Abstract
Measuring local adaptation can provide insights into how coevolution occurs between predators and prey. Specifically, theory predicts that local adaptation in functionally matched traits of predators and prey will not be detected when coevolution is governed by escalating arms races, whereas it will be present when coevolution occurs through an alternate mechanism of phenotype matching. Here, we analyse local adaptation in venom activity and prey resistance across 12 populations of Northern Pacific rattlesnakes and California ground squirrels, an interaction that has often been described as an arms race. Assays of venom function and squirrel resistance show substantial geographical variation (influenced by site elevation) in both venom metalloproteinase activity and resistance factor effectiveness. We demonstrate local adaptation in the effectiveness of rattlesnake venom to overcoming present squirrel resistance, suggesting that phenotype matching plays a role in the coevolution of these molecular traits. Further, the predator was the locally adapted antagonist in this interaction, arguing that rattlesnakes are evolutionarily ahead of their squirrel prey. Phenotype matching needs to be considered as an important mechanism influencing coevolution between venomous animals and resistant prey.
Keywords: coevolution, local adaptation, venom, resistance, molecular variation, predator–prey interactions
1. Background
The concept of an escalating ‘arms race’ has been repeatedly used to account for the evolution of functionally paired sets of morphological, physiological and molecular traits between antagonistic species [1–4]. Yet, theoretical work indicates the arms race concept is one of a number of possible mechanisms that can govern coevolutionary dynamics between predators and their prey [5]. Therefore, a key step in understanding how coevolution has shaped phenotypic variation in offensive traits in predators and defensive traits in prey is assessing the importance of each of these different mechanisms of coevolutionary interaction [6–8].
In an escalating arms race, the winner is determined by ‘phenotype differences’ between antagonists (i.e. which player is bigger, faster, stronger or more toxic [9]). Arms race models can explain trait variation in a number of predator–prey systems in nature: garter snakes have evolved higher resistance to deadly tetrodotoxin when they encounter more toxic newt prey [2,4], and arms races explain coevolution of morphological adaptations in camellia flowers and weevils that eat them [10]. Yet direct assessments of the arms race are absent in other systems where an arms race explanation has been widely suggested (e.g. venomous animals and their prey; [11]) and theory suggests that alternate explanations are possible [5,9].
The chief alternative to antagonistic arms races is the ‘phenotype matching’ model, where one coevolutionary participant benefits from a match to a second participant's phenotype, while this second participant benefits from a mismatch [9]. Therefore, the degree of matching, and not the difference between trait values, decides the fitness outcomes when two individuals meet. This definition of phenotype matching focuses on the degree of ‘fit’ between specific offensive and defensive traits (e.g. [9]), and should be distinguished from ‘trait matching’ at the population level, where the population-mean trait values of two interacting species are positively correlated (e.g. [3,4,12]). Systems where phenotype matching mediates coevolution of traits include egg mimicry and host recognition in avian brood parasites and their hosts [13] and molecular recognition systems in pathogens and their hosts [14].
Coevolutionary theory offers a clear prediction to test for a role of phenotype matching in predator–prey coevolution: local adaptation will be generated when phenotypic matching influences the winner of a predator–prey interaction, whereas it will not be observed when a phenotype differences mechanism drives trait variation [9,15]. The basis of this prediction can be illustrated by considering two hypothetical antagonists with positively correlated values of a key trait among populations that coevolve under an arms race (phenotype differences) mechanism. The populations of predator and prey with the highly exaggerated trait values (e.g. locations with the most toxic venoms and most resistant prey) will always perform better away from home when performance is compared in sympatric and allopatric combinations of predator and prey, whereas predator–prey populations with low trait values will perform poorly away from home. As a result, the average level of local adaptation across all populations will be zero. By contrast, phenotype matching generates populations of predators and prey that occupy different fitness optima, and so local adaptation will be common when comparing sympatric and allopatric pairs of antagonists [9,15].
In venomous animals that feed on resistant prey, the arms race dynamic has been repeatedly invoked to explain the high levels of variation in offensive traits in predators (e.g. venom composition) and defensive traits in prey (e.g. biochemical resistance; [11,16]), while the role phenotype matching might play in these systems remains to be investigated. Phenotype matching can be identified through an analysis of local adaptation in predator venom and prey defences across populations where each coexists. Such analyses are tractable for venomous snakes and their prey through the use of well-developed in vitro tests that assess venom functional activity and prey resistance [17,18]. These functional data can then be analysed using the statistical approaches developed for assessing local adaptation in host–parasite systems [19,20]. Because local adaptation measures the net fit of genotype to environment at a metapopulation level, it can also be used as a measure of the net outcome of coevolutionary interactions, thus estimating the relative success of each partner and revealing which species is evolutionarily ahead in the interaction [19,21].
Here, we focus on the molecular traits that mediate interactions between venomous Northern Pacific rattlesnakes (Crotalus o. oreganus) and their main prey California ground squirrels (Otospermophilus beecheyi). Multiple biological features of this system argue that coevolutionary interactions play a significant role in the evolution of offensive traits in rattlesnakes and defensive traits in squirrels. Where they coexist, O. beecheyi can comprise the majority of the diet of this rattlesnake, and rattlesnakes are the main squirrel predator [22]. Diverse anti-snake behaviours of the ground squirrels [23–25] and behavioural responses by rattlesnakes [26] suggest reciprocity in matched behavioural adaptations of prey and predator. At the molecular level, ground squirrels show counter-adaptations against envenomation through serum-based resistance to the activity of snake venom metalloproteinases (SVMP), major constituents of pitviper venom that break down connective tissue and facilitate the penetration of other venom components to target tissues [16]. Although both resistance factors in squirrels [16] and venom composition in rattlesnakes [27] vary geographically, the extent to which this variation is adaptive and the type of coevolutionary interaction generating it remain to be determined.
The proteins in ground squirrel serum inhibit SVMPs by targeted binding to venom proteins and interactions between these two types of proteins offers an in vitro system to study the evolutionary mechanism driving interactions between offensive and defensive traits at the molecular level [17]. Inhibition can be measured by exposing venom to the serum of ground squirrels, which contains the inhibitors. We measured local adaptation in this predator–prey system by functional comparisons of venom performance among sympatric and allopatric combinations of rattlesnake venom and ground squirrel serum. Our approach is to assume that coevolution has influenced the evolutionary dynamics of venom and resistance based on the biological features of the system and then to use measures of local adaptation to infer the potential role of phenotype matching as a driver of coevolution of venom and resistance.
2. Material and methods
(a). Study sites and sample collection
We collected rattlesnake venom and ground squirrel serum from at least 10 adult rattlesnakes and 10 adult squirrels at each of 12 California locations (figure 1a), under California Department of Fish and Wildlife permit SC-12027 and using approved procedures described in Ohio State University IACUC protocol 2012A00000015. Rattlesnakes were located by visual search and venom was extracted and frozen. Adult ground squirrels were captured with live traps and a blood sample collected and serum frozen.
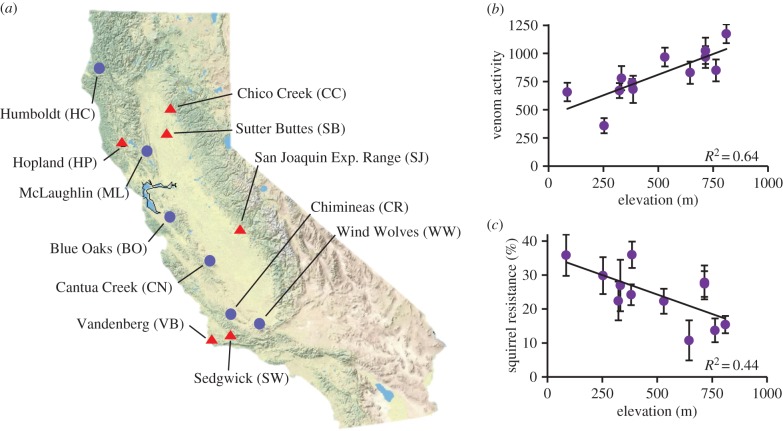
Figure 1.
(a) Map of California marking the 12 sites where rattlesnake venom and ground squirrel blood serum were collected. Circles indicate a site greater than 400 m in elevation, while triangles indicate a site that is less than 400 m in elevation. Scatterplots show site elevation versus the (b) mean baseline snake venom metalloproteinase (SVMP) activity (RFU min−1) of 10 rattlesnakes collected from each site and (c) mean resistance (% inhibition of SVMP activity compared with a snake's baseline activity) of 10 ground squirrels collected from each site. One standard error of the mean is shown for each population. (Online version in colour.)
Ten venom and 10 serum samples from each location were used in our local adaptation tests. Each rattlesnake was randomly paired with 12 ground squirrels; one sympatric squirrel and one squirrel from each of the 11 allopatric sites. Since there were 10 squirrels collected from every population, each of the 10 individual snakes was tested with a unique squirrel from each population, yielding 1440 unique combinations of snake and squirrel in our dataset. We used the method of Biardi et al. [18] to measure SVMP activity and inhibition by O. beecheyi serum (see the electronic supplementary material for detailed protocols). Briefly, metalloproteinase enzymatic activity was measured using the EnzChek Gelatinase Kit (Life Technologies, Carlsbad, CA, USA). We followed product protocols, except that the gelatin substrate was diluted to a concentration of 1 : 100 and 0.3125 ng of venom was used in each test. We measured fluorescence intensity in relative fluorescence units (RFU) every 1.5 min using a FLUOstar Omega microplate reader (BMG Labtech, Ortenberg, Germany). We calculated the slope (RFU min−1) from the linear part of the reaction, which we used as our measure of venom SVMP activity. Larger slopes indicate that the venom is degrading the substrate more quickly, and hence that it is more effective.
Serum-to-venom binding scores calculated during previous work in this system were strongly correlated with venom lethal dose measurements in live ground squirrels [28], suggesting a direct link between levels of serum-based inhibition of venom activity and the fitness outcomes of envenomation for both species. Therefore, determining the reduction in SVMP activity when squirrel venom inhibitors are present provides a functional measure of a snake–squirrel interaction at the molecular level with a probable link to the fitness outcomes of a snake–squirrel interaction in nature.
To do this, we first calculated baseline SVMP activity for each snake by testing the venom by itself. Next, we calculated the difference in SVMP activity between the baseline measure and the SVMP activity when venom was incubated with O. beecheyi serum for 30 min, yielding the amount of SVMP activity lost to an individual squirrel's serum inhibitors for each snake individual. Serum samples were first diluted to 5 mg ml−1 and then incubated with individual venom samples, and their activity measured (see the electronic supplementary material). More negative values of inhibition indicate a larger loss of rattlesnake venom activity when squirrel serum is present. In a biological sense, SVMPs injected into a squirrel are acting on target tissues and avoiding inhibitors, so more positive values (less reduction from baseline activity) would be advantageous to the snake.
(b). Detecting geographical variation in predator and prey performance
We assessed the presence and pattern of geographical variation in the venom and resistance phenotypes using one-way analysis of variance (ANOVA) with population of origin as the factor. Resistance was defined as above and separately as per cent reduction from baseline venom activity for interpretability. Because there are only 10 snakes and squirrels per population, and 66 possible pairwise comparisons between population-mean values for the above variables, we present the post hoc comparisons of population average phenotypes as protected Fisher's least significant differences.
The SVMP activity of C. o. oreganus is hypothesized to be affected by a trade-off between lethal toxicity and general proteolytic activity of venom possibly mediated by temperature or seasonality at a site [27]. Further, O. beecheyi are thought to be less resistant at higher elevations [29]. As such, we explored the possibility that elevation influences both SVMP activity and squirrel resistance. We estimated the elevation of a particular site in ArcMap v. 10.2 (ESRI) as the centroid of a minimum convex polygon encompassing all points from which individual snakes were captured at that site. We used these elevation values in linear regressions on the average baseline venom SVMP activity and average squirrel resistance value at each site; fully factorial multiple regression exploring relationships among SVMP activity, resistance and elevation resulted in all relationships being non-significant due to combined lack of power and collinearity between predictors, so we present these analyses as separate linear models.
(c). Testing for local adaptation
Recent theoretical work on the detection of local adaptation has highlighted the importance of assessing organismal performance in allopatry in the context of relevant environmental variation [19]. Owing to the significant relationships we found between elevation and both SVMP activity and resistance levels (see Results), we partitioned our allopatric combinations of venom and serum based on elevation [19]. We used the median site elevation (422 m.a.s.l.) to designate six high and six low elevation sites. Specific snake–squirrel pairings were then designated as sympatric, allopatric-same elevation (AS) or allopatric-different elevation (AD; figure 2). The AS designation meant the snake–squirrel comparison was between two high elevation sites or two low elevation sites, whereas the AD designation meant the comparison was between a snake and squirrel collected at different elevations (snake low versus squirrel high or snake high versus squirrel low). This ‘comparison type’ term partitions any contribution of local adaptation in SVMP activity into a portion related to local adaptation at high or low elevation, and a portion attributable to other unknown factors [19,30].
Figure 2.
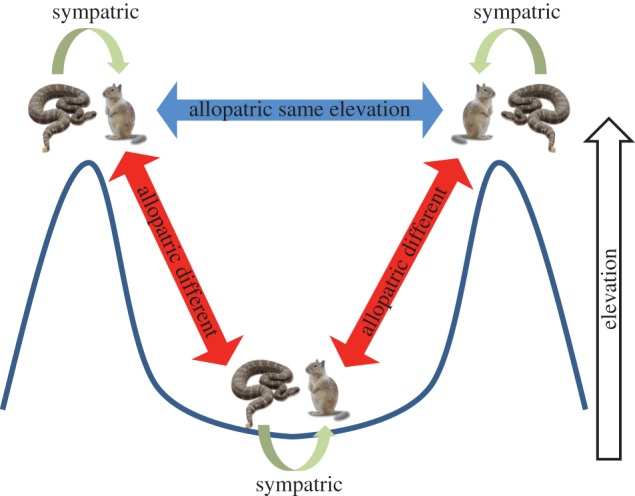
Schematic of the fully crossed experiment where ground squirrel serum was tested for its ability to inhibit the venom of sympatric (n = 120) and allopatric rattlesnakes. Curved lines represent peaks and a valley forming an elevation gradient. Allopatric tests were categorized as same type (low to low or high to high elevation; n = 660) or different type (high to low or low to high elevation; n = 660). (Online version in colour.)
We fit a linear mixed-effects model (LMM) with resistance as the response variable to test for local adaptation [19,20]. Fixed main effects in the full model included snake elevation type (SnElev), squirrel elevation type (SqElev), snake population of origin nested within SnElev (SnPop), squirrel population of origin nested within SqElev (SqPop) and our elevation-partitioned local adaptation term, comparison type (S–AS–AD). In addition, snake individual nested within SnPop, and squirrel individual nested within SqPop, were included as random factors to account for the fact that each snake and squirrel individual was used in 12 comparisons. Dunn–Sidak adjusted p-values were used to compare the model-estimated marginal mean resistance values between S, AS and AD combinations. Larger (more positive) values in sympatric comparisons would show local adaptation of rattlesnakes, whereas smaller (more negative) values in sympatry would indicate local adaptation of ground squirrels.
Finally, we visualized the population-level pattern of performance in sympatric and allopatric populations of snakes or squirrels to determine which sites showed patterns consistent with local adaptation by comparing performance in sympatry versus allopatry (AS and AD) for each population. We took advantage of the fact that the same individual snake or squirrel was used in multiple functional tests, making our study analogous to a paired or repeated measures design and allowing us to derive a ‘venom deviation score’ for each snake–squirrel combination. The venom deviation score was calculated as the amount that a focal squirrel individual caused a particular snake individual to deviate from its average performance against all squirrels. This approach controls for the large main effect differences among individual snakes, and therefore, the populations they are nested within, by measuring performance as the deviation from an individual snake's average performance. Further, since one squirrel population could be analysed at a time, main effects of squirrel population are also factored out, resulting in a value for an individual snake's venom deviation score in sympatry (one value), in all AD comparisons (six values) and in all AS comparisons (five values). For each snake individual, we then subtracted the mean venom deviation scores of all AD and AS comparisons from its sympatric value, generating values for the difference in performance in sympatry versus allopatric sites.
3. Results
(a). Geographical variation in venom activity and resistance
We detected extensive among-population variation in both the venom activity of snakes and the capacity to inhibit venom in squirrels. The baseline SVMP activity of venoms varied among rattlesnake populations, with threefold differences between the least and most enzymatically active populations (one-way ANOVA, F11,119 = 5.49, p < 0.001, R2 = 0.36). Venom activity was reduced by 27%, on average, by the ratio of venom to serum used in our experiments. We found variation in resistance in ground squirrels in both an absolute sense, the average RFU min−1 of SVMP activity a venom lost in the presence of a given serum (F11,119 = 3.08, p < 0.001, R2 = 0.24), and as a percentage reduction in SVMP activity (F11,119 = 2.7, p = 0.004, R2 = 0.22). There was threefold variation in the average amount of SVMP activity lost to particular squirrel populations. On average, squirrels from the mountains of Humboldt County were least resistant, only causing a 10.8% reduction in venom activity, while squirrels from Vandenberg A.F.B. on the coast and the Sutter Buttes in the northern Sacramento River Valley inhibited venoms by nearly 36% (electronic supplementary material, table S1).
A large amount of the site-to-site variation in venom SVMP activity was predicted by linear regression on the elevation of a site, where population average SVMP activity was higher at higher site elevation (T = 4.24, df = 10, p = 0.002, R2 = 64.3%, β = 0.73, figure 1b). The population average resistance values of ground squirrels were also related to elevation, with squirrels from low elevations having serum better at inhibiting venom (T = –2.81, df = 10, p = 0.018, R2 = 44.2%, β = –0.00023, figure 1c). Direct linear regression of average squirrel resistance on average venom SVMP activity among population yields a non-significant relationship (p = 0.12) with a negative slope coefficient. While elevation is clearly an important driver of both overall SVMP activity in snakes and average ability to inhibit SVMPs in squirrels, our test for local adaptation (below) models the change in SVMP activity to test for interactions between snake and squirrel genotypes on venom function (G × G interactions), which applies regardless of baseline venom activity and the apparent environmental gradient. Thus, the population average functional responses we have measured can be correlated with elevation while local adaptation of predator or prey can still exist, especially if different venom or inhibitor isoforms or expression differences lead to the same global average in function.
(b). Detecting local adaptation
We used our linear mixed model approach that factored out population and environmental main effects to test for a difference in performance in sympatric versus allopatric combinations of venom and serum [20]. This model was the best fit to the data based on sample sized-corrected Akaike's information criterion, when compared with a series of other models that treat local adaptation in alternative ways (e.g. comparisons of sympatry versus all allopatric comparisons or consideration of geographical distance as a covariate, electronic supplementary material, table S2). We detected a significant main effect of squirrel elevation (F1,107 = 16.39, p < 0.001) where squirrels originating in low elevation sites (mean change = −199.7 RFU min−1) were approximately 32% more resistant than those from high elevation sites (mean change = −293.3 RFU min−1). The elevation type from which snakes originated had an impact on the amount of SVMP activity they lost (low elevation mean = −228.6, high elevation mean = −264.3, F1,107 = 10.33, p = 0.002). Both snake population and squirrel population of origin had significant main effects on the amount of SVMP activity lost, showing the existence of variation in the outcome of a snake–squirrel interaction even when controlling for the large general effects of site elevation (table 1). After variation in these geographical factors was controlled for, the local adaptation term (S–AS–AD) was a significant predictor of the amount of SVMP activity lost (table 1). Post hoc testing to determine whether snakes or squirrels were locally adapted showed that the S and AS comparisons, where selection pressures may be similar, were comparable in the magnitude of change in baseline venom activity, while snakes lost approximately 10% more venom activity in the AD comparisons than in S or AS trials (figure 3a). This indicates that C. o. oreganus is locally adapted to overcoming the resistance of O. beecheyi in a way that is structured by elevation, where snakes are locally adapted to overcoming squirrel inhibitor proteins when tested against an allopatric squirrel from a site that differs in elevation. Venom deviation scores, the residual difference from an individual snake's average performance across all squirrels, shows this pattern of local adaptation best: high elevation snakes perform best with high elevation squirrels, and low elevation snakes perform best with low elevation squirrels (figure 3b).
Table 1.
Analysis of variance of change in snake venom metalloproteinase activity in the presence of ground squirrel blood serum. Error mean squares and degrees of freedom for elevation and population variables were obtained via a linear combination of several terms using the Satterthwaite approximation. Snake and squirrel population terms were nested within elevation terms. Snake and squirrel ID terms were nested within population terms. The S–AS–AD term is the local adaptation test: S, sympatric; AS, allopatric-same elevation; AD, allopatric-different elevation. Significance values are indicated with asterisks.
| source | d.f. | F-value |
|---|---|---|
| SnElev | 1 | 10.3* |
| SnElev error | 107 | |
| SqElev | 1 | 16.4*** |
| SqElev error | 107 | |
| SnPop (SnElev) | 10 | 7.4*** |
| SnPop error | 107 | |
| SqPop (SqElev) | 10 | 2.0* |
| SqPop error | 107 | |
| snake ID (SnPop) | 108 | 35.7*** |
| squirrel ID (SqPop) | 108 | 22.5*** |
| S–AS–AD | 2 | 13.5*** |
| error | 1199 |
*p < 0.05, p < 0.001, ***p < 0.0001.
Figure 3.
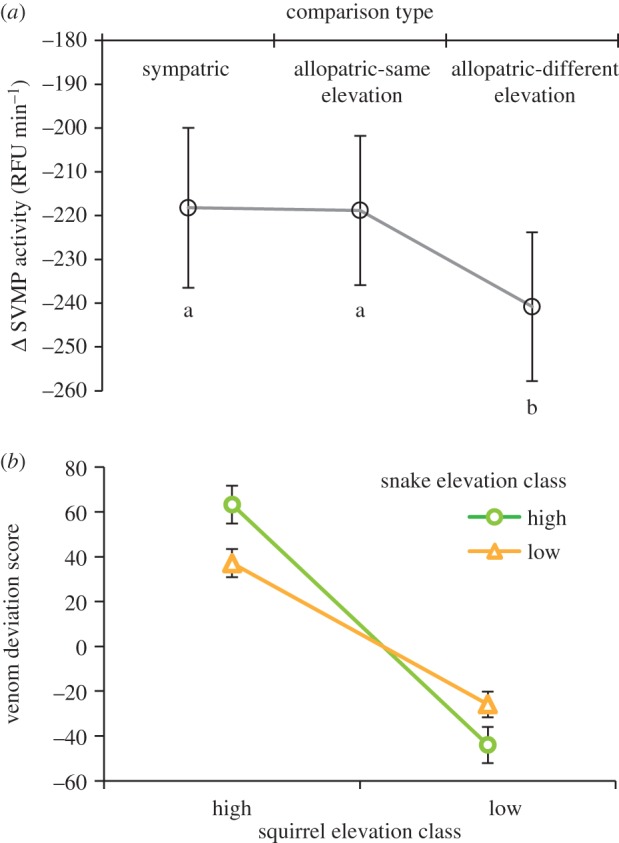
Local adaptation of rattlesnakes to overcoming ground squirrel resistance shown as (a) model-estimated marginal mean changes (±1 s.e.) in SVMP activity in the presence of California ground squirrel serum for sympatric comparisons, allopatric comparisons that involved snakes and squirrels from similar elevations, and allopatric comparisons involving snakes and squirrels from different elevations and (b) venom deviation score (mean ± 1 s.e.) of high (circles) and low (triangles) elevation rattlesnakes based on whether they were paired with ground squirrels from a high or low elevation. Different letters indicate significant differences. (Online version in colour.)
A direct illustration of variation in the outcome of interactions between snakes and squirrels comes from comparisons of the venom deviation scores across populations. Consistent with the result of the overall analysis, in 10 of 12 populations, the mean value of the venom deviation score was more positive when calculated as S minus AD than when calculated as S minus AS, demonstrating environmentally structured local adaptation in rattlesnakes (figure 4). However, three sites (San Joaquin Experimental Range, McLaughlin Reserve and Chico Creek) showed patterns more consistent with local adaptation of ground squirrels to inhibiting snake venoms (figure 4).
Figure 4.
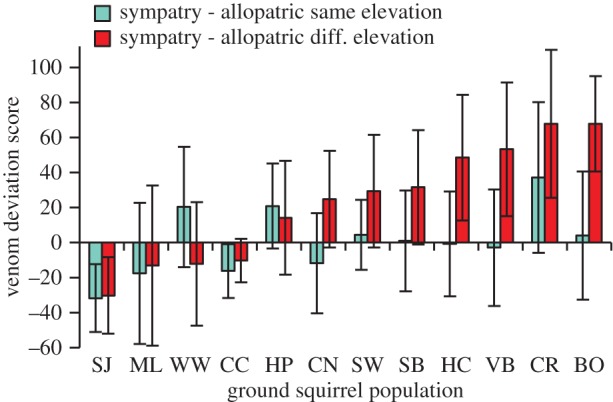
The difference in venom deviation scores (mean ± 1 s.e.) when sympatric versus allopatric rattlesnakes are paired with ground squirrels from a given population. For individual squirrels, the amount they caused a randomly chosen snake to deviate from its average venom activity was calculated and the mean of this score for all allopatric comparisons was subtracted from the sympatric comparison. Comparisons are broken up by whether the allopatric snakes were from the same type of habitat (left bars) or the alternative type of habitat (right bars). (Online version in colour.)
4. Discussion
(a). Coevolution of molecular traits
In addition to the biological features of this system, the threefold variation in rattlesnake venom activity and squirrel resistance across populations and local adaptation of snakes to overcoming squirrel resistance provide support for a coevolutionary interaction between rattlesnakes and squirrels that varies across space due to a selection mosaic [31]. Nonetheless, pattern alone does not provide direct evidence for coevolution since other evolutionary forces can produce similar geographical patterns [8]. For example, local adaptation of rattlesnakes to independently generated geographical variation in ground squirrel resistance is a possible alternative to reciprocal coevolutionary selection. Under this scenario, geographical variation in squirrel resistance would be an evolutionary response to selection independent of rattlesnakes, while rattlesnakes still evolve local adaptation in response to selection from squirrels.
In other systems coevolution is supported when coevolutionary cold spots with no reciprocal selection (e.g. areas where one species is absent) are detected and the phenotype in the present species differs from that in similar locations where both species occur [2,3,9]. Cold spots appear to present in this system: Poran et al. [30] assessed squirrel resistance in the Great Central Valley of California, where snakes are rare. These sites were all low in elevation (less than 100 m.a.s.l.), and so based on the negative correlation between resistance and elevation we have shown, these populations should be highly resistant, if resistance was primarily determined by site elevation. Instead, both venom resistance and the behavioural response to a live rattlesnake [26] are reduced in these Valley floor populations, indicating that the effect of elevation on the squirrel resistance phenotype depends on selection from rattlesnakes. The resistance phenotype in low resistance Central Valley cold spots supports the presence of a geographical mosaic of coevolutionary selection in this system. Taken together the geographical patterns in venom, resistance and behavioural traits all provide strong support for our assumption that coevolution plays an important role in the evolution of offensive and defensive traits in this predator and its prey, while more definitive proof would come from further work which uses methods to partition the strength of selection on venom and resistance traits [10,32].
(b). A role for phenotype matching
Our results identify phenotype matching as an important force governing the coevolutionary interactions of rattlesnakes and ground squirrels, because phenotype matching is the only coevolutionary mechanism that can generate a signature of local adaptation that is detectable in a reciprocal crossing study [5,9,15]. In a statistical sense, among-population comparisons are dominated by population main effects during arms race coevolution, leading to the least escalated populations always appearing locally adapted and the most escalated populations always appearing locally maladapted. On the other hand, phenotype matching through a ‘lock-and-key’ mechanism of target recognition can generate populations of snakes that show local adaptation when comparisons are made between sympatric and allopatric pairs of the interacting predator and prey, because the unique resistance phenotype of each squirrel population represents a distinct adaptive peak for the local snakes [15].
The role of arms race dynamics as a general mechanism for generating spatial variation in snake venom and prey resistance remains unclear. On one hand, some of the best-studied cases of antagonistic arms races have shown that these interactions generate positive correlations between the trait values of the interacting species [2,4,10]. We did not find such a correlation between population-level measures of SVMP activity in snakes and SVMP resistance in squirrels. However, theoretical work suggests that regression analysis of phenotype values is not a strong test of the arms race hypothesis, as there is only a weak expectation for arms races to generate positively correlated trait means under strong coevolutionary selection [33]. Furthermore, our data are based on a functional interaction (enzyme activities and their inhibition) that closely matches the mechanism envisioned for phenotype matching (see below; [16,17]). By contrast, the concentrations of SVMPs and their inhibitors may only be loosely related to our enzymatic measures if various isoforms of these proteins have different activities, a result that has been confirmed for the SVMPs of other pitvipers [34]. Concentrations of venom proteins and their inhibitors may better fit the arms race model and would be a logical next place to search for signs of an arms race mediated by a phenotypic differences mechanism involving venom and resistance.
The implication that phenotype matching influences the fitness outcomes of an interaction between venomous and resistant species is a key step in explaining patterns of diversification in this coevolving system. We suggest that interactions between venom and resistance molecules studied here are analogous to the targeted recognition or gene-for-gene coevolution seen with innate immunity to pathogens, which is a form of coevolution governed by phenotype matching [14]. This explanation is consistent with our understanding of the mechanism by which these proteins interact: resistance molecules in prey bind and inactivate venom protein targets, whereas the venom must bypass these molecules to be effective, which is similar to pathogens evading the immune system [16]. Targeted binding of inhibitors and the evolution of molecular mechanisms by which venom overcomes resistance requires complexity in the molecular underpinnings of both the venom and resistance phenotypes (e.g. allelic variation). Consistent with this idea, multiple SVMP isoforms exist within individual C. o. oreganus [27]. By contrast, the level of variation at O. beecheyi resistance loci is unknown although the small number of resistance proteins identified to date suggests that if genetically based variation is present, it is likely limited [17]. Phenotype matching, compared with arms races, is more likely to promote phenotypic diversification among populations [35], so may better explain the high levels of intraspecific variation observed in snake venoms [27,36].
(c). The predator is ahead
Rattlesnakes appear to be ahead in the coevolving interactions involving SVMPs and their inhibitors, because venom, and not resistance, showed a pattern of local adaptation in our reciprocal crossing study [19]. Predator local adaptation is unexpected given the potential asymmetry in selection intensity and evolvability between predator and prey, as summarized in the Life–Dinner Principle [37]. Prey are expected to be under stronger selection because being eaten entails a higher fitness cost than missing a meal, and because they are in general expected to evolve more rapidly due to higher reproductive rates. The life-history characteristics of rattlesnakes and squirrels match these expectations: O. beecheyi reproduce annually beginning in their second year and live to around 5 years of age [38], while rattlesnakes reproduce semi-annually after their third year, and have a maximum lifespan of 20 years or older [22]. Yet, despite these demographic differences snakes are evolutionarily ahead of their prey at least for the molecular interaction we characterized. This pattern has also been found in other antagonistic interactions: predatory garter snakes are ahead in the arms race dynamic occurring with their toxic newt prey [4], while pipefish hosts are ahead of Vibrio bacterial parasites despite generation times that differ by many orders of magnitude [21]. Our results support Abrams' [39] suggestion that the outcome of antagonistic coevolution in predator–prey systems will be a product of ecological and evolutionary factors specific to each system and cannot be predicted from general concepts such as the Life–Dinner Principle.
Two such factors that theoretical and empirical studies emphasize are relative levels of gene flow of the interacting species and the genetic architecture of the key traits mediating the interaction [40,41]. In host–parasite systems, the ‘winner’ in a coevolutionary interaction is often the species with higher gene flow, as higher migration can facilitate more rapid adaption due to the increased spread of evolutionarily advantageous alleles [40]. Population genetic studies of congeneric species suggest that ground squirrels show higher levels of genetic structure than Crotalus rattlesnakes [42,43], which is consistent with the prediction that the winning partner will experience greater levels of gene flow, but this remains to be documented for the species studied here.
The architecture of key traits at the phenotypic interface of the interaction may also influence whether predators or prey are ahead [44]. In particular, the participant with traits that are less evolutionarily constrained or have a more evolvable genetic architecture may be at an advantage [44]. Both evolvability and constraint were invoked to explain why garter snakes outpaced newts in their tetrodotoxin-based arms race: single-nucleotide substitutions can have large effects on snake resistance, while bio-availability of toxin precursors may constrain newt toxicity levels [4]. Venom is a modular phenotype contained in a secretory venom gland [45], so it may be more evolvable than the blood-based inhibitor proteins of ground squirrels, which could interact pleiotropically with other physiological processes, constraining their ability to respond over evolutionary timescale to changes in venom. We emphasize that our results to date only involve snake SVMP proteins and their inhibitors in squirrels. Whether similar patterns occur for the coevolutionary dynamics of other venom proteins, such as the serine protease and phospholipase A2 proteins to which ground squirrels may be resistant, is an important direction for future work.
(d). Environmental effects on coevolution
Local adaptation can be missed if the geographical location of study sites is not congruent with the scale at which local adaptation occurs, or if variable selection from other sources (e.g. the abiotic environment) among allopatric sites leads to large variance in the outcomes of allopatric tests [19]. In our system, controlling for elevation through comparisons of snake–squirrel populations at the same (AS) versus different (AD) elevations was essential to detecting local adaptation of rattlesnake venom to squirrel resistance. Not doing so leads to much larger variance in allopatric comparisons and a difference between sympatric and allopatric performance of venom that is marginally non-significant (electronic supplementary material, table S2). This result adds to a growing body of work that identifies environmental differences associated with elevation as key influences on the selection mosaic in coevolutionary systems, such as newts and garter snakes [46], and camellias and weevils [3]. More broadly, our study highlights the need to understand the environmental context of variation in traits involved in species interactions when interpreting their evolutionary history.
Hypoxic conditions at higher elevations are one candidate for the environmental cause of a selection mosaic acting on rattlesnakes and ground squirrels. Mammalian serum albumins are non-specific inhibitors of rattlesnake venom activity [47] that are reduced in abundance in some mammals at high elevations to compensate for increased haematocrit [48]. Thus, reduced expression in non-specific protein binding agents like albumin may account for the substantial reduction in the venom-inhibitory capacity of high elevation squirrel serum (the squirrel elevation main effect), with isoforms of alpha-1-antitrypsin involved in the remaining variation to which snakes have become locally adapted. Identifying the physiological underpinnings of elevational differences in resistance, and how these may lead to variable coevolutionary selection between snakes and squirrels, are promising avenues for future work.
5. Conclusion
Our study demonstrates the value of using local adaptation studies to assess the coevolutionary mechanism that is responsible for trait variation at the molecular level in venomous animals and their resistant prey. We used this approach to draw the novel conclusion that phenotype matching is a driver of coevolution between venom and resistance traits involved in a rattlesnake–prey interaction. We also show that despite biological differences between snakes and squirrels that favour squirrels winning the interaction, snakes appear to be evolutionarily ahead of their prey. The evolutionarily labile venom phenotype, combined with phenotype matching and its propensity to promote diversification [35], may provide a mechanism by which coevolution generates biologically and medically important diversity in animal venoms.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank M. Earthman, K. Price, J. Zajdel, E. Taylor, T. Frazier, B. Putman, R. Clark, S. Dorr, A. Branske and R. Denton for assistance in the field, R. Denton, T. Fries, M. Sovic, D. Salazar and S. Smiley for help with planning and analysis, C. Benkman, M. Daly, the Gibbs lab, M. van Baleen and two anonymous reviewers for helpful comments on the manuscript, S. Nuismer for discussion and key insights about the links between coevolution and local adaptation, M. Westphal, H. Hamman, and California Fish and Wildlife, California State Parks and the University of California Natural Reserve System for access to field sites.
Data accessibility
All data associated with this paper are archived in the Dryad repository: http://dx.doi.org/10.5061/dryad.p7p7p.
Authors' contributions
M.L.H., J.E.B. and H.L.G. designed the study, M.L.H. and J.E.B. collected the data, M.L.H. analysed the data, M.L.H. and H.L.G. co-wrote the manuscript, and J.E.B. provided editorial suggestions. All authors gave final approval for submission.
Competing interests
The authors have no competing interests.
Funding
This work was supported by a NSF GRF to M.L.H., research grants from the American Society of Naturalists, Herpetologists’ League, American Museum of Natural History, American Society of Mammalogists, California Bureau of Land Management, Sigma Xi, American Society of Ichthyologists and Herpetologists and the Chicago Herpetological Society (all to M.L.H.), and funds from Ohio State University.
References
- 1.Benkman CW, Parchman TL, Favis A, Siepielski AM. 2003. Reciprocal selection causes a coevolutionary arms race between crossbills and lodgepole pine. Am. Nat. 162, 182–194. ( 10.1086/376580) [DOI] [PubMed] [Google Scholar]
- 2.Brodie ED, Ridenhour BJ, Brodie ED. 2002. The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution 56, 2067–2082. ( 10.1111/j.0014-3820.2002.tb00132.x) [DOI] [PubMed] [Google Scholar]
- 3.Toju H. 2008. Fine-scale local adaptation of weevil mouthpart length and Camellia pericarp thickness: altitudinal gradient of a putative arms race. Evolution 62, 1086–1102. ( 10.1111/j.1558-5646.2008.00341.x) [DOI] [PubMed] [Google Scholar]
- 4.Hanifin CT, Brodie ED Jr, Brodie ED III. 2008. Phenotypic mismatches reveal escape from arms-race coevolution. PLoS Biol. 6, 471–482. ( 10.1371/journal.pbio) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuismer SL, Doebeli M, Browning D. 2005. The coevolutionary dynamics of antagonistic interactions mediated by quantitative traits with evolving variances. Evolution 59, 2073–2082. ( 10.1554/05-141.1.s1) [DOI] [PubMed] [Google Scholar]
- 6.Zangerl AR, Stanley MC, Berenbaum MR. 2008. Selection for chemical trait remixing in an invasive weed after reassociation with a coevolved specialist. Proc. Natl Acad. Sci. USA 105, 4547–4552. ( 10.1073/pnas.0710280105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geffeney S, Brodie ED Jr, Ruben PC, Brodie ED III. 2002. Mechanism of adaptation in a predator–prey arms race: TTX-resistant sodium channels. Science 297, 1336–1339. ( 10.1126/science.1074310) [DOI] [PubMed] [Google Scholar]
- 8.Gomulkiewicz R, Drown DM, Dybdahl MF, Godsoe W, Nuismer SL, Pepin KM, Ridenhour BJ, Smith CI, Yoder JB. 2007. Dos and don'ts of testing the geographic mosaic theory of coevolution. Heredity 98, 249–258. ( 10.1038/sj.hdy.6800949) [DOI] [PubMed] [Google Scholar]
- 9.Nuismer SL, Ridenhour BJ, Oswald BP. 2007. Antagonistic coevolution mediated by phenotypic differences between quantitative traits. Evolution 61, 1823–1834. ( 10.1111/j.1558-5646.2007.00158.x) [DOI] [PubMed] [Google Scholar]
- 10.Toju H, Sota T. 2006. Imbalance of predator and prey armament: geographic clines in phenotypic interface and natural selection. Am. Nat. 167, 105–117. ( 10.1086/498277) [DOI] [PubMed] [Google Scholar]
- 11.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2012. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 28, 219–229. ( 10.1016/j.tree.2012.10.020) [DOI] [PubMed] [Google Scholar]
- 12.Anderson B, Johnson SD. 2008. The geographical mosaic of coevolution in a plant–pollinator mutualism. Evolution 62, 220–225. ( 10.1111/j.1558-5646.2007.00275.x) [DOI] [PubMed] [Google Scholar]
- 13.Davies NB. 2000. Cuckoos, cowbirds, and other cheats. London, UK: T A. D. Poyser. [Google Scholar]
- 14.Dybdahl MF, Jenkins CE, Nuismer SL. 2014. Identifying the molecular basis of host–parasite coevolution: merging models and mechanisms. Am. Nat. 184, 1–13. ( 10.1086/676591) [DOI] [PubMed] [Google Scholar]
- 15.Ridenhour BJ, Nuismer SL. 2007. Polygenic traits and parasite local adaptation. Evolution 61, 368–376. ( 10.1111/j.1558-5646.2007.00029.x) [DOI] [PubMed] [Google Scholar]
- 16.Biardi JE. 2008. The ecological and evolutionary context of mammalian resistance to rattlesnake venoms. In Biology of the rattlesnakes (eds Hayes WK, Beaman KR, Cardwell MD, Bush SP), pp. 557–568. Loma Linda, CA: Loma Linda University Press. [Google Scholar]
- 17.Biardi JE, Ho CYL, Marcinczyk J, Nambiar KP. 2011. Isolation and identification of a snake venom metalloproteinase inhibitor from California ground squirrel (Spermophilus beecheyi) blood sera. Toxicon 58, 486–493. ( 10.1016/j.toxicon.2011.08.009) [DOI] [PubMed] [Google Scholar]
- 18.Biardi JE, Nguyen KT, Lander S, Whitley M, Nambiar KP. 2011. A rapid and sensitive fluorometric method for the quantitative analysis of snake venom metalloproteases and their inhibitors. Toxicon 57, 342–347. ( 10.1016/j.toxicon.2010.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanquart F, Kaltz O, Nuismer S, Gandon S. 2013. A practical guide to measuring local adaptation. Ecol. Lett. 16, 1195–1205. ( 10.1111/ele.12150) [DOI] [PubMed] [Google Scholar]
- 20.Thrall PH, Burdon JJ, Bever JD. 2002. Local adaptation in the Linum marginale–Melampsora lini host–pathogen interaction. Evolution 56, 1340–1351. ( 10.1111/j.0014-3820.2002.tb01448.x) [DOI] [PubMed] [Google Scholar]
- 21.Roth O, Keller I, Landis SH, Salzburger W, Reusch TBH. 2012. Hosts are ahead in a marine host–parasite coevolutionary arms race: innate immune system adaptation in pipefish Syngnathus typhle against Vibrio phylotypes. Evolution 66, 2528–2539. ( 10.1111/j.1558-5646.2012.01614.x) [DOI] [PubMed] [Google Scholar]
- 22.Fitch HS. 1949. Study of snake populations in Central California. Am. Midland Nat. 41, 513–579. ( 10.2307/2421774) [DOI] [Google Scholar]
- 23.Putman B, Coss R, Clark R. 2015. The ontogeny of antipredator behavior: age differences in California ground squirrels (Otospermophilus beecheyi) at multiple stages of rattlesnake encounters. Behav. Ecol. Sociobiol. 69, 1447–1457. ( 10.1007/s00265-015-1957-2) [DOI] [Google Scholar]
- 24.Coss RG, Gusé KL, Poran NS, Smith DG. 1993. Development of antisnake defenses in California ground squirrels (Spermophilus beecheyi): II. Microevolutionary effects of relaxed selection from rattlesnakes. Behaviour 124, 137–164. ( 10.1163/156853993X00542) [DOI] [Google Scholar]
- 25.Rundus AS, Owings DH, Joshi SS, Chinn E, Giannini N. 2007. Ground squirrels use an infrared signal to deter rattlesnake predation. Proc. Natl Acad. Sci. USA 104, 14 372–14 376. ( 10.1073/pnas.0702599104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbour MA, Clark RW. 2012. Ground squirrel tail-flag displays alter both predatory strike and ambush site selection behaviours of rattlesnakes. Proc. R. Soc. B 279, 3827–3833. ( 10.1098/rspb.2012.1112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackessy SP. 2010. Evolutionary trends in venom composition in the western rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon 55, 1463–1474. ( 10.1016/j.toxicon.2010.02.028) [DOI] [PubMed] [Google Scholar]
- 28.Poran NS, Coss RG, Benjamini E. 1987. Resistance of California ground squirrels (Spermophilus beecheyi) to the venom of the northern Pacific rattlesnake (Crotalus viridis oreganus): a study of adaptive variation. Toxicon 25, 767–777. ( 10.1016/0041-0101(87)90127-9) [DOI] [PubMed] [Google Scholar]
- 29.Towers SR, Coss RG. 1990. Confronting snakes in the burrow: snake-species discrimination and antisnake tactics of two California ground squirrel populations. Ethology 84, 177–192. ( 10.1111/j.1439-0310.1990.tb00796.x) [DOI] [Google Scholar]
- 30.Adiba S, Huet M, Kaltz O. 2010. Experimental evolution of local parasite maladaptation. J. Evol. Biol. 23, 1195–1205. ( 10.1111/j.1420-9101.2010.01985.x) [DOI] [PubMed] [Google Scholar]
- 31.Thompson JN. 2005. The geographic mosaic of coevolution. Chicago, IL: The University of Chicago Press. [Google Scholar]
- 32.Ridenhour BJ. 2005. Identification of selective sources: partitioning selection based on interactions. Am. Nat. 166, 12–25. ( 10.1086/430524) [DOI] [PubMed] [Google Scholar]
- 33.Nuismer SL, Gomulkiewicz R, Ridenhour BJ. 2010. When is correlation coevolution? Am. Nat 175, 525–537. ( 10.1086/651591) [DOI] [PubMed] [Google Scholar]
- 34.Bernardoni JL, Sousa LF, Wermelinger LS, Lopes AS, Prezoto BC, Serrano SMT, Zingali RB, Moura-da-Silva AM. 2014. Functional variability of snake venom metalloproteinases: adaptive advantages in targeting different prey and implications for human envenomation. PLoS ONE 9, e109651 ( 10.1371/journal.pone.0109651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoder JB, Nuismer SL. 2010. When does coevolution promote diversification? Am. Nat 176, 802–817. ( 10.1086/657048) [DOI] [PubMed] [Google Scholar]
- 36.Sunagar K, et al. 2014. Intraspecific venom variation in the medically significant southern Pacific rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteomics ( 10.1016/j.jprot.2014.01.013) [DOI] [PubMed] [Google Scholar]
- 37.Dawkins R, Krebs JR. 1979. Arms races between and within species. Proc. R. Soc. Lond. B 205, 489–511. ( 10.1098/rspb.1979.0081) [DOI] [PubMed] [Google Scholar]
- 38.Fitch HS. 1948. Ecology of the California ground squirrels on grazing lands. Am. Midland Nat. 39, 513–596. ( 10.2307/2421524) [DOI] [Google Scholar]
- 39.Abrams PA. 1986. Adaptive responses of predators to prey and prey to predators: the failure of the arms-race analogy. Evolution 40, 1229–1247. ( 10.2307/2408950) [DOI] [PubMed] [Google Scholar]
- 40.Gandon S. 2002. Local adaptation and the geometry of host–parasite coevolution. Ecol. Lett. 5, 246–256. ( 10.1046/j.1461-0248.2002.00305.x) [DOI] [Google Scholar]
- 41.MacPherson A, Hohenlohe PA, Nuismer SL. 2015. Trait dimensionality explains widespread variation in local adaptation. Proc. R. Soc. B 282, 20141570 ( 10.1098/rspb.2014.1570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gavin TA, Sherman PW, Yensen E, May B. 1999. Population genetic structure of the northern Idaho ground squirrel (Spermophilus brunneus brunneus). J. Mammal. 80, 156–168. ( 10.2307/1383216) [DOI] [Google Scholar]
- 43.Bushar LM, Bhatt N, Dunlop MC, Schocklin C, Malloy MA, Reinert HK. 2015. Population isolation and genetic subdivision of timber rattlesnakes (Crotalus horridus) in the New Jersey Pine Barrens. Herpetologica 71, 203–211. ( 10.1655/HERPETOLOGICA-D-14-00030) [DOI] [Google Scholar]
- 44.Brodie ED, Brodie ED. 1999. Predator–prey arms races. Bioscience 49, 557–568. ( 10.2307/1313476) [DOI] [Google Scholar]
- 45.Margres MJ, McGivern JJ, Seavy M, Wray KP, Facente J, Rokyta DR. 2015. Contrasting modes and tempos of venom expression evolution in two snake species. Genetics 199, 165–176. ( 10.1534/genetics.114.172437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stokes AN, Ray AM, Buktenica MW, Gall BG, Paulson E, Paulson D, French SS, Brodie ED, Brodie ED. 2015. Otter predation on Taricha granulosa and variation in tetrodotoxin levels with elevation. Northwest Nat. 96, 13–21. ( 10.1898/NWN13-19.1) [DOI] [Google Scholar]
- 47.Clark WC, Voris HK. 1969. Venom neutralization by rattlesnake serum albumin. Science 164, 1402–1404. ( 10.1126/science.164.3886.1402) [DOI] [PubMed] [Google Scholar]
- 48.Crait JR, Prange HD, Marshall NA, Harlow HJ, Cotton CJ, Ben-David M. 2012. High-altitude diving in river otters: coping with combined hypoxic stresses. J. Exp. Biol. 215, 256–263. ( 10.1242/jeb.059774) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this paper are archived in the Dryad repository: http://dx.doi.org/10.5061/dryad.p7p7p.