Significance
The three major DNA replication fidelity determinants are nucleotide selectivity, proofreading, and mismatch repair. Defects in the two latter determinants are now firmly associated with cancer. Nucleotide selectivity is affected by changes in the absolute or relative concentrations of dNTPs. Here, we show that hemizygous SAMHD1+/− mouse embryos have increased dNTP pools compared with wild-type controls and that heterozygous mutations that inactivate SAMHD1 are frequently found in colon cancers. We infer that such cancer cells have increased dNTP pools and, therefore, higher mutation rates. These observations suggest that changes in dNTP concentrations, which affect nucleotide selectivity, the first major determinant of DNA replication fidelity, are associated with cancer.
Keywords: dNTP pools, colon cancer, DNA replication fidelity
Abstract
Even small variations in dNTP concentrations decrease DNA replication fidelity, and this observation prompted us to analyze genomic cancer data for mutations in enzymes involved in dNTP metabolism. We found that sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1), a deoxyribonucleoside triphosphate triphosphohydrolase that decreases dNTP pools, is frequently mutated in colon cancers, that these mutations negatively affect SAMHD1 activity, and that several SAMHD1 mutations are found in tumors with defective mismatch repair. We show that minor changes in dNTP pools in combination with inactivated mismatch repair dramatically increase mutation rates. Determination of dNTP pools in mouse embryos revealed that inactivation of one SAMHD1 allele is sufficient to elevate dNTP pools. These observations suggest that heterozygous cancer-associated SAMHD1 mutations increase mutation rates in cancer cells.
Recent advances in whole-genome sequencing have revealed that human cancers often contain thousands of subclonal mutations (1–3), lending support to the mutator phenotype hypothesis that postulates that an elevation in spontaneous mutation rate is an early step in cancer evolution (4, 5). The three major determinants of DNA replication fidelity that control the spontaneous mutation rate are nucleotide selectivity by DNA polymerases, proofreading by replicative DNA polymerases, and the mismatch repair (MMR) system (6). Failures in the two latter determinants have now been firmly associated with the development of cancer (7), but they cannot account for the increased spontaneous mutation rates in most cancers (5).
The first determinant, nucleotide selectivity by DNA polymerases, can be affected by changes in the absolute and relative concentrations of the four deoxyribonucleoside triphosphates (dNTPs). We have previously demonstrated that severely imbalanced dNTP pools strongly decrease DNA replication fidelity in Saccharomyces cerevisiae (8, 9) without affecting cell proliferation, as long as none of the dNTPs is limiting for DNA replication (10). An equimolar elevation in dNTP pools also decreases DNA replication fidelity, both in yeast and bacteria, presumably by suppressing the proofreading activity of replicative DNA polymerases and by stimulating lesion bypass by both replicative and translesion DNA polymerases (11–16). Recently, we showed in yeast that even a small elevation of the dNTP pool dramatically decreases the replication fidelity of exonuclease-deficient DNA polymerase ε (Pol ε) and DNA polymerase δ (Pol δ) harboring the cancer-associated R696W mutation (17–19). Based on these observations, we hypothesized that decreased nucleotide selectivity caused by changes in the absolute or relative concentrations of dNTPs could be one of the reasons for the increased mutation rates in cancers.
The absolute and relative concentrations of dNTPs are controlled by several dozen proteins (20), and mutations or a change in abundance in any of these could in principle result in a distortion of the dNTP pool. Ribonucleotide reductase (RNR), dCMP deaminase, dUTPase, dTMP synthase, dTMP kinase, and NDP kinases control dNTP biosynthesis. Purine and pyrimidine de novo synthesis pathways provide substrates for RNR, and multiple (deoxy)nucleoside kinases and 5′ nucleotidases control cellular and mitochondrial dNTP salvage. We analyzed the mutation status of the genes involved in dNTP metabolism in colon cancers using a public dataset from The Cancer Genome Atlas (TCGA) and identified SAMHD1 (sterile alpha motif and histidine-aspartate domain-containing protein 1) as one of the frequently mutated genes.
SAMHD1 is a dual-function enzyme with both nuclease and deoxyribonucleoside triphosphate triphosphohydrolase (dNTPase) activities (21, 22). Germ-line mutations in SAMHD1 have been associated with Aicardi–Goutieres syndrome, a congenital autoimmune disease (23), and more recently SAMHD1 was shown to be an HIV-1 restriction factor operating in nondividing blood cells (24, 25). Initially, the restriction function of SAMHD1 was attributed to its dNTPase activity, which was presumed to decrease the intracellular dNTP concentrations to levels incompatible with viral replication (26). Later, it was suggested that restriction of HIV-1 was primarily caused by the nuclease activity of SAMHD1 degrading viral RNA (27). However, more recently it was proposed that SAMHD1 lacks nuclease activity altogether and that the restriction of HIV-1 is caused by alternating ssRNA-binding and dNTPase activities (28). Franzolin et al. showed that SAMHD1 is expressed in a cell cycle-regulated manner and that loss of SAMHD1 has large effects on dNTP pool composition in vitro in both quiescent and cycling cells (29). SAMHD1 has also been identified as a potential driver gene in chronic lymphatic leukemia, where it is recurrently mutated in early stages of tumor development (30–32). In solid tumors, lower protein and RNA expression of SAMHD1 has been observed, presumably caused by promoter methylations (30, 33, 34). However, it is unknown whether SAMHD1 somatic mutations found in cancers affect its dNTPase activity (35).
Here, we show that colon cancer-associated mutations in SAMHD1 either abolish its dNTPase activity or change its specificity, leading to unequal degradation of individual dNTPs. Importantly, even a hemizygous deletion of SAMHD1 leads to an increase of dNTP pools in mouse embryos, which, similar to tumors, contain actively dividing cells. This result suggests that, although the SAMHD1 mutations identified in cancers are heterozygous, they would still result in an alteration of dNTP pools. Interestingly, several of the identified SAMHD1 mutations were present in MMR-deficient hypermutated cancers. Analysis of MMR-deficient yeast strains and human colorectal carcinoma cells demonstrated that even a small alteration of dNTP pools results in a multiplicative increase of mutation rates. Together, these findings suggest that mutations affecting the activity of SAMHD1 are likely to result in increased mutation rates in cancer.
Results
SAMHD1 Is Frequently Mutated in Colon Cancer.
Mutations in SAMHD1 have previously been associated with chronic lymphatic leukemia, in which SAMHD1 has been suggested to be one of the driver genes, but its role in solid cancers is uncertain (30–32). The Catalogue of Somatic Mutations in Cancer (COSMIC) currently contains 104 SAMHD1 mutations (36), and a quarter of these mutations were found in tumors of the large intestine (26 mutations in 24 samples). With the exception of one 31-nucleotide deletion, all SAMHD1 mutations in tumors of the large intestine were single-nucleotide polymorphisms (Table S1). The majority of the mutations were predicted to be deleterious by using in silico analysis: 87% were predicted to be damaging by at least one of four computational tools (Grantham, SIFT, PolyPhen2, and CADD), 72% by two or more tools, and 50% by three tools (Table S1) (37–40). Four of the mutations (R145, D207, R366, and R451) are located at amino acid residues that have been reported to have functional significance in in vitro studies (27, 41, 42).
Table S1.
Summary of the 26 SAMHD1 mutations present in 24 large intestine tumors from COSMIC
| AA substitution | Grantham | SIFT | PolyPhen | CADD | ConSurf |
| F59C* | 205 | Deleterious | Probably damaging | 11.68 | 3 |
| A76T | 58 | Tolerated | Benign | 6.603 | 5 |
| L78L | Silent | Silent | Silent | 7.774 | 8 |
| L132I | 5 | Deleterious | Possibly damaging | 27.9 | 8 |
| V133I (×2) | 29 | Tolerated | Benign | 15.15 | 7 |
| R145Q | 43 | Deleterious | Probably damaging | 34 | 9 |
| D207Y | 160 | Deleterious | Probably damaging | 20.8 | 9 |
| R226H | 29 | Tolerated | Possibly damaging | 28.8 | 6 |
| T232M | 81 | Deleterious | Possibly damaging | 24.5 | 7 |
| S247Y* | 144 | Deleterious | Benign | 24.1 | 5 |
| K288T* | 78 | Tolerated | Benign | 22.9 | 3 |
| S302Y | 144 | Deleterious | Benign | 25 | 8 |
| R305I | 97 | Tolerated | Benign | 23.7 | 7 |
| A338T | 58 | Deleterious | Possibly damaging | 29.3 | 8 |
| A338V | 64 | Tolerated | Benign | 22.9 | 8 |
| R348C | 180 | Tolerated | Benign | 23.1 | 6 |
| R366H | 29 | Deleterious | Possibly damaging | 34 | 9 |
| R451P | 103 | Deleterious | Probably damaging | 28.1 | 9 |
| D497Y | 160 | Deleterious | Benign | 24.5 | 7 |
| D501Y | 160 | Deleterious | Benign | 26.2 | 4 |
| A525T | 58 | Deleterious | Benign | 8.32 | 5 |
| F578V | 50 | Deleterious | Benign | 13.09 | 5 |
| P581P | Silent | Silent | Silent | 7.263 | 7 |
| K596fs (×2) | fs | fs | fs | fs | fs |
Results are shown from four effect-prediction software programs (Grantham, SIFT, PolyPhen, and CADD) and one conservation analysis software program (ConSurf). The eight mutations originating from the TCGA dataset, hgsc.bcm.edu COAD.IlluminaGA DNASeq.Level 2.1.5.0, are indicated in bold. For Grantham, >60 is nonconservative; for CADD, >15 is damaging; and for ConSurf, 9 is most conserved. fs, frame shift; ×2, the same variant was found in two independent tumors.
All in one individual.
Eight of the 26 SAMHD1 mutations (Table S1) were found in the publicly available colorectal cancer (CRC) TCGA dataset, hgsc.bcm.edu COAD.IlluminaGA DNASeq.Level 2.1.5.0, which was downloaded for further analysis. The dataset contained 114,594 mutations in 217 tumors; thus, 3.7% (8 of 217) of the tumors in this CRC dataset carried a coding mutation in SAMHD1. Approximately 20% of the 217 tumors were hypermutated (>12 mutations per 106 bases) (43), and the eight SAMHD1 mutations (Table S1) were found in these hypermutated tumors. Analysis of the dataset using the Significantly Mutated Genes (SMG) test from the MuSIC suite (44) showed that—when taking into consideration mutation type and gene length—SAMHD1 carried more mutations than expected by chance (P = 0.049, false discovery rate = 0.05, convolution test).
Of the total number of mutations falling within coding regions of all genes in the CRC dataset, 39% were silent, whereas none of the eight mutations in SAMHD1 were silent. Among the 26 SAMHD1 mutations in the current COSMIC database, only 2 (8%) were silent mutations. An increase in the ratio between nonsilent and silent mutations in a gene can indicate that functional mutations within that gene are beneficial for the tumor and are selected for during cancer development. Analysis of a panel of genes of known genetic importance in colon cancer (APC, BRAF, KRAS, PIK3CA, SMAD4, and TP53) (45, 46) also showed a lower frequency of silent mutations (3.4%) compared with the overall frequency of silent mutations (39%).
Collectively, these data indicate that SAMHD1 mutations in colon cancer occur nonrandomly and affect functionally important residues, suggesting that deleterious mutations in this gene can be beneficial for the tumors.
CRC-Specific Mutations Negatively Affect SAMHD1 dNTPase Activity.
To investigate the effect of the CRC-specific amino acid substitutions on SAMHD1 dNTPase activity, we chose four mutations (V133I, A338T, R366H, and D497Y) for recombinant protein purification and characterization (Fig. S1). V133I was selected because two tumors carried this mutation, but it was unclear from the in silico analysis to what extent V133I would affect the activity of SAMHD1. The other three mutations were predicted to be deleterious, and we wanted to confirm this prediction biochemically.
Fig. S1.
Analyses of purified SAMHD1 proteins. A total of 1 µg of purified SAMHD1 (a) and SAMHD1 mutant proteins V133I (b), A338T (c), R366H (d), and D497Y (e) were separated by 12.5% SDS/PAGE and stained with Coomassie Brilliant Blue.
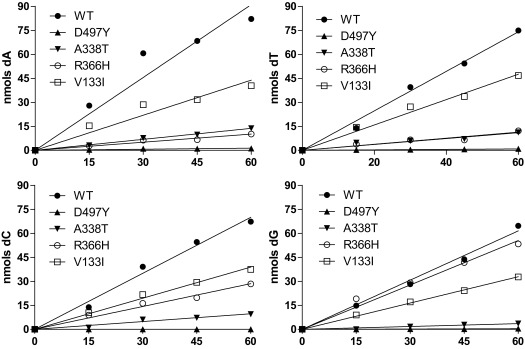
To measure the dNTPase activity, we quantified the deoxyribonucleoside production derived from the hydrolysis of dNTPs by the wild-type (WT) SAMHD1 protein and the four mutants over a time course of 60 min (Fig. S2). Compared with WT, all mutants had reduced specific dNTPase activity toward all four dNTPs (Fig. 1). Interestingly, whereas the D497Y mutation nearly abolished the dNTPase activity, other mutations had varying effects on SAMHD1 activity toward the individual dNTPs. For example, there was a ninefold difference between deoxyadenosine and deoxyguanosine production by the R366H variant of SAMHD1 (Fig. 1D). These data suggest that SAMHD1 mutations might result in imbalanced dNTP pools.
Fig. S2.
In vitro dNTPase activity assays of SAMHD1 protein and the four mutants over a time course of 60 min. All reactions were performed by using the same sample of each protein. Reactions with a total volume of 300 μL [10 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 1 mM GTP, 1 mM dNTP, and 0.5 μM SAMHD1] were incubated at 25 °C. Aliquots were collected at 0, 15, 30, 45, and 60 min, and the dN products were quantified. Lines were fitted by using the linear regression function in GraphPad Prism.
Fig. 1.
In vitro dNTPase activity of SAMHD1 is altered by cancer-associated mutations. Purified SAMHD1 (A) and SAMHD1 mutant proteins V133I (B), A338T (C), R366H (D), and D497Y (E) were incubated with 1 mM dCTP, dTTP, dATP, or dGTP separately with 1 mM GTP as the activator, and the deoxynucleoside products (dC, dT, dA, and dG) were analyzed by HPLC. Numbers indicate detected deoxynucleoside products in mutants compared with WT SAMHD1. Error bars indicate SD. nd, not detectable.
Hemizygous SAMHD1 Deletion Leads to Elevated dNTP Pools in Mouse Embryos.
All eight identified CRC-associated SAMHD1 mutations from the TCGA dataset were heterozygous, and the corresponding methylation data from the TCGA database did not show any SAMHD1 promoter methylation that might indicate inactivation of the WT allele. Thus, although the four tested mutations—V133I, A338T, R366H, and D497Y—negatively affected SAMHD1 dNTPase activity in vitro, it was not clear whether these heterozygous mutations would have any effect on dNTP pools in vivo. To address this question, we compared dNTP pools in WT, SAMHD1+/− hemizygous, and SAMHD1−/− homozygous embryonic day 13.5 (E13.5) mouse embryos. We chose whole embryos because they contain a large number of actively dividing cells, similar to actively proliferating solid tumors. As expected, dNTP pools were significantly increased in homozygous SAMHD1−/− embryos. Importantly, hemizygous SAMHD1+/− embryos also had elevated and imbalanced dNTP pools compared with WT, and dCTP and dTTP were increased by ∼40%, dATP by ∼60%, and dGTP by ∼20% (Fig. 2). These data strongly suggest that CRC-associated heterozygous SAMHD1 mutations that negatively affect its dNTPase activity in vitro should result in an elevation of dNTP pools in vivo.
Fig. 2.
dNTP levels in mouse embryos are affected by SAMHD1 copy number. dNTP levels were measured in E13.5 mouse embryos that were WT (33 embryos), lacking one copy of SAMHD1 (13 embryos), or lacking both copies of SAMHD1 (18 embryos). Results are presented in a boxplot where the central box spans the first to the third quartile, the whiskers represent minimum and maximum values, and the segment inside the box is the median. Outliers are represented by circles. The significance value was calculated by using the Wilcoxon rank sum test.
Small Alteration of dNTP Pools Dramatically Increases Mutation Rates in Combination with MMR Deficiency.
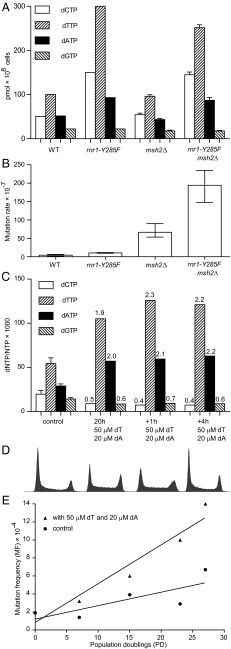
Further analysis of the eight tumors with mutated SAMHD1 showed that six of them had defects in MMR (Table 1 and Fig. S3). To model the effect of minor dNTP pool increases on DNA replication fidelity in the absence of MMR, we used a S. cerevisiae strain containing a deletion of the MSH2 gene and the rnr1-Y285F allele. Y285 is located in the allosteric specificity site of Rnr1, the large subunit of RNR. We have previously demonstrated that rnr1-Y285F alone leads to a slightly imbalanced elevation of dNTPs (3× dCTP, 3× dTTP, 1.8× dATP, and 1× dGTP) that in turn results in a ∼2.5-fold increase in mutation rate compared with WT (10), whereas msh2Δ alone increases the mutation rate ∼15-fold compared with WT. The msh2Δ rnr1-Y285F double mutant tested in this work had a ∼45-fold increased mutation rate compared with WT, demonstrating that minor changes in dNTP pools have a multiplicative effect in combination with the loss of MMR (Fig. 3 A and B).
Table 1.
MMR status in the eight tumors from the TCGA dataset with SAMHD1 mutation
| Amino acid substitution | MMR heterozygous LOF | MLH1 methylation |
| V133I_1 | MLH3 (fs) | MLH1 (d) |
| V133I_2 | MLH1 (fs) | |
| A338T | ||
| A338V | MLH3 (fs) | MLH1 (d) |
| R366H | MLH1 (fs) | MLH1 (s) |
| D497Y | MLH3 (ns) | |
| A525T | MLH1 (fs) | |
| K596fs |
Loss-of-function mutations (LOF) in MLH1, MLH3, MSH2, and MSH6 and promoter methylation of MLH1 were included in the analysis. d, both alleles methylated; fs, frameshift; ns, nonsense; s, one allele methylated.
Fig. S3.
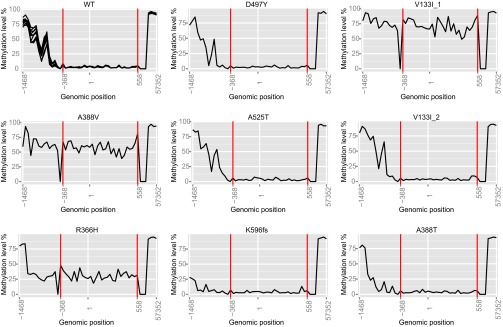
MLH1 promoter methylation in the eight tumors with SAMHD1 mutations. Methylation levels of the MLH1 promoter in percentage are presented as a line plot. The x axis represents 50 CpG sites covering the region 1,468 bp upstream to 57,352 bp downstream of the transcriptional start site (marked by “1”). Red lines specify the variable region where an increased methylation level indicates silencing of the MLH1 gene. WT represents data from nine normal colon samples. The V133I variant is found in two tumors and denoted V133I_1 and V133I_2.
Fig. 3.
Minor alterations of dNTP pools further elevate mutation rates in MMR-deficient cells. (A and B) Amount of each dNTP (A) and mutation rates (B) in the WT, msh2Δ, rnr1-Y285F, and msh2Δ rnr1-Y285F yeast strains. (C) Amount of each dNTP normalized to the total NTP pool in untreated HCT116 cells (control), HCT116 cells incubated in the presence of 50 μM thymidine and 20 μM deoxyadenosine for 20 h (20 h), and in HCT116 cells incubated in the presence of 50 μM thymidine and 20 μM deoxyadenosine for 20 h, after which an additional 50 μM thymidine and 20 μM were added. dNTP pools were measured after 1 h (+1 h) and after 4 h (+4 h). (D) Flow cytometry histograms of the HCT116 cells used for dNTP pool measurements in C. (E) Mutation frequencies and PDs of the HCT116 cells incubated in the presence or absence of 50 μM thymidine (dT) and 20 μM deoxyadenosine (dA).
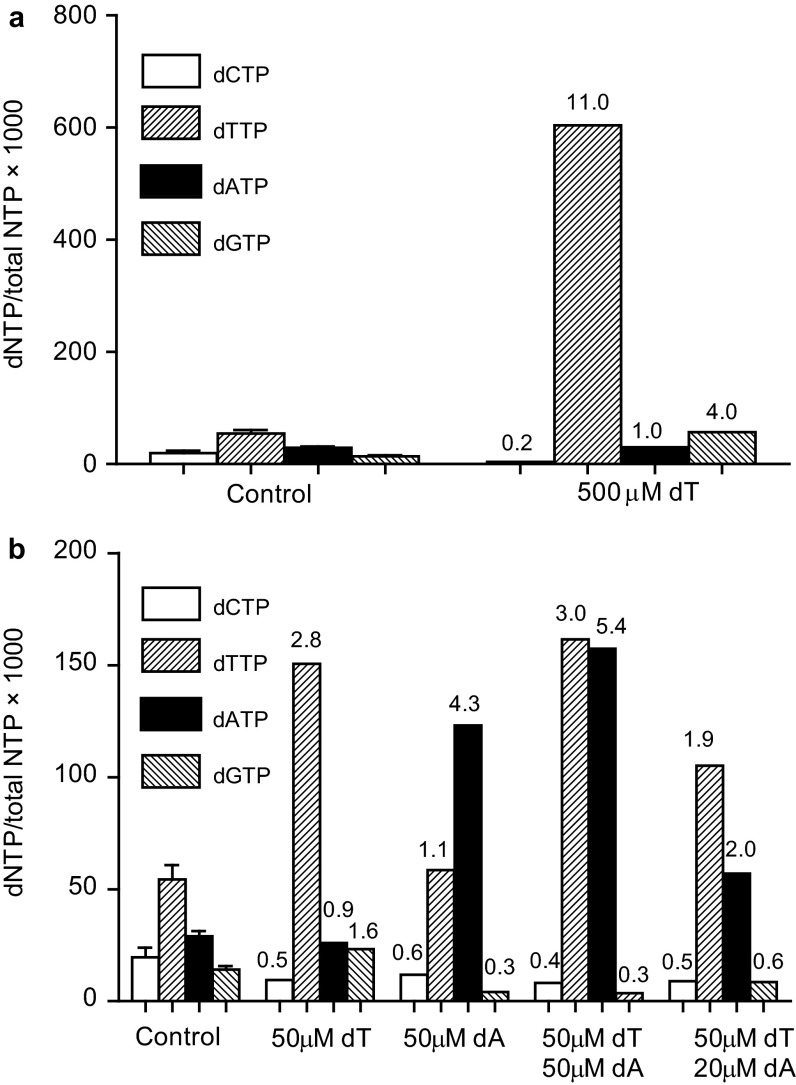
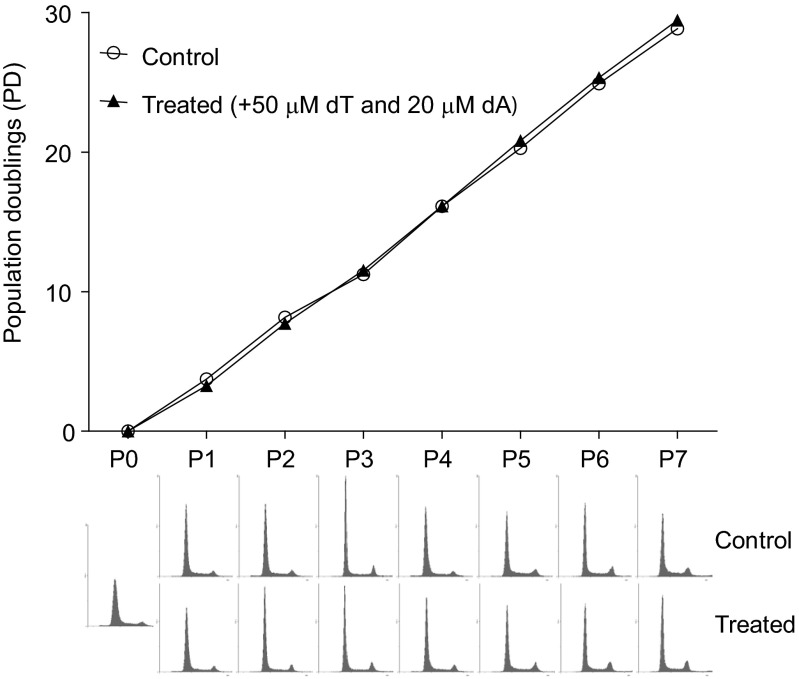
In mammalian cells, severe dNTP pool imbalances are known to be mutagenic (47, 48), but to what extent minor dNTP pool changes increase mutation rates is not well known. To model the effect of minor dNTP pool changes on DNA replication fidelity in human cancer cells, we measured mutation rates at the HPRT locus of the HCT116 colorectal carcinoma cell line that was manipulated to have altered dNTP pools. This cell line lacks functional MMR due to a homozygous mutation of MLH1 (49), and it is commonly used for the analysis of mutation rates under different conditions (50–52). To alter intracellular dNTP pools in this cell line, we supplemented the cell culture medium with deoxyribonucleosides, which are stepwise phosphorylated by deoxynucleoside (dN) kinases and deoxyribonucleotide kinases into dNTPs (20). Addition of one deoxyribonucleoside at high concentrations results in a dramatic increase in one or several dNTPs and a depletion of one or several other dNTPs because of the allosteric regulation of RNR. For example, addition of 1 mM thymidine results in a ∼25-fold increase of dTTP and a ∼10-fold decrease of the dCTP pool and leads to S-phase arrest (53). This effect of thymidine is commonly used for synchronization of cells by the so-called double thymidine block. Therefore, we sought to identify conditions in which the addition of several deoxyribonucleosides at lower concentrations would result in a minor change of dNTP pools without a concomitant S-phase arrest. We first tested thymidine at a concentration of 0.5 mM, which resulted in an 11-fold increase in dTTP and a 5-fold decrease in dCTP (Fig. S4A). Then, we tested thymidine at a concentration of 0.05 mM, which resulted in a 2.8-fold increase in dTTP and a 2-fold decrease in dCTP (Fig. S4B). We then tested the addition of deoxyadenosine alone and in combination with thymidine and found that, in the presence of 50 μM thymidine and 20 μM deoxyadenosine, dTTP and dATP increased approximately twofold and dCTP and dGTP levels decreased approximately twofold (Fig. S4B and Fig. 3C). At these concentrations of dN, flow cytometry analysis did not demonstrate any S-phase arrest (Fig. 3D). Therefore, we decided to proceed with measurements of mutation rates in the presence or absence of 50 μM thymidine and 20 μM deoxyadenosine, according to the schematic in Fig. S5. Mutation frequencies were invariably increased in the cell cultures grown in the presence of 50 μM thymidine and 20 μM deoxyadenosine (Fig. 3E), whereas the population-doubling (PD) time and cell-cycle progression were similar to the control cells grown in the absence of deoxyribonucleosides (Fig. S6). Mutation rates calculated by using mutation frequencies and PD times (Fig. 3E) were approximately threefold higher in the cells grown with exogenous deoxyribonucleosides (4.46 × 10−5 vs. 1.45 × 10−5).
Fig. S4.
dNTP pools in HCT116 cells grown 1 h in the presence of thymidine (dT) and deoxyadenosine (dA) in the following concentrations: 500 μM thymidine (A) and 50 μM thymidine; 50 μM deoxyadenosine; 50 μM thymidine and 50 μM deoxyadenosine; and 50 μM thymidine and 20 μM deoxyadenosine (B).
Fig. S5.
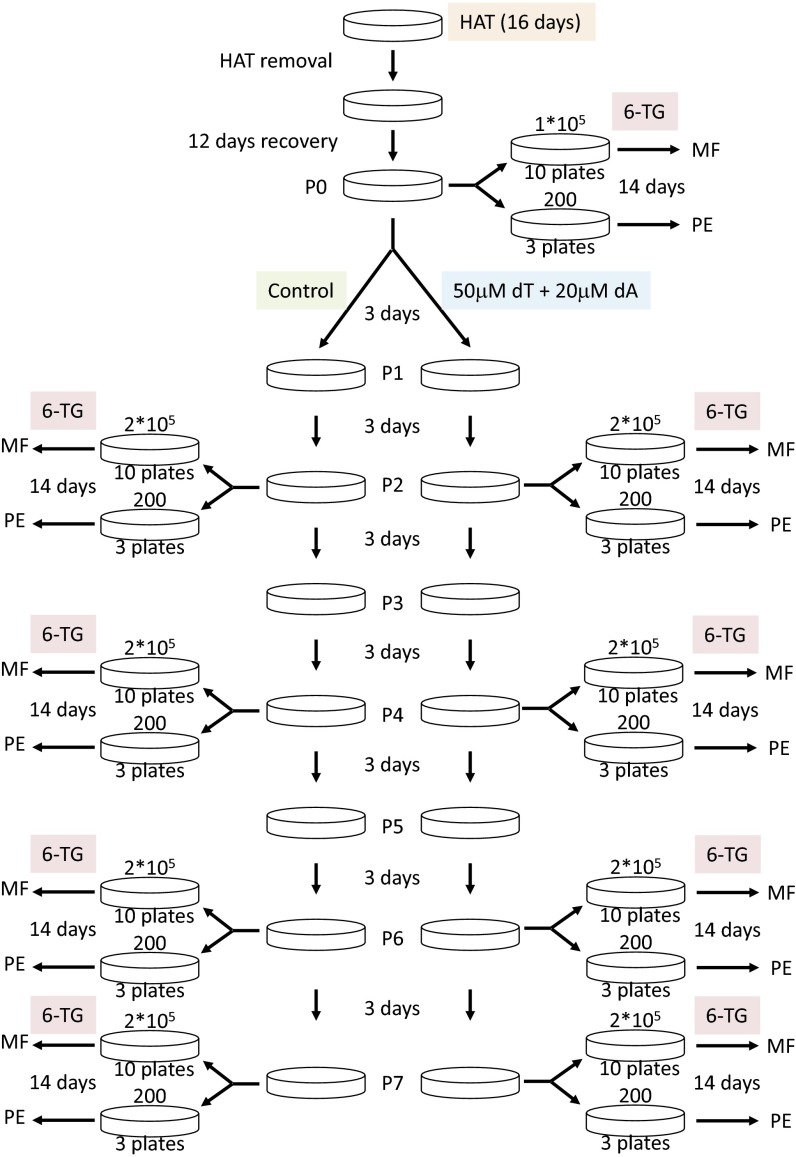
Schematic representation of the workflow of the experiment for calculating the mutation rates of the HPRT locus of the HCT116 cells in the presence or absence of 50 μM thymidine (dT) and 20 μM deoxyadenosine (dA). 6-TG, 6-thioguanine; HAT, hypoxanthine-aminopterin-thymidine; MF, mutation frequency; PE, plating efficiency. For details, see SI Experimental Procedures.
Fig. S6.
Population doubling times and corresponding flow cytometry histograms of the HCT116 cells grown in the absence (control) or presence of 50 μM thymidine and 20 μM deoxyadenosine (treated). Each passage was 3 d.
Discussion
Are dNTP pools altered in cancer cells compared with dNTP pools in normal cells? And, if so, do changes in dNTP pools contribute to the increased mutation rates and development of cancer? These are not easy questions to answer. dNTP pools are orders of magnitude lower than the corresponding NTP pools (54) and are, for that reason, difficult to measure. Publications reporting that cancer tissues have elevated dNTP pools compared with normal surrounding tissues are not informative because dNTPs are produced primarily during S phase. Thus, dNTP pools are high in mitotic cells, such as cancer cells, and low in nondividing cells. Comparisons of dNTP pools in cancerous and normal immortalized cell lines in vitro also have caveats. First, the mitotic index, and thus the proportion of S-phase cells containing elevated dNTP pools, is often higher in cancerous cell lines. Second, the volume of cells in different cell lines might vary. An apparent increase in dNTP pools in cancerous cells will not result in a higher intracellular dNTP concentration if the volume of such cells is also increased compared with normal cells. Finally, dNTP pools in different cell lines in vitro are affected by the nucleosides or bases present in the culture medium (Fig. S4) (55). These dNTP precursors can be taken up with different efficiencies depending on the status of the salvage enzymes, such as thymidine kinase, deoxyguanosine kinase, and deoxyadenosine kinase, in cancerous vs. normal cells.
Here, we propose that some colon cancers have altered intracellular dNTP pools. We base our conclusion on the following observations: (i) These cancers have mutations in SAMHD1, an enzyme responsible for the degradation of dNTPs; (ii) the identified SAMHD1 mutations negatively affect its dNTPase activity in vitro; and (iii) actively dividing hemizygous SAMHD1+/− mouse embryos have increased dNTP pools compared with congenic WT embryos of the same age. Previous dNTP pool measurements were performed in various types of WT and SAMHD1−/− homozygous cells, including E14.5 mouse embryonic fibroblasts (56, 57), but, to our knowledge, ours is the first study to measure dNTP pools in hemizygous SAMHD1 mouse embryos. This observation is important because it demonstrates that cancer cells do not need to inactivate both SAMHD1 alleles to increase the dNTP pools. Of note, the genes encoding Pol ε and δ are examples of two other genes in which heterozygous mutations have recently been associated with human cancers (58).
dNTP pool increases in SAMHD1+/− mouse embryos are modest but significant and range from 20% to 60% (Fig. 2). In fact, even modest changes of dNTP pools above or below normal levels can have a profound effect on cellular physiology or DNA replication fidelity. First, a mutation in the allosteric activity site of yeast RNR (rnr1-D57N) leads to only a ∼1.6- to 2-fold increase in dNTP pools, but a concomitant 3-fold increase in the mutation rate (11). Second, inactivation of Sml1, an inhibitor of yeast RNR, leads to a ∼2.5-fold increase in dNTP pools (59), and this slight increase is enough to rescue the lethality of the deletion of MEC1 (yeast homolog of mammalian ATR) (59) and to increase the speed of replication forks by approximately 2-fold (60). Third, in mice, increased gene dosage of the small RNR subunit Rrm2 elevates RNR activity, but does not lead to elevated dNTP pools (61), presumably because of the strict allosteric dATP feedback inhibition of the mammalian RNR. Interestingly, despite any detectable increase in dNTP levels, increased gene dosage of the small RNR subunit Rrm2 reduces fragile site breakage and prolongs the survival of ATR mutant mice (61). Fourth, the deletion of Dun1, a protein kinase that controls yeast RNR, causes a ∼50% reduction of dNTP pools that, in turn, decreases the mutagenic effect of proofreading-deficient Pol ε (pol2-4) by approximately threefold and results in a mutation rate comparable to WT levels (18). Although the reduction of dNTP levels increases the fidelity of DNA polymerases, it can also lead to fork stalling, the accumulation of single-stranded DNA, and chromosomal rearrangements. Consistent with these outcomes, decreased dNTP pools have been proposed to be a source of genomic instability in early stages of cancer development (62).
Cancer cells that have defects in replicative DNA polymerases and/or MMR will be at further risk of having elevated mutation rates due to minor dNTP pool alterations. We have recently demonstrated that the introduction of the Pol ε-M644G (a mutation that reduces the accuracy of Pol ε) or Pol δ-R696W (a human colon cancer-associated mutation) into budding yeast cells results in replication stress, leading to the activation of the genome integrity checkpoint and concomitant elevation of dNTP pools (17–19). The checkpoint-dependent elevation of dNTP pools was to a large degree responsible for the dramatic elevation of the mutation rates in these polymerase-defective yeast strains. Here, we show that minor increases of dNTP pools also have a profound effect on the mutation rate in an MMR-deficient yeast strain. The onefold to threefold increase in dNTP pools caused by the rnr1-Y285F substitution elevates the mutation rate from ∼65 × 10−7 in msh2Δ yeast to ∼200 × 10−7 in msh2Δ rnr1-Y285F yeast (Fig. 3B). To model the effect of minor dNTP pool alterations on the mutation rates in cancer cells, we perturbed dNTP pools in the HCT116 colorectal carcinoma cell line by adding low concentrations of thymidine and deoxyadenosine to the growth medium. Such treatment resulted in an approximately twofold increase in dTTP and dATP, an approximately twofold decrease in dCTP and dGTP, and a concomitant approximately threefold elevation of mutation rates at the HPRT locus. Because the HCT116 cells lack functional MMR, they already have ∼12-fold elevated mutation rates, compared with the HCT116 cells with normal MMR function (52). Our results indicate that minor alterations of dNTP pools further elevate mutation rates in MMR-deficient cells.
Replication stress-dependent activation of the genome integrity checkpoint has been shown to lead to elevated dNTP pools and to higher mutation rates in budding yeast (11). In contrast to yeast, genome integrity checkpoint activation in mammalian cells does not result in a similar global expansion of the dNTP pools (63). It is possible that, in response to checkpoint activation, there are local increases of dNTP levels near the sites of DNA damage. For example, colocalization of RNR and dTMP kinase at the sites of DNA damage has been reported (64, 65), but it is not known whether RNR and dTMP kinase colocalize at stalled replication forks. It is also not clear whether the rest of the machinery required for the production of dNTPs—including dUTPase, dCMP deaminase, dTMP synthase, dTMP kinase, and NDP kinase—colocalize at the sites of DNA damage. Even if there is no local checkpoint-dependent increase in dNTP pools in the mammalian cells in response to replication stress, it is possible that dNTP pools increase due to mutation or misregulation of one or more of the many enzymes involved in nucleotide metabolism.
In this study, we demonstrate that mutations in SAMHD1 that alter its dNTPase activity are associated with colon cancer. We speculate that other mutations that elevate or imbalance dNTP pools will also be identified in cancer cells, and we propose that minor dNTP pool disturbances in combination with defects in proofreading or MMR might enhance the mutator phenotype of cancer cells. Recently, the minidriver model of polygenic cancer evolution has been put forward (66). This model proposes that many mutations found in cancer might not be major drivers or “passenger” mutations, but instead might have relatively weak tumor-promoting effects and are referred to as “mini drivers.” It has been suggested that multiple mini drivers can substitute for a major driver. We believe that mutations in SAMHD1 that are predicted to result in mutagenic dNTP pool alterations fall into the category of mini drivers.
The findings presented here might have implications for the treatment of mutator-driven cancers. Chemotherapeutic reduction of dNTP pools could decrease mutation rates and slow down cancer progression, whereas an additional elevation of dNTP pools by treatment with exogenous deoxyribonucleosides might further increase mutation rates and kill cancer cells through mutation overload.
Experimental Procedures
Public Databases.
The cancer mutation database COSMIC was used to get an overview of reported SAMHD1 mutations in solid cancers. For detailed studies of tumors with SAMHD1 mutations, the publicly available mutation dataset (hgsc.bcm.edu COAD.IlluminaGA DNASeq.Level 2.1.5.0), consisting of data from 217 CRC tumors analyzed by using Illumina exome sequencing technology, was downloaded from TCGA. Only publicly available datasets were used for all analyses.
In Silico Analysis of Mutations.
The deleteriousness of the SAMHD1 mutations in the tumors of the large intestine from COSMIC was analyzed by using four different computational tools: Grantham, SIFT, PolyPhen2, and CADD. Because these are prediction tools, we increased our confidence in their results by taking into consideration the overlap between them when evaluating whether mutations were likely to affect protein function. ConSurf was used to generate conservation scores from 1 to 9, with 9 being the most conserved amino acids within the protein. The TCGA mutation dataset was analyzed by using the SMG test from the MuSIC suite with default settings in all cases except for background mutation rate groups, which was set to 2 based on the fact that colon cancers can be divided into hypermutated and non-hypermutated.
SAMHD1 Expression and Purification.
The pGEX-6P-1 plasmids encoding N-terminal GST-tagged WT and mutant SAMHD1 were expressed in Escherichia coli (BL21) grown in LB medium by using 0.4 mM isopropyl β-d-1-thiogalactopyranoside for induction at 18 °C for 16 h, and the proteins were purified essentially as described (67). After batch loading on 1.5 mL of glutathione–Sepharose (GE Healthcare), the beads were collected in a column and washed twice with 10 mL of buffer A [50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, and 10% (vol/vol) glycerol]. The GST affinity tag was removed by overnight enzymatic cleavage with PreScission Protease. SAMHD1 was eluted and stored in buffer A.
SAMHD1 in Vitro dNTPase Assay.
dNTPase assays were performed essentially as described (68). Reactions in a total volume of 300 μL [10 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 1 mM GTP, 1 mM dNTP, and 0.5 μM SAMHD1] were incubated at 25 °C. Aliquots collected at 0, 15, 30, 45, and 60 min were diluted in nine volumes of ice-cold PBS to stop the reaction and spun through a 0.5-mL Nanosep 3-kDa filter (PALL) at 14,000 × g for 10 min. The deproteinized samples were analyzed by HPLC using an UltraCore SuperC18 50-mm × 2.1-mm column (ACE) equilibrated in buffer (7% MeOH and 17 mM KH2PO4, pH 6.0). The dN products were quantified in buffer (7% MeOH and 17 mM KH2PO4, pH 6.0) by using the peak integration of the mV response. The specific activity of SAMHD1 was defined as nanomoles of dN product per hour (1 nmol of dN per hour = 1 unit) per milligram of protein [(nanomoles of dN per hour) per milligram].
dNTP Pool Measurements in Mouse Embryos.
All animal experiments were approved by the Animal Review Board at the Court of Appeal of Northern Norrland (Umea). Homozygous SAMHD1−/− knockout mice in the C57BL/6 background (57) were kindly provided by Jan Rehwinkel (University of Oxford, Oxford) and mated with WT C57BL/6 mice. From these crosses, E13.5 embryos were isolated, and after their tails were removed for genotyping, the embryos were snap-frozen in Eppendorf tubes in liquid nitrogen. After the addition of ice-cold 12% (wt/vol) TCA, 15 mM MgCl2 solution, and glass beads, the embryos were thawed on ice and homogenized on a BeadBeater (BioSpec) for 30 s at 4 °C in a cold room. The supernatant was collected by centrifugation at 14,000 × g for 5 min at 4 °C and processed as described (69).
dNTP Pool and Mutation Rate Measurements in S. cerevisiae.
All yeast culturing was carried out at 30 °C in YPAD (1% yeast extract, 2% bacto-peptone, and 20 mg/L adenine) liquid cultures in a shaking incubator at 160 rpm. For plates, the YPAD contained 2% agar. dNTP pools were measured in asynchronous cultures as described (69). The canavanine resistance assay was used to calculate mutation rates as described (10).
SI Experimental Procedures
Methylation Analysis.
To analyze whether SAHMD1 and/or MLH1 were transcriptionally silenced by methylation in the eight tumors carrying heterozygous SAMHD1 mutations, the corresponding Illumina HumanMethylation450 open-access data files for these individuals, available in TCGA, were downloaded. Nine additional datasets from normal colon tissue were downloaded for comparison. For MLH1, the HumanMethylation450 chip contained 51 CpG sites from 1,468 bp upstream to 57,352 bp downstream of the transcriptional start site. An increase in methylation level between healthy tissue and tumors was noticed in the region −368 to +558 bp relative to the transcriptional start site. An average methylation level of between 10% and 50% in this region was used as a cutoff for concluding that at least one copy of MLH1 was methylated, whereas an average methylation level of >50% was considered to indicate that the tumor contained cells where both copies were methylated (Table 1). The one individual who had an average methylation level between 10% and 50% also carried an MLH1 frameshift mutation. For SAMHD1, the HumanMethylation450 chip contained 16 CpG sites from 309 bp upstream to 59,469 bp downstream of the transcriptional start site. No difference in methylation level between normal tissue and tumor tissue was found for SAMHD1.
HPRT Mutation Rates in HCT116 Cells.
HPRT mutation rates in HCT116 cells (ATCC CCL-247) were determined by measuring the accumulation of mutant cells, with or without altered dNTP pools, during serial passaging as described (52). The cells were grown in 10% FBS and Dulbecco's Modified Eagle Medium (Sigma-Aldrich). dNTP pool and flow cytometry analyses in HCT116 cells were performed as described (63). To alter the dNTP pool, cells were grown in medium supplemented with 50 μM thymidine, 20 μM deoxyadenosine, and 1 μM deoxycoformycin (DCF). DCF was added to inhibit deamination of deoxyadenosine by adenosine deaminase. The cells were cleansed of existing HPRT mutations before the experiment by growth in 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine medium (Sigma-Aldrich) for approximately nine PDs. HPRT mutant frequency was determined at passage 0 (P0) and at several later passages as illustrated in Fig. S5. A total of 1–2 × 106 cells were plated in 30 μM 6-thioguanine (6-TG) medium at a density of 1–2 × 105 cells per 10-cm dish. Additionally, 2 × 102 cells per 10-cm dish were plated in triplicate in normal culture medium to obtain the plating efficiency (PE) at the time of selection. Cells were incubated for 14 d, and colonies were visualized by staining with 0.5% crystal violet in 50% methanol. The mutation frequency (MF) was calculated by using the following equation: MF = C6-TG/(N6-TG × PE) where C6-TG is the number of 6-TG–resistant colonies and N6-TG is the total number of cells plated (∼2 × 106). PE was calculated by dividing the number of colonies in the nonselective dishes by the total number of cells plated (∼6 × 102). PE and the exact number of subcultured cells were used to calculate PD by using the following equation: PD = [ln(total number of cells) − ln(number of cells plated × PE)]/ln2. The mutation rate was estimated by plotting the observed mutant frequencies as a function of PD and calculating the slope by linear regression using the LINEST function in Microsoft Excel. This slope yields the mutation rate (the number of mutations per cell in each generation) (52).
Acknowledgments
We thank Dr. Jan Rehwinkel for the SAMHD1−/− mice; Dr. Kim Baek for the pGEX-6P-1-SAMHD1 plasmid; Drs. Polina Shcherbakova and Pelle Håkansson for the HCT116 cells; Dr. Anders Hofer and Ms. Farahnaz Ranjbarian for the assistance with nucleoside measurements; and Dr. Jörgen Johansson for 6-thioguanine. This work was supported by a Knut and Alice Wallenberg Foundation grant (to J.T., B.M., A.C., and E.J.). Additional support was provided by the Swedish Research Council (E.J. and A.C.) and the Swedish Cancer Society (E.J. and A.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519128113/-/DCSupplemental.
References
- 1.Schmitt MW, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods. 2015;12(5):423–425. doi: 10.1038/nmeth.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox EJ, Loeb LA. Cancer: One cell at a time. Nature. 2014;512(7513):143–144. doi: 10.1038/nature13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34(9):2311–2321. [PubMed] [Google Scholar]
- 5.Schmitt MW, Prindle MJ, Loeb LA. Implications of genetic heterogeneity in cancer. Ann N Y Acad Sci. 2012;1267:110–116. doi: 10.1111/j.1749-6632.2012.06590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hewish M, Lord CJ, Martin SA, Cunningham D, Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol. 2010;7(4):197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- 8.Buckland RJ, et al. Increased and imbalanced dNTP pools symmetrically promote both leading and lagging strand replication infidelity. PLoS Genet. 2014;10(12):e1004846. doi: 10.1371/journal.pgen.1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watt DL, Buckland RJ, Lujan SA, Kunkel TA, Chabes A. Genome-wide analysis of the specificity and mechanisms of replication infidelity driven by imbalanced dNTP pools. Nucleic Acids Res. 2016;44(4):1669–1680. doi: 10.1093/nar/gkv1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38(12):3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabes A, et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112(3):391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 12.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36(17):5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone JE, et al. Lesion bypass by S. cerevisiae Pol ζ alone. DNA Repair (Amst) 2011;10(8):826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(48):19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunz BA, et al. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: A critical factor in the maintenance of genetic stability. Mutat Res. 1994;318(1):1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 16.Ahluwalia D, Schaaper RM. Hypermutability and error catastrophe due to defects in ribonucleotide reductase. Proc Natl Acad Sci USA. 2013;110(46):18596–18601. doi: 10.1073/pnas.1310849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertz TM, Sharma S, Chabes A, Shcherbakova PV. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc Natl Acad Sci USA. 2015;112(19):E2467–E2476. doi: 10.1073/pnas.1422934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams LN, et al. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc Natl Acad Sci USA. 2015;112(19):E2457–E2466. doi: 10.1073/pnas.1422948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohl CD, Ray S, Sweasy JB. Pools and Pols: Mechanism of a mutator phenotype. Proc Natl Acad Sci USA. 2015;112(19):5864–5865. doi: 10.1073/pnas.1505169112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews CK. Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer. 2015;15(9):528–539. doi: 10.1038/nrc3981. [DOI] [PubMed] [Google Scholar]
- 21.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 22.Beloglazova N, et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem. 2013;288(12):8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice GI, et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laguette N, Benkirane M. How SAMHD1 changes our view of viral restriction. Trends Immunol. 2012;33(1):26–33. doi: 10.1016/j.it.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryoo J, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20(8):936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seamon KJ, Sun Z, Shlyakhtenko LS, Lyubchenko YL, Stivers JT. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015;43(13):6486–6499. doi: 10.1093/nar/gkv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franzolin E, et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci USA. 2013;110(35):14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clifford R, et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123(7):1021–1031. doi: 10.1182/blood-2013-04-490847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuh A, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120(20):4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- 32.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JL, Lu FZ, Shen XY, Wu Y, Zhao LT. SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochem Biophys Res Commun. 2014;455(3-4):229–233. doi: 10.1016/j.bbrc.2014.10.153. [DOI] [PubMed] [Google Scholar]
- 34.de Silva S, et al. Downregulation of SAMHD1 expression correlates with promoter DNA methylation in Sézary syndrome patients. J Invest Dermatol. 2014;134(2):562–565. doi: 10.1038/jid.2013.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohnken R, Kodigepalli KM, Wu L. Regulation of deoxynucleotide metabolism in cancer: Novel mechanisms and therapeutic implications. Mol Cancer. 2015;14:176. doi: 10.1186/s12943-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbes SA, et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 38.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 39.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji X, Tang C, Zhao Q, Wang W, Xiong Y. Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci USA. 2014;111(41):E4305–E4314. doi: 10.1073/pnas.1412289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu C, et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat Commun. 2013;4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]
- 43.Donehower LA, et al. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J Pathol. 2013;229(1):99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dees ND, et al. MuSiC: Identifying mutational significance in cancer genomes. Genome Res. 2012;22(8):1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esplin ED, Snyder MP. Genomic era diagnosis and management of hereditary and sporadic colon cancer. World J Clin Oncol. 2014;5(5):1036–1047. doi: 10.5306/wjco.v5.i5.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oikonomou E, Pintzas A. Cancer genetics of sporadic colorectal cancer: BRAF and PI3KCA mutations, their impact on signaling and novel targeted therapies. Anticancer Res. 2006;26(2A):1077–1084. [PubMed] [Google Scholar]
- 47.Kunz BA. Mutagenesis and deoxyribonucleotide pool imbalance. Mutat Res. 1988;200(1-2):133–147. doi: 10.1016/0027-5107(88)90076-0. [DOI] [PubMed] [Google Scholar]
- 48.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181(2):305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 49.Papadopoulos N, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263(5153):1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 50.Tili E, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci USA. 2011;108(12):4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agbor AA, Göksenin AY, LeCompte KG, Hans SH, Pursell ZF. Human Pol ε-dependent replication errors and the influence of mismatch repair on their correction. DNA Repair (Amst) 2013;12(11):954–963. doi: 10.1016/j.dnarep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glaab WE, Tindall KR. Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis. 1997;18(1):1–8. doi: 10.1093/carcin/18.1.1. [DOI] [PubMed] [Google Scholar]
- 53.Bjursell G, Reichard P. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J Biol Chem. 1973;248(11):3904–3909. [PubMed] [Google Scholar]
- 54.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107(11):4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 56.Behrendt R, et al. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Reports. 2013;4(4):689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rehwinkel J, et al. SAMHD1-dependent retroviral control and escape in mice. EMBO J. 2013;32(18):2454–2462. doi: 10.1038/emboj.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rayner E, et al. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16(2):71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 59.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2(3):329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 60.Poli J, et al. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31(4):883–894. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Contreras AJ, et al. Increased Rrm2 gene dosage reduces fragile site breakage and prolongs survival of ATR mutant mice. Genes Dev. 2015;29(7):690–695. doi: 10.1101/gad.256958.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145(3):435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281(12):7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 64.Niida H, et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24(4):333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu CM, et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer Cell. 2012;22(1):36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 66.Castro-Giner F, Ratcliffe P, Tomlinson I. The mini-driver model of polygenic cancer evolution. Nat Rev Cancer. 2015;15(11):680–685. doi: 10.1038/nrc3999. [DOI] [PubMed] [Google Scholar]
- 67.Amie SM, et al. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J Biol Chem. 2013;288(28):20683–20691. doi: 10.1074/jbc.M113.472159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan J, et al. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem. 2013;288(15):10406–10417. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jia S, Marjavaara L, Buckland R, Sharma S, Chabes A. Determination of deoxyribonucleoside triphosphate concentrations in yeast cells by strong anion-exchange high-performance liquid chromatography coupled with ultraviolet detection. Methods Mol Biol. 2015;1300:113–121. doi: 10.1007/978-1-4939-2596-4_8. [DOI] [PubMed] [Google Scholar]