Abstract
The social amoeba Dictyostelium discoideum is unusual among eukaryotes in having both unicellular and multicellular stages. In the multicellular stage, some cells, called sentinels, ingest toxins, waste and bacteria. The sentinel cells ultimately fall away from the back of the migrating slug, thus removing these substances from the slug. However, some D. discoideum clones (called farmers) carry commensal bacteria through the multicellular stage, while others (called non-farmers) do not. Farmers profit from their beneficial bacteria. To prevent the loss of these bacteria, we hypothesize that sentinel cell numbers may be reduced in farmers, and thus farmers may have a diminished capacity to respond to pathogenic bacteria or toxins. In support, we found that farmers have fewer sentinel cells compared with non-farmers. However, farmers produced no fewer viable spores when challenged with a toxin. These results are consistent with the beneficial bacteria Burkholderia providing protection against toxins. The farmers did not vary in spore production with and without a toxin challenge the way the non-farmers did, which suggests the costs of Burkholderia may be fixed while sentinel cells may be inducible. Therefore, the costs for non-farmers are only paid in the presence of the toxin. When the farmers were cured of their symbiotic bacteria with antibiotics, they behaved just like non-farmers in response to a toxin challenge. Thus, the advantages farmers gain from carrying bacteria include not just food and protection against competitors, but also protection against toxins.
Keywords: toxin resistance, Dictyostelium, bacteria, symbiosis, farmers, innate immunity
1. Introduction
Many hosts derive a defensive benefit from symbiotic associations. These beneficial interactions between symbionts are called ‘defensive mutualisms’, a term first coined to describe protection from predation by herbivores on plants in fungus–plant mutualisms [1]. Since then, examples of microbial symbionts protecting hosts from predation and pathogens to enhance fitness have been reported in many systems. For example, the facultative bacterial symbiont Hamiltonella protects aphids against parasitoid wasps [2]. Antibiotic producing bacteria carried by fungus-growing ants protect their fungal crop from attack by the parasitic fungus Escovopsis [3]. Streptomyces bacteria protect digger wasp larvae from fungal attack [4]. Other examples include triterpene glycosides produced by Caribbean reef sponges to discourage predation by reef fish [5], and toxic secondary compounds produced by lichens to avoid predation by beetles [6]. Symbionts protecting hosts against toxins have been reported less often, but examples include detoxification of specific compounds by mammalian gut microbiota [7,8], and Burkholderia symbionts detoxifying compounds for both insect and fungal hosts [9,10].
In the soil environment, many different organisms are close together, so host defence may be particularly important. Under such dense conditions, organisms are at great risk of encountering threats such as toxins and pathogens. One organism found in soil and leaf detritus is the eukaryote social amoeba Dictyostelium discoideum. In favourable conditions, D. discoideum amoebae consume prey bacteria and increase in number by binary fission. However, if prey bacteria become scarce, D. discoideum amoebae enter a multicellular social stage, and aggregate by the tens of thousands [11]. This aggregate becomes a motile slug that moves towards heat and light. During this migration, exposure to toxins or bacterial pathogens can occur and compromise survival by limiting spore production. After migration, the slug reorganizes and ultimately becomes a structure known as a fruiting body. The fruiting body consists of a sorus containing fertile spores held aloft by a dead stalk composed of cells that have died to support the others. Spores formed during this stage are more resistant to environmental threats than the amoebae are [11].
Recent research into how D. discoideum slugs protect themselves against toxins and pathogens discovered a new kind of cell that has immune-like phagocyte activity [12]. These specialized cells are called sentinel cells, and can sequester harmful toxins and pathogens in large vesicles within the sentinel cell. After collecting toxins and/or pathogens, these cells are left behind in the discarded slug sheath as the slugs move. As sentinel cells are sloughed off in the slug trail, new ones take their place in the slug, so sentinel cell numbers appear to remain constant in the slug at about 1% of the cell population. Sentinel cells do not increase in number even with increased toxic assault [12]. Sentinel cells thus function to sequester and remove harmful contaminants before final culmination as a fruiting body. These cells carry away contaminants, both toxins and pathogens, from the presumptive spore population, and can be compared with mammalian immune cells, such as neutrophils and macrophages [13], or liver cells [14].
In most wild D. discoideum clones, the multicellular fruiting body is effectively purged of all bacteria, presumably by the sentinel cell system. However, about one-third of the clones retain some bacteria in the final fruiting body stage and exhibit a primitive form of agriculture [15]. These clones, called farmers, engage in husbandry of their farmer-associated bacteria by not consuming all available preferred prey bacteria, but instead saving some to carry through the dispersal stage to seed new food populations. This ability provides a distinct advantage if edible bacteria are lacking at the new site. However, it comes with the cost of reduced spore production compared with clones that do not carry bacteria (non-farmers), so they are less successful if edible bacteria are already abundant at the new site [15].
Farmers also carry non-food bacteria such as Burkholderia and Pseudomonas with defensive capabilities against non-farmer competitors [16,17]. Non-farmers (but not farmers) are harmed when farmers and non-farmers are mixed as amoebae and allowed to complete the social stage as evidenced by reduced spore production. We showed the inhibition (or harm) of non-farmer spore production is due to molecules secreted by farmer-associated Burkholderia [16]. Stallforth et al. also reported one strain of farmer-associated Pseudomonas fluorescens produces a novel small molecule, chromene, a polycyclic aromatic compound broadly similar to ethidium bromide (EtBr), which again decreases non-farmer spore production and dramatically increases host farmer spore production [17]. Additionally, two clades of the non-food bacterium Burkholderia appear to induce farming; farmers cured of Burkholderia no longer carry food bacteria, and non-farmers exposed to certain clones of Burkholderia can carry food bacteria [18].
Sentinel cells function to clear toxins and bacteria during the multicellular social stage. However, farmers carry host-associated bacteria during this same stage. This suggests a possible conflict between the need to carry beneficial bacteria and the mechanisms for clearing detrimental bacteria. Consequently, we hypothesize that in order to carry bacteria, farmers have a diminished ability to respond to pathogens or toxins compared with non-farmers, which would be shown by fewer sentinel cells. To test this, we investigated features of sentinel cells in farmers and non-farmers. We found that farmers have fewer sentinel cells than non-farmers and that bacteria in farmers augment the function of sentinel cells.
2. Material and methods
(a). Wild Dictyostelium discoideum isolates and culture conditions
We used farmer and non-farmer D. discoideum clones collected at either Mountain Lake Biological Station in Virginia (N 37°21′, W 80°31′), Houston Arboretum in Texas (N 29°46′, W 95°27′) or Lake Itasca in Minnesota (N 47°22′, W 95°21′) for our study. Electronic supplementary material, table S1 details specific clones used in each assay. We plated wild clones from spores on nutrient agar Petri plates (2 g glucose, 2 g Oxoid bactopeptone, 2 g Oxoid yeast extract, 0.2 g MgSO4, 1.9 g KH2PO4, 1 g K2HPO4 and 15.5 g agar per litre DDH2O) in association with Klebsiella pneumoniae at room temperature (22°C). We prepared our bacterial food stock by growing overnight cultures of K. pneumoniae in liquid Luria broth (10 g tryptone, 5 g Oxoid yeast extract and 10 g NaCl per litre DDH2O) shaking in a 25°C incubator.
(b). Preparation of ethidium bromide-stained sentinel cells
We adapted our methods from Chen et al. [12] to prepare and produce sentinel cells stained with EtBr. We varied the concentration of EtBr from the original method and determined that non-nutrient agar Petri plates (0.198 g KH2PO4, 0.0356 g Na2HPO4 and 15.5 g agar per litre DDH2O) containing 1.5 µg ml−1 EtBr allowed migration across the Petri plate with minimal amoebae death while sufficiently staining the sentinel cells for quantification. We used 10 farmers and 10 non-farmers for these two assays (electronic supplementary material, table S1). The 10 farmers carried five different host-associated bacteria species as well as the laboratory food bacterium K. pneumoniae (electronic supplementary material, table S2). To prepare the migration plates, we embedded glass slides under a thin layer of EtBr non-nutrient agar on top of partially filled and solidified EtBr non-nutrient agar in a 150 × 15 mm Petri plate. To set up the migration plates, we first prepared a concentrated stationary-phase liquid bacteria suspension by centrifuging an overnight culture of K. pneumoniae at 10 000g for 5 min at 4°C. We resuspended the bacterial pellet in a small amount of non-nutrient buffer by vortexing. Next, we determined its optical density (OD) with a BioPhotometer (Eppendorf, New York) and diluted the solution to an OD A600 of 35.0 in starvation buffer so that the bacterial concentration would be constant across treatments. We collected D. discoideum spores from each clone in starvation buffer, and determined spore concentration with dilution using a haemacytometer and a light microscope. We mixed 1.0 × 105 spores in 50 µl of the prepared stationary-phase liquid bacteria suspension. We deposited the spore suspensions in a line on one side of each plate and allowed the liquid in the spore suspension to absorb into the agar (figure 1). Next, we wrapped the plates in foil with a small hole in the foil directly opposite the line of deposited spores and placed the plates in a lighted incubator (22°C). Under these conditions, spores hatch and proliferate until the bacteria are consumed, and then starvation induces the start of the social stage. During the social stage, slugs can move. They are phototrophic and migrate across the agar towards the pinhole of light. We allowed the slugs to migrate towards the light for about 72 h before data collection.
Figure 1.
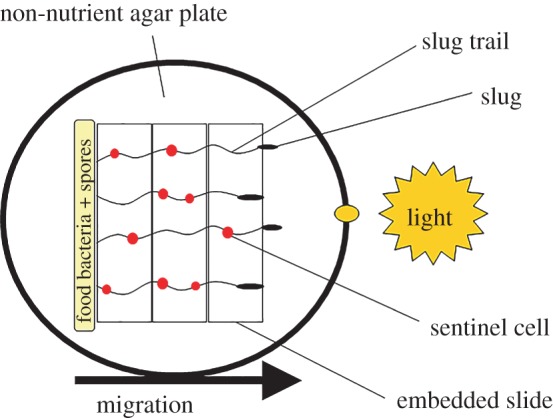
Cartoon of D. discoideum slug migration. For each clone, we deposited spore and bacteria suspensions in a line on one side of each non-nutrient agar plate containing EtBr, a toxin that is taken up by sentinel cells. We allowed phototropic slugs to form and migrate towards a light source across embedded slides to facilitate imaging of slug trails. (Online version in colour.)
(c). Visualizing sentinel cells
After the slugs migrated, we picked up the slides containing the slug trails that they had moved across. We used a Nikon A1Si laser scanning confocal microscope (Nikon, Tokyo) to image the slides under UV light (Texas Red filter) and 10× magnification. Individual images (5–6) of each slug trail were taken and used to construct a composite image of the slug trails using the image-manipulation program Gimp (v. 2.6; GNU Image-Manipulation Program, Public Domain). We counted the number of EtBr-stained sentinel cells (the cells which fluoresced most brightly and appeared rounded) found along five individual slug trails for each clone. Next, we used ImageJ64 (NIH, Maryland) to measure the length of each selected trail. The sentinel cell assay was conducted blind in the sense that farmer/non-farmer status for each wild isolate was revealed only after data collection was completed. Then, we compared the number of sentinel cells sloughed off per millimetre of slug trail by farmer slugs to the number sloughed off by non-farmers. The number of sentinel cells counted in individual slug trails and slug trail length are tabulated in electronic supplementary material, table S3.
(d). Spore production assay and spore viability under toxic conditions
We tested spore production in the presence of EtBr to examine the impact of toxic conditions during the social stage. We used seven farmer clones and seven non-farmers (electronic supplementary material, table S1). The seven farmers carried four different host-associated bacteria species as well as the laboratory food bacterium K. pneumoniae (electronic supplementary material, table S2). To obtain log-growth amoebae for the assay, we individually collected spores in starvation buffer from stock plates of our test clones. Then we plated 2 × 105 spores in 200 µl K. pneumoniae suspension in starvation buffer at an OD of 1.5 A600 on nutrient agar plates. We previously determined that spore germination and amoebae log growth occurs at about 32–36 h after plating with this plating regime. When clones reached log-phase growth, we added ice-cold starvation buffer to the plate to collect the amoebae. Next, we centrifuged the collected suspension of amoebae and bacteria at 1500g for 3 min to wash the amoebae free from bacteria. We washed the pelleted amoebae again using an excess volume of ice-cold starvation buffer three to four times depending on the amount of uneaten bacteria still present. We determined the density of washed amoebae with dilution using a haemacytometer.
To test spore production in a toxic environment, we used two conditions: (i) starvation agar plates containing 15 µg ml−1 EtBr as our toxic environment or (ii) starvation agar plates without EtBr as a control. We laid 13 mm AABP 04700 black filter squares equidistant on agar plates in duplicate for each clone in each condition. Next, we spotted the filters individually with 1.25 × 106 amoebae in starvation buffer. We allowed the clones to hatch, grow and develop under direct light to limit potential movement of slugs before final culmination to fruiting bodies. The social stage was complete for all clones after about 24 h. We allowed the spores to mature in the fruiting bodies for an additional 24–48 h before collection. At that point, we collected each filter by placing the filter in a 1.5 ml conical Eppendorf tube containing 1 ml starvation buffer + 0.1% NP-40 alternative. We vortexed each Eppendorf Tube briefly to evenly disperse the spores and counted without dilution to determine density by counting spores using a haemacytometer and a light microscope. We performed two technical replicates.
After determining spore density as above, we made serial dilutions of the EtBr-treated spores in non-nutrient buffer to determine spore viability under toxic conditions. We then plated 100 spores in association with K. pneumoniae on 10 nutrient agar plates. We determined the total number of hatched spores by counting plaques 2–3 days after plating.
(e). Preparation of cured farmers and non-farmers and spore production assay under toxic conditions
The five farmers carry farmer-associated Burkholderia Clade 2 bacteria [18] similar by 16S rDNA to Burkholderia xenovorans LB400 (98–99% identity; electronic supplementary material, table S2) as well as food bacteria K. pneumoniae. We removed (cured) farmer-associated bacteria from five farmers and five non-farmers (electronic supplementary material, table S1). Although non-farmers do not carry bacteria, we included the non-farmers with the farmers when treating with antibiotics to remove symbiotic bacteria (curing) as a control for the curing process. We treated all 10 clones by growing them on nutrient agar containing 0.1 g ampicillin and 0.3 g streptomycin sulfate per litre of nutrient agar and used heat-killed bacteria as food as reported by Brock et al. [15], with two exceptions: (i) K. pneumoniae was substituted for Escherichia coli for the dead food bacteria; and (ii) we performed one round of treatment with antibiotics instead of two rounds [15]. After the social stage was complete, we affirmed that the D. discoideum clones no longer carried bacteria by placing 10 individual sori (spore masses), collected with a sterile, filtered pipette tip, from each treated clone on nutrient agar, and subsequently observed no bacterial growth (spot tests) [15]. Only the five untreated farmers contained bacteria in their sori and were 100% positive for bacteria in the tested sori (electronic supplementary material, figure S1). We also performed PCR using Burkholderia-specific 16S primers on sori from the cured and uncured farmer and non-farmer clones. All treated clones and untreated non-farmers were negative for Burkholderia; all untreated farmers were positive [19]. As an additional control, we plated K. pneumoniae and the farmer-associated Burkholderia Clade 2 strains on nutrient agar Petri plates with or without ampicillin/streptomycin to verify that these antibiotics killed the bacteria. We observed no bacterial growth on the nutrient agar plates with antibiotics while bacterial growth was normal on the control plates. We therefore had five untreated farmers, five cured farmers, five untreated non-farmers and five non-farmers that had gone through the curing process. We performed the spore production assay as above with one replicate.
(f). Statistical analyses
We analysed our data using a generalized linear mixed model with fixed effects (farmer and non-farmer) and a random effect (clone). Standard error and F-statistics were Kenward–Roger-corrected so that degrees of freedom are approximated using variances and correlations in the observed data [20]. Specifics of each analysis are found in the results section. We used SAS software (v. 9.2 for Windows) to analyse our data.
3. Results
In order to determine the differences in immune-like phagocyte activity between farmers and non-farmers, we compared the numbers of sentinel cells visible in the slug trails generated by farmer and non-farmer clones. For this experiment, we used a population of 10 farmers carrying five different species of farmer-associated bacteria (based on 16S rDNA sequence) as well as the laboratory food bacteria, and 10 non-farmers (electronic supplementary material, tables S1 and S2). Figure 1 shows a schematic cartoon of the experimental design. Cells left behind as the slugs moved were clearly visible in the slug trails under high magnification (figure 2a). Sentinel cells readily took up the fluorescent toxin EtBr [12] and had a distinctly rounded appearance. Thus, the brightness of these cells, when imaged under an ultraviolet light filter, allowed them to be distinguished from other cells present in slug trails.
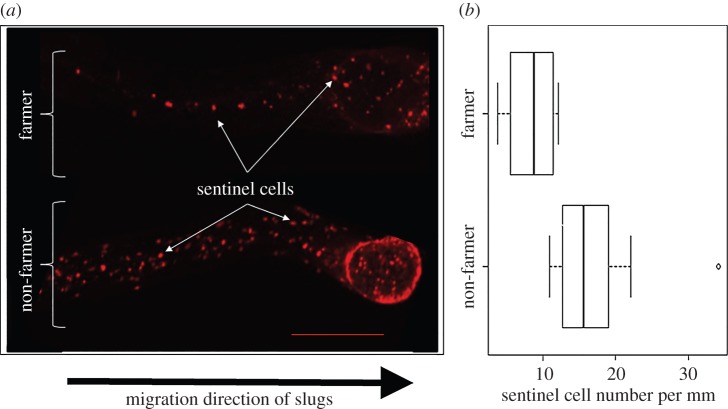
Figure 2.
Sentinel cell number varies with farmer/non-farmer status. (a) Fluorescent images of sentinel cells in D. discoideum slug trails. Sentinel cells are visible as bright red to orange round spots when stained with ethidium bromide (EtBr) and illuminated for imaging. Representative examples of EtBr-stained slug trails from one random farmer and one random non-farmer. Scale bar is 250 µm. (b) Farmers have fewer sentinel cells compared with non-farmers. We counted and averaged the number of sentinel cells found in five slug trails for each of 10 farmers and 10 non-farmers. We found farmers have significantly fewer sentinel cells than non-farmers (F1,18 = 13.04, p = 0.002), as displayed in this box plot.
We counted the number of sentinel cells found in five slug trails for each farmer and non-farmer clone, and then we calculated the average number of sentinel cells per millimetre of slug trail for each clone (electronic supplementary material, table S3). We tested whether sentinel cell number varies by farmer status by analysing these overall average numbers of sentinel cells per millimetre for each farmer and each non-farmer clone using a generalized linear mixed model with fixed effects (farmer and non-farmer) and random effect (clone). We found farmers have significantly fewer sentinel cells compared with non-farmers (farmer F1,18 = 13.04, p = 0.002). We found the overall average numbers of sentinel cells per millimetre for farmers and non-farmers to be 8.78 ± 0.97 cells mm−1 and 17.3 ± 2.16 cells mm−1, respectively (figure 2b; for individual clones, see electronic supplementary material, figure S2).
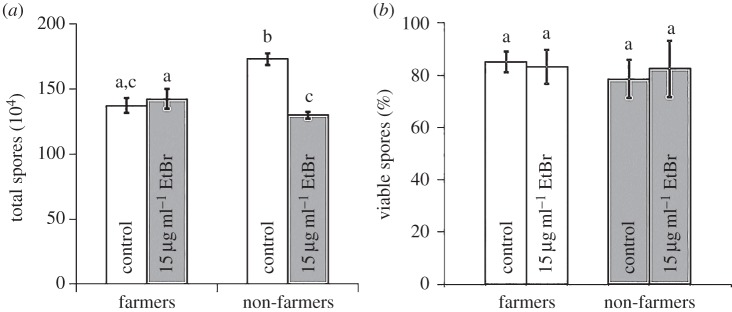
Fewer sentinel cells may impair a slug's ability to defend against toxins and/or pathogens, generating a fitness cost. To address this possibility, we determined spore production and spore viability. For these tests, we used seven farmers with four different species of farmer-associated bacteria as well as the laboratory food bacterium, and seven non-farmers (electronic supplementary material, tables S1 and S2). We determined farmer and non-farmer spore production by allowing clones to complete the social stage in the presence of EtBr as our selected toxin or in non-nutrient buffer as our control, repeating the entire experiment with the same clones at two different times. We tested whether spore production varies by farmer status and/or the presence of EtBr. We found significant main and interaction effects (generalized linear mixed model: farmer F1,12 = 10.36, p = 0.0074; EtBr F1,12 = 51.41, p≤ 0.0001; farmer × EtBr F1,12 = 93.31, p ≤ 0.0001). Differences among the four treatments are indicated by letters found in figure 3a, which reflect results of a post hoc Tukey's HSD test. Control non-farmers produced more spores than control farmers, as previously reported [15]. We assume this reflects a cost to the farmers of carrying and supporting bacteria. From these baselines, EtBr had different effects. We found non-farmer spore production was reduced from their baseline under toxic conditions, while, surprisingly, spore production for farmers did not change from their baseline, despite their lower number of sentinel cells.
Figure 3.
Farmers are unharmed under toxic conditions and spores produced remain equally viable as under control conditions. (a) Non-farmer spore production is significantly reduced in toxic conditions, but farmer spore production is unchanged. (b) Importantly, farmer and non-farmer spores, whether untreated or treated with a toxin during development, remain equally viable. Significant differences in spore production or spore viability are indicated by different letters, which reflect results of a post hoc Tukey's HSD test. Error bars show s.e.m.
Although farmer spore production is unchanged, potential harm from the toxin could be masked if the farmer spores are less viable than spores produced by non-farmers. We tested for this and found it was not the case. Spore viability did not differ between farmers and non-farmers whether grown in toxic (EtBr) conditions or under control (non-nutrient) conditions (generalized linear mixed model: farmer F1,12 = 0.10, p = 0.7613; EtBr F1,12 = 0.01, p = 0.9070; farmer × EtBr F1,12 = 0.24, p = 0.6344), as shown in figure 3b.
To determine if the host-associated farmer bacteria could be augmenting the function of sentinel cells with regard to protection against toxic chemicals for farmers, we removed host-associated bacteria from farmers by treating with antibiotics (curing). We used five uncured farmers, each carrying a farmer-associated bacteria isolate most similar to B. xenovorans LB400 by 16S rDNA (the Burkholderia Clade 2 of DiSalvo et al. [18]); the same five farmers, but deprived of their host-associated bacteria by curing; five uncured non-farmers; and the same five non-farmers also treated with antibiotics as a control for the curing process (electronic supplementary material, table S1). We tested whether spore production varies by farmer status and/or antibiotic treatment (curing) when clones are exposed to a toxin (EtBr). The analysis of variance showed strongly significant effects for EtBr treatment and most interactions, and no effect on spore production of farmer status or curing (generalized linear mixed model: farmer F1,24 = 0.30, p = 0.5970; EtBr F1,24 = 111.28, p ≤ 0.0001; farmer × EtBr F1,24 = 30.98, p ≤ 0.0001; cured F1,24 = 1.11, p = 0.3021; farmer × cured F1,24 = 0.44, p = 0.5147; cured × EtBr F1,24 = 8.80, p = 0.0067; farmer × cured × EtBr F1,24 = 8.14, p = 0.0088). Interestingly, we found the farmer's protection from toxins is lost when Burkholderia Clade 2 farmer-associated bacteria are removed (figure 4). As before, uncured farmer spore production remains unchanged in toxic versus non-toxic conditions, but cured farmers produce fewer spores in toxic conditions, like the uncured or cured non-farmers.
Figure 4.
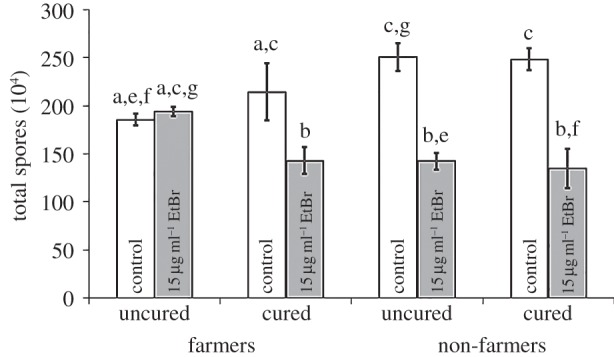
Protection against the toxic effect of EtBr is lost when farmer-associated bacteria are removed. We compared spore production of five farmer clones either with (uncured) or without (cured) their farmer-associated bacteria to five non-farmers treated under the same conditions as a control. These clones were grown with or without EtBr (toxin). Spore production for cured farmers is reduced to the same level of harm as seen for non-farmers (either cured or uncured). The curing process had no significant effect on non-farmer spore production in either condition. Significant differences in spore production between conditions are indicated by different letters, which reflect results of a post hoc Tukey's HSD test. Error bars show s.e.m.
4. Discussion
Dictyostelium discoideum's sentinel cells sequester and remove pathogenic bacteria and toxins in the multicellular social stage, performing functions analogous to vertebrate macrophages and liver cells. This trait could be disadvantageous to farmers because it might compromise their ability to retain the helpful bacteria that they need to carry through the social stage before colonizing a new site [15]. We therefore predicted farmers would have fewer sentinel cells than non-farmers. We confirmed this prediction, so we further predicted that the reduction in sentinel cells would cause farmers to have reduced ability to cope with toxins. But surprisingly, their deficit in sentinel cell production does not appear to impair or impact the ability of farmers to produce viable spores in a toxic environment. This is in marked contrast to non-farmers, who suffer significant harm in the same environment despite having more sentinel cells. Thus, something is preventing the farmers from being harmed by EtBr, so we hypothesized that the farmer-associated bacteria compensate (and in fact over-compensate) for the fact that farmers have fewer sentinel cells, at least with respect to this toxin. In support of this hypothesis, we found that removal of farmer-associated bacteria caused the farmers to lose their protection against toxins, evidenced by reduced spore production, just like non-farmers. Their spore production increased in the absence of toxin to the level of non-farmers (presumably because they no longer pay a cost of supporting the bacteria), but their spore production was now harmed by EtBr. We have previously reported that non-food bacteria carried by farmers benefit their host by protecting against competition from non-farmers [16,17]. Here our findings suggest an additional role for non-food farmer-associated bacteria: enhancing the capacity of farmers to protect against toxins.
How could farmer-associated bacteria supplement the sentinel cell protection role of removing toxins and pathogenic bacteria during the multicellular, social stage? In this study, we used EtBr as our toxin to challenge farmer and non-farmer clones. EtBr is a polycyclic, aromatic compound with a phenanthridine core [21]. EtBr binds nucleic acids, and one hypothesis is that the carried bacteria simply bind up the EtBr. An alternative is that the EtBr is degraded and even used as a carbon source, as has been reported for other members of Betaproteobacteria and Gammaproteobacteria [22], to which the carried bacteria studied here belong. The farmers used in the spore production and viability assays carry four different species of farmer-associated bacteria, each of which is capable of degrading polycyclic aromatic hydrocarbons and using these reduced compounds as potential sources of carbon and/or energy: Burkholderia Clade 2 [18] similar by 16S rDNA to B. xenovorans [23], Burkholderia Clade 1 [18] similar by 16S rDNA to B. fungorum [24], Stenotrophomonas maltophilia [25] and P. fluorescens [26]. Therefore, the ability of these farmer-associated bacteria to degrade aromatic compounds taken together with the evidence presented here suggests that farmer-associated bacteria may be augmenting the protection role of sentinel cells for farmers.
Cases of symbiotic bacteria detoxifying compounds for their hosts have been reported across other taxa and may represent a general phenomenon. In one example, the rumen microbiota of some cattle, sheep and goats enabled them to forage successfully on a legume, Leucaena leucocephala, containing the toxic acid mimosine [7]. The microbiota of these ruminants degrade toxic intermediates (dihydroxypyridines) from mimosine metabolism and interestingly can be transferred to naive ruminants to confer resistance. In another report using mice with and without antibiotic treatment to suppress gut flora, the author found bacteria in the gut can rapidly degrade methylmercury, a potent neurotoxin, by demethylation to elemental mercury and mercuric ions [8]. Inorganic mercuric mercury is poorly absorbed by the body and quickly eliminated by faeces, relieving the body burden by greatly reducing neurotoxicity.
Two recent studies report a beneficial interaction between hosts and their symbiont Burkholderia bacteria similar to the detoxification we reported in our system. One of these studies describes insecticide-degrading bacterial symbionts of stinkbugs [9]. These mutualistic gut symbionts of the genus Burkholderia can confer resistance to Fenitrothion, a common organophosphorus insecticide used worldwide, immediately upon infection of the nymphal host. These symbiotic bacteria break down the insecticide and use the final metabolite as a carbon source for their growth while simultaneously enhancing the growth and size of their host. In another study, Nazir et al. found that the soil bacterium Burkholderia terrae BS001 can protect several types of colonized soil fungi partners from antifungal agents such as the metabolite cyclohexamide produced by P. fluorescens strain CHAO [10]. Protection provided by the bacteria to the fungal mycelia could be from detoxification of antifungal metabolites, physical shielding of the fungal hyphae, or both.
Although detoxification mutualisms do not seem to have been very widely reported, they could nevertheless be very important. It has been suggested that the endosymbiosis leading to mitochondria might have originated as a detoxification mutualism [27]. With the rise in environmental O2, anaerobes to which it is toxic could gain protection by harbouring aerobic bacteria that mop up oxygen.
How toxic the soil environment is for D. discoideum or other protists seems to have been little studied. However, bacteria, including soil bacteria, are notorious for production of secreted compounds that are toxic to other bacteria, including various antibiotics and bacteriocins. It would be very surprising if they did not sometimes use parallel means to protect themselves from common predators, and toxin release has been proposed as a major mechanism of defence against protists [28]. This probably includes toxins encoded by phage carried by bacteria, including shigatoxin and diphtheria toxin [29–31]. An unusually well-studied system is P. fluorescens, which is one of the four species we studied. It secretes a variety of extracellular compounds, including the antibiotics 2,4-diacetylphloroglucinol, pyoluteorin and pyrrolnitrin, an extracellular protease (AprA), and the volatile compound hydrogen cyanide, which have profound effects on several protists [32,33]. Fungi also produce many toxic secondary metabolites [34]. Soil, and particular forest soil, also contains polycyclic aromatic compounds from both human (e.g. fossil fuels) and natural sources (e.g. wildfires [35,36]), and various bacteria are good at breaking them down [37]. Finally, that some such compounds are natural dangers to D. discoideum is supported by the fact that it has evolved a complex mechanism—sentinel cells—for their removal. Sentinel cells appear to be a somewhat general defence, removing EtBr, acridine orange and Hoecht 3, three polycyclic aromatic compounds with rather different structures [12].
Here, we showed farmer-associated bacteria protect the host farmer from deleterious effects caused by the toxin EtBr. These farmer-associated bacteria may behave similarly towards other toxins, particularly those with polycyclic aromatic hydrocarbons. But this needs further testing, particularly given the possibility that protection from EtBr, but not most other toxins, could come from binding to bacterial nucleic acids. In this study, we have not tested whether the farmer-associated bacteria enhance immune function against pathogens, although examples of bacteria aiding the immune function of their hosts have been reported in other systems [38,39]. Using this eukaryote Dictyostelium farmer/host-associated bacteria system could provide a simple platform to elucidate the precise mechanisms of defence and immune action. Overall, the data we describe here broaden the scope of advantages farmers gain from carrying bacteria to include not just food and protection against competitors, but also a new function: protection against toxins.
Supplementary Material
Acknowledgements
Many thanks to the Strassmann/Queller laboratory group for useful discussions. Special thanks to Dianne Duncan at the Washington University Core Biology Imaging Facility for invaluable help.
Data accessibility
The sequences reported in this paper have been deposited in the GenBank database (accession nos. KR607503–KR607504, KR607510–KR607512) and data of figure 2 are found in electronic supplementary material, table S3. Data from figures 3 and 4 can be found in the Dryad digital depository (http://dx.doi.org/10.5061/dryad.1cc85).
Authors' contributions
D.A.B., W.É.C., D.C.Q. and J.E.S. designed the experiments, discussed the results, analysed the data and wrote the manuscript. D.A.B. and W.É.C. performed the experiments.
Competing interests
We have no competing interests.
Funding
This material is based upon work supported by the National Science Foundation under grant nos. NSF DEB1146375 and NSF IOS 1256416, and the John Templeton Foundation grant no. 43667.
References
- 1.Clay K. 1988. Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology 69, 10–16. ( 10.2307/1943155) [DOI] [Google Scholar]
- 2.Oliver KM, Moran NA, Hunter MS. 2006. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. Lond. B 273, 1273–1280. ( 10.1098/rspb.2005.3436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie CR, Scott JA, Summerbell RC, Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398, 701–704. ( 10.1038/19519) [DOI] [Google Scholar]
- 4.Kaltenpoth M, Göttler W, Herzner G, Strohm E. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15, 475–479. ( 10.1016/j.cub.2004.12.084) [DOI] [PubMed] [Google Scholar]
- 5.Kubanek J, Pawlik JR, Eve TM, Fenical W. 2000. Triterpene glycosides defend the Caribbean reef sponge Erylus formosus from predatory fishes. Mar. Ecol. Progr. Ser. 207, 69–77. ( 10.3354/meps207069) [DOI] [Google Scholar]
- 6.Nimis PL, Skert N. 2006. Lichen chemistry and selective grazing by the coleopteran Lasioderma serricorne. Environ. Exp. Bot. 55, 175–182. ( 10.1016/j.envexpbot.2004.10.011) [DOI] [Google Scholar]
- 7.Allison MJ, Hammond AC, Jones RJ. 1990. Detection of ruminal bacteria that degrade toxic dihydroxypyridine compounds produced from mimosine. Appl. Environ. Microbiol. 56, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowland IR. 1988. Interactions of the gut microflora and the host in toxicology. Toxicol. Pathol. 16, 147–153. ( 10.1177/019262338801600207) [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 2012. Symbiont-mediated insecticide resistance. Proc. Natl Acad. Sci. USA 109, 8618–8622. ( 10.1073/pnas.1200231109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazir R, Tazetdinova DI, van Elsas JD. 2014. Burkholderia terrae BS001 migrates proficiently with diverse fungal hosts through soil and provides protection from antifungal agents. Front. Microbiol. 5, 598 ( 10.3389/fmicb.2014.00598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessin RH. 2001. Dictyostelium—evolution, cell biology, and the development of multicellularity. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 12.Chen G, Zhuchenko O, Kuspa A. 2007. Immune-like phagocyte activity in the social amoeba. Science 317, 678–681. ( 10.1126/science.1143991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124, 783–801. ( 10.1016/j.cell.2006.02.015) [DOI] [PubMed] [Google Scholar]
- 14.Racanelli V, Rehermann B. 2006. The liver as an immunological organ. Hepatology 43, S54–S62. ( 10.1002/hep.21060) [DOI] [PubMed] [Google Scholar]
- 15.Brock DA, Douglas TE, Queller DC, Strassmann JE. 2011. Primitive agriculture in a social amoeba. Nature 469, 393–396. ( 10.1038/nature09668) [DOI] [PubMed] [Google Scholar]
- 16.Brock DA, Read S, Bozhchenko A, Queller DC, Strassmann JE. 2013. Social amoeba farmers carry defensive symbionts to protect and privatize their crops. Nat. Commun. 4, 2385 ( 10.1038/ncomms3385) [DOI] [PubMed] [Google Scholar]
- 17.Stallforth P, Brock DA, Cantley AM, Tian X, Queller DC, Strassmann JE, Clardy J. 2013. A bacterial symbiont is converted from an inedible producer of beneficial molecules into food by a single mutation in the gacA gene. Proc. Natl Acad. Sci. USA 110, 14 528–14 533. ( 10.1073/pnas.1308199110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiSalvo S, Haselkorn TS, Bashir U, Jimenez D, Brock DA, Queller DC, Strassmann JE. 2015. Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. Proc. Natl Acad. Sci. USA 112, E5029–E5037. ( 10.1073/pnas.1511878112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brock DA, Jones K, Queller DC, Strassmann JE. 2016. Which phenotypic traits of Dictyostelium discoideum farmers are conferred by their bacterial symbionts? Symbiosis 68, 39–48. ( 10.1007/s13199-015-0352-0) [DOI] [Google Scholar]
- 20.Kenward MG, Roger JH. 1997. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53, 983–997. ( 10.2307/2533558) [DOI] [PubMed] [Google Scholar]
- 21.Sabnis RW, Wiley I. 2010. Handbook of biological dyes and stains synthesis and industrial applications. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 22.Patil SM, Berde CV. 2015. Biodegradation of carcinogenic dye, ethidium bromide, by soil microorganisms. World J. Pharm. Pharm. Sci. 4, 1210–1219. [Google Scholar]
- 23.Chain PSG, et al. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl Acad. Sci. USA 103, 15 280–15 287. ( 10.1073/pnas.0606924103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreolli M, Lampis S, Zenaro E, Salkinoja-Salonen M, Vallini G. 2011. Burkholderia fungorum DBT1: a promising bacterial strain for bioremediation of PAHs-contaminated soils. FEMS Microbiol. Lett. 319, 11–18. ( 10.1111/j.1574-6968.2011.02259.x) [DOI] [PubMed] [Google Scholar]
- 25.Juhasz AL, Stanley GA, Britz ML. 2000. Microbial degradation and detoxification of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia strain VUN 10 003. Lett. Appl. Microbiol. 30, 396–401. ( 10.1046/j.1472-765x.2000.00733.x) [DOI] [PubMed] [Google Scholar]
- 26.Cerniglia CE. 1993. Biodegradation of polycyclic aromatic hydrocarbons. Curr. Opin. Biotechnol. 4, 331–338. ( 10.1016/0958-1669(93)90104-5) [DOI] [Google Scholar]
- 27.Andersson SGE, Kurland CG. 1999. Origins of mitochondria and hydrogenosomes. Curr. Opin. Microbiol. 2, 535–541. ( 10.1016/S1369-5274(99)00013-2) [DOI] [PubMed] [Google Scholar]
- 28.Matz C, Kjelleberg S. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. ( 10.1016/j.tim.2005.05.009) [DOI] [PubMed] [Google Scholar]
- 29.Lainhart W, Stolfa G, Koudelka GB. 2009. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J. Bacteriol. 191, 5116–5122. ( 10.1128/jb.00508-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold JW, Koudelka GB. 2014. The trojan horse of the microbiological arms race: phage-encoded toxins as a defence against eukaryotic predators. Environ. Microbiol. 16, 454–466. ( 10.1111/1462-2920.12232) [DOI] [PubMed] [Google Scholar]
- 31.Mauro SA, Koudelka GB. 2011. Shiga toxin: Expression, distribution, and its role in the environment. Toxins 3, 608–625. ( 10.3390/toxins3060608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jousset A, Lara E, Wall LG, Valverde C. 2006. Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl. Environ. Microbiol. 72, 7083–7090. ( 10.1128/aem.00557-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen AL, Winding A, Altenburger A, Ekelund F. 2011. Protozoan growth rates on secondary-metabolite-producing Pseudomonas spp. correlate with high-level protozoan taxonomy. FEMS Microbiol. Lett. 316, 16–22. ( 10.1111/j.1574-6968.2010.02182.x) [DOI] [PubMed] [Google Scholar]
- 34.Fox EM, Howlett BJ. 2008. Secondary metabolism: regulation and role in fungal biology. Curr. Opin. Microbiol. 11, 481–487. ( 10.1016/j.mib.2008.10.007) [DOI] [PubMed] [Google Scholar]
- 35.Wilcke W. 2000. SYNOPSIS polycyclic promatic hydrocarbons (PAHs) in soil—a review. J. Plant Nutr. Soil Sci. 163, 229–248. () [DOI] [Google Scholar]
- 36.Youngblood WW, Blumer M. 1975. Polycyclic aromatic hydrocarbons in the environment: homologous series in soils and recent marine sediments. Geochim. Cosmochim. Acta 39, 1303–1314. ( 10.1016/0016-7037(75)90137-4) [DOI] [Google Scholar]
- 37.Kanaly RA, Harayama S. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182, 2059–2067. ( 10.1128/jb.182.8.2059-2067.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. 2010. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231. ( 10.1038/nm.2087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA 108, 5354–5359. ( 10.1073/pnas.1019378108) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences reported in this paper have been deposited in the GenBank database (accession nos. KR607503–KR607504, KR607510–KR607512) and data of figure 2 are found in electronic supplementary material, table S3. Data from figures 3 and 4 can be found in the Dryad digital depository (http://dx.doi.org/10.5061/dryad.1cc85).