Abstract
Farnesoid X receptor (FXR) is a bile acid-activated transcription factor belonging to the nuclear receptor superfamily. FXR is highly expressed in liver and intestine and crosstalk mediated by FXR in these two organs is critical in maintaining bile acid homeostasis. FXR deficiency has been implicated in many liver and intestine diseases. However, regulation of transcription by FXR at the genomic level is not known. This study analyzed genome-wide FXR binding in liver and intestine of mice treated with a synthetic FXR ligand (GW4064) by chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq). The results showed a large degree of tissue-specific FXR binding, with only 11% of total sites shared between liver and intestine. The sites were widely distributed between intergenic, upstream, intragenic, and downstream of genes, with novel sites identified within even known FXR target genes. Motif analysis revealed a half nuclear receptor binding site, normally bound by a few orphan nuclear receptors, adjacent to the FXR response elements, indicating possible involvement of some orphan nuclear receptors in modulating FXR function. Furthermore, pathway analysis indicated that FXR may be extensively involved in multiple cellular metabolic pathways.
Conclusion
This study reports genome-wide FXR binding in vivo and the results clearly demonstrate tissue-specific FXR/gene interaction. In addition, FXR may be involved in regulating broader biological pathways in maintaining hepatic and intestinal homeostasis.
Farnesoid X receptor (FXR), highly expressed in liver and intestine, is a ligand-activated transcription factor belonging to the nuclear receptor superfamily. FXR has been adopted when bile acids are identified as its endogenous ligands.1,2 FXR is essential in maintaining bile acid homeostasis and is important for energy balance through regulating lipid and glucose metabolism.3–5 FXR deficiency in mice has been implicated not only in hepatic and gastroenterological diseases, such as cholestasis, gallstones, nonalcoholic fatty liver diseases, liver and intestinal carcinogenesis, but also in systemic metabolic abnormalities such as atherosclerosis.6–10
As a transcription factor (TF), FXR induces gene transcription by directly binding to an IR1, as a heterodimer with the retinoid X receptor α (RXRα), in promoters of target genes including Nr0b2, Abcb11, Ibabp, Osta, and Ostb.11 However, direct binding of FXR homodimer to promoter of Apoa1, encoding apolipoprotein A-I, suppresses gene transcription.12 FXR also suppresses gene transcription indirectly by way of induction of small heterodimer partner (SHP; encoded by Nr0b2), an orphan nuclear receptor with only a ligand, but not a DNA, binding domain. SHP suppresses transcription through interactions with other TFs.13,14 FXR has emerged as a critical factor in mediating crosstalk between the liver and intestine to maintain bile acid, lipid, and glucose homeostasis. Activation of FXR in the intestine induces fibroblast growth factor 15/19 (Fgf 15/FGF 19) that travels to the liver to suppress the transcriptional activation of Cyp7a1 that encodes a rate-limiting enzyme in bile acid synthesis.15–17
Despite the importance of FXR in regulating liver and intestine pathophysiology, a complete understanding of FXR-DNA interaction at the genomic level is not known. Several studies have identified transcriptional profiles in liver and intestine with FXR activation and/or deletion by microarray analysis.18–20 However, this approach cannot elucidate direct FXR binding. In addition, many in vitro FXR binding studies were performed in cell lines not of hepatic or intestinal origin. Instead, FXR was overexpressed in an artificial cellular environment and the promoters used to investigate FXR binding activity were outside of their natural chromatin context. Thus, many binding sites for a TF in living cells could be masked by the presence of a nonpermissive chromatin environment and, therefore, the assays would reveal a false-positive binding at these sites. New techniques have been developed to identify genomic binding sites of TFs. These techniques are chromatin immunoprecipitation (ChIP) followed by either the hybridization of the immunoprecipitated DNA pool to a tiling array (ChIP-chip) or by end-sequencing of millions of immunoprecipitated DNA fragments (ChIP-seq). ChIP-chip data tend to have low resolution and are often quite noisy,21 and therefore, even with certain challenges, ChIP-seq is a better tool for analyzing genome-wide binding of TFs.22 ChIP-seq has already been applied for quantitatively detecting nuclear receptor binding sites in a genome-wide manner.23
In the current study we used ChIP-seq analysis to determine genome-wide FXR binding sites, in both the mouse liver and intestine. The results not only revealed novel binding sites for FXR, but also implicated new patterns of transcriptional regulation. This work highly suggests novel mechanism(s) by which FXR regulates tissue-specific gene expression in vivo and reveals potential pathways to be regulated by FXR.
Materials and Methods
Animals and Treatment
Ten-week-old fasted C57BL/6 male mice (n = 4 per group) were gavaged with vehicle (1% methocellulose, 1% Triton X-100 in phosphate-buffered saline [PBS]) or GW4064 (75 mg/kg). GW4064 was synthesized by the Chemical Discovery Laboratory at the University of Kansas, Lawrence, KS. Liver and intestines (ileum and colon) were collected 2, 4, or 8 hours later. Animal protocols and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Kansas Medical Center.
Chromatin Immunoprecipitation for Sequencing
This procedure was performed by Genpathway (San Diego, CA). Briefly, flash-frozen tissues were fixed in formaldehyde before being quenched with glycine. The nuclei were extracted and sonicated to yield 500–1,000 basepair (bp) DNA fragments. Chromatin were precleared with blocked Staph A cells (Pansorbin, CalBiochem, San Diego, CA) before incubation with aChIP-quality anti-FXR antibody (H-130x, Santa Cruz Biotechnology, Santa Cruz, CA). Antibody specificity for mouse FXR was validated and is presented in supporting data (Supporting Fig. 1a,b). Samples were incubated with prepared Staph A cells to extract antibody-chromatin complexes, followed by washing and elution. DNA fragments associated with the FXR antibody were released and purified. The purified DNA fragments were first analyzed by quantitative PCR (qPCR) with primers amplifying known FXRREs of target genes. The primers are presented in the supporting data (Supporting Table 1).
ChIP-Seq
ChIP DNA fragments associated with the FXR antibody were amplified using the Illumina ChIPSeq DNA Sample Prep Kit (San Diego, CA). The DNA libraries generated were further tested by qPCR at the same specific genomic regions as the original ChIP DNA to assess quality of the amplification reactions. DNA libraries were sequenced on a Genome Analyzer II by Illumina Sequencing Services. After sequencing, data files were processed through Genome Analyzer Pipeline Software (Illumina) and mapped to the genome. Enriched intervals, referred to as peak values, were defined when a given region appeared more than 20 times (a conservative threshold was arbitrarily set at >20). A genomic region containing more than one enriched interval overlapping by at least one basepair was defined as an active region. BED files are stored in the UCSC database (http://genome.ucsc.edu/) and can be downloaded using UCSC genome browser (custom track number will be added).
The distribution of FXR binding sites relative to the transcription start site (TSS) was analyzed by JMP 7.0. Analysis was done to determine the average peak value of FXR binding as well as to determine the total number of FXR binding events in relation to distance of site from a gene TSS.
Motif Identification
ChIP-seq peaks unique to liver, intestine, or both were used to define tissue-specific FXR-DNA binding motifs. Using the midpoint from the 500 highest peak values for each tissue as a reference, 48 bp sequences flanking either side were retrieved from the UCSC Browser.24 Each file was then independently run using MEME (Multiple Em for Motif Elicitation).25 MEME uses a position-dependent letter probability matrix that describes the probability of each possible letter at each position in the pattern. Individual MEME motifs do not contain gaps. Patterns with variable-length gaps were split by MEME into two or more separate motifs. Motifs containing or resembling the canonical “AGGTCA” motif (which were also the highest scoring motifs) were cross-compared between tissues.
Pathway Analysis
Peaks identified in ChIP-seq data, in both liver and intestine, that were located 0–2 kb upstream from TSSs were analyzed using the Functional Annotation Tool in DAVID (DAVID; http://www.david.niaid.nih.gov).26 For a pathway or process to be defined, the threshold count was set at 2 with a minimum EASE (Expression Analysis Systematic Explorer) score, a modified Fisher Exact Test, of 0.1. Only Bonferroni-corrected P values with false-discovery rates (FDRs) less than or equal to 0.1 were accepted.
ChIP-qPCR
ChIP assays were performed on mice livers and intestines (n = 4) using EZ CHIP KIT (Millipore, Temecula, CA) following a similar protocol as described above. qPCR reactions were carried out using Maxima SYBR Green (Fermentas, Glen Burnie, MD).
Statistical Analysis
For ChIP-qPCR data, the difference between two groups was analyzed by Student’s t test. P < 0.05 was considered statistically significant.
Results
Determining the Optimal Timepoint of FXR-DNA Binding In Vivo Following Ligand Treatment
We first determined the optimal timepoint for FXR-DNA binding following treatment with GW4064 for 2, 4, or 8 hours.
In liver, FXR is known to bind to the promoter of the Nr0b2 gene.27 Conversely, in intestine, FXR binds to FXRREs in the promoter regions of Osta and Ostb genes.28 Therefore, initial ChIP-qPCR assays were performed to amplify known FXRREs in Nr0b2 (−320 to −220 bp upstream of TSS) for liver samples and Osta (−1,245 to −1,145 bp upstream of TSS) and Ostb (−220 to −150 bp upstream of TSS) for intestine samples. In both liver and intestine samples, binding of FXR to known FXRREs was strong and independent of GW4064 treatment when compared to untranscribed regions (Untr6), which serves as a negative control (Supporting Fig. 2a,b). This confirms the specificity of FXR binding. Based on these results, only samples from GW4064-treated mice were used for ChIP-seq analysis. Liver and intestine samples with 4- and 2-hour GW4064 treatment, respectively, showed relatively stronger FXR binding, thereby they were used for subsequent detection of FXR genome-wide binding sites by ChIP-seq.
Validation of FXR Binding Sites Discovered by ChIP-seq
To validate the ChIP-seq results, novel FXR binding sites were randomly chosen for confirmation by ChIP-qPCR assay. In the ChIP-seq dataset, the intensity of FXR binding to DNA was reported as peak value. Many known FXRREs previously identified were detected with relatively high peak values (Table 1). Based on peak values reported in ChIP-seq data, the sites listed in Table 1, together with other randomly selected novel sites, were categorized as having high (>300), medium (100–300), or low (<100) peak values and were tested by ChIP-qPCR, with gene names listed in Supporting Tables 2 and 3. These sites were tested in liver only (20 sites), intestine only (5 sites), or both tissues (27 sites). The results showed 100% of the sites with high, 84.4% with medium and 70% with low peak values were confirmed (Table 2). Altogether, 86% (68/79) of the sites were verified by ChIP-qPCR as true FXR binding sites with examples shown in Supporting Fig. 3a – c. These results led us to conclude the ChIP-seq results are accurate and we proceeded with more detailed analysis of the ChIP-seq results.
Table 1.
Previously Reported FXR Binding Sites Detected by ChIP-Seq
| Gene | Binding Site | Peak Value | Tissue |
|---|---|---|---|
| Nr0b2 | −320 to −220 bp upstream of TSS | 258; 124 | Liver; intestine |
| Abcb11 | −240 to −140 bp upstream of TSS | 308 | Liver |
| Fgf15 | 1880–1980 bp with in an intron | 43; 273 | Liver; intestine |
| Osta | −1245 to −1145 bp upstream of TSS | 502 | Intestine |
| Ostb | −220 to −150 bp upstream of TSS | 271; 572 | Liver; intestine |
| Fabp6 | site 1: −220 to −120; site 2: −2990 to −2890; and site 3: −7600 to −7500 bp upstream |
540, 638, 93 | Intestine |
Table 2.
Validation of ChIP-seq: Percentage of ChIP-seq Binding Sites Confirmed by ChIP-qPCR
| Peak Value | % Confirmation | ||
|---|---|---|---|
| 27 genes | >300 | 100.0 | (27/27) |
| 32 genes | 100–300 | 84.4 | (27/32) |
| 20 genes | <100 | 70.0 | (14/20) |
Analysis of Genome-Wide FXR Binding Sites in Liver and Intestine
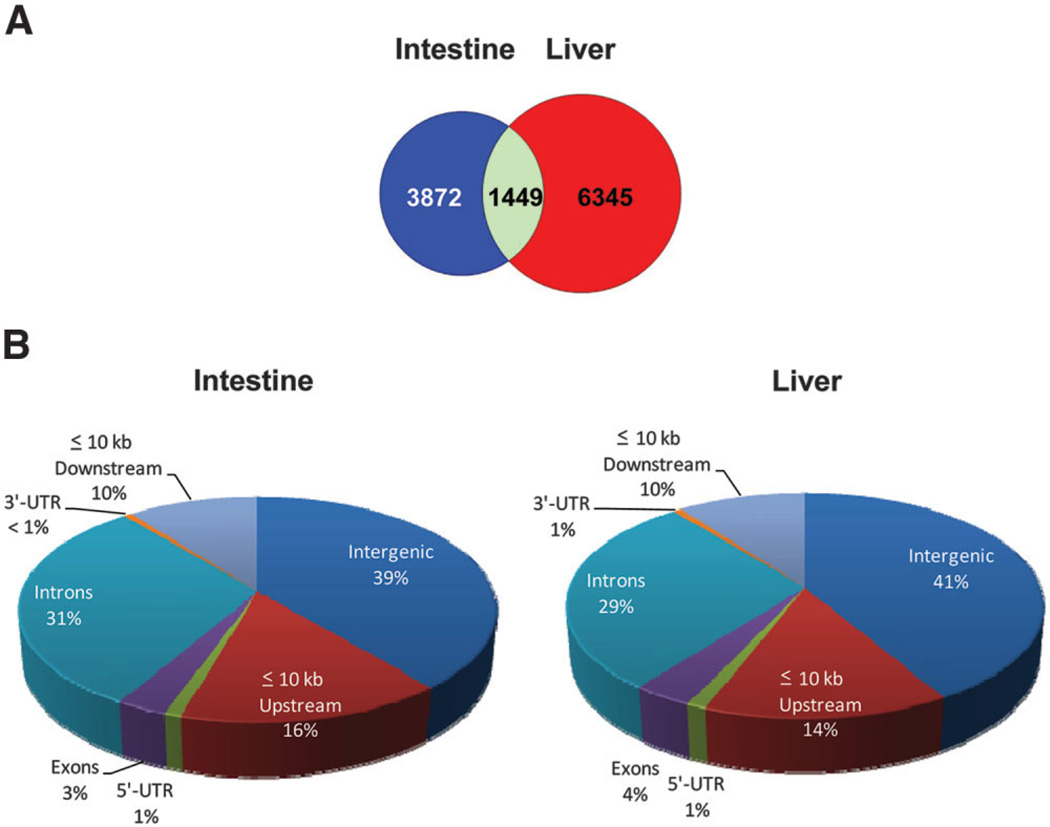
There were a total of 7794 FXR binding sites in liver and 5321 in intestine. However, 6345 sites were found specifically in liver, 3872 specifically in intestine, and 1449, representing 11% of total sites, were bound by FXR in both tissues (Fig. 1A). The FXR binding sites were widely distributed throughout the mouse genome. Forty-one percent of total FXR binding sites in liver and 39% in intestine were located more than 10 kb upstream of a RefSeq gene (intergenic regions). Conversely, 59% of the total sites in liver and 61% in intestine were found directly associated with a gene, indicating that FXR binding in both liver and intestine is more highly concentrated within coding regions of the mouse genome. Of these binding sites: 1% in liver and intestine overlap with 5′-untranslated regions (5′UTRs); 4% in liver and 3% in intestine were located in exon regions; 29% in liver and 31% in intestine were located in intron regions; less than 1% in liver and intestine overlap with 3′UTRs; and 10% in liver and intestine were located within 10 kb downstream of genes (Fig. 1B).
Fig. 1.
Total FXR binding sites in liver and intestine. (A) A Venn diagram of total FXR binding in liver (red), intestine (blue), or both (green). (B) Percentage of FXR binding sites in liver and intestine that were distributed to >10 kb from genes (intergenic), <10 kb upstream of genes, introns, exons, 5′UTRs, 3′UTRs, and <10 kb downstream of genes.
Many genes were bound by FXR at multiple regions, including upstream, in-gene, and downstream of genes, including known FXR target genes previously identified to have only one FXRRE. For example, multiple FXR binding sites were identified for Nr0b2 and Ostb (Fig. 2A,B). Therefore, the total number of genes to which FXR bound was less than the total number of binding sites. There were 4248 genes in liver and 3406 genes in intestine to which FXR was shown to bind at a single or multiple regions. Among them, 1713 genes were shared by both liver and intestine.
Fig. 2.
Histogram of novel FXR binding sites within known FXR target genes, Nr0b2 and Ostb. The y-axis displays the peak value and the x-axis shows the chromosomal location of the gene. The top panel (displayed in red) represents FXR binding in liver, and the bottom panel (displayed in blue) represents FXR binding in intestine. Genes that are displayed above the chromosome scale are oriented in the sense directions (right arrow), and genes that are displayed below the chromosome scale are oriented in the antisense direction (left arrow). The threshold is set at peak value >20. (A) The Nr0b2 gene is located on chromosome 4 (Chr4): 133109305-133112451 and is oriented in the sense direction in this figure. FXR binds at two locations around the Nr0b2 gene in intestine, promoter region (peak value = 124) and 3′ end of the gene (peak value = 381). FXR binds at four locations of the Nr0b2 gene in liver, two in the promoter and two at the 3′ end (peak values are 69, 258, 498, and 49, respectively). (B) The Ostb gene is located at Chr9: 65260560-65270580 and is oriented in the antisense direction. In intestine, FXR binds at four locations near the Ostb gene, with three sites within the promoter (peak value = 572, 27, and 210) and one located at the 3′ end (peak value = 132). In liver, FXR binds once within the promoter (peak value = 271) and once at the 3′ end (peak value = 44). Novel FXR sites found at the 3′ end of both Nr0b2 and Ostb gene have not been reported and are indicated by an asterisk.
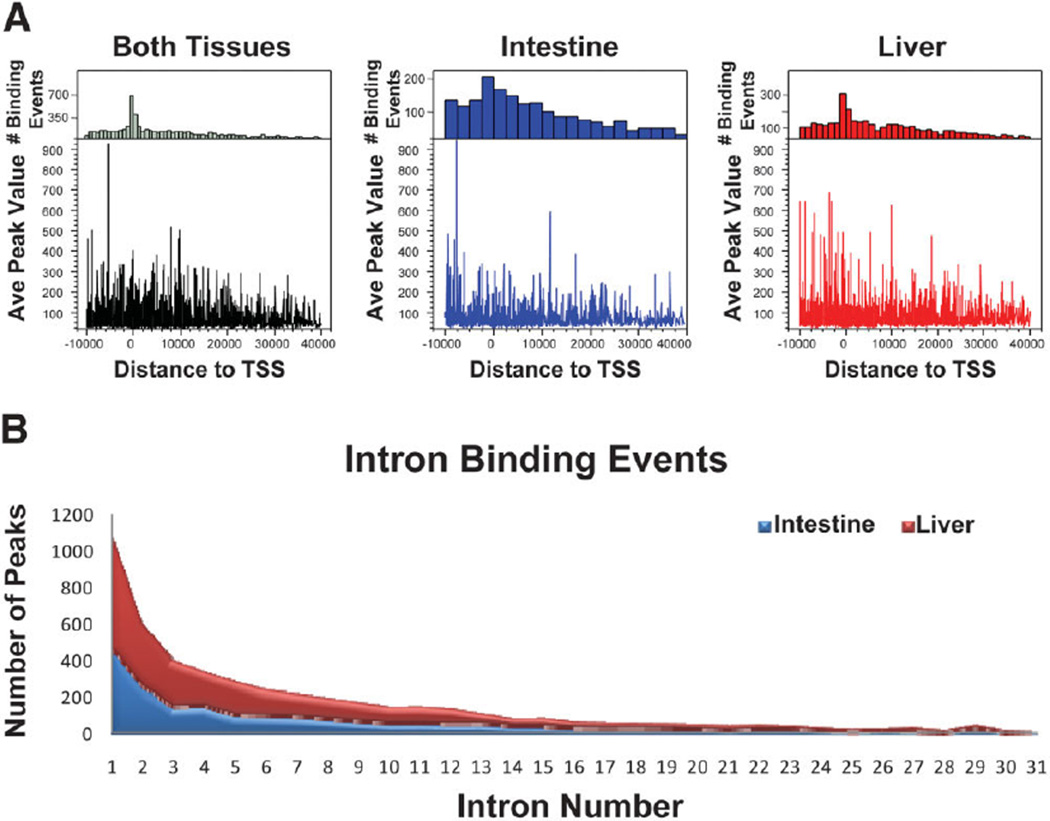
The average peak value of FXR binding sites was higher for sites within 10 kb upstream of TSSs, and the frequency of binding (number of binding events) was higher for binding sites located near the TSS of target genes in both liver and intestine (Fig. 3A). The average peak value decreased with distance from proximal promoter and frequency of binding decreased with the distance from the TSSs. As reported earlier, there was a high percentage of intron binding of FXR. Therefore, the genome-wide intron binding of FXR was further evaluated for cumulative binding events of FXR within intron regions in the liver and intestines (Fig. 3B). Most of the intron binding of FXR was in close proximity to TSSs of target genes, with 1107 peaks from both tissues falling within the first intron of a gene (640 peaks in the liver [red] and 467 peaks in the intestine [blue]).
Fig. 3.
Distribution of total FXR binding sites relative to TSSs and intron binding of FXR. (A) The top panel is the frequency distribution of FXR binding, showing the number of binding events (y-axis) at each distance from TSSs (x-axis). The bottom panel displays average peak value of binding sites at each distance from TSSs. The y-axis displays the average peak value and the x-axis shows the distance of binding site from the TSS. The highest number of FXR binding events was greatest at the TSSs of genes, and the average peak value of FXR binding sites was greatest within 10 kb upstream of TSSs of genes. (B) The cumulative binding events of FXR distributed only to introns of genes in the liver (red) and intestine (blue). The graph displays the total number of FXR binding peaks (y-axis) in the liver and intestine located within intron 1–31 of genes (x-axis). There were a higher number of peaks within the first intron of genes, with 1107 total peaks within first intron for both liver and intestine.
Novel FXR Binding Sites Revealed by ChIP-Seq
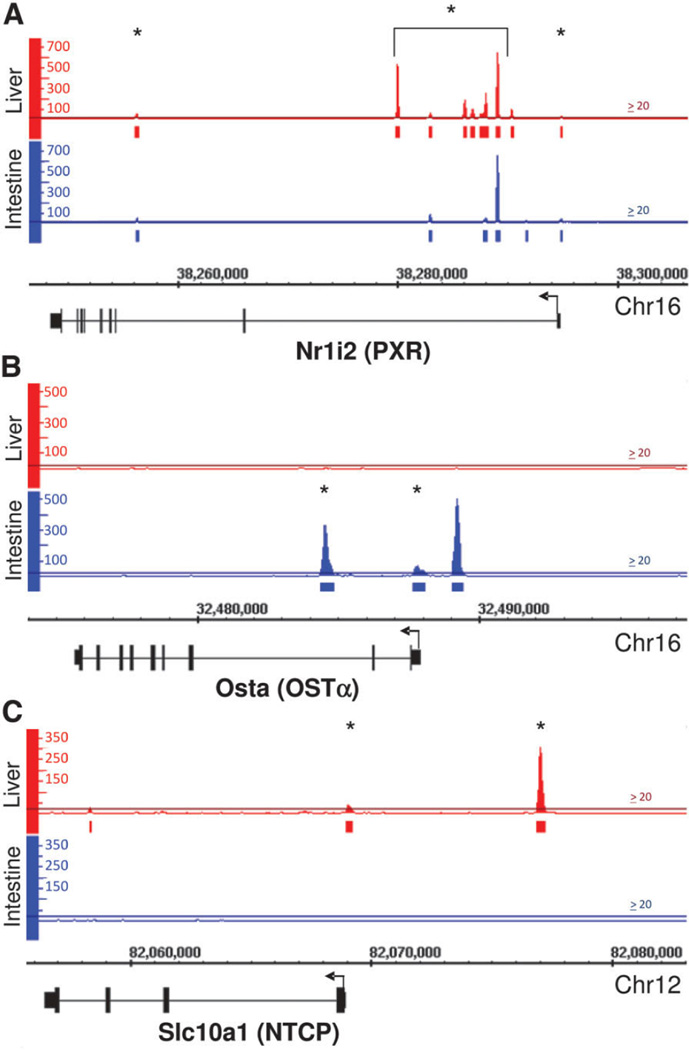
FXR was found to bind to many novel sites within the mouse genome. For example, FXR bound to several sites within the first intron of the Nr1i2 gene, which encodes pregnane X receptor (PXR; highest peak value: 654 in liver and intestine, indicated by an asterisk; Fig. 4A). A novel FXR binding site was also discovered at a 3′ regulatory region of the Nr0b2 gene (peak value: 498 in liver and 381 in intestine, indicated by an asterisk; Fig. 2A).
Fig. 4.
Histogram of novel FXR binding sites within Nr1i2, Osta, and Slc10a1. (A) The Nr1i2 gene is located on Chr16: 38248437-38294909 and is oriented in the antisense direction. Previously uncharacterized FXR binding sites were found at multiple regions within the Nrli2 gene, indicated by an asterisk (the brackets include all FXR binding sites in the 1st intron of the gene). These binding sites were located within the promoter region and introns 1 and 2 of the gene (highest peak value was 646 in liver and 654 in intestine). (B) The Osta gene is located on Chr16: 32475664-32487965 and is oriented in antisense direction. FXR binds to Osta in the intestine but not in the liver, displaying tissue-specific binding. Binding of FXR to promoter region (−1245 to −1145 upstream of TSS) of Osta in the intestine has been previously characterized, and serves as a positive control for our analysis (peak value = 502). However, binding of FXR within −100 bp upstream of TSS (peak value = 70) and within intron 2 of Osta in the intestine are novel findings (peak value = 333; indicated by an asterisk). (C) The Slc10a1 gene is located on Chr12: 82056479-82068971 and is oriented in the antisense direction. FXR binds at two sites in the Slc10a1 promoter (around −100 bp and −8,000 bp upstream of TSS, peak value = 38 and 304, respectively; indicated by an asterisk). Binding of FXR to these sites were confirmed by ChIPqPCR (data not shown).
FXR has been shown to up-regulate both organic solute transporter α and β (Ostα and Ostβ, encoded by Osta and Ostb, respectively) in liver and intestine.28,29 This has been previously recorded as being a result of direct FXR binding to regulatory regions of Osta and Ostb genes in both liver and intestine.30 The current study confirmed the binding of FXR in intestine to upstream regions of Osta and Ostb (Fig. 4B: Osta and Fig. 2B: Ostb). In addition, in liver FXR bound to upstream regions of Ostb (Fig. 2B). However, these results clearly showed that FXR did not bind to regulatory regions of Osta in liver (Fig. 4B). Furthermore, binding of FXR to 3′ end of Ostb and to the proximal promoter and intron region of Osta in the intestine has not been previously reported (indicated by an asterisk). The present results also showed FXR bound directly to two upstream regulatory regions of Slc10a1, which encodes Na+-taurocholate cotransporting polypeptide (Ntcp), in liver with relatively high intensity (peak value: 50 at −150 to −50 bp and 300 at −8,100 to −8,000 bp), which are uncharacterized FXR binding sites (indicated by an asterisk, Fig. 4C). Ntcp is a bile acid uptake transporter essential for hepatic uptake of conjugated bile acids. ChIP-seq results on FXR binding to Osta, Ostb, and Slc10a1 were confirmed by ChIP-qPCR (data not shown). The histograms of Osta and Slc10a1 clearly show tissue-specific DNA binding of FXR, with Osta only being bound in intestine (Fig. 4B) and Slc10a1 only being bound in liver (Fig. 4C).
Motif Analysis of FXR Binding Sites
As mentioned above, the most characterized FXRRE is an IR1. This motif has been found within many known FXR target genes, including Nr0b2, Fabp6 (encodes intestine bile acid binding protein, Ibabp) and Fgf15/FGF19. In the current study the most common motif identified was an IR1 (Fig. 5). Interestingly, a half nuclear receptor binding site, AGGTCA, was found adjacent to the IR1 at the 3′ end in the liver. Conversely, the most commonly occurring sequences for intestine FXR binding sites were an IR1 and an everted repeat separated by two nucleotides (ER2). As with the liver samples, there was a half-site adjacent to the FXR binding sites in intestine. However, the half-site could be at the 3′ end (TGACCT) if FXR binding site was an ER2 or at the 5′ end (TGACCT) if FXR binding site was an IR1. Nevertheless, the motif analysis suggests tissue-specific sequence motifs to which FXR recognizes and binds.
Fig. 5.
Motif analysis. The three most commonly identified sequence motifs of the top 500 FXR binding sites in the liver (top panel) and intestine (bottom panel) are shown according to MEME analysis. In the liver an IR1 was the most common sequence motif found. Two of the three motifs identified (left and right motif) in the liver showed the presence of a half nuclear receptor binding site at the 3′ end of the IR1. In the intestine, an IR1 (left motif) or an ER2 (middle motif) were motifs commonly found within the top 500 FXR binding sites. The motifs found in the intestine are arbitrarily aligned to match up with the sequence motifs in the liver to show sequence similarity. In the intestine, a half nuclear receptor binding site was also associated with the identified FXRRE either at the 5′ end (half-site 1) or at the 3′ end (half-site 2), depending on which motif is recognized as an FXRRE. E-values, a measure of significance, were between 3.8e-037 to 9.3e-085.
Biological Pathway Analysis
FXR is essential in regulating pathways important for bile acid, lipid, and glucose homeostasis. However, FXR is likely involved in other biological pathways that have not been previously recognized. To further determine biological pathways potentially regulated by FXR, genes bound by FXR less than 2 kb upstream of their TSSs were selected for biological pathway analysis (Table 3). Functional annotation analysis revealed FXR may be involved in several metabolic pathways in liver. In fact, 90% of genes categorized into the cellular metabolism process were bound by FXR in the liver. Furthermore, FXR may also be significantly involved in regulating other pathways that are not known to be regulated by FXR, such as amino acid and nitrogen compound metabolism.
Table 3.
Pathways Enriched with FXR Binding 2 kb Upstream of Genes
| Tissue | Biological Process | Genes* | % Bound by FXR |
P Value | Bonferroni |
|---|---|---|---|---|---|
| Liver | Primary metabolic process | 92 | 93.9 | 1.70E-25 | 8.80E-22 |
| Cellular metabolic process | 88 | 89.8 | 1.90E-20 | 9.60E-17 | |
| Biosynthetic process | 38 | 38.8 | 7.60E-16 | 4.00E-12 | |
| Cellular biosynthetic process | 25 | 25.5 | 5.60E-09 | 2.90E-05 | |
| Nitrogen compound metabolic process | 25 | 25.5 | 4.90E-17 | 2.60E-13 | |
| Amine metabolic process | 24 | 24.5 | 1.40E-16 | 5.80E-13 | |
| Amino acid and derivative metabolic process | 24 | 24.5 | 3.80E-18 | 2.00E-14 | |
| Amino acid metabolic process | 23 | 23.5 | 3.20E-19 | 1.70E-15 | |
| Lipid biosynthetic process | 21 | 21.4 | 8.90E-17 | 5.80E-13 | |
| Steroid metabolic process | 18 | 18.4 | 7.50E-17 | 5.80E-13 | |
| Monosaccharide metabolic process | 11 | 11.2 | 3.80E-08 | 1.90E-04 | |
| Hexose metabolic process | 11 | 11.2 | 3.10E-08 | 1.60E-04 | |
| Sterol metabolic process | 11 | 11.2 | 4.50E-11 | 2.30E-07 | |
| Cholesterol metabolic process | 11 | 11.2 | 1.70E-11 | 8.80E-08 | |
| Glucose metabolic process | 10 | 10.2 | 2.80E-08 | 1.50E-04 | |
| Lipid catabolic process | 10 | 10.2 | 2.40E-08 | 1.20E-04 | |
| Fatty acid oxidation | 8 | 8.2 | 1.30E-10 | 7.00E-07 | |
| Steroid biosynthetic process | 7 | 7.1 | 6.30E-06 | 3.20E-02 | |
| Glycine, serine and threonine metabolism | 7 | 7.1 | 1.20E-04 | 2.30E-02 | |
| Isoprenoid metabolic process | 7 | 7.1 | 8.70E-08 | 4.50E-04 | |
| Pyruvate metabolic process | 7 | 7.1 | 8.40E-09 | 4.30E-05 | |
| Pyruvate metabolism | 6 | 6.1 | 5.60E-04 | 1.00E-01 | |
| Amino acid biosynthetic process | 6 | 6.1 | 7.80E-06 | 4.00E-02 | |
| Glycerol ether metabolic process | 6 | 6.1 | 1.30E-06 | 6.80E-03 | |
| Glycerolipid metabolic process | 6 | 6.1 | 1.10E-06 | 5.70E-03 | |
| Neutral lipid metabolic process | 6 | 6.1 | 1.10E-06 | 5.70E-03 | |
| Acylglycerol metabolic process | 6 | 6.1 | 9.20E-07 | 4.80E-03 | |
| Sterol transport | 6 | 6.1 | 9.50E-08 | 4.90E-04 | |
| Cholesterol transport | 6 | 6.1 | 7.00E-08 | 3.60E-04 | |
| Triacylglycerol metabolic process | 5 | 5.1 | 9.80E-06 | 4.90E-02 | |
| Gluconeogenesis | 5 | 5.1 | 6.50E-06 | 3.30E-02 | |
| Cholesterol homeostasis | 5 | 5.1 | 5.20E-06 | 2.70E-02 | |
| Lipid homeostasis | 5 | 5.1 | 5.20E-06 | 2.70E-02 | |
| Sterol homeostasis | 5 | 5.1 | 5.20E-06 | 2.70E-02 | |
| Intestine | Catalytic activity | 283 | 40.8 | 2.20E-16 | 6.00E-13 |
| Oxidoreductase activity | 64 | 9.2 | 2.30E-07 | 6.20E-04 | |
| Generation of precursor metabolites and energy | 46 | 6.6 | 2.40E-07 | 1.20E-03 | |
| Electron transport | 36 | 5.2 | 9.90E-06 | 5.00E-02 | |
| Iron ion binding | 30 | 4.3 | 3.10E-06 | 8.40E-03 | |
| Cofactor binding | 24 | 3.5 | 1.30E-07 | 3.40E-04 | |
| Vitamin binding | 19 | 2.7 | 1.70E-08 | 4.60E-05 | |
| Monooxygenase activity | 17 | 2.4 | 1.00E-05 | 2.80E-02 | |
| Complement and coagulation cascades | 14 | 2 | 9.70E-05 | 1.90E-02 | |
| Unspecific monooxygenase activity | 9 | 1.3 | 2.10E-05 | 5.50E-02 |
The number of genes bound by FXR in this pathway.
In intestine, however, FXR seems to regulate different pathways than in liver. For example, FXR binds to 40% of the genes categorized in the catalytic pathway processes, suggesting FXR may be highly involved in regulating catalytic pathways in the intestine. In addition, pathways involved in oxidoreductase activity, monooxygenase activity, and cofactor binding seemed to be highly enriched with FXR binding in intestine (Table 3). Interestingly, pathways involved in intestinal inflammation, such as complement and coagulation cascades, are also enriched with FXR binding.
Discussion
In the current study, genome-wide FXR binding sites were analyzed by ChIP-seq analysis in both liver and intestine following treatment of mice with a potent synthetic FXR ligand. The current study not only summarizes binding sites for FXR within the mouse genome in liver and intestine, but also suggests potential tissue-specific patterns of transcriptional regulation mediated by FXR.
The initial validation study by regular ChIP-qPCR assay showed that in vivo FXR was already bound to its response element in the regulatory region of FXR target genes, presumably due to activation by endogenous ligands (bile acids). The binding intensity was enhanced upon treatment with a potent synthetic ligand, GW4064. Thereby treatment with GW4064, a ligand with higher affinity for FXR than bile acids,31 may result in an endogenous-to-synthetic ligand switch. In agreement with this notion, previous in vitro studies using HepG2 cells, which are known to synthesize bile acids, have shown that FXR binds to its response element weakly in the presence of control vehicle, and treatment with various synthetic ligands enhanced the binding.32,33 Furthermore, we also observed a weak FXR interaction with its response element in the mouse hepatoma cell line, Hepa1c1c cells, and this interaction was enhanced upon treatment with GW4064 (data not shown). Collectively these results may indicate a ligand switch for FXR.
In order to address the possibility of false-positive binding, the ChIP-seq data were further validated by applying a second peak finding algorithm, MACS, to our dataset.34 This program looks for local enrichments of alignments that show the correct orientation that would be expected for a ChIP peak (peak modeling). This program thus eliminates at least some false positives. The analysis from the MACS showed that of the 7,796 liver peaks (intervals) in the original analysis, 6,942 (89.05%) were also identified by MACS, and of the 5,324 original ileum peaks, 5,023 (94.35%) were also identified by MACS.
Using the ChIP-seq discovery approach, DNA regions bound by FXR have been identified throughout the entire mouse genome. This study shows a novel FXR binding pattern with novel FXR binding sites located large distances upstream (intergenic), downstream, or within genes (intragenic). In fact, this study shows a large portion of FXR binding sites are located more than 10 kb away from a RefSeq gene. This indicates long-distance chromatin interactions as a possible mechanism for FXR-mediated regulation of gene transcription.35 However, the majority of FXR binding sites are concentrated within coding regions of the mouse genome. Of these sites, FXR is highly associated with the proximal promoter and first intron. Recent ChIP-tiling array and ChIP-seq reports on other nuclear receptors and TFs have also revealed a high percentage of intergenic binding.23,36,37 In addition, a previous report of genome-wide estrogen receptor α (ERα) binding has also shown a higher intensity of binding near the TSSs of genes, which is consistent with this study.36
This study exhibits a high degree of tissue-specific binding sites for FXR as well as a tissue-specific binding pattern, with a relatively low portion of total sites shared between liver and intestine. For example, Nr0b2, a classical FXR target gene encoding SHP, is expressed at a much higher level in liver than in intestine.38 In both tissues, SHP expression levels can be strongly induced through activation of FXR. The current study reveals that FXR binds with high intensity to a downstream IR1 of the Nr0b2 gene in both liver and intestine, but shows a tissue-specific binding pattern. Furthermore, FXR also binds to other known target genes, including Osta, Ibabp, and Slc10a1 in a tissue-specific manner. Ibabp and Osta are bound by FXR in intestine but not in liver, which, for Osta, is in contrast with a previous report that FXR also binds to the promoter region of Osta in liver.29 Likewise, the Slc10a1 gene, encoding Ntcp, has two FXR binding sites within promoter regions in liver. Ntcp has been shown to be suppressed by high concentrations of bile acids under cholestatic conditions through an FXR-SHP-mediated mechanism.39 However, the finding of FXR binding sites in the Slc10a1 promoter region suggests that FXR may directly regulate the transcription of the Slc10a1 gene. Furthermore, tissue-specific FXR regulation of these genes may be mediated by tissue-specific cofactors or chromatin modification.
The motif analysis indicates the most represented FXR binding motif in mouse liver is an IR1, which has been identified in many FXR target genes. In mouse intestine, an IR1 was also the most commonly represented motif; however, an ER2 motif was also represented as a possible FXR binding sequence. In addition, there was a half nuclear receptor binding site adjacent to the FXR binding site in both liver and intestine. The motif analysis suggests there are tissue-specific sequence motifs to which FXR recognizes and binds. In addition, this new finding indicates a potential interaction between FXR and other orphan nuclear receptors/TFs known to bind to this half-site.40 These cofactors may be important for modulating transcriptional activation of FXR target genes in a tissue-specific manner. The interaction between hormone nuclear receptors and other TFs have been previously reported. For example, ERα has been shown to interact with FoxA1 (hepatocyte nuclear factor 3β) for optimal DNA binding and transcriptional activation.37
The current study also suggests that FXR may directly regulate more diverse biological pathways. For example, FXR binds to Rara and Rorc and may regulate transcriptional activation of these genes as well as retinol metabolism and hormone metabolism (data not shown). This means that FXR not only regulates bile acid and lipid homeostasis, but may also be directly involved in other cellular functions. In addition, FXR may regulate bile acid and lipid homeostasis to a greater extent than previously determined. Further studies are needed to characterize the significance of FXR’s involvement in these pathways to clarify how FXR maintains cellular and organ homeostasis.
In summary, this study analyzed genome-wide FXR-DNA binding in both the mouse liver and intestine following ligand treatment. The identification of novel FXR binding sites, as well as unique binding patterns, enable a more profound understanding of FXR-regulated biological and disease pathways in a tissue-specific manner.
Supplementary Material
Acknowledgments
We thank Ms. Noriko Esterly for help with collecting animal tissues and Ms. Kaori Nakamoto for help with cell culture. We also thank Dr. Curtis Klaassen and Ms. Yue Cui for discussion in the Department of Pharmacology, Toxicology, and Therapeutics, and Dr. Kenneth Peterson and his group in the Department of Biochemistry at the University of Kansas Medical Center. We thank Dr. Jeff Aube and laboratory in the Department of Medicinal Chemistry, University of Kansas, Lawrence, KS for synthesizing GW 4064 for use in the animal studies.
Supported by DK031343-01 (to G.L.G.); 5P20-RR021940 (to G.L.G., X.B.Z.); T32ES007079 (to A.M.T.); Madison and Lila Self Graduate Fellowship from the University of Kansas (to S.N.H.).
Abbreviations
- ChIP-seq
chromatin immunoprecipitation followed by massively parallel sequencing
- ER2
everted hexanucleotide repeat separated by 2 nucleotides
- FXR
farnesoid X receptor
- IR1
inverted hexanucleotide repeat separated by 1 nucleotide
- Ntcp
sodium taurocholate cotransporting polypeptide
- Ostα/β
organic solute transporter alpha/beta
- PXR
pregnane X receptor
- RE
response element
- RXRα
retinoid X receptor α
- SHP
small heterodimer partner
- TF
transcription factor
- TSS
transcription start site.
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 2.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 3.Lambert G, Amar MJ, Guo G, Brewer HB, Jr, Gonzalez FJ, Sinal CJ. The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J Biol Chem. 2003;278:2563–2570. doi: 10.1074/jbc.M209525200. [DOI] [PubMed] [Google Scholar]
- 4.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 6.Guo GL, Santamarina-Fojo S, Akiyama TE, Amar MJ, Paigen BJ, Brewer B, Jr, et al. Effects of FXR in foam-cell formation and atherosclerosis development. Biochim Biophys Acta. 2006;1761:1401–1409. doi: 10.1016/j.bbalip.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 8.Moschetta A, Bookout AL, Mangelsdorf DJ. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med. 2004;10:1352–1358. doi: 10.1038/nm1138. [DOI] [PubMed] [Google Scholar]
- 9.Maran RRM, Thomas A, Roth M, Sheng Z, Esterly N, Pinson D, et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J Pharmacol Exp Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong B, Luyendyk JP, Tawfik O, Guo GL. Farnesoid X receptor deficiency induces nonalcoholic steatohepatitis in low-density lipoprotein receptor-knockout mice fed a high-fat diet. J Pharmacol Exp Ther. 2009;328:116–122. doi: 10.1124/jpet.108.144600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- 12.Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, et al. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest. 2002;109:961–971. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 14.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim I, Ahn S-H, Inagaki T, Choi M, Ito S, Guo GL, et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, et al. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing X, Burgermeister E, Geisler F, Einwachter H, Fan L, Hiber M, et al. Hematopoietically expressed homeobox is a target gene of farnesoid X receptor in chenodeoxycholic acid-induced liver hypertrophy. Hepatology. 2009;49:979–988. doi: 10.1002/hep.22712. [DOI] [PubMed] [Google Scholar]
- 21.Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 26.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 27.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res. 2006;47:201–214. doi: 10.1194/jlr.M500417-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Boyer JL, Trauner M, Mennone A, Soroka CJ, Cai SY, Moustafa T, et al. Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1124–G1130. doi: 10.1152/ajpgi.00539.2005. [DOI] [PubMed] [Google Scholar]
- 30.Landrier J-F, Eloranta JJ, Vavricka SR, Kullak-Ublick GA. The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-{ alpha} and -beta genes. Am J Physiol Gastrointest Liver Physiol. 2006;290:G476–G485. doi: 10.1152/ajpgi.00430.2005. [DOI] [PubMed] [Google Scholar]
- 31.Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, Plunket KD, et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem. 2000;43:2971–2974. doi: 10.1021/jm0002127. [DOI] [PubMed] [Google Scholar]
- 32.Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol Pharmacol. 2005;68:551–558. doi: 10.1124/mol.105.012104. [DOI] [PubMed] [Google Scholar]
- 33.Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, et al. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem. 2008;283:35086–35095. doi: 10.1074/jbc.M803531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li YJ, Fu XH, Liu DP, Liang CC. Opening the chromatin for transcription. Int J Biochem Cell Biol. 2004;36:1411–1423. doi: 10.1016/j.biocel.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Gao H, Falt S, Sandelin A, Gustafsson JA, Dahlman-Wright K. Genome-wide identification of estrogen receptor alpha-binding sites in mouse liver. Mol Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamisako T, Ogawa H, Yamamoto K. Effect of cholesterol, cholic acid and cholestyramine administration on the intestinal mRNA expressions related to cholesterol and bile acid metabolism in the rat. J Gastroenterol Hepatol. 2007;22:1832–1837. doi: 10.1111/j.1440-1746.2007.04910.x. [DOI] [PubMed] [Google Scholar]
- 39.Zollner G, Wagner M, Fickert P, Geier A, Fuchsbichler A, Silbert D, et al. Role of nuclear receptors and hepatocyte-enriched transcription factors for Ntcp repression in biliary obstruction in mouse liver. Am J Physiol Gastrointest Liver Physiol. 2005;289:G798–G805. doi: 10.1152/ajpgi.00319.2004. [DOI] [PubMed] [Google Scholar]
- 40.Ito M, Achermann JC, Jameson JL. A naturally occurring steroidogenic factor-1 mutation exhibits differential binding and activation of target genes. J Biol Chem. 2000;275:31708–31714. doi: 10.1074/jbc.M002892200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.