Significance
How exactly protein motions facilitate substrate recognition and catalysis has remained largely unanswered. Characterization of protein dynamics at atomistic level is essential to understanding function. Molecular dynamics and NMR are helpful in this regard; however, analyzing multidimensional data from very long molecular dynamics (MD) simulations to elucidate key dynamical features observed in NMR remains very challenging. We present results from an approach for data analysis in which dynamics is defined in terms of interresidue contact formation and breaking. Analyzing simulation data on a therapeutically important Cyclophilin A and carrying out NMR experiments, we uncovered remarkable and unprecedented changes in its motions at a site over 15 Å from the active site upon substrate binding and how mutation in this distal site affects catalysis.
Keywords: allostery, enzyme dynamics, residue–residue contacts, Cyclophilin A, molecular dynamics
Abstract
Detailed understanding of how conformational dynamics orchestrates function in allosteric regulation of recognition and catalysis remains ambiguous. Here, we simulate CypA using multiple-microsecond-long atomistic molecular dynamics in explicit solvent and carry out NMR experiments. We analyze a large amount of time-dependent multidimensional data with a coarse-grained approach and map key dynamical features within individual macrostates by defining dynamics in terms of residue–residue contacts. The effects of substrate binding are observed to be largely sensed at a location over 15 Å from the active site, implying its importance in allostery. Using NMR experiments, we confirm that a dynamic cluster of residues in this distal region is directly coupled to the active site. Furthermore, the dynamical network of interresidue contacts is found to be coupled and temporally dispersed, ranging over 4 to 5 orders of magnitude. Finally, using network centrality measures we demonstrate the changes in the communication network, connectivity, and influence of CypA residues upon substrate binding, mutation, and during catalysis. We identify key residues that potentially act as a bottleneck in the communication flow through the distinct regions in CypA and, therefore, as targets for future mutational studies. Mapping these dynamical features and the coupling of dynamics to function has crucial ramifications in understanding allosteric regulation in enzymes and proteins, in general.
As a biomolecule samples various conformations governed by its free energy landscape, it undergoes a wide spectrum of motions that range over a broad timescale and length scale (1). Except for certain cases in which large-scale displacements are observed upon ligand binding (2), these motions, in general, account for modest fluctuations around an average native structure (3). Despite extensive studies, detailed understanding of how conformational dynamics lead to or facilitate function remains formidable (4–6). It has been frequently observed that enzymes in their free unliganded state sample 3D conformations that consist of those visited in the presence of the ligand (7). Differences in intrinsic conformational dynamics in the wild type and the mutant maltose-binding proteins have been shown to be related to association and dissociation of ligand and thereby to affect dissociation constants (8). Enzyme dynamics in Cyclophilin A (CypA), a peptidyl prolyl cis–trans isomerase, have been demonstrated to occur on the same millisecond timescale as the catalytic turnover (9). Mutants of dihydrofolate reductase that lead to suppressed conformational dynamics in the active site have exhibited concomitant loss in enzymatic activity (10).
These exemplary studies are suggestive of a key role for conformational dynamics in recognition and catalysis. This notion has been controversial (10–13) due to limited or, in some cases, a complete lack of microscopic analysis of experimental observations. The suppressed catalytic activity in mutants may be a result of an increase in activation energy and not decreased dynamics (14, 15). Although enzyme dynamics may not be responsible for catalytic speed up relative to the uncatalyzed reaction, CypA dynamics has been shown to be coupled to catalytic function (16). Allosteric regulation, i.e., modification of binding or catalysis at the active site due to binding of a ligand at a distal nonoverlapping site, is widespread in biochemical signaling (17, 18). It is natural that allosteric regulation depends on modulation of protein motions, because substrate binding and catalysis are linked to conformational dynamics (19, 20). Unlike static X-ray structures, solution NMR relaxation dispersion techniques have been instrumental in providing high-resolution conformational exchange information using site-specific isotope labeling (21). In NMR studies of CypA, a dynamic continuum has been identified such that the relaxation profiles cannot be globally fit to one or two exchange phenomena and are instead indicative of more localized motions (22). Exchange rates coalesce somewhat during turnover, perhaps suggesting an increase in coordination throughout the protein, but appear to still consist of localized motions that are not fully coherent (23). However, dynamical signals during catalytic turnover could be affected by substrate binding and unbinding, especially if the substrate binding affinity is low, thereby leading to ambiguity in the interpretation of NMR analysis (23).
Complementary to experiments, long-timescale molecular dynamics (MD) simulations can be greatly instrumental in providing a microscopic picture of biomolecular dynamics and establishing its exact linkage to function. However, the challenge at hand is to elucidate the key dynamical features and correlations between different parts of the protein from a vast amount of multidimensional time-dependent data from MD. Principal component analysis (PCA) is often used to reduce the dimensionality of conformational space and map the differences in conformational ensembles. PCA, which is usually performed on Cartesian coordinates, may sometimes mask certain important long-range dynamical relations and complex features of biomolecular conformational dynamics. Comparing residue–residue contacts in various isoforms from difference contact maps built from simulation trajectories has aided in determining certain similar structural properties and some unique to a particular isoform (24). Another analysis that involves monitoring the time evolution of residue–residue contact formation and breaking has been proven useful in identifying certain characteristic events during conformational transitions (25). Correlated motions between different biomolecular segments have been identified using cross-correlation analysis and building dynamical networks (25–29). However, interpretation of the results of such analysis in case of CypA, which exhibit subtle changes upon substrate binding and catalysis, has been ambiguous.
Here, we characterize the conformational dynamics of CypA using very long atomistic standard MD simulations in explicit solvent, and the results are validated using NMR experiments. CypA is an archetypal and extensively studied enzyme belonging to the family of peptidyl prolyl isomerases (PPIases), speeding the cis–trans isomerization of peptidyl prolyl ω-bond in its protein substrates by more than 105 times (30–34). In five independent microsecond-long simulations, we monitored the dynamics in wild-type CypA, V29L variant of CypA, CypA bound to a substrate analogue in the trans, transition state, and cis configurations. When we apply our method of analyzing the trajectories at a coarse-grained level, i.e., interresidue contact interactions, and use PCA in contact space, the specific differences in CypA dynamics upon association to its substrate, during the catalytic process and upon alteration of a single residue distant from the active site are revealed.
Results and Discussion
Analyzing Standard Molecular Dynamics Trajectories at the Residue-Level Interactions.
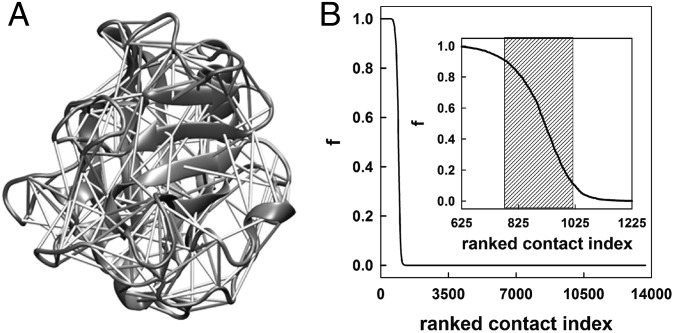
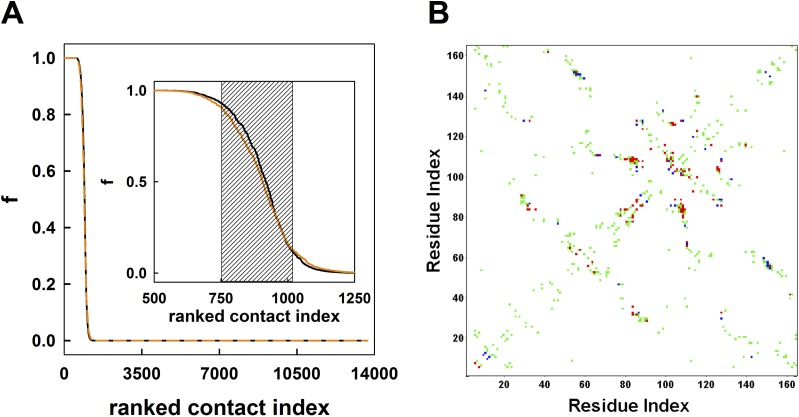
We analyzed the trajectories of the free wild-type CypA (Cwt) and CypA in complex with the substrate (Ctrans, Cts, and Ccis) to reveal the differences in the conformational ensembles of the free and substrate-bound enzyme and map the dynamical features in each ensemble (Figs. S1A and S2). Mining the time evolution data of 3N atomic coordinates (n = 2,505 atoms of CypA) for key features presents a “big data” problem. To simplify the complexity of the high-dimensional data, we established a data reduction scheme similar to the Computation of Allosteric Mechanism by Evaluating Residue–Residue Associations approach (35). The 3D Cartesian coordinates of the trajectory is condensed to a binary trajectory (consisting of 0s and 1s) of Nres*(Nres-1)/2 residue–residue contacts. A contact is defined to have formed between two residues if the distance between any of their heavy atoms is within a cutoff of 4.5 Å during the simulation. It is common to use such cutoff values in coarse-grained representation of protein structures (36–38) and evaluation of protein contacts (39, 40). A contact receives a value of either 1 when formed or 0 when the contact is broken. Although some contacts that are essential for the structural integrity of CypA will be present throughout the simulation and have an f value near 1.0, others may never be formed (f close to 0) (Fig. 1). The region around the transition midpoint of the sigmoidal contact probability curve represents those contacts that are formed and broken as time evolves as a consequence of conformational dynamics. These interesting contacts are potentially relevant for ligand recognition, allosteric communication, conformational transitions, and function, as they can form and break and are easily perturbed upon ligand binding or during catalysis. From the average contact probability curve (Fig. 1B, Inset), we selected the contacts in the transition region between 10% and 90% for further analysis (Fig. 1 and Fig. S1). This cutoff was decided based on an error analysis between the first and the second half of the simulation (Fig. S3). We refer to these contacts as dynamic contacts (cdyn) and the residues that are involved in formation of these contacts as dynamic residues (rdyn). As these contacts represented less than 2% of the unique interresidue contacts, not only the size of the binary trajectory but also the noise in the multidimensional data were drastically reduced.
Fig. S1.
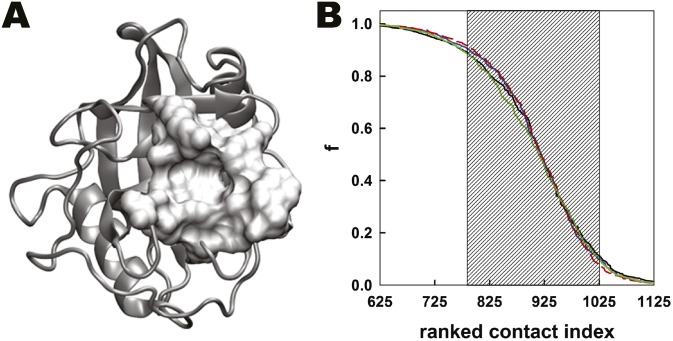
Structure of CypA and the dynamical network of residue-residue contacts in CypA. (A) The 3D structure of CypA (gray) with its substrate-binding cavity (Van der Waals surface) formed by residues Arg55, Phe60, Met61, Gln63, Ala101, Asn102, Ala103, Phe113, Leu122, and His126. (B) Individually ranked contact curves obtained from trajectories of substrate-free CypA (black), substrate-bound CypA in the trans (blue), transition state (dashed red), and cis (green) configurations.
Fig. S2.
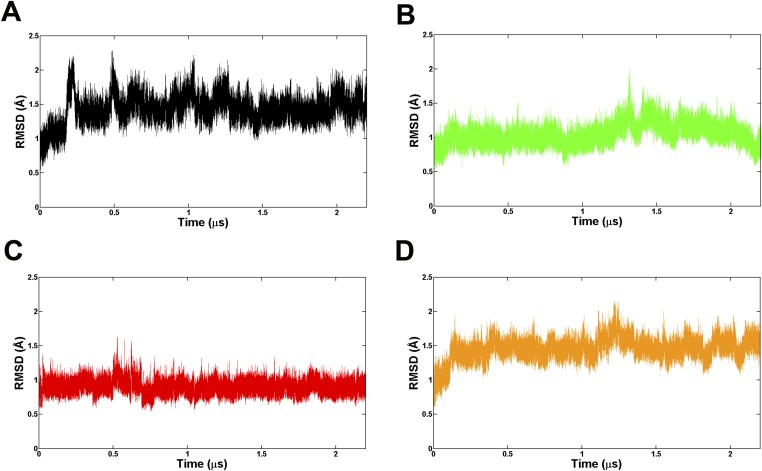
Root mean square deviation from the starting structure as a function of time from the trajectories of (A) free CypA (black), CypA bound to the substrate in the (B) transition state (green) and (C) cis (red) configurations, and (D) V29L mutant (orange).
Fig. 1.
Structure of CypA and the dynamical network of residue–residue contacts in CypA. (A) The selected contacts (shaded area in B) are represented as cylinders connecting the residue pairs on the CypA structure. (B) Residue–residue contacts within CypA are ranked according to their probability of formation, f (i.e., fraction of the simulation time when a contact is formed). Contact probabilities are plotted against their respective ranked contact index. This curve represents the average from individual simulations of free Cwt, Ctrans, Cts, and Ccis. Inset shows a magnified portion of the curve with the shaded area depicting contacts that are formed between 10% and 90% of the simulation time.
Fig. S3.
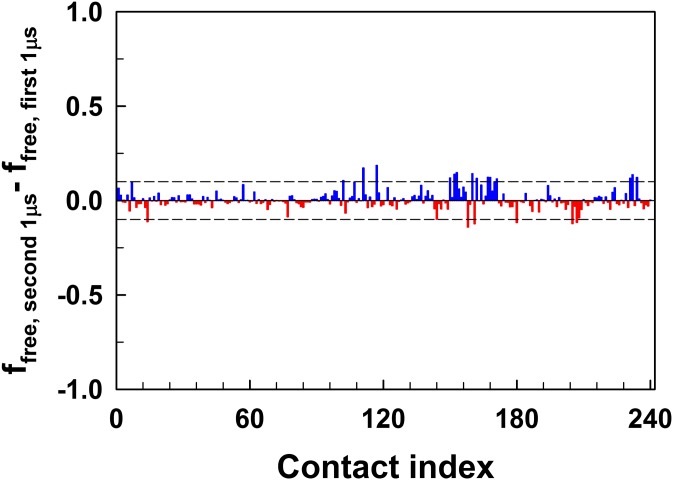
Error analysis of MD simulations. The plot shows the difference in contact probabilities in the first and the second halves of the MD trajectory of the free wild-type CypA. Those contacts that are formed between 10% and 90% of the simulation time are chosen. Blue and red bars represent the contacts that are formed and broken, respectively in the second 1 μs of the trajectory. The dotted lines mark the deviation of 0.1, i.e., a difference of 10% in contact probabilities in two parts of the trajectory within which majority of the contacts fall.
Validating our Coarse-Grained Approach to Analysis with NMR Chemical Shift Data.
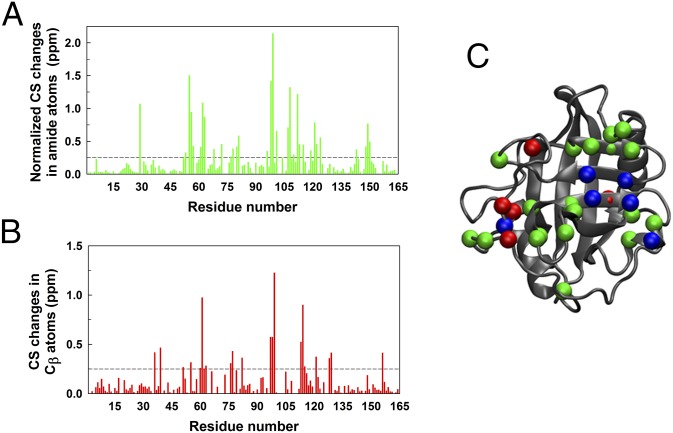
We identified 40 residues that exhibited significant NMR chemical shift changes in either their backbone amide or side-chain Cβ atoms or both after association with the substrate (Fig. 2 A and B). For the contacts selected according to Fig. 1B, we computed the difference in contact probabilities obtained from simulations of the free and substrate-bound CypA. Out of 40 residues, we observed 30 residues that belonged to rdyn (i.e., involved in forming one or more contacts that exhibited more than 10% changes in contact probabilities) in simulations of CypA-substrate complexes (Fig. 2C). From the remaining 10 residues that showed no changes upon substrate binding in our simulations, the immediate neighbors of 8 residues were found to be in rdyn when the substrate was present in the active site. These additional residues were most likely affected by the change in chemical environment, rather than the dynamics, upon binding the substrate. It should be noted that a longer peptide substrate, Gly-Ser-Phe-Gly-Pro-Asp-Leu-Arg-Ala-Gly-Asp, was used in NMR experiments (41) than the one used in the simulations. Majority of the residues that undergo chemical shift changes in the presence of the substrate were observed in our simulations to be perturbed due to dynamical changes and not due to direct interaction with the substrate. Such comparisons between our simulations and chemical shift data thus validated the conformational ensembles obtained from our microsecond-long standard MD simulations as well as our coarse-graining approach to analyzing the trajectories.
Fig. 2.
Experimental validation of dynamical change in CypA upon substrate binding. (A) Normalized changes in chemical shift in amide atoms (green) and (B) chemical shift changes in Cβ atoms (red) of CypA upon binding the substrate, as obtained from NMR experiments, as previously described (41). (C) Residues that exhibit chemical shift changes of greater than 0.25 ppm and are involved in contacts that show >10% variation in the probabilities of formation (and are a subset of contacts with f between 10% and 90%, i.e., cdyn) upon substrate binding are shown as spheres (Cα atoms) on the CypA structure. The color code is consistent with A and B. Blue spheres represent those residues that are involved in formation of those contacts that have >10% change in their probability of formation upon substrate binding and with chemical shift changes of greater than 0.25 ppm in both the amide and Cβ atoms.
Mapping the Dynamical Changes in CypA upon Substrate Binding and Along the Reaction Pathway.
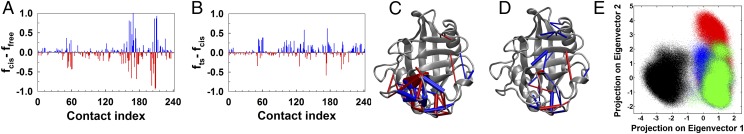
NMR chemical shift changes in a protein upon association of a ligand report on the changes in the chemical environment of its residues. It is hard to decipher whether these chemical shift changes are due to ligand binding or the dynamical changes associated with ligand binding. However, whether these active site residues also undergo changes in their dynamics cannot be determined from ensemble-averaged chemical shift data. This issue can be easily addressed from MD trajectories of the free CypA and substrate-bound CypA. We computed the difference in the probabilities of contact formation of cdyn between the ensembles obtained from MD simulations of the free CypA and CypA bound to the cis substrate (Fig. 3A). A value of fcis–ffree close to +1.0 represents a contact that is formed in the cis-bound CypA but rarely or never formed in the free enzyme. A value close to −1.0 signifies a contact formed with higher probability in the free CypA and broken when the cis substrate is present in the active site. Mapping these contacts that are formed and broken with varied probabilities upon substrate binding on the CypA structure revealed the involved residues that were predominantly clustered (termed “dynamic cluster”) in a region distinct from the active site (Fig. 3C). Notably, the active site residues were involved in very few contacts that were formed and broken at low probabilities and through which they communicated with the distant cluster of dynamic residues. Indeed most of these dynamic cluster residues also exhibited NMR chemical shift changes, indicating the long-range effects of substrate binding.
Fig. 3.
Dynamical changes in CypA following substrate binding and during catalytic function. (A) Differences in the fraction of the time the selected contacts (cdyn) are formed in trajectories of free CypA and CypA bound to the substrate in the cis configuration. Positive (blue) and negative (red) values signify contacts that are formed and broken, respectively, in CypA upon binding the cis substrate. (B) Differences in the fraction of the time the selected contacts are formed in trajectories of CypA bound to substrate in the transition state and the cis state. Positive values signify contacts that have higher probability of forming in the ensemble of CypA structures when bound to the substrate in the transition state. Negative values signify contacts that have higher probability of forming in CypA when bound to the cis substrate. (C) For CypA contacts that show a difference of more than 10% in the probability of contact formation or breaking upon substrate binding [i.e., abs(fcis-ffree) ≥ 0.1] are shown as blue and red cylinders, respectively, on the CypA structure (gray). The cylinders are shown to join the Cα atoms of the pair of residues. The radius of the cylinder is proportional to the absolute values of fcis-ffree. (D) CypA (gray) with red and blue cylinders showing contacts broken and formed, according to the criterion mentioned in C. The radii of the cylinders are proportional to the absolute values of fts-fcis. (E) Projection of contact space on the first two principal components—cdyn contacts that are formed and broken in each snapshot of the trajectories of Cwt (black), Ctrans (blue), and Cts (green), and Ccis (red) are shown.
Comparison of the contact probabilities of cdyn in the cis- and transition-state–bound ensembles of CypA (Fig. 3 B and D) revealed the enzyme residues that exhibited dynamical changes during catalysis. The differences in the contact formation probabilities were relatively less drastic compared with the differences observed upon substrate binding. However, the contacts that are formed and broken in the ensemble of transition-state–bound CypA were distributed throughout the protein. We further performed principal component analysis in contact space (i.e., cdyn) by combining the binary trajectories of Cwt, Ctrans, Cts, and Ccis. Although the conformational ensemble of the free CypA was primarily distinct from those of substrate-bound CypA, there was significant overlap between conformational ensembles of CypA bound to the substrate in different configurations along the reaction pathway. This indicated that overall a new set of contacts were formed and broken upon substrate association. We would like to note that PCA performed on contact space reports on features that are distinct from PCA of fluctuations in Cartesian coordinates (16, 42). PCA in contact space defines conformational ensemble in terms of contacts formed and broken and thus yields more information of dynamic changes that take place upon substrate binding or catalysis.
We further computed the average time it took to form and break each cdyn contact from the trajectories of Cwt, Ctrans, Cts, and Ccis. The time constants (τ) exhibited an overall distribution ranging from picoseconds to microseconds (Fig. S4) that altered upon substrate binding (i.e., comparing Fig. S4A with Fig. S4 B–D) and when the bound substrate was in a different configuration (i.e., comparing Fig. S4 B, C, and D). Interestingly, contacts with slower dynamics than the picosecond timescale were primarily clustered in the region that was distant from the active site. It was only in the Cts that a higher number of active site residues were involved in the formation and breakage of contacts on a relatively slower timescale. Furthermore, certain contacts that took more than 100 ns to form and break in Cwt, Ctrans, and Ccis were completely absent when the substrate was in the transition-state configuration, thereby narrowing the range of contact dynamics timescale in Cts. These results suggested noncoherent contact dynamics that was altered upon association of the substrate and during the various stages of catalysis.
Fig. S4.
Temporally dispersed contact dynamics in CypA. Shown are the contacts from individual molecular dynamics trajectories of CypA (A) in the absence of a substrate, CypA bound to the substrate in the (B) trans, (C) transition state, and (D) cis configurations. Contacts are color coded according to their time constants (τ) of formation and breakage—τ < 0.1 ns (white), 0.1 ns ≤ τ < 1 ns (red), 1 ns ≤ τ <10 ns (orange), 10 ns ≤ τ < 100 ns (green), τ ≥ 100 ns (blue). The time constant, τ = τf + τb where τf and τb are the average times of contact formation and breaking, respectively. The radii of the cylinders between the Cα atoms are proportional to the magnitude of τ.
Effects of a Distal Mutation within the Dynamic Cluster of Residues on CypA Catalysis.
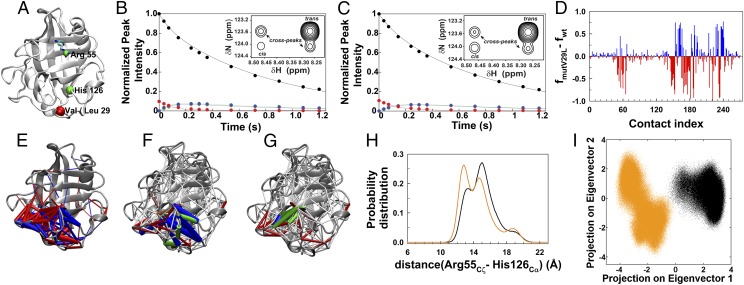
Interestingly, it was observed in previous NMR studies by Schlegel et al. that Valine 29, which is part of the dynamic cluster, exhibited significant variation in dynamical motion upon substrate binding to CypA compared with unbound CypA (22). Due to the observed dynamic coupling between the active site and the dynamic cluster from the previous NMR studies, we speculated that the effects of mutating Valine 29 in this distal region with a residue that did not significantly perturb the dynamics and maintained the fold and integrity of CypA should be transmitted back to the active site of CypA. The question was whether a mutation in the dynamic cluster that was over 15 Å away from the active site would exert any effect on the active site residues or the activity of CypA. We carried out a conservative mutation within this region by mutating Valine 29 (Fig. 4A) to Leucine and performed NMR studies to determine the effects of the mutation on the catalytic isomerization rate of CypA. The isomerization rates were determined by NMR ZZ exchange, as previously described (41) (Fig. 4 B and C). This single conserved mutation resulted in a decrease in the catalytic isomerization rate from 10.4 ± 0.4 s−1 for the wild-type CypA to 6.9 ± 0.3 s−1 for the V29L mutant i.e., the V29L mutant CypA catalyzed at ∼70% the rate of wild-type CypA. These results provided a direct test of the significance of the cluster of residues that are perturbed on substrate binding and further raised the question as to how the effects of modification of a single residue distant from the active site are communicated to the active site.
Fig. 4.
Dynamical changes in CypA upon mutation. (A) On the CypA structure are shown the Cα atoms of the mutated residue Val29 (red sphere) and those of active site residues, Arg55 and His126 (green spheres). Decay of ZZ-exchange peak intensity spectra of residue Leu7 in the substrate in the presence of (B) wild-type CypA and (C) the V29L mutant. Data points are shown for the trans (black) and cis (red) peaks, trans-to-cis (green), and cis-to-trans (blue) cross-peaks. Continuous lines are best fits to the data. Representative ZZ-exchange spectra collected on 15N-labeled substrate at 144 ms in the presence of wild-type CypA and the V29L mutant are shown as Inset to B and C, respectively. Arrows indicate the exchange peaks in the presence of the enzyme. (D) Differences in the fraction of the time the selected contacts (cdyn′) are formed in trajectories of wild-type CypA and V29L mutant of CypA. Positive (blue) and negative (red) values signify contacts that are formed and broken, respectively, with higher probabilities in the mutant than the wild type. (E) Contacts having an absolute difference in contact probabilities between the wild-type and the mutant (i.e.fmutV29L-fwt) > 0.1 are shown as cylinders connecting the Cα atoms of residues with the same color code as in D. The radii of the cylinders are proportional to the abs(fmutV29L-fwt). The selected contacts from the trajectories of (F) wild-type CypA and (G) V29L mutant are color coded according to their time constants (τ) of formation and breakage: τ < 0.1 ns (white), 0.1 ns ≤ τ < 1 ns (red), 1 ns ≤ τ <10 ns (orange), 10 ns ≤ τ < 100 ns (green), and τ ≥ 100 ns (blue). The radii of the cylinders between the Cα atoms are proportional to the magnitude of τ. (H) Probability distribution of the distance between the guanidinium carbon of the side chain of Arg55 and backbone Cα atom of His126 in the wild-type (black) and the V29L mutant CypA (orange). (I) Projection of contact space on to the first two principal components. Each data point represents a configuration of either the wild-type (black) CypA or its mutant (orange) in contact space.
We simulated V29L CypA (CmutV29L) for 2.2 μs with standard MD to address this question. Similarly, we selected the set of contacts (Cdyn′) with probability of formation between 10% and 90% from an average ranked contact probability curve (computed from the ensembles of Cwt and CmutV29L; Fig. S5A). Comparing the contact trajectories of Cwt and CmutV29L showed that most of the contacts that were formed and broken with probability of >10% were expectedly found in the region around Val29 (Fig. 4 D and E). However, the cluster of contacts around V29 was dynamically connected or coupled to the active site through few contacts that were formed and broken with relatively lower probabilities upon mutation (Fig. 4E).
Fig. S5.
Ranked contact probability curve for the wild-type CypA and its V29L mutant and the selected contacts (A) Contact probabilities from the trajectories of wild-type CypA (black) and its V29L mutant (orange) are plotted against their respective ranked contact index. Inset shows magnified portion of the curves with the shaded area depicting contacts that are formed between 10% and 90% of the simulation time. This cutoff is decided from an average ranked contact probability curve calculated from the probability curves of wild-type CypA and V29L mutant. (B) Shown are residue–residue contacts selected exclusively from (A), i.e., cdyn′ (red) and Fig. 1A for the ensembles of free and substrate-bound CypA, i.e., cdyn (blue). Contacts that are common in cdyn and cdyn′ are shown in green.
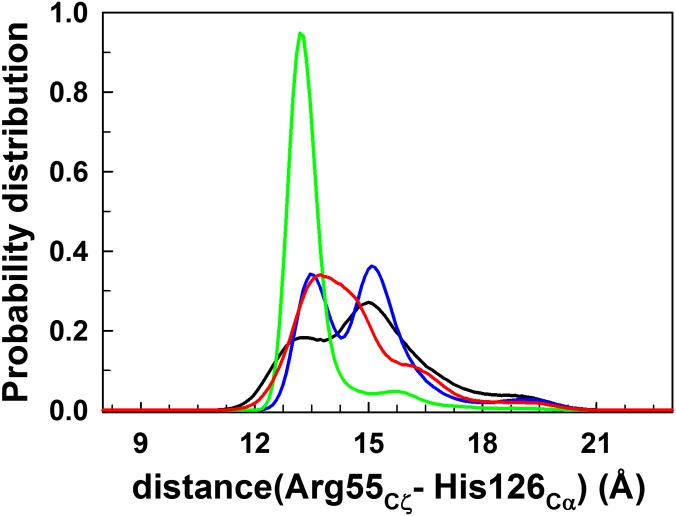
Structural studies have demonstrated that a conserved active site residue, Arg55, is well positioned to stabilize the transition state of the substrate and thereby facilitates catalysis (43). Mutating Arg55 to Ala or Lys has been shown to result in drastic loss of catalytic efficiency (7, 44). Previous quantum mechanics/molecular mechanics (QM/MM) (45) and MD studies (46, 47) have shown that Arg55 is involved in key stabilizing hydrogen bonds with the substrate in the transition state. Given the importance of Arg55 in binding and catalysis, we monitored its side chain motion by computing the distance between the guanidinium carbon of the side chain of Arg55 and backbone Cα of His126 (Figs. 4A and S6). In the Cts ensemble, the predominantly unimodal probability distribution of this distance peaked at 13.5 Å (Fig. S6), indicating a relatively fixed and less dynamic backbone at the active site’s key residue. In Cwt, however, the distribution exhibited a second peak centered on ∼15.5 Å that constituted the major population (Figs. 4H and S6). Such a multimodal distribution suggested the motions of the side chain of Arg55, moving away and approaching the active site and substrate. Interestingly, in the V29L mutant, there was a redistribution such that the major population was shifted to 13.5 Å (Fig. 4H), reminiscent of the well-organized and less dynamic active site in the transition-state–bound CypA (Cts; Fig. S6). These results suggested that the active site of the V29L mutant is preorganized—a characteristic that might confer slightly higher substrate binding affinity to V29L mutant over the wild-type CypA. As Cts binds better than Ctrans or Ccis, the V29L mutant is expected to bind the substrate better as well. In CypA, trans and cis states are both substrates and products, because CypA catalyzes cis–trans isomerization reversibly. Assuming the mutant CypA interacts with the transition state in a manner similar to the wild type, the increased binding affinity would confer relatively decreased transition-state stabilization by the mutant CypA than the wild-type CypA and thereby relatively increase the activation barrier for catalysis. Similarly, it has been shown that substrates of CypA with optimal binding do not necessarily have the highest catalytic rates (43). Also, substrates that bind optimally may not dissociate from the enzyme with the same ease as those with lower binding affinity, thereby resulting in lower catalytic efficiency. Therefore, relatively stronger binding of the substrate is not always ideal for catalysis.
Fig. S6.
Probability distributions of the distance between the guanidinium carbon of the side chain of Arg55 and backbone Cα atom of His126 calculated from the trajectories of free CypA (black), CypA bound to the substrate in the trans (blue), transition state (green), and cis (red) configurations.
We further projected the configurations from each snapshot on to the two most dominant principal components obtained from PCA of Cwt and CmutV29L contact trajectories (Fig. 4I). The two ensembles were remarkably well separated in contact space, despite minimal differences in contact probabilities in the active site region (Fig. 4 E and I). Marked dissimilarities between the dynamics of wild-type CypA and the V29L mutant were noticed in the average times of formation and breakage for cdyn’s contacts (Fig. 4 F and G). Majority of the contacts that were formed and broken in the nanosecond timescale (green and blue cylinders in Fig. 4F) in the Cwt ensemble took less than 100 ps in the mutant, indicating the speedup in dynamics around the site of mutation.
Using Graph Theory to Analyze the Alteration in CypA Dynamics upon Substrate Binding, Mutation, and During Catalysis.
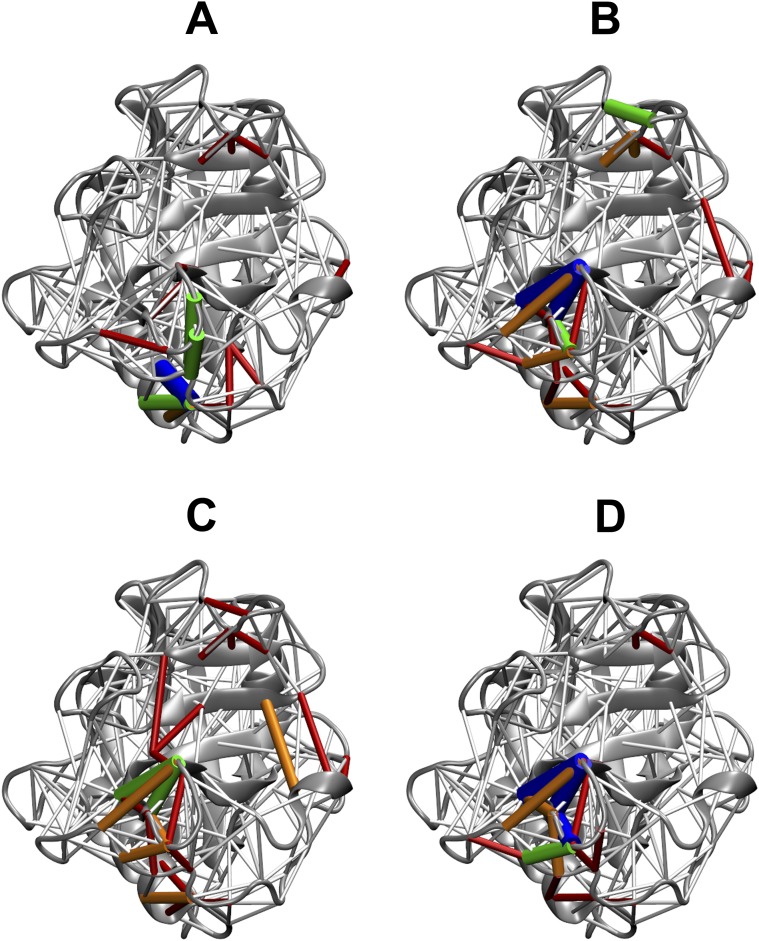
We used vertex centrality measures to identify the relative importance of CypA residues in the network of residue–residue contacts as shown in Figs. S7 and S8 and Fig. 5. We calculated eigenvector centrality to gauge the relative influence of each residue in the network. The idea behind eigenvector centrality is that a residue connected to other residues that are well connected makes this residue influential. We computed the percent change in eigenvector centrality in CypA ensembles when bound to the substrate and upon mutation with respect to the free wild-type ensemble (Fig. S8). The influence of Ala103, an active site residue, markedly decreased upon binding the substrate, irrespective of the substrate configuration (Fig. S8, Upper). On the other hand, Pro105 and Asn106 exhibited noticeably large increase in eigenvector centrality upon association with the substrate. Intriguingly, certain residues in the vicinity of Val29 whose degree centrality (Fig. S7) had significantly increased upon mutation were found to have a large decrease in eigenvector centrality. These results indicated that although more number of new interactions was formed upon mutation, the influence of those residues diminished (red spheres in Fig. S8D).
Fig. S7.
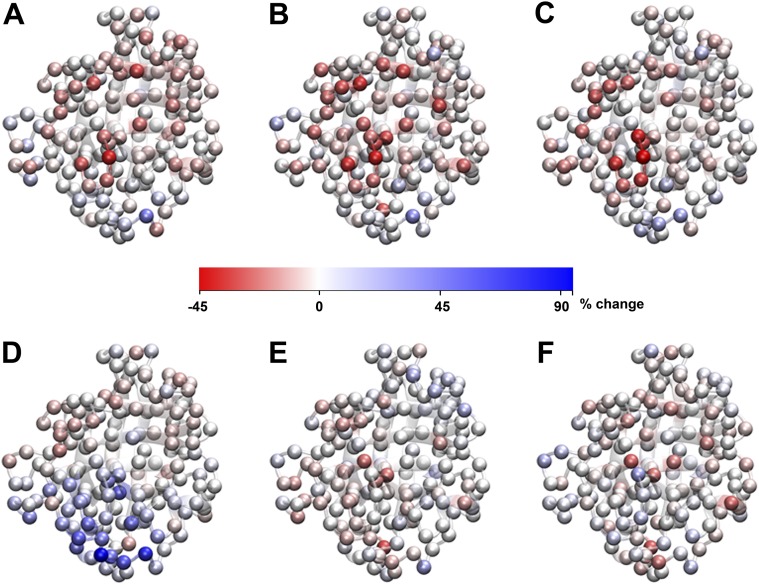
Changes in degree centrality of CypA residues upon substrate recognition, catalysis, and mutation. The percent change in degree centrality, i.e., , where is the degree centrality of residue v calculated from the reference ensemble, is computed for every CypA residue and depicted as a color-coded sphere on its 3D structure. Each sphere represents a Cα atom. The color scale extends from −45% to +94%. Here, ΔCD is shown for CypA bound to the substrate in the (A) trans, (B) transition state, (C) cis configurations, and (D) CypA V29L mutant with respect to the free wild-type CypA. Also shown are ΔCD when the substrate undergoes transition from (E) the trans to the transition state, and from (F) the cis to the transition state.
Fig. S8.
Changes in eigenvector centrality of CypA residues upon substrate recognition, catalysis, and mutation. Each residue is represented as a sphere on the CypA structure and color coded according to the percent change in eigenvector centrality, . Here, Δxv is calculated for ensembles of CypA bound to the substrate in the (A) trans, (B) the transition state, and (C) the cis configurations as well as for (D) CypA V29L mutant, with respect to the free wild-type CypA. E and F shows Δxv in the transition-state–bound CypA with respect to CypA bound to the ground-states, i.e., trans and cis, respectively. The color scale extends from −72% to 104%.
Fig. 5.
Distribution of shortest-path betweenness in the free, the substrate-bound, and mutant CypA. Betweenness centrality is depicted for each CypA residue calculated for the (A) free wild-type enzyme, (B) V29L mutant, CypA bound to the substrate in the (C) trans, (D) the transition state, and (E) the cis configurations. The Cα atoms that are represented as spheres are colored according to the residue betweenness: 0 < CB(v) ≤ 0.05 (blue), 0.05 < CB(v) ≤ 0.1 (cyan), 0.1 < CB(v) ≤ 0.3 (green), 0.3 < CB(v) ≤ 0.5 (yellow), 0.5 < CB(v) ≤ 0.7 (orange), and 0.7 < CB(v) ≤ 0.9 (red).
Communication between two long-range residues takes place via paths of interresidue contacts that ultimately connect them. If a residue falls on the shortest paths between many other pairs of residues, it can control the information flow and act as a bottleneck in the network. Betweenness centrality of a residue is the fraction of the shortest paths, enumerated over all pairs of residues that pass through it. We calculated betweenness centrality for each CypA residue from the ensembles of Cwt, CmutV29L, Ctrans, Cts, and Ccis. If a contact was formed with high probability, a shorter distance was assigned between the contacting residues. Fig. 5 highlights remarkable differences in the manner in which information was passed when wild-type CypA was unbound versus when it was in complex with the substrate or it was mutated. The disparity in betweenness for Phe112 and Val29 in the mutant compared with the wild-type CypA was quite pronounced (Fig. 5 A and B). The betweenness score for Phe112 was decreased in the mutant; whereas, information flow through the mutated residue Leu29 was significantly enhanced compared with that through the wild-type Val29. Not only Leu29 but also Pro30 and Lys31 exhibited higher betweenness in the mutant, almost creating an alternate information channel that extended from the site of mutation to the active site. The betweenness of residues in the three substrate-bound states exhibited differences that were spread throughout the enzyme, indicating the changes in information flow as the substrate passed from the trans or the cis to the transition state (Fig. 5, Lower). Interestingly, all of the residues having relatively higher scores (spheres colored in yellow, orange, and red) in each ensemble were identified as hydrophobic. Moreover, Phe112 stood out with the highest score in each ensemble, which suggested that it could be the key bottleneck residue controlling the information flow. Furthermore, in Cts, there was a striking increase in the number of residues with betweenness scores of >0.3, suggesting an enhanced cross-talk over long range that may have important implication for the catalytic function.
Conclusions
We defined the dynamics of CypA in terms of contact formation and breaking. The approach to analyzing the MD trajectories significantly reduced the noise in the high-dimensional data, and we could reveal unprecedented key dynamical features in CypA that generally show subtle conformational changes. We demonstrated that variation in contact dynamics and communication across CypA upon substrate binding, mutation, and during catalysis can be captured. Surprisingly, the effects of substrate binding on contact dynamics were maximally felt in a region about 15 Å distal from the active site and very limited in the active site region. We used NMR experiments to validate the simulation results and to confirm the atomistic description of the residue–residue contact analysis. These results implied the allosteric role of the distal region in substrate binding. Our NMR studies and analysis of contact dynamics of the MD simulations in the V29L mutant revealed the underlying reason for its slightly lower catalytic isomerization rate than the wild type. The ensemble in contact space of conformations in the free unbound CypA was largely distinct from those of CypA in complex with the substrate. The ensemble of CypA conformations when in complex with the substrate in the transition state exhibited formation of more number of contacts relative to CypA ensemble bound to the substrate in the ground-state configurations. Moreover, contact dynamics in Cts were found to be in the relatively faster regime with a greater number of residues involved in information passage between distant residues. We found that contact dynamics is dispersed over a timescale that ranges from picoseconds to longer than hundreds of nanoseconds. Our results suggest that these dynamical features have crucial importance in the allosteric regulation of biological systems by revealing the coupling of protein dynamics to function. Mapping these dynamical features has ramifications in fully understanding the relationship between dynamics and function in biomolecular systems.
Materials and Methods
Molecular Dynamics Simulations.
For each system—Cwt, Ctrans, Cts, Ccis, and CmutV29L—the initial structures of CypA were solvated in individual TIP3P (48) water boxes. Standard molecular dynamics were carried out for each system at 300 K for a total simulation time of 11 μs. Operational details on molecular dynamics are included in the Supporting Information. Contact analysis and calculation of centrality measures are described in detail in the Supporting Information.
NMR.
The NMR chemical shifts were determined as previously described (41). The catalytic isomerization rates were determined by ZZ exchange as previously described (41). More specifically, ZZ-exchange data were collected on 1 mM 15N-labeled peptide with 20 µM unlabeled wild-type CypA or V29L CypA mutant at 10 °C on a Varian 600 MHz spectrometer with a cryogenically cooled probe. Data were fit according to Farrow et al. (49).
SI Text and Materials and Methods
Molecular Dynamics Simulations.
The assisted model building with energy refinement (AMBER) 14 suite of programs (50) was used along with the AMBER ff14SB (51, 52) force field and reoptimized parameters for the ω-bond torsional angle (53). The starting simulation structure for CypA was based on the PDB structure with PDB ID 1M9F. The following modifications were performed on the structure: for the substrate-bound CypA, the substrate in the PDB was modified to Ac-Ser-Phe-Gly-Pro-Asp-Leu-Nme by keeping the common atoms and using the xleap program in AMBER to add the missing atoms. Ctrans, Cts, and Ccis were modeled as described previously (16). The PDB file was modified by changing the residue name from Val to Leu for residue 29 and further regenerated using xleap that ensured correct number of atoms for Leu. For each simulation, the protein (or the protein–substrate complex) was solvated in a periodic octahedron TIP3P water box with a spacing distance of at least 10 Å between the protein and any edge of the box. About 5,000 water molecules were used. Minimization and equilibration were performed for each system according to the protocol outlined in a previous study. Production runs were carried out at constant pressure (1 bar) and constant temperature (300 K). Standard MD simulations were carried out for 2.2 μs each for Cwt, Ctrans, Cts, Ccis, and CmutV29L. The SHAKE (54) algorithm was used to constrain all of the covalent bonds involving hydrogen atoms. Long-range electrostatics was evaluated using particle mesh Ewald (55) summation method. Truncation of short-range nonbonded interactions was performed using a cutoff of 9 Å. Newton’s equations of motion were integrated using a time step of 2 fs. Data were saved every 1 ps (i.e., 500 steps).
Starting with a minimized and equilibrated structure of CypA solvated in TIP3P water, we performed standard MD at 300 K for 2.2 μs. In three independent runs of over 2 μs each, CypA was simulated with the substrate Ace-Ser-Phe-Gly-Pro-Ala-Leu-Nme bound at the active site and the peptidyl prolyl (i.e., Gly-Pro ω-bond) bond in the trans, constrained in the transition state, and in the cis configurations. The substrate used here is a shorter analog of the high binding affinity peptide construct that was used in the recent NMR studies on CypA by Holliday et al. (41). This set of simulations modeled CypA associated with its substrate as well as the three states along the cis–trans isomerization pathway. The reversible cis–trans isomerization catalyzed by CypA occurs in the millisecond timescale (31). Standard MD implemented on general-purpose computers is still not amenable to model such long timescale processes and sample sufficient number of cis–trans transitions as well as within each well (i.e., trans and cis). Because the three distinct states along the reaction pathway are reasonably well defined, i.e., regions around ω = 180°, ω = 90°, ω = 0° define the trans, transition state, and cis states, respectively, we sampled each state in an independent trajectory for a very long time, without applying any constraints to the entire system (except for the transition-state–bound CypA system. The peptidyl prolyl bond was restrained to ω = 90° using a harmonic potential and a force constant of 1,000 kcal/rad2). Root mean square deviations from the starting structure (Fig. S2) suggested that the overall fold of the enzyme was preserved throughout the length of the simulation for each system.
Contact Analysis.
A contact was defined to have formed between two residues if any of their heavy atoms approaches a distance less than 4.5 Å. Contact probability, fi, for a contact i is computed as , where ti is the fraction of the total simulation time T when the contact was observed to have been formed. An average curve was calculated from the ranked contact probability curves obtained from the trajectories of free CypA, CypA in complex with the substrate in the trans, transition state, and cis configurations. Similarly, another average curve was calculated from the ranked contact probability curves of the wild-type CypA and its V29L mutant. Contacts in the transition region of the average sigmoidal curve with 0.1 ≤ f < 0.9 were selected where as those with f < 0.1 and f > 0.9 were omitted from further analysis. Contact trajectories with binary data were generated for the selected 241 contacts for each system, resulting in N X 241 matrix of 0s and 1s, where N is the total number of snapshots. PCA was performed on the combined binary trajectories of the free and substrate-bound CypA. In an independent analysis, PCA was carried out on the combined binary trajectories of the free and the mutant CypA. We used Matlab for the calculation and diagonalization of the covariance matrix as well as for projection of the binary data onto the first two dominant modes (i.e., with the highest eigenvalues and contributing more than 95% to the overall motions) in contact space.
Centrality Analysis.
If G(V,E) is a graph (with V vertices and E edges) representing a network of contacts, its adjacency matrix can be given by a(v,t). Each vertex v represents a residue and each edge e a contact. If vertex v forms a contact with vertex t, a(v,t) = 1, otherwise a(v,t) = 0.
Degree Centrality.
The degree centrality of a vertex v is calculated as where n is the number of residues (165 in case of CypA). For calculating degree centrality, no cutoffs based on contact probabilities were used.
Degree centrality of a vertex (a residue here) is defined by the number of immediate neighbors that it is connected to (or makes contacts with). This centrality measure reports on the local connectivity in a network. Residues that are involved in contacts that are formed to maintain the secondary and tertiary structure of CypA will have more or less identical degree centrality in the free and substrate-bound CypA. Therefore, to identify the residues that undergo dynamical changes, we calculated the percent change in degree centrality upon substrate binding and mutation as well as during catalysis (Fig. S7). Fig. S7, Upper that represents changes in degree centrality upon substrate association showed an overall decrease in degree centrality as the intra-CypA contacts are broken in the free enzyme (spheres in shades of red) and new interactions are formed with the substrate. The effects of binding the substrate in different configurations were not as striking as those associated with the modification of a single residue, Val29, to Leu. In correspondence to Fig. 4E there was a marked increase in degree centrality in the region around the site of mutation (spheres in blue gradation in Fig. S7D). Comparing the ensembles of Cts with respect to those bound to the substrate in the ground-state configurations (i.e., Ctrans and Ccis), we noticed degree centrality changes that were distributed throughout the protein but of very small magnitudes (Fig. S7 E and F). A residue with a higher degree may have limited potential in communicating with a larger part of the network if it is connected to other residues only in its vicinity that may or may not themselves be well connected.
Eigenvector Centrality.
A weighted adjacency matrix aw(v,t) is built with its elements equal to the edge weights, i.e., aw(v,t) = contact probability between vertices (or residues) v and t. The eigenvector centrality of a vertex, xv, is proportional to the sum of the centralities of its neighbors, i.e., , where λ is a constant. Rewriting this equation in a matrix–vector notation gives the eigenvector equation: , where x and λ are the eigenvectors and eigenvalues of the adjacency matrix A. The eigenvector corresponding to the largest eigenvalue is generally preferred because it ensures that all of the elements of the eigenvector are positive.
Shortest-Path Betweenness Centrality.
The weighted adjacency matrix is modified as follows: aw(v,t) = 1 − aw(v,t), when aw(v,t) ≈ 0, otherwise aw(v,t) = 106. This treatment represents the weights as distances between vertices such that edges with higher contact probabilities are assigned lower distances. Vertices that are not connected, i.e., with contact probability of 0, are assigned an arbitrarily large number to represent a very large distance between them. Using this adjacency matrix, the number of shortest paths for each pair of vertices is calculated using the Dijkstra’s algorithm. Betweenness of a vertex is then computed from the fraction of the total number of shortest paths that pass through that vertex: . Here σst is the total number of shortest paths between vertices s and t and σst(v) is the number of shortest paths between vertices s and t that pass through v. We used Matlab to calculate the above-mentioned centrality measures.
Error Analysis.
We compared the first 1-μs and the second 1-μs parts of the trajectory of free CypA. Fig. S3 shows the difference in the contact probabilities of the selected 241 contacts between the first and the second half of the trajectory. Due to stochasticity, each contact is formed with a different contact probability in two parts of the simulation. However, majority of the contact probability differences are within 10%. Therefore, while analyzing the trajectories of free CypA, its mutant and the substrate-bound CypA, we considered any difference in contact probabilities that is less than 10% as error.
Acknowledgments
We will like to thank Prof. Tongye Shen for insightful conversations and discussion about his CAMERRA method. This research was supported by the National Science Foundation (MCB-1517617). We also acknowledge support from Georgia State University and the Georgia Research Alliance. A portion of the research was performed using Environmental Molecular Sciences Laboratory, a Department of Energy Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. The Information System and Technology Center of Georgia State University is acknowledged for allocating computational time on the IBM System x3850 X5 Servers.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523573113/-/DCSupplemental.
References
- 1.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Henzler-Wildman KA, et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450(7171):913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 3.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 4.Kamerlin SC, Warshel A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins. 2010;78(6):1339–1375. doi: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doshi U, Hamelberg D. The dilemma of conformational dynamics in enzyme catalysis: Perspectives from theory and experiment. Adv Exp Med Biol. 2014;805:221–243. doi: 10.1007/978-3-319-02970-2_10. [DOI] [PubMed] [Google Scholar]
- 6.Klinman JP. Importance of protein dynamics during enzymatic C-H bond cleavage catalysis. Biochemistry. 2013;52(12):2068–2077. doi: 10.1021/bi301504m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 8.Seo M-H, Park J, Kim E, Hohng S, Kim H-S. Protein conformational dynamics dictate the binding affinity for a ligand. Nat Commun. 2014;5:3724. doi: 10.1038/ncomms4724. [DOI] [PubMed] [Google Scholar]
- 9.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295(5559):1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 10.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332(6026):234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamczyk AJ, Cao J, Kamerlin SC, Warshel A. Catalysis by dihydrofolate reductase and other enzymes arises from electrostatic preorganization, not conformational motions. Proc Natl Acad Sci USA. 2011;108(34):14115–14120. doi: 10.1073/pnas.1111252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loveridge EJ, Behiry EM, Guo J, Allemann RK. Evidence that a ‘dynamic knockout’ in Escherichia coli dihydrofolate reductase does not affect the chemical step of catalysis. Nat Chem. 2012;4(4):292–297. doi: 10.1038/nchem.1296. [DOI] [PubMed] [Google Scholar]
- 13.Kohen A. Enzyme dynamics: Consensus and controversy. J Biocat Biotrans. 2012;1(1):1–2. [Google Scholar]
- 14.Liu H, Warshel A. The catalytic effect of dihydrofolate reductase and its mutants is determined by reorganization energies. Biochemistry. 2007;46(20):6011–6025. doi: 10.1021/bi700201w. [DOI] [PubMed] [Google Scholar]
- 15.Pisliakov AV, Cao J, Kamerlin SC, Warshel A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc Natl Acad Sci USA. 2009;106(41):17359–17364. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doshi U, McGowan LC, Ladani ST, Hamelberg D. Resolving the complex role of enzyme conformational dynamics in catalytic function. Proc Natl Acad Sci USA. 2012;109(15):5699–5704. doi: 10.1073/pnas.1117060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57(3):433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 18.Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: A classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5(3):207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nussinov R, Ma B, Tsai CJ. Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys Chem. 2014;186:22–30. doi: 10.1016/j.bpc.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508(7496):331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta. 2011;1814(8):942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlegel J, Armstrong GS, Redzic JS, Zhang F, Eisenmesser EZ. Characterizing and controlling the inherent dynamics of cyclophilin-A. Protein Sci. 2009;18(4):811–824. doi: 10.1002/pro.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holliday MJ, Armstrong GS, Eisenmesser EZ. Determination of the full catalytic cycle among multiple Cyclophilin family members and limitations on the application of CPMG-RD in reversible catalytic systems. Biochemistry. 2015;54(38):5815–5827. doi: 10.1021/acs.biochem.5b00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorfe AA, Grant BJ, McCammon JA. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16(6):885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarabelli G, Grant BJ. Mapping the structural and dynamical features of kinesin motor domains. PLOS Comput Biol. 2013;9(11):e1003329. doi: 10.1371/journal.pcbi.1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci USA. 2009;106(16):6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander RW, Eargle J, Luthey-Schulten Z. Experimental and computational determination of tRNA dynamics. FEBS Lett. 2010;584(2):376–386. doi: 10.1016/j.febslet.2009.11.061. [DOI] [PubMed] [Google Scholar]
- 28.Rivalta I, et al. Allosteric pathways in imidazole glycerol phosphate synthase. Proc Natl Acad Sci USA. 2012;109(22):E1428–E1436. doi: 10.1073/pnas.1120536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasper PM, Fuglestad B, Komives EA, Markwick PR, McCammon JA. Allosteric networks in thrombin distinguish procoagulant vs. anticoagulant activities. Proc Natl Acad Sci USA. 2012;109(52):21216–21222. doi: 10.1073/pnas.1218414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fanghänel J, Fischer G. Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front Biosci. 2004;9:3453–3478. doi: 10.2741/1494. [DOI] [PubMed] [Google Scholar]
- 31.Kern D, Kern G, Scherer G, Fischer G, Drakenberg T. Kinetic analysis of cyclophilin-catalyzed prolyl cis/trans isomerization by dynamic NMR spectroscopy. Biochemistry. 1995;34(41):13594–13602. doi: 10.1021/bi00041a039. [DOI] [PubMed] [Google Scholar]
- 32.Dugave C, Demange L. Cis-trans isomerization of organic molecules and biomolecules: Implications and applications. Chem Rev. 2003;103(7):2475–2532. doi: 10.1021/cr0104375. [DOI] [PubMed] [Google Scholar]
- 33.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3(10):619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 34.Wedemeyer WJ, Welker E, Scheraga HA. Proline cis-trans isomerization and protein folding. Biochemistry. 2002;41(50):14637–14644. doi: 10.1021/bi020574b. [DOI] [PubMed] [Google Scholar]
- 35.Johnson QR, Lindsay RJ, Nellas RB, Fernandez EJ, Shen T. Mapping allostery through computational glycine scanning and correlation analysis of residue-residue contacts. Biochemistry. 2015;54(7):1534–1541. doi: 10.1021/bi501152d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karanicolas J, Brooks CL., 3rd The structural basis for biphasic kinetics in the folding of the WW domain from a formin-binding protein: Lessons for protein design? Proc Natl Acad Sci USA. 2003;100(7):3954–3959. doi: 10.1073/pnas.0731771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shehu A, Kavraki LE, Clementi C. Multiscale characterization of protein conformational ensembles. Proteins. 2009;76(4):837–851. doi: 10.1002/prot.22390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noel JK, Whitford PC, Onuchic JN. The shadow map: A general contact definition for capturing the dynamics of biomolecular folding and function. J Phys Chem B. 2012;116(29):8692–8702. doi: 10.1021/jp300852d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinda KV, Vishveshwara S. A network representation of protein structures: Implications for protein stability. Biophys J. 2005;89(6):4159–4170. doi: 10.1529/biophysj.105.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethi A, Tian J, Derdeyn CA, Korber B, Gnanakaran S. A mechanistic understanding of allosteric immune escape pathways in the HIV-1 envelope glycoprotein. PLOS Comput Biol. 2013;9(5):e1003046. doi: 10.1371/journal.pcbi.1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holliday MJ, et al. Structure and dynamics of GeoCyp: A thermophilic Cyclophilin with a novel substrate binding mechanism that functions efficiently at low temperatures. Biochemistry. 2015;54(20):3207–3217. doi: 10.1021/acs.biochem.5b00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaraju M, McGowan LC, Hamelberg D. Cyclophilin A inhibition: Targeting transition-state-bound enzyme conformations for structure-based drug design. J Chem Inf Model. 2013;53(2):403–410. doi: 10.1021/ci300432w. [DOI] [PubMed] [Google Scholar]
- 43.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol. 2003;10(6):475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J, Koblinski JE, Dutson LV, Feeney YB, Clevenger CV. Prolyl isomerase cyclophilin A regulation of Janus-activated kinase 2 and the progression of human breast cancer. Cancer Res. 2008;68(19):7769–7778. doi: 10.1158/0008-5472.CAN-08-0639. [DOI] [PubMed] [Google Scholar]
- 45.Li G, Cui Q. What is so special about Arg 55 in the catalysis of cyclophilin A? Insights from hybrid QM/MM simulations. J Am Chem Soc. 2003;125(49):15028–15038. doi: 10.1021/ja0367851. [DOI] [PubMed] [Google Scholar]
- 46.Hamelberg D, McCammon JA. Mechanistic insight into the role of transition-state stabilization in cyclophilin A. J Am Chem Soc. 2009;131(1):147–152. doi: 10.1021/ja806146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGowan LC, Hamelberg D. Conformational plasticity of an enzyme during catalysis: Intricate coupling between cyclophilin A dynamics and substrate turnover. Biophys J. 2013;104(1):216–226. doi: 10.1016/j.bpj.2012.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 49.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4(5):727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 50.Case DA, et al. 2015. AMBER 2015 (University of California, San Francisco)
- 51.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier JA, et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doshi U, Hamelberg D. Reoptimization of the AMBER force field parameters for peptide bond (Omega) torsions using accelerated molecular dynamics. J Phys Chem B. 2009;113(52):16590–16595. doi: 10.1021/jp907388m. [DOI] [PubMed] [Google Scholar]
- 54.Ryckaert J, Cicotti G, Berendsen H. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 55.Darden T, York D, Pedersen L. Particle mesh Ewald-an N Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]