Significance
This study defines a molecular mechanism by which clock- and light-signaling pathways converge in Arabidopsis. The data reveal that Timing of CAB expression 1 (TOC1), an essential core component of the central oscillator, binds to and represses Phytochrome-interacting factor (PIF) transcriptional activators, which also are the direct molecular signaling partners of the phytochrome photosensory receptors. This finding shows that TOC1 functions as a clock output transducer, directly linking the core oscillator to a pleiotropically acting transcriptional network through repression of target genes. Collectively, in the plant, these components comprise a transcriptionally centered signaling hub that provides clock-imposed gating of PIF-mediated, photosensory-regulated diurnal growth patterns. These results provide a framework for future research aimed at understanding how circadian dynamics are integrated with other plant physiological processes important for optimal plant fitness.
Keywords: PIFs, photoperiod, TOC1, circadian clock, gating of growth
Abstract
A mechanism for integrating light perception and the endogenous circadian clock is central to a plant’s capacity to coordinate its growth and development with the prevailing daily light/dark cycles. Under short-day (SD) photocycles, hypocotyl elongation is maximal at dawn, being promoted by the collective activity of a quartet of transcription factors, called PIF1, PIF3, PIF4, and PIF5 (phytochrome-interacting factors). PIF protein abundance in SDs oscillates as a balance between synthesis and photoactivated-phytochrome–imposed degradation, with maximum levels accumulating at the end of the long night. Previous evidence shows that elongation under diurnal conditions (as well as in shade) is also subjected to circadian gating. However, the mechanism underlying these phenomena is incompletely understood. Here we show that the PIFs and the core clock component Timing of CAB expression 1 (TOC1) display coincident cobinding to the promoters of predawn-phased, growth-related genes under SD conditions. TOC1 interacts with the PIFs and represses their transcriptional activation activity, antagonizing PIF-induced growth. Given the dynamics of TOC1 abundance (displaying high postdusk levels that progressively decline during the long night), our data suggest that TOC1 functions to provide a direct output from the core clock that transiently constrains the growth-promoting activity of the accumulating PIFs early postdusk, thereby gating growth to predawn, when conditions for cell elongation are optimal. These findings unveil a previously unrecognized mechanism whereby a core circadian clock output signal converges immediately with the phytochrome photosensory pathway to coregulate directly the activity of the PIF transcription factors positioned at the apex of a transcriptional network that regulates a diversity of downstream morphogenic responses.
Given the importance of solar energy to plants, they have evolved sophisticated photosensory-response systems to monitor and adapt to the diurnal photoperiod (1). This environmental parameter provides a precise index of the progression of the earth’s seasons and the time of the day and thereby a signal that regulates a spectrum of growth and developmental responses (such as elongation growth, flowering, and dormancy) appropriate to the prevailing conditions.
The photoreceptors in the phytochrome family (phyA–E in Arabidopsis) are the primary sensors of this signal (2, 3). These chromoproteins regulate two pathways in parallel that converge to control the morphogenic response: (i) the phytochrome-interacting factor (PIF) pathway, whereby the photoactivated-phytochrome molecules bind to and induce the degradation of the PIF proteins (notably the PIF1, PIF3, PIF4, and PIF5 quartet, a subfamily of basic helix–loop–helix transcription factors), thereby altering the expression of the PIF direct-target genes and the cognate downstream transcriptional network (4, 5), and (ii) the circadian clock, whereby the phytochromes entrain the circadian oscillations of the core clock components by sensing the dark-to-light transition at dawn each day (6). Much has been learned about these two pathways, but the mechanism by which their activities are integrated is not well understood.
A central consequence of light-regulated phytochrome activity is that PIF protein abundance oscillates diurnally over each 24-h cycle, with low PIF levels during the light hours (when the phytochromes are photoactivated) and progressive accumulation during the long dark period (as the levels of the active Pfr form of the phytochromes declines) (7–9). This oscillation of PIF proteins controls rhythmic growth under short photoperiods by collectively promoting increased elongation rates in the predawn hours when they are most abundant (7, 8, 10, 11). In parallel, transcription of PIF4 and PIF5 genes is regulated by the circadian clock, most likely in a direct fashion by several central clock components (4), which drive an internal rhythm whose periodicity is also set by the external photoperiodic information. In contrast, PIF1 and PIF3 transcription is maintained at a constant level during the diurnal cycle (8, 11).
Of particular biological relevance to phytochrome and circadian clock integration is circadian gating of light signaling, whereby the circadian clock limits the timing of maximum responsiveness to light to specific times of day (6). Elongation growth is subject to permissive gating during shade avoidance (12) and diurnal growth (7, 10, 13), and there is evidence that this behavior is founded on the phasing of downstream effector transcript abundance through the interaction of the light and circadian clock signaling networks (13). However, despite the importance of temporal gating in the control of the elongation activity in plants, a fundamental understanding of the underlying mechanism is still incomplete.
Here we provide evidence that the core clock oscillator component Timing of CAB expression 1 (TOC1) directly represses the transcriptional-activator activity of the PIF protein when TOC1 is most abundant in the circadian cycle. Specifically, we show that in short days (SDs) TOC1 constrains PIF growth-promoting activity in early postdusk darkness despite rising PIF levels, thereby reducing the extent of the PIF-induced growth that would have accrued otherwise.
Results
PIF3 and TOC1 Display Coincident Cobinding to Dawn-Phased Genes Under SD Diurnal Conditions.
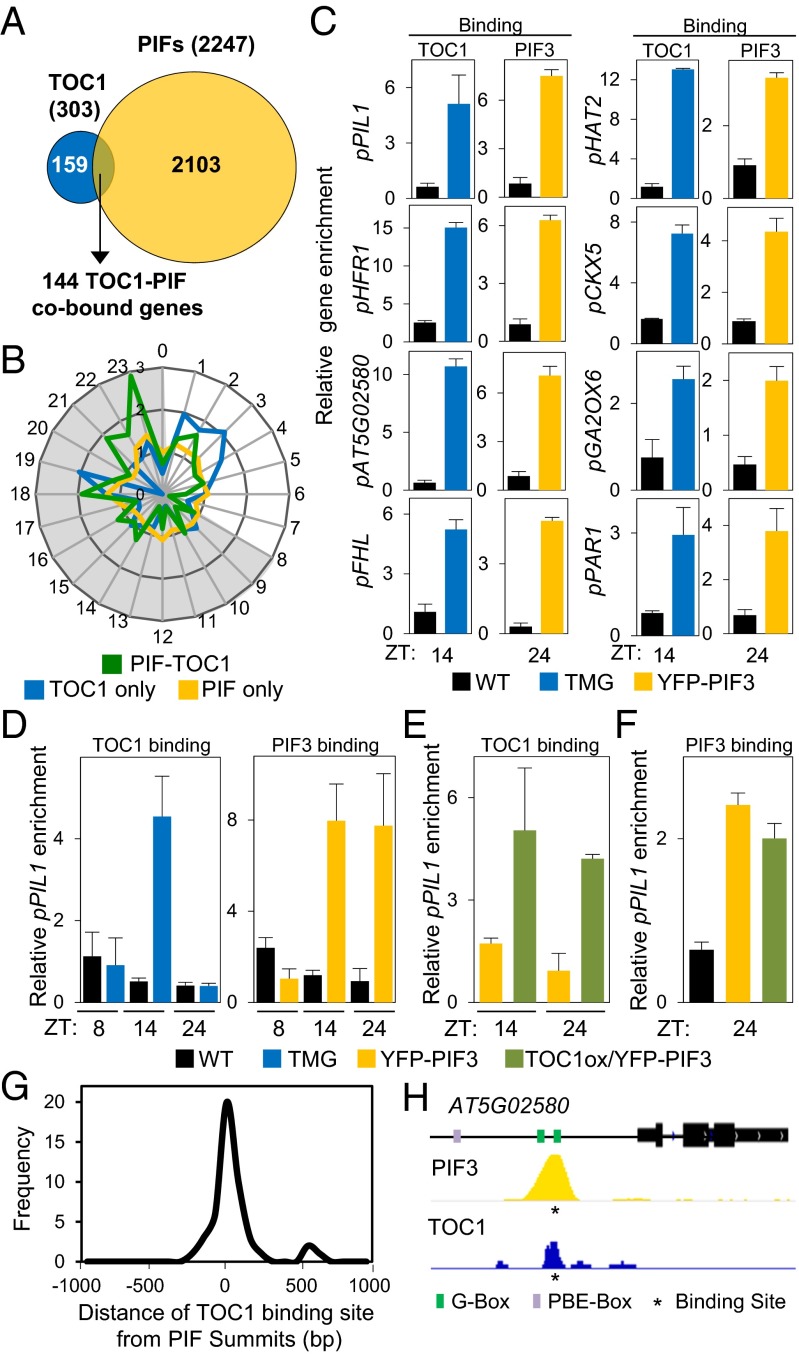
Genome-wide reanalysis of ChIP sequencing (ChIP-seq) data for PIF-associated (5) and TOC1-associated (14) loci, using identical criteria for defining both (see SI Appendix, SI Expanded Results for details), revealed an overlap of 144 shared genes, representing 48% and 7% of the redefined TOC1- and PIF-bound loci (the PIF-TOC1 gene set), respectively (Fig. 1A). Although the two ChIP-seq analyses were performed under different conditions (5, 14), the overlap that emerges suggests that the PIFs and TOC1 might bind a common set of genes in conditions in which their combined function is concomitantly relevant. Because both light and the clock regulate responses in diurnal light/dark cycles, and the PIFs have been shown to accumulate progressively during the long nights of SD photoperiods (8 h light:16 h dark) (7–9), we hypothesized that these genes might be targeted directly by both TOC1 and PIFs under SD conditions. Consistent with this possibility, time-of-day-expression enrichment analysis of these genes using the available data at the PHASER website (phaser.mocklerlab.org/) showed that the 144 cobound PIF–TOC genes displayed an overrepresented phase of expression under SD photocycles at the end of the dark period (Fig. 1B), with 49 of these genes phased between 18 and 23 h (the predawn-specific PIF–TOC1 set) (Dataset S1), when PIF abundance is maximum. Notably, this phase-overrepresentation pattern was absent from the 159 TOC1-only and the 2,103 PIF-only genes (Fig. 1 A and B and Dataset S1) and was specific for SD versus long-day (LD) conditions (SI Appendix, Fig. S1). These data suggest that the predawn-specific PIF–TOC1 genes might be targeted directly by both TOC1 and PIFs to drive a SD-specific expression pattern. ChIP-quantitative PCR assays confirmed the direct binding of TOC1 and PIF3 to the promoters of selected predawn-specific PIF–TOC1 genes at postdusk [zeitgeber time (ZT)14] and dawn (ZT24), respectively (Fig. 1C), when each protein is most abundant in the SD diurnal cycle, respectively (SI Appendix, Fig. S2 A and B and see SI Appendix, SI Expanded Results for details) (5, 14).
Fig. 1.
PIF3 and TOC1 display coincident cobinding to dawn-phased genes under diurnal SD conditions. (A) Comparison of TOC1-bound (14) and PIF-bound (5) genes using identical criteria for defining binding. (B) Expression phases in SD of gene sets defined in A. The 144 PIF–TOC1 genes are shown in green, the 159 TOC1-only genes in blue, and the 2,103 PIF-only genes in yellow. Phases as defined by PHASER (phaser.mocklerlab.org) are indicated on the circumference, and fold-change phase enrichment of genes (count/expected) is shown on the radius. Day is shown in white; night is shown in gray. (C–F) ChIP-qPCR analysis. Samples of 3-d-old SD-grown pTOC1::TOC1:YFP (TMG) (23) and pPIF3::YFP:PIF3 (YFP-PIF3) (32) seedlings (SI Appendix, SI Materials and Methods) were harvested at the indicated times during the third day and were immunoprecipitated using anti-GFP (C, D, and F) or anti-MYC (E) antibodies. Data are from two independent ChIP experiments. Error bars indicate SEM. (C) TOC1 and PIF3 binding to the promoters of selected dawn-phased genes at ZT14 and ZT24 in TMG and YFP-PIF3 seedlings, respectively. WT controls were Col-0 for YFP-PIF3 and C24 for TMG seedlings. (D–F) TOC1 and PIF3 binding to the PIL1 promoter at ZT8, ZT14, and ZT24 in TMG, YFP-PIF3, and TOC1ox/YFP-PIF3 seedlings as indicated. (G) Frequency distribution of the pairwise distance in base pairs (bp) between the TOC1-binding (14) and PIF-binding (5) sites in each of the 49 dawn-phased cobound genes. (H) Visualization of PIF3 and TOC1 ChIP-seq data in the genomic region encompassing the AT5G02580 locus cobound by PIF3 and TOC1. The statistically significant binding sites identified are indicated by an asterisk below the ChIP-seq pile-up tracks. G-box and PBE-box motifs in the promoter are indicated.
Consistent with this pattern, time-course analysis of TOC1 and PIF3 binding to the promoters of three of these dawn-phased genes [Phytochrome interacting factor 3-like 1 (PIL1), Long hypocotyl in far red 1 (HFR1), and AT5G02580] through the night (ZT8, ZT14, and ZT24) showed maximum enrichment of TOC1 at ZT14 and of PIF3 at ZT14 and ZT24 (Fig. 1D and SI Appendix, Fig. S2C). Using double-transgenic lines that constitutively overexpress constant levels of TOC1-MYC in the YFP-PIF3 background (TOC1ox/YFP-PIF3) throughout the night (SI Appendix, Fig. S3A) (14), we found a significant enrichment of promoter binding at ZT24, similar to the levels at ZT14 (Fig. 1E and SI Appendix, Fig. S3B), in contrast to the TOC1 minigene (TMG) lines, in which TOC1 levels are reduced by ZT24. This result affirms that TOC1 binding to its target promoters is dictated by its protein abundance (14). The overexpression of TOC1 did not significantly affect the abundance of YFP-PIF3 (SI Appendix, Fig. S3C) or the promoter binding of PIF3 at ZT24 (Fig. 1F and SI Appendix, Fig. S3D), indicating that the binding of TOC1 and PIF3 to these promoters is likely simultaneous rather than competitive.
To gain insight into the topology of DNA occupancy by TOC1 and PIF3, we examined the binding distance between the PIFs and TOC1 on the promoters of their cobound predawn-specific PIF–TOC1 genes using the available ChIP-seq data (5, 14). The data show that the PIF and TOC1 binding sites lie within 120 bp for 74% of the cobound genes and within 40 bp for 40% of these genes (Fig. 1G). These distances are consistent with concurrent, closely coincident DNA binding of the PIF and TOC1 proteins. A visual example of the highly spatially coincident binding peaks for PIF3 and TOC1 is shown for AT5G02580 in Fig. 1H. See SI Appendix, SI Expanded Results for the DNA motifs associated with PIF- and TOC1-bound genes.
PIF3 and TOC1 Interact and Colocalize in the Nucleus in Planta.
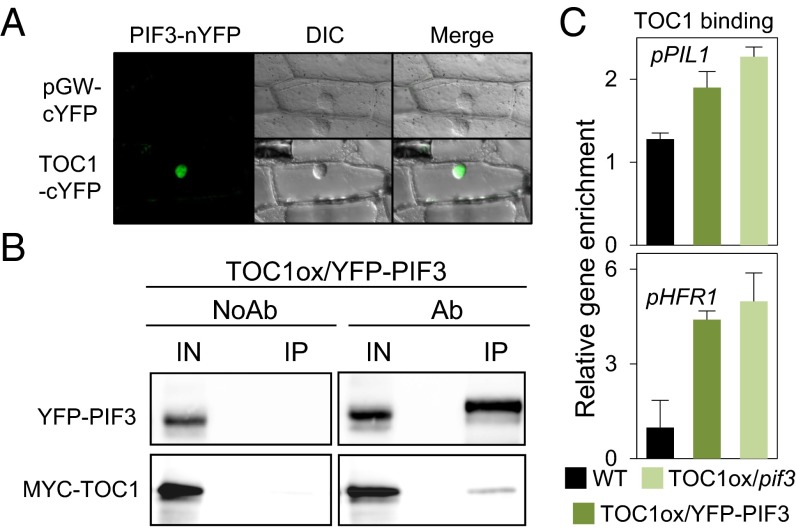
A previous study showed that PIF3 and TOC1 can interact in yeast (15). To determine if the two proteins directly interact in planta, we performed bimolecular fluorescence complementation (BiFC) assays. The data show direct PIF3–TOC1 interaction in the nucleus (Fig. 2A). Furthermore, we observed coimmunoprecipitation of PIF3 and TOC1 from extracts of transgenic TOC1ox/YFP-PIF3 seedlings (Fig. 2B). Together, these results indicate that PIF3 and TOC1 can interact directly with each other in the nucleus under SD conditions. Binding-domain mapping shows that the C-terminal half of PIF3 is predominantly necessary for TOC1 binding (SI Appendix, SI Expanded Results and Fig. S4).
Fig. 2.
PIF3 and TOC1 interact and colocalize in the nucleus in planta. (A) BiFC assay of PIF3 and TOC1 fusions to N- and C-terminal fragments of YFP, respectively, in transfected onion cells. cYFP was used as control. (Left) YFP fluorescence image. (Center) Bright-field image. (Right) Merge of YFP fluorescence and bright-field image. (B) Coimmunoprecipitation of TOC1-MYC and YFP-PIF3 proteins from 3-d SD-grown Arabidopsis seedlings. Samples were harvested under green safelight at ZT14, and extracts were immunoprecipitated with anti-GFP antibody and detected by Western blot using anti-GFP and anti-MYC antibodies. (C) TOC1 binds to target promoters in the absence of PIF3. ChIP-qPCR analysis as in Fig. 1 shows TOC1 binding to the PIL1 and HFR1 promoters at ZT24 in 3-d-old SD-grown TOC1ox/YFP-PIF3 and TOC1ox/pif3 seedlings. Data are from two independent ChIP experiments. Error bars indicate SEM.
It has been reported that TOC1 can associate with DNA both directly through its CONSTANS, CO-like, and TOC1 (CCT) domain (16) and indirectly through interaction with DNA-binding factors (17). We examined the possibility that PIF3 might be necessary to recruit TOC1 to the DNA, comparing TOC1-MYC–overexpressing seedlings in a pif3 background (TOC1ox/pif3) with TOC1ox/YFP-PIF3 seedlings, also in a pif3 background. The data (Fig. 2C and SI Appendix, Fig. S3 D and E) suggest that TOC1 likely binds DNA independently of PIF3, but the possibility that TOC1 binds through a different member of the PIF quartet cannot be discarded. Conversely, as described above for PIF3 promoter binding (Fig. 1F and SI Appendix, Fig. S3D), the data suggest that the interaction of TOC1 with PIF3 does not significantly affect PIF3 binding to DNA (SI Appendix, SI Expanded Results).
TOC1 Represses PIF3 Transcriptional Activity in Regulating Predawn-Phased Growth-Related Genes.
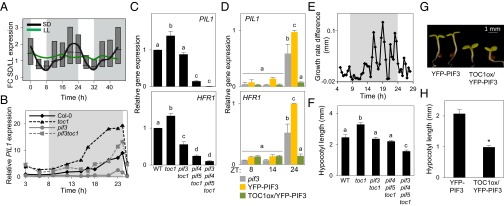
Under SD photoperiods, PIFs directly promote a progressive increase in the expression of genes such as PIL1 and HFR1 during the second half of the night to peak at dawn (7, 8, 10, 11). Consistent with this pattern, the average expression of the dawn-specific PIF–TOC1 gene set under SD conditions shows an oscillatory pattern with maximum expression at the end of the night (Fig. 3A), suggesting that the PIFs directly target these genes to promote their expression at dawn. Strikingly, in contrast, under free-running conditions the average expression of this gene set is almost constant (Fig. 3A), a pattern that is not a classical clock-output pattern. We confirmed directly here that the dawn-specific PIF–TOC1 genes PIL1, HFR1, and AT5G02580 lose rhythmicity and are maintained at low levels across the day and subjective night in seedlings grown for 2 d under SD conditions and then released into constant light (LL), in contrast to the oscillation of clock outputs such as Chlorophyll A/B-binding protein 2 (CAB2) (SI Appendix, Fig. S5).
Fig. 3.
TOC1 and PIF3 antagonistically regulate dawn-phased growth-related genes in controlling early growth in diurnal SD conditions. (A) Median values of expression data (diurnal.mocklerlab.org) of the 49 dawn-specific PIF-TOC1 genes under SD and LL conditions are indicated by colored lines. Boxes indicate upper and lower quartiles of distributions of fold-change (FC) expression for all genes. (B–D) Seedlings grown for 2 d in SD conditions were harvested during the third day at the indicated times. Expression was analyzed by qRT-PCR, and values were normalized to PP2A. Data in C and D are from three independent biological replicates. Error bars indicate SEM. (B) PIL1 expression in 3-d-old SD-grown WT (Col-0) and mutant seedlings. Data are the average of three technical replicates of one representative biological experiment. (C) PIL1 and HFR1 expression in 3-d-old SD-grown WT and mutant seedlings at ZT23. (D) PIL1 and HFR1 expression in 3-d-old SD-grown pif3, YFP-PIF3, and TOC1ox/YFP-PIF3 seedlings. (E) The difference in hypocotyl elongation rate in toc1 and WT seedlings grown under SD conditions. Seedling growth was monitored by infrared imaging (n = 7) every 30 min from day 2 onwards. The growth rate (per 30 min) of WT seedlings was subtracted from the growth rate of toc1 seedlings at each time point. (F) Hypocotyl length of 3-d-old SD-grown WT and mutant seedlings. (G) Visible phenotype of 3-d-old SD-grown YFP-PIF3 and TOC1ox/YFP-PIF3 seedlings. (H) Hypocotyl length of seedlings shown in G. Error bars in F and H indicate SEM of three independent studies with at least 25 seedlings each. In C, D, and F, different letters denote statistically significant differences among means by Tukey’s b test. The asterisk in H indicates statistically significant differences between mean values by Student’s t test.
Previous evidence indicates that TOC1 can act as a transcriptional repressor (SI Appendix, SI Expanded Results) (14, 16). To begin to assess potential TOC1 repression of PIF activity under SD conditions, we examined whether TOC1 levels affect the diurnal pattern of dawn-phased rising expression of their cobound target genes in these conditions. The transcript levels of these genes begin rising at ZT14–ZT16 in the TOC1-deficient toc1-101 mutant (18), several hours earlier than in Col-0 (WT) seedlings, and continues to increase at this elevated level throughout the night, peaking at dawn (Fig. 3B and SI Appendix, Fig. S6). This window of early expression in toc1 coincides with the time of highest TOC1 protein abundance in WT plants (SI Appendix, Fig. S2B). In contrast to the clock-output gene CAB2, this pattern cannot be attributed to toc1 being a short-period mutant (SI Appendix, Fig. S7A) (19). Together, these data indicate that TOC1 prevents early postdusk PIF-induced expression of predawn-phased direct-target genes when PIF3 first begins to accumulate in the middle of the dark period (ZT12–ZT16) in SD conditions. In strong support of this suggestion, we found that the early (ZT12–ZT16) PIL1 expression in toc1 seedlings compared with WT plants was suppressed in a pif3toc1 mutant (Fig. 3B). Also, removal of PIF4 and PIF5 in the pif4pif5toc1 and pif3pif4pif5toc1 mutants partially suppressed the expression of PIL1 and HFR1 (Fig. 3C and SI Appendix, Fig. S8A). Although potentially complicated by higher PIF4 and PIF5 levels in toc1 seedlings (SI Appendix, Fig. S9A) (14), this result suggests that TOC1 represses PIF4 and PIF5 as well as PIF3 activity. It also is notable that TOC1 repression of PIL1 and HFR1 expression occurred under LD as well as SD conditions (SI Appendix, Fig. S8A) and that, conversely to toc1 seedlings, constitutive overexpression of high levels of TOC1 throughout the night completely suppressed dark-induced expression of PIF3 target genes, not only at ZT14 but also at ZT24 (Fig. 3D). Because PIF3 transcript and protein levels are not affected in toc1 seedlings (SI Appendix, Fig. S9 B–D), the data indicate that TOC1 acts directly as a transcriptional repressor of PIF3, which itself acts intrinsically as a transcriptional activator (4), and thus that PIF3 and TOC1 act antagonistically in regulating the expression of their cotarget genes.
Under SD conditions, hypocotyl elongation is rhythmic and peaks at the end of the night (7, 8, 20). To determine whether the apparent antagonistic activities of the PIFs and TOC1 affect this phenotype, we initially compared the growth rates of WT and the toc1 mutant seedlings under our SD conditions. In agreement with previous reports (7), the data show that toc1 mutants elongate more rapidly than WT seedlings through the middle of the night (Fig. 3E and SI Appendix, Fig. S9E) and therefore are taller than WT seedlings (Fig. 3F). This tall phenotype persists under a light/dark cycle of 21 h (T21) (SI Appendix, Fig. S7 B and C), consistent with the conclusion that it is not a consequence of toc1 being a short-period mutant. However the phenotype is strongly suppressed in the pif3toc1 double mutant (Fig. 3F), indicating that PIF3 is necessary for the long toc1 hypocotyls and that PIF3 and TOC1 act antagonistically in regulating growth under diurnal conditions. Similarly, the pif4pif5toc1 triple mutant partially suppresses the tall toc1 phenotype, and PIF3 removal further suppresses the hypocotyl elongation in the pif4pif5toc1 triple mutant (Fig. 3F and SI Appendix, Fig. S8C). This effect was stronger in SD than in LD conditions (SI Appendix, Fig. S8 B and C). Overall, these results mirror the PIF direct target gene-expression data presented above. Conversely, TOC1 overexpression in TOC1oxYFP-PIF3 lines resulted in a strong inhibition of hypocotyl length (Fig. 3 G and H), as also is consistent with the repression of predawn-specific PIF-TOC1 genes when TOC1 is overexpressed (Fig. 3D). Consistent with a role of these genes in growth, gene ontology (GO) analysis shows enrichment for genes responsive to the growth-regulating hormones auxin, brassinosteroids, cytokinin, and gibberellin (SI Appendix, SI Expanded Results and Fig. S10).
TOC1 Can Repress PIF Activity During Skotomorphogenesis.
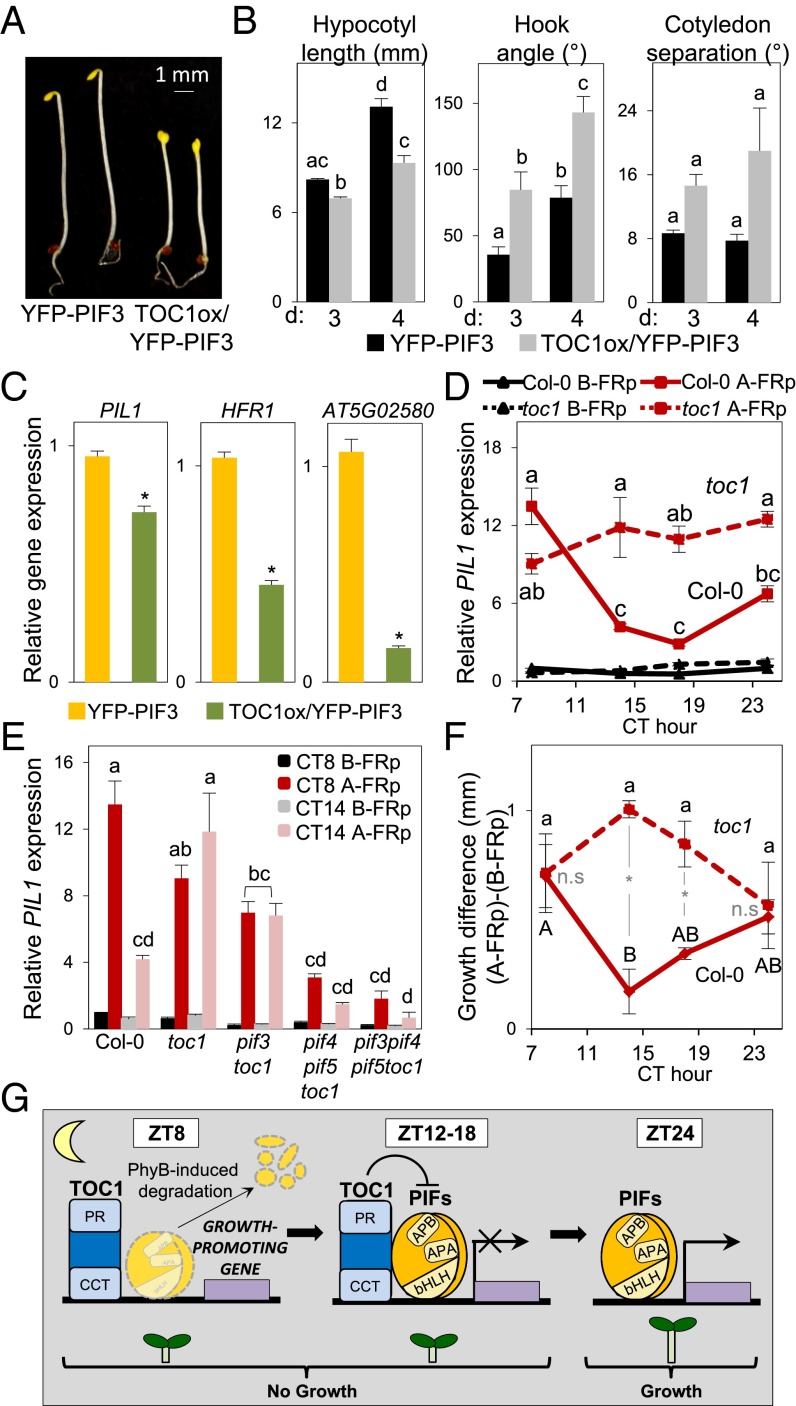
PIFs accumulate to maximum levels in postgerminative seedlings in the dark, thereby promoting skotomorphogenesis, a developmental stage in which TOC1 levels are low and constant (18). Comparison of dark-grown YFP-PIF3 and TOC1ox/YFP-PIF3 seedlings shows that TOC1 overexpression induces partial photomorphogenic development in darkness (hypocotyl-length inhibition, open hooks, and partially separated cotyledons) (Fig. 4 A and B) suggestive of TOC1 repression of PIF activity under these conditions (21). Indeed, expression analysis confirms that TOC1 overexpression suppresses the full expression of PIF3 target genes (Fig. 4C).
Fig. 4.
The transcriptional activity of PIF3 is repressed by TOC1. (A) Visible phenotype of 3-d-old dark-grown YFP-PIF3 and TOC1ox/PIF3-YFP seedlings. (B) Quantification of hypocotyl length, hook angle, and cotyledon separation in YFP-PIF3 and TOC1ox/PIF3-YFP seedlings. The x axis shows the number of days in the dark. Error bars indicate SEM. (C) Gene expression in 3-d-old dark-grown YFP-PIF3 and TOC1ox/PIF3-YFP seedlings. (D) PIL1 expression in Col-0 and toc1 seedlings grown for 2 d in SD and released into continuous white light until exposure to a 15-min FRp at CT8, CT14, CT18, and CT24, followed by 15 min of darkness. Samples were collected either before (B-FRp) (black lines) or after (A-FRp) (red lines) the FRp-plus-dark treatment, as specified in SI Appendix, Fig. S11A. Values are shown relative to Col-0 B-FRp at CT 7 set at 1. (E) PIL1 expression at CT8 and CT14 in Col-0 and mutant seedlings before (B-FRp) (black and gray bars) and after (A-FRp) (red and pink bars) the FRp-plus-dark treatment described in D. Expression in C–E was analyzed by qRT-PCR, and values were normalized to PP2A. Data are for three independent experiments. Error bars indicate SEM. (F) Growth difference induced by a 15-min FRp given at CT8, CT14, CT18, and CT24 to Col-0 and toc1 seedlings, followed by 8 h of darkness (A-FRp) (SI Appendix, Fig. S10C), compared with samples collected before the FRp (B-FRp). In B–E and F, different letters denote statistically significant differences among means by Tukey’s b test. In F, Col-0 (uppercase) and toc1 (lowercase) data were processed independently. Asterisks in C and F indicate statistically significant differences between mean values by Student’s t test. n.s., not significant. (G) Model of the proposed role of TOC1 as a repressor of PIF transcriptional regulatory activity in gating growth to the predawn hours. (Left) TOC1 binds directly or indirectly to the promoters of growth-promoting genes as it accumulates during the postdusk hours. (Center) PIFs accumulate progressively during the night and bind to the same promoters. TOC1 directly interacts with PIFs and represses their transcriptional activity. (Right) As night proceeds, TOC1 abundance declines and PIFs accumulate. At predawn, TOC1 is no longer present, repression is relieved, and PIFs induce growth-promoting gene expression.
TOC1 Gates Shade-Stimulated PIF Activity.
The data reported above suggest that the growth rate is determined by the balance between PIF and TOC1 abundance. We reasoned that this concept might provide mechanistic insight into the permissive gating of growth by the clock as previously reported under seasonal and shade-avoidance conditions (12, 13). To test this possibility, we artificially induced accumulation of PIFs at different time points during a subjective night in SD-grown seedlings released into LL conditions (SI Appendix, Fig. S11 A and B). Under these conditions, TOC1 oscillations persist (diurnal.mocklerlab.org), but PIF3 levels remain low because of phytochrome-imposed degradation (8, 11). By giving a far-red light pulse (FRp) followed by darkness at different time points during the subjective night [circadian time (CT)8, CT14, CT18, and CT24], we induced rapid PIF3 accumulation that was able to induce rapid PIL1 expression (detected within 15 min) to high levels at the beginning and at the end of the subjective night (CT8 and CT24), when TOC1 levels are low, but only to much lower levels at CT14 and CT18, when TOC1 levels are high (Fig. 4D). This result strongly suggests that PIF3-induced expression of target genes is indeed gated by high TOC1 levels. Consistent with this suggestion, this repression was absent in the toc1 mutant (Fig. 4D), confirming that TOC1 is essential to gate PIF-dependent growth-promoting activity. In addition, PIL1 expression in toc1 and piftoc1 mutants at CT8 and CT14 (time points with low and high TOC1 levels, respectively, in WT plants) shows that PIF removal suppresses expression in toc1 mutants after a FRp both at CT14 and CT18 (Fig. 4E). To test whether the TOC1-imposed permissive or restrictive gene-expression pattern correlates with growth, we submitted WT and toc1 seedlings to 8 h of darkness after the FRp given during a subjective night at CT8, CT14, CT18, and CT24 (SI Appendix, Fig. S11C) and measured the hypocotyl elongation that took place during this time. The difference in hypocotyl length before and after the FRp plus 8 h of darkness was low in the WT seedlings at CT14 and CT18, when TOC1 levels are high, and was significantly greater at CT8 and CT24 (the beginning and end of the subjective night, respectively), when TOC1 levels are low (Fig. 4F). By contrast, the repression of growth at CT14 and CT18 was absent in the toc1 mutant (Fig. 4F). This pattern mirrors the data for marker gene expression (Fig. 4 D and E), strongly supporting the conclusion that the transcriptional repressor activity of TOC1 toward the PIFs mediates the gating of PIF-promoted growth by the clock. Together, these data support our hypothesis and provide a direct mechanism explaining the permissive gating of growth by TOC1 to time the maximum PIF3-promoted hypocotyl elongation precisely to the predawn period.
Discussion
We show that TOC1 directly interacts with and acts to repress the transcriptional activating activity of PIF3 (and, by extension, likely that of the other PIFs) (SI Appendix, SI Discussion) on the promoters of their cotargeted genes. Given the different dynamics of TOC1 and PIF3 protein levels during SD photocycles, we propose a model in which TOC1 binds, directly or indirectly, to the promoters of predawn-phased PIF and TOC1 cotarget genes during the early postdusk hours (Fig. 4G). Then, as the PIFs accumulate during the night, they are initially subjected to the transcriptional repressing action of TOC1, a repression that is lifted toward the end of the dark period when TOC1 levels decline coincident with maximum PIF levels. The cotargeted genes include growth-related and hormone-associated genes (8, 13, 20), which are PIF induced predawn, thereby promoting an increase in hypocotyl elongation rates (Fig. 4G).
These data indicate that the net transcriptional activation activity of the PIFs is determined by a dynamic balance in the relative abundance of the PIF and TOC1 proteins. We propose that this antagonistic interaction is potentially operative throughout the life cycle. In fully dark-grown, etiolated seedlings, the PIFs are at high levels that appear to be saturating for promotion of skotomorphogenesis, because the absence of any single member of the quartet in monogenic pif mutants has little or no effect on the phenotype (22). Under these conditions, the absence of native levels of TOC1 in the toc1 mutant has a minimal, albeit promotive, effect (SI Appendix, Fig. S12) (23). Exposure to light induces a precipitous reduction in PIF abundance through degradation to levels that become susceptible to significant repression by TOC1. We suggest that this repression explains the gene-expression patterns observed in de-etiolated seedlings under two different conditions: during the early night of diurnal photocycles as shown in Fig. 3B and during the light period in seedlings exposed to vegetative shade, as shown in Fig. 4D (12). The latter conclusion was suggested by the report by Salter et al. (12) that rapid shade-induced increases in PIL1 expression are gated in circadianly entrained seedlings released into LL conditions.
Although previous evidence has established TOC1 (also known as “Pseudo-response regulator 1,” PRR1) as a general transcriptional repressor (14, 16), our identification of the PIF transcriptional activators as direct molecular targets of TOC1 repression reveals a molecular mechanism by which that activity is exerted. Moreover, given the evidence that other members of the PRR protein family, i.e., PRR5, PRR7, and PRR9, impose transcriptional repression on target genes by recruiting the corepressor TOPLESS (TPL) (24), we speculate that TOC1 may invoke a similar mechanism to repress PIF activity, albeit using a different corepressor, because Wang et al. (24) failed to detect any direct interaction of TPL with TOC1. The question of the topology of PIF–TOC1 co-occupancy of target promoters remains open. The recruitment of TOC1 to G-box–containing promoter regions (Fig. 1 G and H) (14, 16) is consistent with either direct or indirect interaction with these genomic sites. The interaction could be the result of binding only to DNA-bound PIFs or indirectly to the pervasive TGTG DNA motifs, as reported by Gendron et al. (16), accompanied by interaction with neighboring PIFs (Fig. 4G).
One consequence of this general mechanism of TOC1 as a repressor of PIF transcriptional activation activity is that, although core clock-generated oscillations in TOC1 abundance have the potential to generate sustained, circadianly entrained oscillations in direct target-gene transcription in subsequent constant darkness (DD), where PIF levels are high, they lose this capacity in LL conditions, where PIF levels are too low to activate those genes (SI Appendix, Fig. S13). The initially surprising lack of sustained oscillations in LL conditions for the predawn-specific PIF-TOC1 genes in Fig. 3A and SI Appendix, Fig. S5 support the generality of this notion.
An additional ramification of the present data is that the functionally antagonistic interaction between the PIF and TOC1 proteins provides insight into the mechanism underlying the anticipated convergence of the light- and clock-regulated pathways in controlling common facets of plant morphogenesis (SI Appendix, Fig. S13) (4, 7, 12, 13, 25). Evidence continues to accumulate that, in addition to implementing this specific convergence, the PIFs function to integrate the activities of an increasing number of other signaling pathways, including the gibberellin, ethylene, and brassinosteroid hormones, sugar, and temperature (4, 25, 26). Many of the outputs from these pathways, in addition to diurnal growth, such as cellular metabolism and responses to temperature and biotic and abiotic stress (25), are subjected to permissive gating by the clock. At the transcriptome level, a striking feature of circadian activity is the large number of expressed genes that are regulated by the clock (27). Our present findings indicate that a significant fraction of this regulation is channeled through modulation of the PIF transcriptional network, which is known to control a broad range of biological processes, from seed germination and seedling development, through vegetative-shade avoidance and temperature responsiveness, to flowering. Thus, more generally, our data provide evidence that a core clock component functions as an output transducer that directly links the plant central oscillator to the regulatory machinery of a transcriptionally centered signaling hub that pleiotropically controls a diversity of plant growth and developmental responses to multiple inputs throughout the life cycle.
Materials and Methods
Available online tools were used to analyze and visualize the ChIP-seq data. Arabidopsis thaliana lines were in Columbia and C24 ecotypes. See SI Appendix, SI Material and Methods for transgenic and mutant line references, seedling growth conditions, and hypocotyl measurements. For gene-expression analysis, RNA extraction, cDNA synthesis, and quantitative RT-PCR (qRT-PCR) were done as described (28). PP2A was used for normalization. Primer details can be found in SI Appendix, Table S2. Protein extracts were prepared from seedlings grown under SD conditions as described (29). ChIP assays were performed as previously described (8) using SD-grown seedlings during the third day of growth at the indicated times. Primers used in the detection of each gene by qRT-PCR can be found in SI Appendix, Table S2. Coimmunoprecipitation assays were performed using SD-grown seedlings at ZT16 during the third day of growth as described (30), with modifications specified in SI Appendix, SI Materials and Methods. For BiFC the coding regions of PIF3 and TOC1 were PCR-amplified and cloned into pGWnY and pGWcY vectors (31). Details of all reagents and procedures are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Pfeiffer for early sharing of data and P. Más for materials and comments on the manuscript. This work was supported by Spanish Ministerio de Ciencia e Innovación Grant BIO2009-07675, Ministerio de Economía Grant BIO2012-31672, and Generalitat de Catalunya Grant 2009-SGR-206 (to E.M.); by a Comissionat per a Universitats i Recerca del Departament d’Innovació, Universitats i Empresa Fellowship from the Generalitat de Catalunya (Beatriu de Pinós program) and by Marie Curie International Reintegration Grant PIRG06-GA-2009-256420 and Generalitat de Catalunya Grant 2014-SGR-00268 (to P.L.); by Marie Curie Career Integration Grant 10-GA-2011-304008 (to C.D.); and by NIH Grant 2R01 GM-047475 and US Department of Agriculture Agricultural Research Service Current Research Information System Grant 5335-21000-032-00D (to P.H.Q.). J.S. was supported by a Junta para la Ampliación de Estudios Predoctoral Fellowship, and C.D. was supported by a Postdoctoral Fellowship from the Center for Research in Agricultural Genomics.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603745113/-/DCSupplemental.
References
- 1.Galvão VC, Fankhauser C. Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr Opin Neurobiol. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8(3):217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 3.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leivar P, Monte E. PIFs: Systems integrators in plant development. Plant Cell. 2014;26(1):56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant. 2014;7(11):1598–1618. doi: 10.1093/mp/ssu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen T, et al. Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell. 2006;18(10):2506–2516. doi: 10.1105/tpc.105.037358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448(7151):358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 8.Soy J, et al. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 2012;71(3):390–401. doi: 10.1111/j.1365-313X.2012.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashino T, et al. Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal Behav. 2013;8(3):e23390. doi: 10.4161/psb.23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomoto Y, Kubozono S, Yamashino T, Nakamichi N, Mizuno T. Circadian clock- and PIF4-controlled plant growth: A coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53(11):1950–1964. doi: 10.1093/pcp/pcs137. [DOI] [PubMed] [Google Scholar]
- 11.Soy J, Leivar P, Monte E. PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J Exp Bot. 2014;65(11):2925–2936. doi: 10.1093/jxb/ert465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426(6967):680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- 13.Michael TP, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6(9):e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336(6077):75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 15.Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002;43(1):58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- 16.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109(8):3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323(5920):1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44(2):300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- 19.Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125(3):485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 20.Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011;156(1):357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18(23):1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leivar P, et al. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24(4):1398–1419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15(1):223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA. 2013;110(2):761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 2015;16(10):598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 26.Shin J, Anwer MU, Davis SJ. Phytochrome-interacting factors (PIFs) as bridges between environmental signals and the circadian clock: Diurnal regulation of growth and development. Mol Plant. 2013;6(3):592–595. doi: 10.1093/mp/sst060. [DOI] [PubMed] [Google Scholar]
- 27.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sentandreu M, et al. Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell. 2011;23(11):3974–3991. doi: 10.1105/tpc.111.088161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leivar P, et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20(2):337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieto C, López-Salmerón V, Davière JM, Prat S. ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol. 2015;25(2):187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka Y, et al. Gateway Vectors for Plant Genetic Engineering: Overview of Plant Vectors, Application for Bimolecular Fluorescence Complementation (BiFC) and Multigene Construction. InTech; Rijeka, Croatia: 2012. [Google Scholar]
- 32.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23(3):439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.