. expansum analysis-and normalized against the calibration sample to generate the ∆∆CT values. Each experiment was performed in triplicate, and three different biological experiments were conducted. One representative set of results is presented as mean values of 2-∆∆CT ± SE for each treatment.
Availability of data and material
All data presented in this manuscript were deposited in the NCBI Sequence Read Archive (SRA) database under accession number SRP071104.
Acknowledgements
We acknowledge the financial support for D.P. from the US/Israel Binational Agricultural R&D (BARD) fund; project I-IS-4773-14. We appreciate the contributions of the Bioinformatics units of the Weizmann Institute and the Technion – Israel Institute of Technology, in preparing libraries and providing sequencing services.
Funding
We acknowledge funding by the US/Israel Binational Agricultural Research Fund, BARD, IS-4773-14. SB was funded by a scholarship of the Volcani Center, Israel.
Additional files
GO-enriched terms of genes in cluster 1. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 231 kb)
GO-enriched terms of genes in cluster 2. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 159 kb)
GO-enriched terms of genes in cluster 3. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 212 kb)
GO-enriched terms of genes in cluster 6. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 159 kb)
GO-enriched terms of genes in cluster 7. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 164 kb)
GO-enriched terms of genes in cluster 5. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 159 kb)
GO-enriched terms of genes in cluster 8. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 164 kb)
GO-enriched terms of genes in cluster 9. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 196 kb)
GO-enriched terms of genes in cluster 4. GO enrichment was calculated by using Fisher's Exact Test with blast2go software [18]. All Penicillium expansum genes served as background for the calculations. The percentages of the GO term in all P. expansum genes are marked in red, and the percentages of the GO terms in the gene clusters are marked in blue. (JPG 123 kb)
Fungal primers used in this study. (DOCX 13 kb)
Apple primers used in this research. (DOCX 15 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SB and NS analyzed the data and contributed to the chemical analyses; DV, AD, and NG contributed to specific experiments; AS and DP contributed to writing the manuscript and final analysis of the data. All authors read and approved the final manuscript.
References
- 1.Pitt JI, Hocking AD. Fungi and food spoilage. New York: Springer; 2009. Penicillium and related genera; pp. 169–273. [Google Scholar]
- 2.Jurick WM, Janisiewicz WJ, Saftner RA, Vico I, Gaskins VL, Park E, Forsline PL, Fazio G, Conway WS. Identification of wild apple germplasm (Malus spp.) accessions with resistance to the postharvest decay pathogens Penicillium expansum and Colletotrichum acutatum. Plant Breed. 2011;130(4):481–6. doi: 10.1111/j.1439-0523.2011.01849.x. [DOI] [Google Scholar]
- 3.Prusky D, McEvoy JL, Saftner R, Conway WS, Jones R. Relationship between host acidification and virulence of Penicillium spp. on apple and citrus fruit. Phytopathology. 2004;94(1):44–51. doi: 10.1094/PHYTO.2004.94.1.44. [DOI] [PubMed] [Google Scholar]
- 4.Hadas Y, Goldberg I, Pines O, Prusky D. Involvement of gluconic acid and glucose oxidase in the pathogenicity of Penicillium expansum in apples. Phytopathology. 2007;97(3):384–90. doi: 10.1094/PHYTO-97-3-0384. [DOI] [PubMed] [Google Scholar]
- 5.Yao C, Conway WS, Sams CE. Purification and characterization of a polygalacturonase produced by Penicillium expansum in apple fruit. Phytopathology. 1996;86:1160–6. doi: 10.1094/Phyto-86-1160. [DOI] [Google Scholar]
- 6.Sanchez-Torres P, Gonzalez-Candelas L. Isolation and characterization of genes differentially expressed during the interaction between apple fruit and Penicillium expansum. Mol Plant Pathol. 2003;4(6):447–57. doi: 10.1046/j.1364-3703.2003.00190.x. [DOI] [PubMed] [Google Scholar]
- 7.Barad S, Horowitz SB, Moskovitch O, Lichter A, Sherman A, Prusky D. A Penicillium expansum glucose oxidase-encoding gene, GOX2, Is essential for gluconic acid production and acidification during colonization of deciduous fruit. Mol Plant-Microbe Interact. 2012;25(6):779–88. doi: 10.1094/MPMI-01-12-0002. [DOI] [PubMed] [Google Scholar]
- 8.Barad S, Espeso EA, Sherman A, Prusky D. Ammonia activates pacC and patulin accumulation in acidic environment during apple colonization by Penicillium expansum. Mol Plant Pathol. 2015. [DOI] [PMC free article] [PubMed]
- 9.Yang C, Sudderth J, Dang T, Bachoo RG, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–93. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi F, Barad S, Ment D, Luria N, Dubey A, Casado V, et al. Carbon regulation of environmental pH by secreted small molecules that modulate pathogenicity in phytopathogenic fungi. Mol Plant Pathol. 2015. [DOI] [PMC free article] [PubMed]
- 11.Penalva MA, Lucena-Agell D, Arst HN., Jr Liaison alcaline: Pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol. 2014;22:49–59. doi: 10.1016/j.mib.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Ballester AR, Marcet-Houben M, Levin E, Sela N, Selma-Lázaro C, Carmona L, Wisniewski M, Droby S, González-Candelas L, Gabaldón T. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol Plant Microbe Interact. 2015;28(3):232–48. doi: 10.1094/MPMI-09-14-0261-FI. [DOI] [PubMed] [Google Scholar]
- 13.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
- 16.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 17.Routledge R. Fisher's exact test. 2005. [Google Scholar]
- 18.Li B, Zong Y, Du Z, Chen Y, Zhang Z, Qin G, Zhao W, Tian S. Genomic characterization reveals insights into patulin biosynthesis and pathogenicity in Penicillium species. Mol Plant-Microbe Interact. 2015;28(6):635–47. doi: 10.1094/MPMI-12-14-0398-FI. [DOI] [PubMed] [Google Scholar]
- 19.Zong Y, Li B, Tian S. Effects of carbon, nitrogen and ambient pH on patulin production and related gene expression in Penicillium expansum. Int J Food Microbiol. 2015;206:102–8. doi: 10.1016/j.ijfoodmicro.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Snini SP, Tannous J, Heuillard P, Bailly S, Lippi Y, Zehraoui E, Barreau C, Oswald IP, Puel O. The patulin is a cultivar dependent aggressiveness factor favoring the colonization of apples by Penicillium expansum. Mol Plant Pathol. 2015. [DOI] [PMC free article] [PubMed]
- 21.Prusky D, Yakoby N. Pathogenic fungi: leading or led by ambient pH? Mol Plant Pathol. 2003;4(6):509–16. doi: 10.1046/j.1364-3703.2003.00196.x. [DOI] [PubMed] [Google Scholar]
- 22.Yadav S, Yadav PK, Yadav D, Yadav KDS. Pectin lyase: a review. Process Biochem. 2009;44(1):1–10. doi: 10.1016/j.procbio.2008.09.012. [DOI] [Google Scholar]
- 23.López‐Pérez M, Ballester AR, González‐Candelas L. Identification and functional analysis of Penicillium digitatum genes putatively involved in virulence towards citrus fruit. Mol Plant Pathol. 2015;16(3):262–75. doi: 10.1111/mpp.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naumann TA, Price NP. Truncation of class IV chitinases from Arabidopsis by secreted fungal proteases. Mol Plant Pathol. 2012;13(9):1135–9. doi: 10.1111/j.1364-3703.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Punekar NS. The metabolism of 4-aminobutyrate (GABA) in fungi. Mycol Res. 1997;101(04):403–9. doi: 10.1017/S0953756296002742. [DOI] [Google Scholar]
- 26.Mead O, Thynne E, Winterberg B. Characterising the role of GABA and its metabolism in the wheat pathogen Stagonospora nodorum. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyara I, Shafran H, Kramer Haimovich H, Rollins J, Sherman A, Prusky D. Multi-factor regulation of pectate lyase secretion by Colletotrichum gloeosporioides pathogenic on avocado fruits. Mol Plant Pathol. 2008;9(3):281–91. doi: 10.1111/j.1364-3703.2007.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barad S, Horowitz SB, Kobiler I, Sherman A, Prusky D. Accumulation of the mycotoxin patulin in the presence of gluconic acid contributes to pathogenicity of Penicillium expansum. Mol Plant-Microbe Interact. 2014;27(1):66–77. doi: 10.1094/MPMI-05-13-0138-R. [DOI] [PubMed] [Google Scholar]
- 29.Iida K, Cox-Foster DL, Yang X, Ko WY, Cavener DR. Expansion and evolution of insect GMC oxidoreductases. BMC Evol Biol. 2007;7(1):75. doi: 10.1186/1471-2148-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alkan N, Davydov O, Sagi M, Fluhr R, Prusky D. Ammonium secretion by Colletotrichum coccodes activates host NADPH oxidase activity enhancing host cell death and fungal virulence in tomato fruits. Mol Plant-Microbe Interact. 2009;22(12):1484–91. doi: 10.1094/MPMI-22-12-1484. [DOI] [PubMed] [Google Scholar]
- 31.Alkan N, Meng X, Friedlander G, Reuveni E, Sukno S, Sherman A, Thon M, Fluhr R, Prusky D. Global aspects of pacC regulation of pathogenicity genes in Colletotrichum gloeosporioides as revealed by transcriptome analysis. Mol Plant Microbe Interact. 2013;26(11):1345–58. doi: 10.1094/MPMI-03-13-0080-R. [DOI] [PubMed] [Google Scholar]
- 32.Song X, She X, Yue M, Liu Y, Wang Y, Zhu X, Huang A. Involvement of copper amine oxidase (CuAO)-dependent hydrogen peroxide synthesis in ethylene-induced stomatal closure in Vicia faba. Russ J Plant Physiol. 2014;61(3):390–6. doi: 10.1134/S1021443714020150. [DOI] [Google Scholar]
- 33.Cona A, Rea G, Angelini R, Federico R, Tavladoraki P. Functions of amine oxidases in plant development and defence. Trends Plant Sci. 2006;11(2):80–8. doi: 10.1016/j.tplants.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Rea G, Metoui O, Infantino A, Federico R, Angelini R. Copper amine oxidase expression in defense responses to wounding and Ascochyta rabiei invasion. Plant Physiol. 2002;128(3):865–75. doi: 10.1104/pp.010646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pongpom P, Cooper CR, Jr, Vanittanakom N. Isolation and characterization of a catalase-peroxidase gene from the pathogenic fungus, Penicillium marneffei. Med Mycol. 2005;43(5):403–11. doi: 10.1080/13693780400007144. [DOI] [PubMed] [Google Scholar]
- 36.Miyara I, Shnaiderman C, Meng X, Vargas WA, Diaz-Minguez JM, Sherman A, Thon M, Prusky D. Role of nitrogen-metabolism genes expressed during pathogenicity of the alkalinizing Colletotrichum gloeosporioides and their differential expression in acidifying pathogens. Mol Plant-Microbe Interact. 2012;25(9):1251–63. doi: 10.1094/MPMI-01-12-0017-R. [DOI] [PubMed] [Google Scholar]
- 37.Hammond-Kosack KE, Parker JE. Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol. 2003;14(2):177–93. doi: 10.1016/S0958-1669(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 38.Chalutz E, Lieberman M. Methionine-induced Ethylene Production by Penicillium digitatum. Plant Physiol. 1977;60(3):402–6. doi: 10.1104/pp.60.3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia YJ, Kakuta Y, Sugawara M, Igarashi T, Oki N, KisAKi M, Shoji T, Kanetuna Y, Horita T, Matsui H. Synthesis and degradation of 1-aminocyclopropane-1-carboxylic acid by Penicillium citrinum. Biosci Biotechnol Biochem. 1999;63(3):542–9. doi: 10.1271/bbb.63.542. [DOI] [PubMed] [Google Scholar]
- 40.Eliasson E, Mkrtchian S, Ingelman-Sundberg M. Hormone-and substrate-regulated intracellular degradation of cytochrome P450 (2E1) involving MgATP-activated rapid proteolysis in the endoplasmic reticulum membranes. J Biol Chem. 1992;267(22):15765–9. [PubMed] [Google Scholar]
- 41.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Z, Liu H, Wang C, Xu JR. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14(1):274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcet-Houben M, Ballester AR, de la Fuente B, Harries E, Marcos JF, González-Candelas L, Gabaldón T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics. 2012;13(1):646. doi: 10.1186/1471-2164-13-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010;20(12):1574–84. doi: 10.1093/glycob/cwq106. [DOI] [PubMed] [Google Scholar]
- 46.O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Gen. 2012;44(9):1060–5. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z. Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 2009;151(4):1758–68. doi: 10.1104/pp.109.145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Biol. 1997;48(1):355–81. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- 49.Schaller F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot. 2001;52(354):11–23. doi: 10.1093/jexbot/52.354.11. [DOI] [PubMed] [Google Scholar]
- 50.Droby S, Porat R, Cohen L, Weiss B, Shapiro B, Philosoph-Hadas S, Meir S. Suppressing green mold decay in grapefruit with postharvest jasmonate application. J Am Soc Hortic Sci. 1999;124(2):184–8. [Google Scholar]
- 51.Rose USR, Manukian A, Heath RR, Tumlinson JH. Volatile semiochemicals released from undamaged cotton leaves (a systemic response of living plants to caterpillar damage) Plant Physiol. 1996;111(2):487–95. doi: 10.1104/pp.111.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134(3):1017–26. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falcone Ferreyra ML, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouche N, Fromm H. GABA in plants: just a metabolite? Trends Plant Sci. 2004;9(3):110–5. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Oliver RP, Solomon PS. Does the oxidative stress used by plants for defence provide a source of nutrients for pathogenic fungi? Trends Plant Sci. 2004;9(10):472–3. doi: 10.1016/j.tplants.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Solomon PS, Oliver RP. The nitrogen content of the tomato leaf apoplast increases during infection by Cladosporium fulvum. Planta. 2001;213(2):241–9. doi: 10.1007/s004250000500. [DOI] [PubMed] [Google Scholar]
- 57.Ferreira P, Hernandez-Ortega A, Herguedas B, Martinez AT, Medina M. Aryl-alcohol oxidase involved in lignin degradation: a mechanistic study based on steady and pre-steady state kinetics and primary and solvent isotope effects with two alcohol substrates. J Biol Chem. 2009;284(37):24840–7. doi: 10.1074/jbc.M109.011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hallberg BM, Henriksson G, Pettersson G, Divne C. Crystal structure of the flavoprotein domain of the extracellular flavocytochrome cellobiose dehydrogenase. J Mol Biol. 2002;315(3):421–34. doi: 10.1006/jmbi.2001.5246. [DOI] [PubMed] [Google Scholar]
- 59.Yang G, Zhou R, Tang T, Shi S. Simple and efficient isolation of high-quality total RNA from Hibiscus tiliaceus, a mangrove associate and its relatives. Prep Biochem Biotechnol. 2008;38(3):257–64. doi: 10.1080/10826060802164991. [DOI] [PubMed] [Google Scholar]
- 60.Jung S, Staton M, Lee T, Blenda A, Svancara R, Abbott A, Main D. GDR (Genome Database for Rosaceae): integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res. 2008;36(suppl 1):D1034–40. doi: 10.1093/nar/gkm803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Consortium GO. Gene Ontology annotations and resources. Nucleic Acids Res. 2013;41(D1):D530–5. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37(suppl 1):D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Team RDC . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 67.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kolde R. Pheatmap: pretty heatmaps. 2012. p. 61. [Google Scholar]
- 69.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki R, Shimodaira H. Hierarchical clustering with P-values via multiscale bootstrap resampling. 2013. [Google Scholar]
- 71.Dahl DB. Model-based clustering for expression data via a Dirichlet process mixture model. 2006. pp. 201–18. [Google Scholar]
- 72.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19(2):295–6. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]


