Significance
Host plant specialization is a major cause of diversification in insects. The specialization of the fly Drosophila sechellia on the toxic fruits of noni has been a source of great scientific value, but selection is old enough that genetic variation does not seem useful in mapping the causative genes. On the island of Mayotte, we discovered a population of the related species Drosophila yakuba that is strongly associated with noni compared with generalist mainland populations. We then leveraged genomic variation to reconstruct the recent divergence history of this population and identify the potential targets of selection. Our top candidates included genes that confer tolerance to noni's toxin in D. sechellia. These findings establish a new model for recurrent ecological specialization.
Keywords: host plant adaptation, ecological genomics, parallel evolution, island speciation, Drosophila yakuba
Abstract
Recurrent specialization on similar host plants offers a unique opportunity to unravel the evolutionary and genetic mechanisms underlying dietary shifts. Recent studies have focused on ecological races belonging to the same species, but it is hard in many cases to untangle the role of adaptive introgression versus distinct mutations in facilitating recurrent evolution. We discovered on the island of Mayotte a population of the generalist fly Drosophila yakuba that is strictly associated with noni (Morinda citrifolia). This case strongly resembles Drosophila sechellia, a genetically isolated insular relative of D. yakuba whose intensely studied specialization on toxic noni fruits has always been considered a unique event in insect evolution. Experiments revealed that unlike mainland D. yakuba strains, Mayotte flies showed strong olfactory attraction and significant toxin tolerance to noni. Island females strongly discriminated against mainland males, suggesting that dietary adaptation has been accompanied by partial reproductive isolation. Population genomic analysis indicated a recent colonization (∼29 kya), at a time when year-round noni fruits may have presented a predictable resource on the small island, with ongoing migration after colonization. This relatively recent time scale allowed us to search for putatively adaptive loci based on genetic variation. Strong signals of genetic differentiation were found for several detoxification genes, including a major toxin tolerance locus in D. sechellia. Our results suggest that recurrent evolution on a toxic resource can involve similar historical events and common genetic bases, and they establish an important genetic system for the study of early stages of ecological specialization and speciation.
Ever since Darwin’s (1) description of finch diversity on the Galapagos archipelago, dietary specialization has been considered a major drive of speciation by means of natural selection. Adaptation to similar diets have led to the parallel evolution of beak morphology in some species inhabiting different islands, but genome analyses revealed that this was most likely due to the adaptive introgression of the underlying loci between species (2). In herbivorous insects, host plant specialization also plays a major role in diversification (3), and spectacular examples of convergent evolution both in plant resistant toxins and insect toxin resistances spanning hundred million of years of divergence have been observed. For example, several unrelated flowering plants produce cardenolides that block activity of the ion gradient regulating enzyme (Na++K+)ATPase in insects, but identical cardenolide-resistant amino acid substitutions in this enzyme have independently arisen in beetles, butterflies, flies, and aphids specializing on such plants (4). Recent genomic studies have focused on parallel dietary shifts in early-diverging ecological races, pointing to a substantial degree of common molecular mechanisms (5–8). However, at this evolutionary scale, detailed population genetic analyses would be required to distinguish convergent evolution due to adaptive introgression versus selection on independent mutations.
With few exceptions, most drosophilid species are detritivorous, scraping decomposing plant parts (mostly fruits) for yeasts and bacteria. However, ecological differences in preference for the degree of ripeness/decay exist, with species using less decaying material evolving capabilities to resist plant defensive toxins, digest living plant material, and detect the appropriate host and ripening stage (9–12). Such an “ecological gradient” makes drosophilids ideal for the study of the genetic basis of dietary shifts. A peculiar example is the specialization of Drosophila sechellia, which is endemic on the Seychelles islands in the Indian Ocean, on noni (Morinda citrifolia) fruits that are highly toxic to other drosophilids (13–22). The partial reproductive isolation between D. sechellia and its close relatives from the melanogaster subgroup retaining the ancestral decaying habitat has facilitated the investigation of the genetics of some phenotypes related to herbivory (18, 19, 21). However, the order by which these adaptations appeared, their effect on reproductive isolation, and the reproducibility of the genetic mechanisms underlying them remain largely unclear.
Here, we present a previously unidentified case of noni specialization involving a population of Drosophila yakuba on Mayotte Island (Comoros archipelago in the Indian Ocean), which was accompanied by partial reproductive isolation. D. yakuba is very abundant on mainland Africa and surrounding oceanic islands such as Madagascar (Fig. 1A), where it breeds on rotten fruits of more than 28 identified plant species (23). The ancestors of D. yakuba and D. sechellia diverged on the order of 10 million years ago (24) and these species are completely reproductively and geographically isolated (23). Consequently, any common genetic basis between them should be the result of parallel natural selection, rather than introgression. Interestingly, the insular case of D. yakuba occurred recently enough that genetic variation facilitated our pursuit of genes important in the specialist population’s evolution, with a genome-wide scan of genetic differentiation identifying regions associated with parallel noni specialization in D. sechellia. Our results indicate that some common genetic routes of host shift could be targeted by natural selection in isolated species and offer an ideal model to unravel these routes.
Fig. 1.
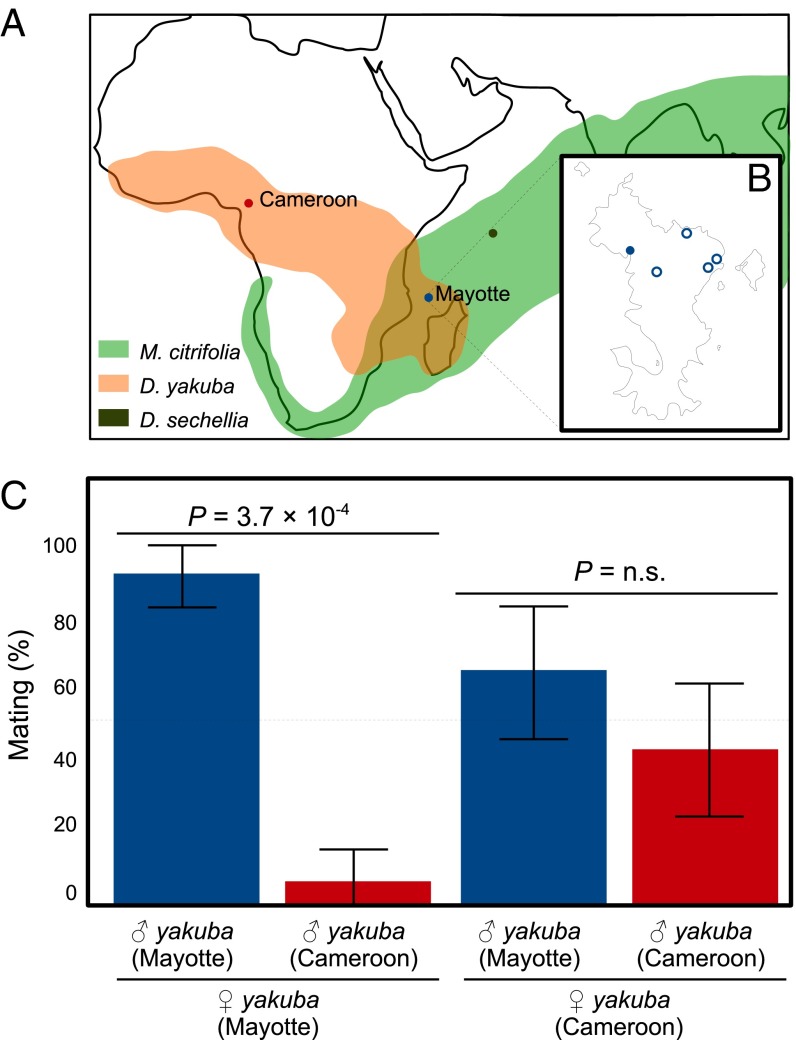
Association of D. yakuba with noni (Morinda citrifolia) on Mayotte and assortative mating with mainland flies. (A) Geographical distribution of D. yakuba (orange) (www.taxodros.uzh.ch/) and noni (light green) (28). Colors indicate location of strains used in subsequent experiments: D. sechellia (green), Mayotte D. yakuba (blue), and Cameroon D. yakuba (red). (B) Map of the island of Mayotte showing the location where D. yakuba was collected on noni (solid circle) and other prospected areas where D. yakuba was not found (open circles). (C) Proportion of homo- and heterogamic copulations between Mayotte and Cameroon D. yakuba. Fisher’s exact test P values are given by comparing outcomes of each cross to parity. Error bars indicate 95% confidence interval.
Results
Strong Association of D. yakuba with Noni on Mayotte.
During a recent collection of drosophilids on Mayotte Island (Comoros archipelago, between Madagascar and mainland Africa) we made a serendipitous discovery: the presence of numerous D. yakuba that are strictly associated with noni fruits. We collected flies, with banana traps or nets, in various habitats, ranging from forests to cultivated areas. Such a protocol, on the African mainland or Madagascar, would have produced many D. yakuba adults (refs. 25 and 26 and J.R.D., personal observations). In Mayotte, D. yakuba was found exclusively in two small areas at the Bay of Soulou, in which noni trees with ripe and fallen fruits were abundant (Fig. 1B). D. yakuba comprised the majority of the drosophilids collected by net sweeping at each noni site (Table 1). Strikingly, when we swept flies from rotten jackfruit (Artocarpus) adjacent to the second site, D. yakuba was virtually absent. From fallen noni fruits brought back to the laboratory, numerous adults emerged, with a predominance of D. yakuba (Table 1).
Table 1.
Number of flies belonging to different species collected in the Bay of Soulou (Mayotte)
| Species | Site A | Site B | Emergence from noni | |
| Noni | Noni | Jackfruit | ||
| D. yakuba | 94 | 331 | 2 | 44 |
| D. melanogaster | 1 | 0 | 0 | 0 |
| Drosophila malerkotliana | 71 | 129 | 151 | 30 |
| Drosophila nasuta | 5 | 39 | 11 | 12 |
| Scaptodrosophila latifasciaeformis | 0 | 10 | 227 | 0 |
| Zaprionus indianus | 0 | 8 | 1 | 0 |
| Zaprionus tuberculatus | 5 | 105 | 24 | 8 |
Asymmetric Prezygotic Isolation Between Mayotte and Mainland Flies.
We crossed Mayotte flies in the laboratory with flies from a mainland population from Cameroon. The flies readily breed in no-choice experiments, producing vigorous and fully fertile F1 males and females regardless of the cross direction, suggesting that Mayotte flies do belong to the species D. yakuba. However, when females from Mayotte or the mainland had to choose between males, we found an almost absolute homogamy for the Mayotte females but no preference for the mainland females (Fig. 1C). Among 30 copulations observed for Mayotte females (out of 46 experiments; see Methods), 28 (i.e., 93.3%) were homogamic, a highly significant difference from parity (Fisher’s exact test P < 0.0004). However, 14 out of 32 copulating mainland females (i.e., 43.8%) mated with their homogamic male (Fisher’s exact test P = 0.8025). Such asymmetric, prezygotic isolation is common in drosophilids and has been suggested to be an early step in speciation (27).
Higher Preference for and Performance on Noni of Mayotte Flies.
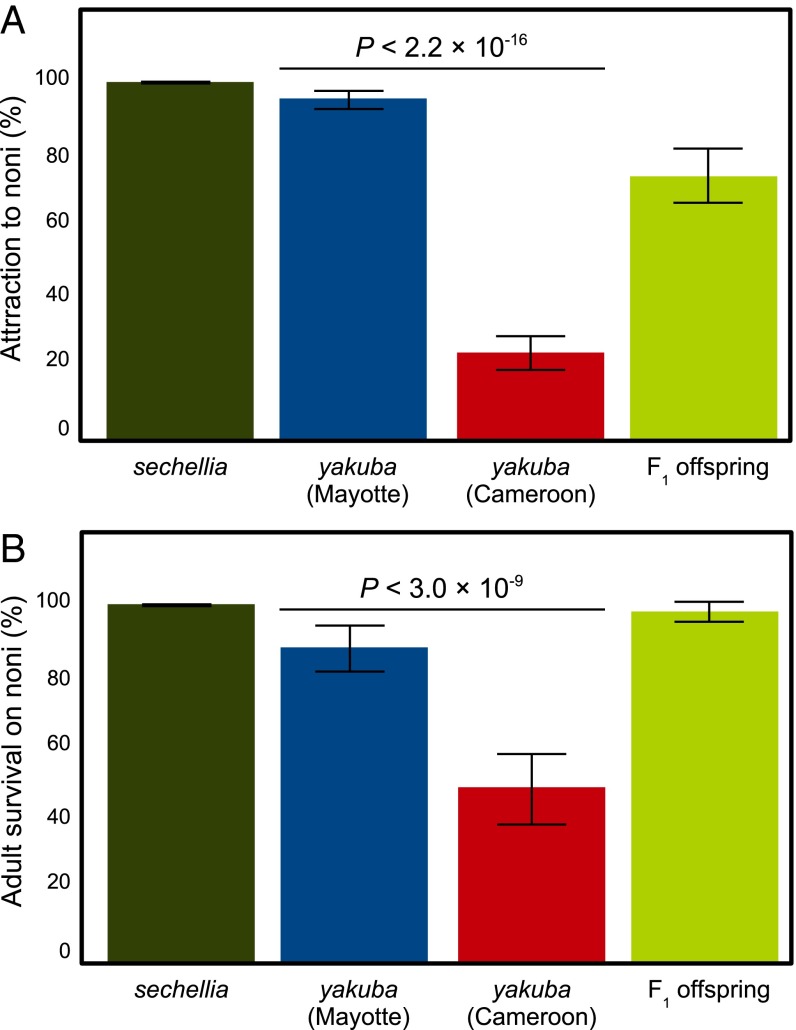
We conducted laboratory experiments to compare the degree of noni specialization of Mayotte flies with that of D. sechellia and of mainland D. yakuba. D. sechellia exhibits absolute olfactory attraction to noni when given a choice between noni and banana and nearly complete resistance to its toxin after exposed to ripe fruits for 1 d (Fig. 2). Unlike mainland D. yakuba, the Mayotte flies showed nearly absolute attraction to noni (Fisher’s exact test P < 2.2 × 10−16, Fig. 2A). The large majority of Mayotte flies attracted to the noni trap over the banana trap is consistent with noni’s being a primary food source for this population. Nonetheless, a minority of the mainland flies were still attracted to noni, despite the lack of records of noni in Cameroon (28), which could reflect stochastic variability in the fruit choice experiment or else genetic variation for odor preference. The attraction of the two D. yakuba populations is higher than that of the remaining seven species of the melanogaster subgroup (Dataset S1).
Fig. 2.
Specialization of D. yakuba from Mayotte on noni. Proportion of flies (A) recaptured from noni traps and (B) surviving on noni after 24-h exposure. Fisher’s exact test P values are given between flies from Mayotte and a mainland population from Cameroon. Error bars indicate 95% confidence interval.
When kept with a fresh noni fruit in a vial for 24 h, Mayotte flies showed a significant degree of tolerance to noni toxin compared with mainland flies (Fisher’s exact test P < 2.2 × 10−16, Fig. 2B). However, fewer flies survived than D. sechellia (Fisher’s exact test P = 0.0013). These results might reflect a preference for relatively more ripe noni fruit than offered here, or that the evolution of noni specialization is not yet complete.
We also tested attraction to and survival on noni in F1 offspring between Mayotte and mainland flies. In agreement with previous results on hybrids between D. sechellia and Drosophila simulans (13), attraction in F1 offspring in D. yakuba was intermediate between the two parental strains (73.6% attracted to noni, Fig. 2A) and significantly different from Mayotte and mainland parents (Fisher’s exact test P = 4.81 × 10−6 and < 2.2 × 10−16, respectively). F1 offspring also significantly differed from Mayotte and mainland parents in tolerance (Fisher’s exact test P = 0.0006 and < 2.2 × 10−16, respectively; Fig. 2B), but with a much higher odds ratio (O.R.) against the mainland parent (O.R. = 50.17) than with Mayotte (O.R. = 6.64), suggesting a possible dominance effect on tolerance similar to the one previously described in hybrids between D. sechellia and D. simulans (13). This result suggests that these shared features with D. sechellia may reflect common evolutionary and molecular processes.
Relatively Recent Colonization of Mayotte with Evidence for a Bottleneck.
We sequenced the genomes of a pooled sample of 22 isofemale lines from Mayotte D. yakuba and estimated its genetic diversity and differentiation from two mainland populations from Kenya and Cameroon (29). Mayotte was more distant from both mainland populations (FST = 0.153 and 0.162, respectively) than were the latter two populations from each other (FST = 0.068). On average, nucleotide diversity of the Mayotte population (π = 0.0089) was also slightly lower than that of the Kenya (π = 0.0095; Mann–Whitney U P < 4.61 × 10−6) and Cameroon populations (π = 0.0100; Mann–Whitney U P = 6.33 × 10−10) (Dataset S2).
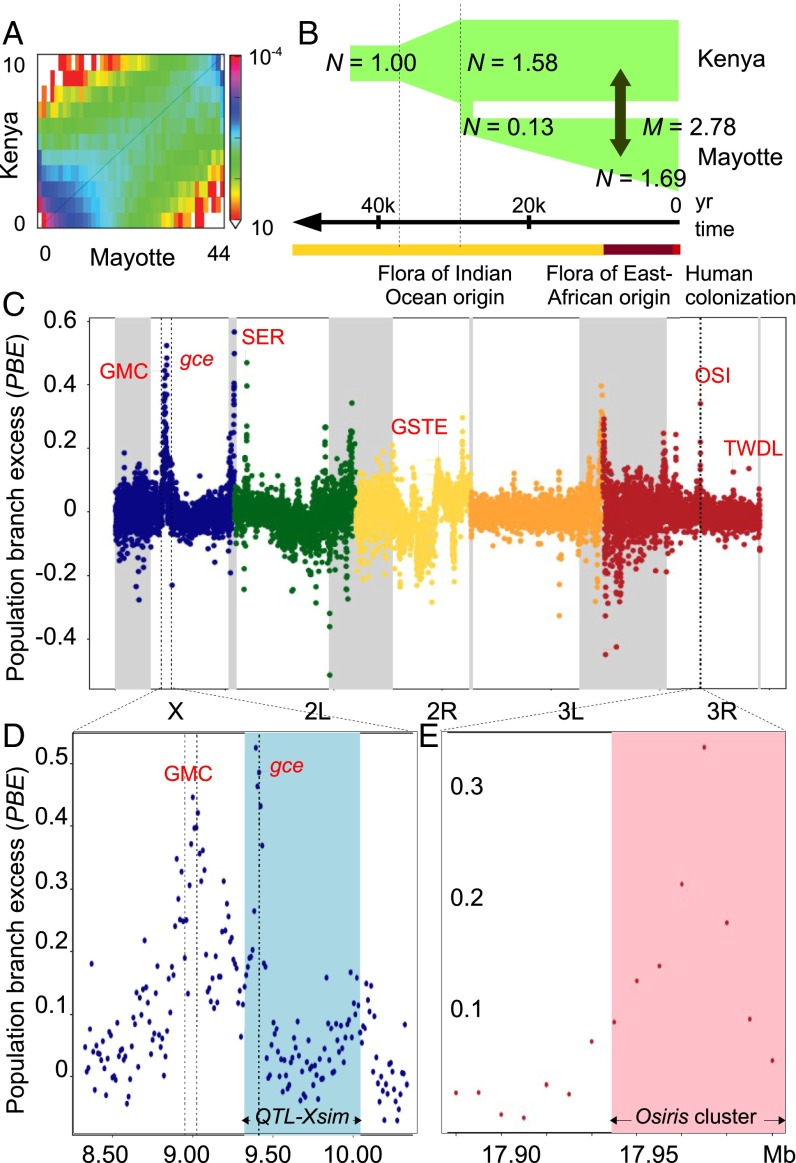
To estimate the demographic history of Mayotte flies, we compared the allele frequency spectrum of Mayotte flies with that of the closest mainland population from Kenya (29). Based on parameter estimates and model comparisons from δαδι (30), the best-fit model involved a relatively recent colonization of Mayotte (about 29,210 ± 1,024 y ago; see SI Methods) with some subsequent migration between populations (Fig. 3 A and B and Dataset S3; P = 0.0027 rejecting a model with no migration). The colonization event was accompanied by a moderate bottleneck, which may explain the low π of the Mayotte population. Nonetheless, genetic differentiation between mainland and island populations is relatively low. We therefore have an ideal scenario to detect genomic regions with elevated genetic differentiation due to local evolutionary processes such as noni specialization and/or reproductive isolation.
Fig. 3.
Demographic history of and selection traces on the genome of Mayotte D. yakuba. (A) Plot of allele frequency spectrum of D. yakuba from Mayotte and Kenya. (B) Illustration of the best-fit demographic model showing estimates of changes in size (N), the time of the split between Mayotte and Kenya and migration rate (M), compared with the floristic history of Mayotte (41). (C) Plot of average PBE per 10-kb nonoverlapping window in the Mayotte population. Note that PBE is positive when Mayotte-specific genetic differentiation exceeds its predicted value and may be negative if selection has acted in another population. Low recombining centromeric regions are highlighted in gray. (D) PBE values at the most differentiated region on the X chromosome with the major D. sechellia larval noni toxin tolerance QTL_simX (21) highlighted in blue. (E) PBE values at the major D. sechellia adult noni toxin tolerance QTL (19) with the Osiris genes cluster highlighted in pink. Major gene or gene cluster at the top of each divergence peak that are discussed in the text are given in red with clusters abbreviated: GMC, glucose–methanol–choline oxidoreductases; GSTE, GST E; OSI, Osiris proteins; SER, serine proteases; TWDL, Tweedle proteins.
Genome-Wide Scan of Targets of Natural Selection in Mayotte Population.
To identify genes that have been selected in Mayotte, we used a modification of the Population Branch Statistic (PBS) (31), which uses FST values among three populations to quantify genetic differentiation specific to one of them. To increase our focus on loci that were under selection in the Mayotte population specifically, rather than evolving adaptively in all populations, we quantified the “Population Branch Excess” (PBE). PBE quantifies the degree to which PBS exceeds its predicted value, based on differentiation between the other two populations at this locus, and in light of the typical patterns observed at other loci (Methods). We used the pair of mainland populations (Cameroon and Kenya) to quantify genetic differentiation that is specific to the Mayotte population. Our scan revealed strong, localized peaks of genetic differentiation in Mayotte, resembling “genomic islands of divergence” (Fig. 3C and Dataset S4).
The locus with the strongest allele frequency shift was found on the X chromosome and falls in the middle of a major quantitative trait locus (QTL) region (∼1 Mb) recently identified in D. sechellia for larval resistance against octanoic acid, the main toxin of noni (21). Our most differentiated windows correspond to the peak of this QTL, which includes the juvenile hormone receptor gene gce, an essential regulator of molting (32) (Fig. 3D). A closer look to this locus also reveals a secondary shift of PBE values falling only 0.4 Mb upstream from the peak. It centers on a cluster of glucose–methanol–choline oxidoreductases, with the highest PBE values falling near Ecdysone oxidase, which is another essential regulator of molting (33), and neighboring paralogs.
The second-highest peak of differentiation in normally recombining regions is detected on chromosome arm 2L (Fig. 3C). It contains a cluster of digestive serine proteases that have rapidly evolved in D. sechellia (34) and expanded in cactophilic Drosophila (35) and are associated with detoxification (36) and food choice (37) in D. melanogaster. Interestingly, the third-highest peak in these regions falls in the middle of a major QTL region on chromosome arm 3R that confers adult resistance against octanoic acid in D. sechellia (19). The ∼170-kb-long QTL was found from interspecific introgression mapping and contains 18 genes, with “none … show[ing] a strong signature of positive selection that may be expected for a gene contributing to D. sechellia adaptation to its host” (10). We found evidence for such strong selection in Mayotte D. yakuba, with the highest PBE values falling amid a cluster of Osiris genes at a window containing Osiris 4 and Osiris 5 (Fig. 3E).
Among loci with moderately elevated PBE values, there is a locus containing a cluster of GST E genes (Fig. 3C and Dataset S5), which significantly differ in expression upon exposure to noni in D. sechellia (17). A similar PBE signal includes a family of larval cuticular proteins, Tweedle, which corresponds to a noni tolerance QTL in D. sechellia larvae (21) (Fig. 3C and Dataset S5). These genes are also differentially expressed when larvae of the herbivorous drosophilid Scaptomyza flava are exposed to Arabidopsis toxin (38).
Genetically Parallel Adaptation to Noni Toxin.
To more formally test for a signal of genetically parallel evolution, we compared the observed concordance of D. yakuba PBE outliers with D. sechellia QTLs to that expected by chance, using genomic permutations. This analysis included tolerance QTLs (refs. 19 and 21 and Dataset S6) and attraction QTLs (ref. 18 and C. D. Jones, personal communication) present outside of putative low recombination regions. We found that just 3 of 13 attraction QTLs overlapped a window with PBE greater than 0.125 (P = 0.200). However, four of nine tolerance QTLs matched this criterion (P = 0.013). By the same process, we confirmed the unlikelihood that the single finely mapped QTL for adult tolerance (encompassing the Osiris cluster) should overlap a PBE value as high as 0.34 by chance (P = 0.008). As expected, no significant overlap was observed for attraction or tolerance QTLs if PBE outliers from Cameroon or Kenya were used instead (P > 0.37 for all four tests). These results suggest that some of the same genes (or paralogous gene clusters) may contribute to noni tolerance in D. sechellia and D. yakuba. In contrast, the lack of D. yakuba PBE outliers corresponding to D. sechellia attraction QTLs could reflect a distinct genetic basis of noni preference, or else natural selection in D. yakuba that did not produce strong effects on linked variation (e.g., soft sweeps, as discussed below). Thus, in addition to demonstrating the parallel phenotypic evolution of noni attraction and tolerance in D. sechellia and D. yakuba, our results support genetic parallelism in the evolution of tolerance, which could include some of the most strongly differentiated loci in the Mayotte population (i.e., the X-linked gce and the Osiris cluster on 3R).
SI Methods
Preparation of Genome Libraries.
We sequenced two pooled samples each consisting of four F1 females of 11 different isofemale lines (44 flies per pool). Genomic DNA was obtained using chloroform extraction and ethanol precipitation. DNA was then fragmented using Bioruptor sonicator (Diagenode), and paired-end libraries with ∼300-bp inserts were prepared using NEBNext DNA Library Prep Reagent Set for Illumina (E6000L; New England Biolabs). Libraries’ concentration and quality were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). They were then sequenced at the University of Wisconsin–Madison Biotechnology Center on the Illumina HiSEq. 2000 platform with 100-bp paired-end reads.
Genome Alignment.
We obtained BAM files for 20 isofemale lines from two mainland populations of D. yakuba that have recently been published from Nairobi, Kenya and Nguti, Cameroon [Sequence Read Archive (SRA) project SRP029453] (29). For comparative purposes we followed the same alignment protocol that was used to generate these 20 BAM files in aligning the reads of our two libraries from Mayotte. We used the command line bwa aln −l 13 −m 50000000 −R 5000 to align our reads to the D. yakuba reference genome r.1.3 obtained from Flybase (flybase.org/), resolved paired-end mappings using the command line bwa sampe −a 5000 −N 5000 −n 500 as implemented in the BWA ver. 0.6.2-r126 software package (55), and sorted and converted the output into a BAM file using SAMtools ver. 0.1.18 (56). After merging the two libraries, the average sequencing depth was about 77×. Sequence data are available on SRA (project SRP063516). We also generated a BAM file for a genome of Drosophila santomea, the closest relative to D. yakuba that is endemic on the island of Sao Tomé, which has also been recently published (SRA project SRR1774234) (57), by aligning it to the D. yakuba reference. The PoPoolation2 ver. 1.201 software package (58) was used to generate a synchronized mpileup file for the 23 BAM files. We then used a custom Perl script to pool files belonging to the same population resulting into a synchronized mpileup file with four population columns (Mayotte, Kenya, Cameroon, and D. santomea) aligned to the D. yakuba reference. To avoid discrepancies in allele frequency estimates due to different sequencing depth in Kenya and Cameroon, we conservatively counted the major allele for each line leading to sample sizes of 10 for each of these populations, because each consisted of 10 inbred isofemale lines maintained in the laboratory for a long time (29).
Inference of Demographic History.
We used the allele frequency spectrum (AFS) from 102,477 SNPs having a minimum sequencing depth of 44× in Mayotte and a called allele in at least five genomes each from Kenya and Cameroon. These SNPs were from short autosomal intronic segments (< 86 bps, with 16 and 6 bps removed from the intron start and end, respectively), because these SNPs are presumably neutral (59). We note that local adaptation or background selection at linked loci could still increase genetic differentiation at neutral sites, potentially decreasing and increasing estimates of gene flow and divergence time, respectively. However, the small proportion of windows that clearly depart from the genomic distribution of PBE (Fig. S1) provides hope that such biases may be minor. Because pooled sequences of F1 flies from 22 isofemale lines were used for Mayotte (potentially representing 88 or more parental alleles per locus), we estimated allele counts by conservatively assuming the presence of just 44 alleles and rounding the counts to the nearest integer 0–44.
Fig. S1.
Histogram of PBE values in the Mayotte population estimated for nonoverlapping 10-kb genomic windows.
We fit the “prior_onegrow_mig” demographic model to our observed AFS of Mayotte and Kenya populations using a diffusion approximation approach implemented in the δαδι ver. 1.7 software package (30). This model implies two stages of growth/decline, wherein the ancestral population grows before splitting, and a bottleneck took place in one of the two populations (Mayotte) with ongoing migration. Parameters were optimized using δαδι and several runs were conducted with the most optimized parameters until convergent fit log-likelihood estimates were obtained. We then used nonparametric bootstrapping to infer parameter uncertainties. For this, we generated 100 bootstrapped AFS using a custom Perl script, and parameter SDs were estimated using the Godambe information matrix (GIM) approach as implemented in δαδι ver 1.7. GIM was also used in δαδι to test the likelihood of a model implying no migration compared with our best-fit model with migration using the adjusted D statistic.
To translate δαδι parameters into their corresponding proper units, we needed to estimate the ancestral effective population size (N). For this we estimated the level of nucleotide diversity (π) for the most diverse population (Cameroon) in 100-kb windows using a custom Perl script and estimated N by dividing π over four times the mutation rate [using an estimate from Drosophila melanogaster of 3.27 × 10−9 (60)]. Divergence times, given in terms of 2 N, were then estimated assuming 15 generations per year (61).
Discussion
Our results provide insights on a recurrent shift from generalism toward specialism. Recent studies on phenotypic evolution have illustrated that similar selective pressures often favor common genes in different taxa (39). However, ecological specialization is a complex phenotype, involving multiple, interacting selection targets that affect behavioral, physiological, and morphological phenotypes (40). We show that attraction, tolerance, and prezygotic barriers can evolve relatively quickly. Our estimated divergence time for the Mayotte D. yakuba population (∼29 kya) could reflect a marine crossing facilitated by lower sea levels during the last glaciation. However, in light of uncertainties such as generation time (SI Methods) and the precise source population for the Mayotte dispersal, we cannot firmly rule out a more recent crossing corresponding to the Holocene marine transgression (10 kya), when other plant species of African origin colonized the island (41). Before the introduction of nontoxic fruits by humans much later, year-round fleshy noni fruits were probably among few predictable native resources, which may have favored the evolution of specialization.
The only other known drosophilid case (i.e., D. sechellia) of noni specialization occurred in the nearby Seychelles archipelago. However, this case is roughly eight times older than ours (∼250,000 y) (42), and despite evidence for recent and ongoing introgression between D. sechellia and its relative D. simulans (42, 43), it is unknown whether the initial phases of noni specialization have also involved gene flow. The geographic parallelism between the two species supports a major role of islands in facilitating specialization on otherwise nonpreferred resources by reducing gene flow (44).
In D. sechellia, attraction to noni and survival thereon seem to have different genetic bases, but the order by which these adaptations appeared remains unclear. Hungate et al. (19) hypothesized that olfactory attraction might have evolved first from recessive alleles segregating in the ancestral range, such as loss-of-function mutations at olfaction-related genes (7, 12, 15, 45). Such attraction would yield strong selection for tolerance alleles, which may often be dominant (13, 14) and may originate from new mutations or rare variants in the ancestral populations. Our results support some of these predictions. First, we found that, similar to D. sechellia (13), tolerance in F1 offspring was dominant whereas attraction was intermediate. Second, our most extreme population genetic differentiation signals did not center on obvious attraction-related genes such as olfactory or gustatory receptors, which might be expected if selection acted on standing genetic variation present in the colonizing population (46, 47). Indeed, none of the largest peaks contained any member of the chemosensory gene families whose rapid evolution and turnover are associated with ecological specialization in drosophilids and other insects (7, 12, 16, 22, 45). Because a minority of mainland flies visited noni in our experiment, it may be worth testing whether standing genetic variation exists for this trait (Fig. 2A and Dataset S1). Further study will be needed to test whether, in addition to common genes potentially being targeted by similar selective pressures, a similar order of adaptive events may have been repeated during parallel ecological specialization.
The outlier regions that appear in our analysis provide excellent candidates for studies on the molecular convergence of toxin resistance, not only between D. yakuba and D. sechellia, which are both genetically and geographically isolated, but also with other herbivorous drosophilids and insects. The two peaks within the highly differentiated X-linked locus include genes controlling juvenile hormone and ecdysteroid signaling whose perturbation is a common target of plant defensive toxins (48, 49). Mutations in genes affecting the ecdysteroid metabolism pathway have enhanced D. sechellia adaptation on toxic noni (20) and helped make Drosophila pachea an obligatory specialist on a toxic cactus (9). Another particularly intriguing candidate is the Osiris cluster associated with resistance to octanoic acid in D. sechellia (19). Although the exact function of these transmembrane proteins remains unknown, they are differentially expressed upon exposure to Arabidopsis glucosinolates in the herbivorous drosophilid S. flava (38) and they have expanded in herbivorous silkworm and pea aphid (50). Other candidates for convergence with herbivorous Scaptomyza include the above-mentioned Tweedle family (38) and cluster D GST genes (51). It is therefore possible that aspects of the parallel evolution between D. yakuba and D. sechellia may extend to other distantly related drosophilids and perhaps beyond. Future functional dissection of the nucleotide differences at these genes in these species and other insects will unravel the molecular basis of recurrent host shift and distinguish between toxin-specific and generalist detoxification mechanisms.
Ecological specialization may contribute to speciation, but the link between genes involved in both processes has long remained elusive (52). Along with D. sechellia, the Mayotte population of D. yakuba may represent two points along an “ecological speciation continuum” involving specialization on toxic noni and the evolution of reproductive isolation. However, it is unclear whether reproductive isolation might have a similar biological basis in these taxa, or how much it is associated with genes driving specialization. The evolution of prezygotic isolation is known to correlate with genetic divergence between allopatric Drosophila species (53), but further research will be needed to relate DNA sequence divergence to reproductive isolation for multiple allopatric species pairs, to determine whether prezygotic isolation in Mayotte D. yakuba has evolved unexpectedly quickly. The lack of postzygotic incompatibilities between Mayotte and mainland flies, together with the molecular and genomic tool kit of Drosophila, provides a unique opportunity to reveal the genetic basis of prezygotic isolation. Our discovery and results reflect important steps toward understanding the mechanisms leading to ecological specialization and insular speciation.
Methods
Fly Collection and Establishment of Isofemale Lines.
Five locations on the northern part of the larger island of Mayotte (Grande Terre) were surveyed for drosophilids in January 2013: Kangani, Mamudzu, La Maison du Gouverneur, Combani, and Bay of Soulou. The locations presented a diversity of habitats, ranging from sea levels to mountainous elevations and from urban regions to primary forests. We collected flies using standard fermenting fruit baits (i.e., placing fermented bananas in a plastic bottle that is hung from a tree branch), or by net sweeping or aspirating over fallen ripe fruits. The taxonomic inventory of this collection is given in David et al. (54). For D. yakuba, 22 isofemale lines were established from flies collected on noni by aspirating over ripe fruits or net sweeping over fallen fruits on costal forest. A proportion of F1 progeny of each line was mixed to establish a mass laboratory culture that was later used for further behavioral and physiological experiments. The remaining progeny were separately preserved in absolute ethanol for subsequent genomic analyses. Comparisons were conducted with a mass culture of a mainland population (from Kunden, Cameroon, collected in 1967, and maintained at ∼2,000 individuals per generation) and laboratory strains of various species of the melanogaster subgroup including D. sechellia (Dataset S1). Strains were kept at 21–24 °C on standard Drosophila medium.
Hybridization and Mate Choice Experiments.
No choice experiments were conducted by placing 10 virgin females and 10 males in the same vial from Mayotte and Kunden in both directions of crosses and examining vials for F2 progeny indicating fertile F1 offspring. For mate choice experiments, we placed without anesthesia 46 virgin 4-d-old females from each population in separate vials with two males each from one of the two populations. Males were marked by clipping the tip of the wing of one of them. Half Mayotte and half Kunden males were clipped to assess the possible effect of clipping on female preferences but no difference was detected. Preference itself was assessed by 1.5-h survey and once a pair was established the noncopulating male was aspirated and identified under a binocular. Experiments were conducted in the morning at room temperature (∼24 °C). Statistical significance for each population females was assigned by comparing the counts of homogamic versus heterogamic pairs using a Fisher’s exact test as implemented in the R software package (https://www.r-project.org/).
Olfactory Attraction Experiment.
We used R’kha et al.’s (13) experimental protocol to investigate olfactory attraction in the field. In summary, three bottles containing ∼1,000, 3- to 4-d-old laboratory-grown adults were released 20 m away from traps containing either a piece of ripe but not rotten noni or crushed banana. Unripe noni fruits were obtained from Mayotte, courtesy of T. Claverie, Centre Universitaire de Mayotte, Dembéni. Fruits were left at room temperature until ripening (changing to grayish color) and then frozen at −20 °C. One day before an experiment, one fruit was removed from the freezer and left to thaw overnight. Pieces of ∼100 g were cut and set with a piece of absorbent paper in the trap bottles. For each experiment four recapture sites were set, each containing one noni and one banana trap placed 1.5 m apart. Flies were then recaptured on three successive days, and the total number of recaptured flies was compared between the two kinds of traps. We also compared the behavior of eight species of the melanogaster subgroup in a total of eight successive experiments (Dataset S1), as well as for F1 offspring from reciprocal crosses between the Mayotte and Kunden populations of D. yakuba. Flies for the above experiments were obtained from outbred mass cultures with the location and date of collection given in Dataset S1. Statistical significance for each population/species was assigned by comparing the counts of recaptured flies in noni versus banana traps using a Fisher’s exact test as implemented in R.
Tolerance to Noni Experiments.
To test for tolerance to noni toxin, we introduced groups of 20 adults in air-tight Falcon 50-mL conical centrifuge tubes containing 2 g of noni pulp and scored the number of dead flies after 1 d. For each experiment, a single ripe, frozen fruit was left to thaw overnight, and then the pulp was separated from the skin and seeds using a forceps and a scalpel, cut into small pieces, and weighted to 2 g on a balance. These pieces were placed on the surface of a piece of absorbent paper soaked with 2 mL of a 3% (wt/vol) sucrose solution and introduced to the tube. For each species or strain five replicates were tested. We conducted the same test simultaneously on F1 offspring using five replicates for each cross direction. Statistical significance for each population/species was assigned by comparing the counts of live versus dead flies after 24 h using a Fisher’s exact test in R.
Quantifying Genetic Differentiation.
We used PoPoolation2 to estimate pairwise genetic differentiation (FST) between the three populations of D. yakuba, using a minimum allele count of two and a minimum depth of five. We estimated FST for nonoverlapping 10-kb windows. To avoid any potential bias due to low recombination regions potentially having a greater variance of FST, we excluded from our analyses centromere- and telomere-proximal intervals showing reduced diversity. The included intervals for each chromosome arm, in megabases, were X:6.32–20.72, 2L:0.35–17.39, 2R:6.67–20.96, 3L:0.24–20.02, and 3R:11.56–28.65.
To search for natural selection that took place in Mayotte, we included the two continental populations (Cameroon and Kenya) and used the three FST estimates to calculate a modified version of the PBS (31). We define PBE as the degree to which PBS exceeds its expected value based on (i) the degree of locus-specific genetic differentiation between the two nonfocal populations and (ii) the median values of that quantity and PBS across all windows on the chromosome arm. Formally,
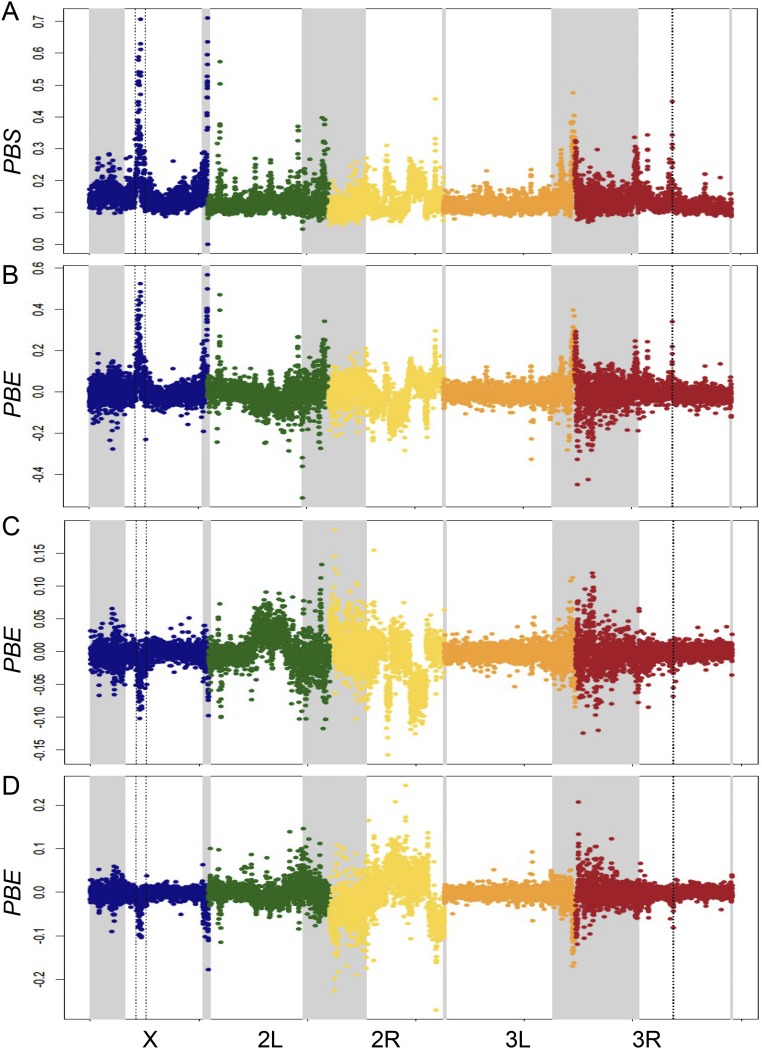
TBC represents the branch length between populations B and C (the nonfocal populations) and is equal to –log(1 – FST), whereas PBS is the branch length specific to the focal population A (31). Here, TBC serves to scale our locus-specific expectations for genetic differentiation, whereas the ratio term in the PBE equation indicates the typical relationship between PBS and TBC. PBE should therefore be centered around zero (as observed in our data; Fig. S1), it will be positive to the degree that PBS exceeds its predicted value, and it will be negative if there is elevated genetic differentiation specific to population B or C. Because PBS simply measures the branch length specific to population A, it should be large whenever positive selection has occurred in population A, regardless of whether it has also occurred in population B or C. In contrast, PBE will be strongly positive if selection is specific to population A but closer to zero if widespread positive selection or background selection has increased the lengths of all branches similarly. Hence, PBE is intended to focus more directly on loci under positive selection in the focal population only. However, we note that our PBE peaks are all detected by PBS as well (Fig. S2).
Fig. S2.
Comparison between PBS values in Mayotte and PBE values in the three populations. (A) PBS in Mayotte. PBE values in (B) Mayotte, (C) Kenya, and (D) Cameroon.
Test for Parallel Adaptation Between D. yakuba and D. sechellia.
We implemented a genomic permutation approach to test for a significant enrichment of genomic windows that had both elevated PBE in Mayotte D. yakuba and mapped QTL for D. sechellia. QTLs were drawn from mapping studies of noni attraction (ref. 18 and C. D. Jones, personal communication) and tolerance to noni or octanoic acid (19, 21). QTL positions were obtained with respect to the D. yakuba genome. For larval tolerance, we used a logarithm of the odds difference of 0.5 from the QTL peak to define its boundary (Dataset S6). We chose a PBE threshold of 0.125 for this analysis; above this value, the genomic distribution formed a long tail encompassing ∼1.5% of all windows (Fig. S1). We used a shift-based permutation scheme, sliding the full genomic landscape of PBE by set increments. This practice maintains the native landscape of PBE from our data, including the proximity of closely linked regions in which PBE may bounce above and below our threshold. To increase the independence of each shift replicate, PBE values were shifted five windows (500 kb) at a time. Only shift increments at least 500 kb displaced from the empirical landscape were used, yielding 1,633 total shift replicates. A P value was defined by the proportion of shift replicates in which at least as many unique QTLs overlapped PBE outliers as observed in the empirical data.
Supplementary Material
Acknowledgments
We thank C. B. Jones for updated data on the D. sechellia olfactory quantitative trait locus data, D. Erezyilmaz for sharing D. sechellia larval toxin tolerance mapping data, T. Claverie for noni supplied from Mayotte, J. B. Lack for bioinformatic assistance, and M. J. Monette for laboratory assistance. Funding was provided by the Richard Lounsbery Foundation (J.E.P. and J.R.D.), NIH Grant R01 GM111797 (to J.E.P.), and Terres Australes et Antarctiques Françaises (TAAF) Grant EPARDROS (to V.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive database (accession nos. SRX1212819–SRX1212838).
See Commentary on page 4558.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522559113/-/DCSupplemental.
References
- 1.Darwin C. On the Origin of the Species by Means of Natural Selection: Or, the Preservation of Favoured Races in the Struggle for Life. John Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Lamichhaney S, et al. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 3.Wiens JJ, Lapoint RT, Whiteman NK. Herbivory increases diversification across insect clades. Nat Commun. 2015;6:8370. doi: 10.1038/ncomms9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobler S, Dalla S, Wagschal V, Agrawal AA. Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proc Natl Acad Sci USA. 2012;109(32):13040–13045. doi: 10.1073/pnas.1202111109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan SP, Nosil P, Funk DJ. Selection and genomic differentiation during ecological speciation: Isolating the contributions of host association via a comparative genome scan of Neochlamisus bebbianae leaf beetles. Evolution. 2008;62(5):1162–1181. doi: 10.1111/j.1558-5646.2008.00352.x. [DOI] [PubMed] [Google Scholar]
- 6.Soria-Carrasco V, et al. Stick insect genomes reveal natural selection’s role in parallel speciation. Science. 2014;344(6185):738–742. doi: 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- 7.Duvaux L, et al. Dynamics of copy number variation in host races of the pea aphid. Mol Biol Evol. 2015;32(1):63–80. doi: 10.1093/molbev/msu266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan SP, et al. Experimental evidence of genome-wide impact of ecological selection during early stages of speciation-with-gene-flow. Ecol Lett. 2015;18(8):817–825. doi: 10.1111/ele.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang M, et al. Mutations in the neverland gene turned Drosophila pachea into an obligate specialist species. Science. 2012;337(6102):1658–1661. doi: 10.1126/science.1224829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzkin LM. Population transcriptomics of cactus host shifts in Drosophila mojavensis. Mol Ecol. 2012;21(10):2428–2439. doi: 10.1111/j.1365-294X.2012.05549.x. [DOI] [PubMed] [Google Scholar]
- 11.Linz J, et al. Host plant-driven sensory specialization in Drosophila erecta. Proc R Soc London B Biol Sci. 2013;280(1760):20130626. doi: 10.1098/rspb.2013.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman-Huertas B, et al. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc Natl Acad Sci USA. 2015;112(10):3026–3031. doi: 10.1073/pnas.1424656112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R’Kha S, Capy P, David JR. Host-plant specialization in the Drosophila melanogaster species complex: A physiological, behavioral, and genetical analysis. Proc Natl Acad Sci USA. 1991;88(5):1835–1839. doi: 10.1073/pnas.88.5.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones CD. The genetic basis of Drosophila sechellia’s resistance to a host plant toxin. Genetics. 1998;149(4):1899–1908. doi: 10.1093/genetics/149.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5(5):e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopp A, et al. Evolution of gene expression in the Drosophila olfactory system. Mol Biol Evol. 2008;25(6):1081–1092. doi: 10.1093/molbev/msn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dworkin I, Jones CD. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics. 2009;181(2):721–736. doi: 10.1534/genetics.108.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earley EJ, Jones CD. Next-generation mapping of complex traits with phenotype-based selection and introgression. Genetics. 2011;189(4):1203–1209. doi: 10.1534/genetics.111.129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hungate EA, et al. A locus in Drosophila sechellia affecting tolerance of a host plant toxin. Genetics. 2013;195(3):1063–1075. doi: 10.1534/genetics.113.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavista-Llanos S, et al. Dopamine drives Drosophila sechellia adaptation to its toxic host. eLife. 2014;3:e03785. doi: 10.7554/eLife.03785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Erezyilmaz D. The genetics of resistance to Morinda fruit toxin during the postembryonic stages in Drosophila sechellia. G3(Bethesda) 2015;5(10):1973–1981. doi: 10.1534/g3.114.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiao M-S, et al. Expression divergence of chemosensory genes between Drosophila sechellia and its sibling species and its implications for host shift. Genome Biol Evol. 2015;7(10):2843–2858. doi: 10.1093/gbe/evv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachaise D, et al. Historical biogeography of the Drosophila melanogaster species subgroup. In: Hecht MK, Wallace B, Prance GT, editors. Evolutionary Biology. Springer; New York: 1988. pp. 159–225. [Google Scholar]
- 24.Obbard DJ, et al. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol Biol Evol. 2012;29(11):3459–3473. doi: 10.1093/molbev/mss150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escher SA, Eriksson K, Bächli G. Collection of Drosophilidae (Diptera) along a transect in Kenya. Mitt Schweiz Entomol Ges. 1997;70(1/2):1–14. [Google Scholar]
- 26.Prigent SR, Le Gall P, Mbunda SW, Veuille M. Seasonal and altitudinal structure of drosophilid communities on Mt Oku (Cameroon volcanic line) C R Geosci. 2013;345(7–8):316–326. [Google Scholar]
- 27.Yukilevich R. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution. 2012;66(5):1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x. [DOI] [PubMed] [Google Scholar]
- 28.Razafimandimbison SG, McDowell TD, Halford DA, Bremer B. Origin of the pantropical and nutriceutical Morinda citrifolia L. (Rubiaceae): Comments on its distribution range and circumscription. J Biogeogr. 2010;37(3):520–529. [Google Scholar]
- 29.Rogers RL, et al. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol. 2014;31(7):1750–1766. doi: 10.1093/molbev/msu124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jindra M, Uhlirova M, Charles J-P, Smykal V, Hill RJ. Genetic evidence for function of the bHLH-PAS protein Gce/Met as a juvenile hormone receptor. PLoS Genet. 2015;11(7):e1005394. doi: 10.1371/journal.pgen.1005394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi H, Rigden DJ, Ebrahimi B, Turner PC, Rees HH. Regulation of ecdysteroid signalling during Drosophila development: Identification, characterization and modelling of ecdysone oxidase, an enzyme involved in control of ligand concentration. Biochem J. 2005;389(Pt 3):637–645. doi: 10.1042/BJ20050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prokupek A, et al. An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution. 2008;62(11):2936–2947. doi: 10.1111/j.1558-5646.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- 35.Kelleher ES, Pennington JE. Protease gene duplication and proteolytic activity in Drosophila female reproductive tracts. Mol Biol Evol. 2009;26(9):2125–2134. doi: 10.1093/molbev/msp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedra JHF, McIntyre LM, Scharf ME, Pittendrigh BR. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc Natl Acad Sci USA. 2004;101(18):7034–7039. doi: 10.1073/pnas.0400580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toshima N, Hara C, Scholz C-J, Tanimura T. Genetic variation in food choice behaviour of amino acid-deprived Drosophila. J Insect Physiol. 2014;69:89–94. doi: 10.1016/j.jinsphys.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 38.Whiteman NK, et al. Genes involved in the evolution of herbivory by a leaf-mining, Drosophilid fly. Genome Biol Evol. 2012;4(9):900–916. doi: 10.1093/gbe/evs063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin A, Orgogozo V. The Loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67(5):1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 40.Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu Rev Ecol Syst. 1988;19:207–233. [Google Scholar]
- 41.Pascal O, Labat J-N, Pignal M, Soumille O. Diversité, affinités phytogéographiques et origines présumées de la flore de Mayotte (Archipel des Comores) Syst Geogr Plants. 2001;71(2):1101–1123. [Google Scholar]
- 42.Garrigan D, et al. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 2012;22(8):1499–1511. doi: 10.1101/gr.130922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matute DR, Ayroles JF. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J Evol Biol. 2014;27(6):1057–1068. doi: 10.1111/jeb.12391. [DOI] [PubMed] [Google Scholar]
- 44.Warren BH, et al. Islands as model systems in ecology and evolution: Prospects fifty years after MacArthur-Wilson. Ecol Lett. 2015;18(2):200–217. doi: 10.1111/ele.12398. [DOI] [PubMed] [Google Scholar]
- 45.Almeida FC, Sánchez-Gracia A, Campos JL, Rozas J. Family size evolution in Drosophila chemosensory gene families: a comparative analysis with a critical appraisal of methods. Genome Biol Evol. 2014;6(7):1669–1682. doi: 10.1093/gbe/evu130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pennings PS, Hermisson J. Soft sweeps III: The signature of positive selection from recurrent mutation. PLoS Genet. 2006;2(12):e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messer PW, Petrov DA. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol. 2013;28(11):659–669. doi: 10.1016/j.tree.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mithöfer A, Boland W. Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol. 2012;63(1):431–450. doi: 10.1146/annurev-arplant-042110-103854. [DOI] [PubMed] [Google Scholar]
- 49.Lee S-H, et al. Identification of plant compounds that disrupt the insect juvenile hormone receptor complex. Proc Natl Acad Sci USA. 2015;112(6):1733–1738. doi: 10.1073/pnas.1424386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah N, Dorer DR, Moriyama EN, Christensen AC. Evolution of a large, conserved, and syntenic gene family in insects. G3 (Bethesda) 2012;2(2):313–319. doi: 10.1534/g3.111.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gloss AD, et al. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the drosophilidae. Mol Biol Evol. 2014;31(9):2441–2456. doi: 10.1093/molbev/msu201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nosil P. Ecological Speciation. Oxford Univ Press; Oxford: 2012. [Google Scholar]
- 53.Coyne JA, Orr HA. “Patterns of Speciation in Drosophila” revisited. Evolution. 1997;51(1):295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- 54.David JR, Yassin A, Gidaszewski N, Debat V. Drosophilids (Diptera) from Mayotte island: An annotated list of species collected in 2013 and comments on the colonisation of Indian Ocean Islands. Annales de la Société entomologique de France (NS) 2014;50(3–4):336–342. [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turissini DA, Liu G, David JR, Matute DR. The evolution of reproductive isolation in the Drosophila yakuba complex of species. J Evol Biol. 2015;28(3):557–575. doi: 10.1111/jeb.12588. [DOI] [PubMed] [Google Scholar]
- 58.Kofler R, Pandey RV, Schlötterer C. PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27(24):3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garud NR, Messer PW, Buzbas EO, Petrov DA. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 2015;11(2):e1005004. doi: 10.1371/journal.pgen.1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrider DR, Houle D, Lynch M, Hahn MW. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics. 2013;194(4):937–954. doi: 10.1534/genetics.113.151670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pool JE. The mosaic ancestry of the Drosophila genetic reference panel and the D. melanogaster reference genome reveals a network of epistatic fitness interactions. Mol Biol Evol. 2015;32(12):3236–3251. doi: 10.1093/molbev/msv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.